CSDE1 Intracellular Distribution as a Biomarker of Melanoma Prognosis
Abstract
1. Introduction
2. Results
2.1. Design of the Study
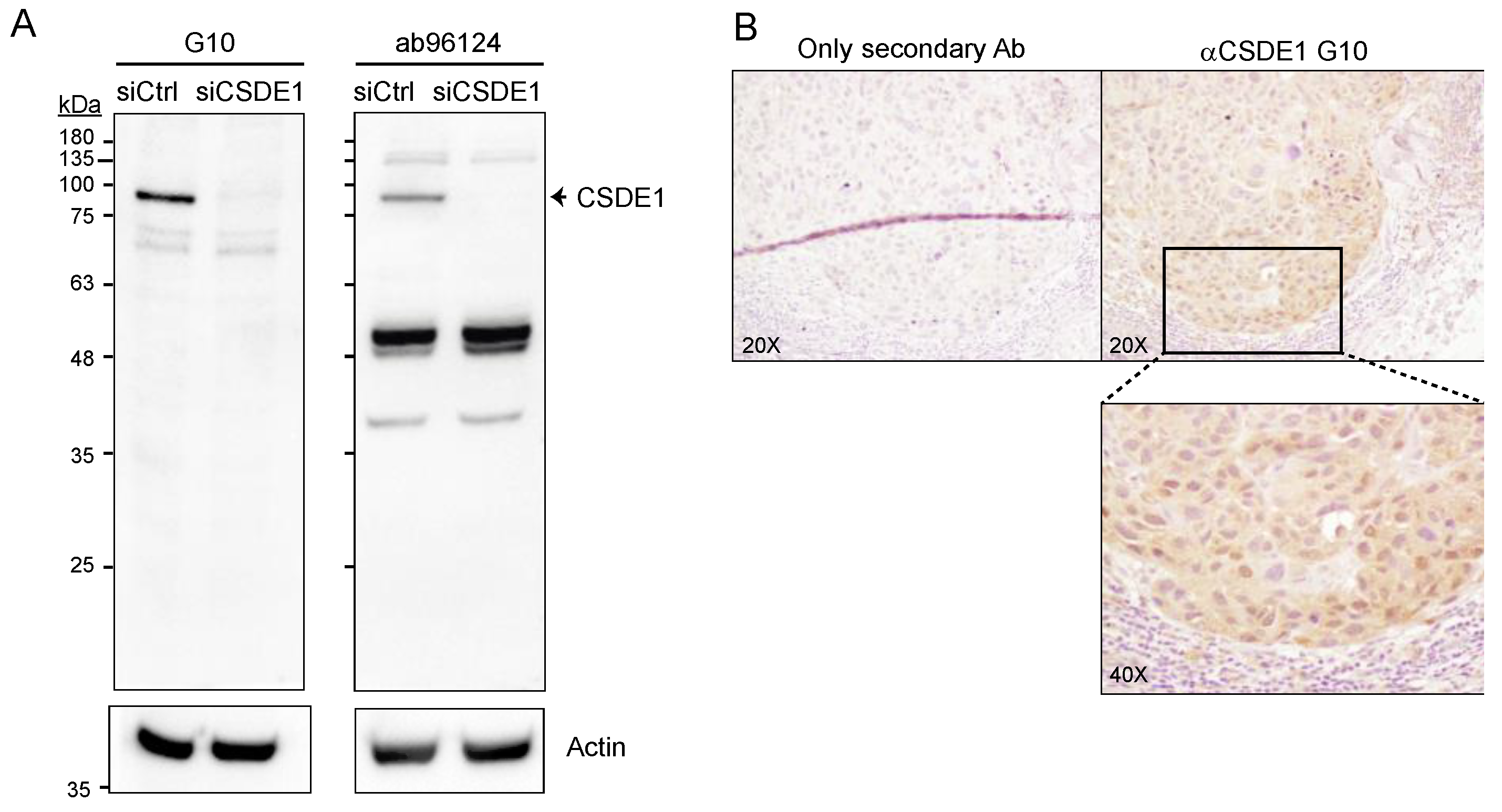
2.2. Generation of a Suitable anti-CSDE1 Monoclonal Antibody
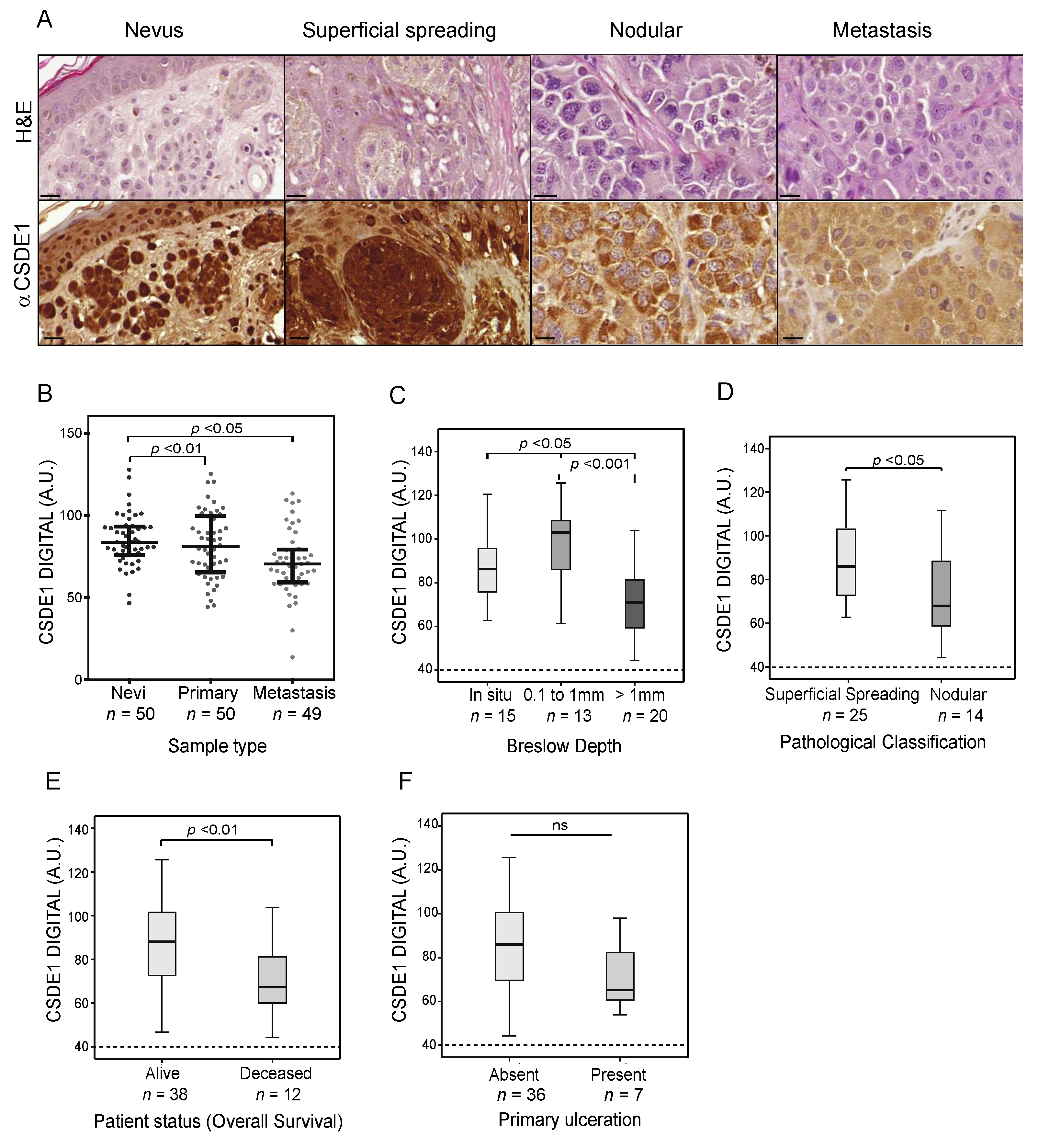
2.3. Global CSDE1 Levels Decrease with Melanoma Malignancy
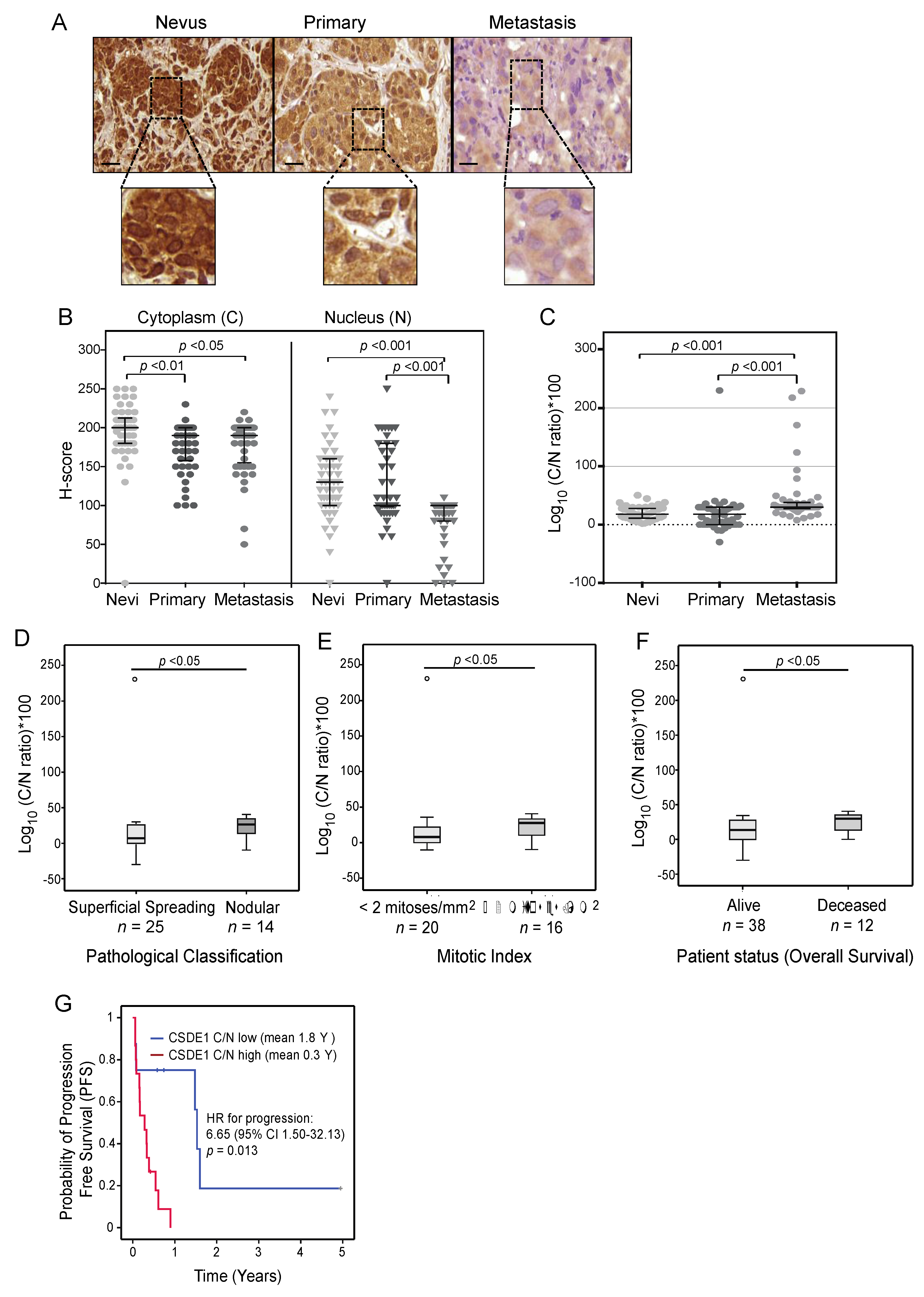
2.4. CSDE1 Cytoplasmic/Nuclear Ratio Correlates with Disease-Specific Survival
3. Discussion
4. Materials and Methods
4.1. Ethical Statement
4.2. Immunohistochemistry (IHC) of Human Tissue Specimens
4.3. Immunocytochemistry (ICC)
4.4. Visual and Digital Quantification
4.5. Statistical Analysis
4.6. Cell Culture and Depletion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schadendorf, D.; van Akkooi, A.C.J.; Berking, C.; Griewank, K.G.; Gutzmer, R.; Hauschild, A.; Stang, A.; Roesch, A.; Ugurel, S. Melanoma. Lancet 2018, 392, 971. [Google Scholar] [CrossRef]
- Swe, T.; Kim, K.B. Update on systemic therapy for advanced cutaneous melanoma and recent development of novel drugs. Clin. Exp. Metastasis 2018, 35, 503–520. [Google Scholar] [CrossRef] [PubMed]
- Olbryt, M.; Rajczykowski, M.; Widłak, W. Biological Factors behind Melanoma Response to Immune Checkpoint Inhibitors. Int. J. Mol. Sci. 2020, 21, 4071. [Google Scholar] [CrossRef]
- Ng, M.F.; Simmons, J.L.; Boyle, G.M. Heterogeneity in Melanoma. Cancers 2022, 14, 3030. [Google Scholar] [CrossRef] [PubMed]
- Eisenstein, A.; Gonzalez, E.C.; Raghunathan, R.; Xu, X.; Wu, M.; McLean, E.O.; McGee, J.; Ryu, B.; Alani, R.M. Emerging Bi-omarkers in Cutaneous Melanoma. Mol. Diagn. Ther. 2018, 22, 203–218. [Google Scholar] [CrossRef]
- Rabbie, R.; Ferguson, P.; Molina-Aguilar, C.; Adams, D.J.; Robles-Espinoza, C.D. Melanoma subtypes: Genomic profiles, prognostic molecular markers and therapeutic possibilities. J. Pathol. 2019, 247, 539–551. [Google Scholar] [CrossRef]
- Tonella, L.; Pala, V.; Ponti, R.; Rubatto, M.; Gallo, G.; Mastorino, L.; Avallone, G.; Merli, M.; Agostini, A.; Fava, P.; et al. Prog-nostic and Predictive Biomarkers in Stage III Melanoma: Current Insights and Clinical Implications. Int. J. Mol. Sci. 2021, 22, 4561. [Google Scholar] [CrossRef]
- Seyhan, A.A.; Carini, C. Insights and Strategies of Melanoma Immunotherapy: Predictive Biomarkers of Response and Resistance and Strategies to Improve Response Rates. Int. J. Mol. Sci. 2022, 24, 41. [Google Scholar] [CrossRef] [PubMed]
- Hentze, M.W.; Castello, A.; Schwarzl, T.; Preiss, T. A brave new world of RNA-binding proteins. Nat. Rev. Mol. Cell Biol. 2018, 19, 327–341. [Google Scholar] [CrossRef]
- Pereira, B.; Billaud, M.; Almeida, R. RNA-Binding Proteins in Cancer: Old Players and New Actors. Trends Cancer 2017, 3, 506–528. [Google Scholar] [CrossRef]
- Kang, D.; Lee, Y.; Lee, J.-S. RNA-Binding Proteins in Cancer: Functional and Therapeutic Perspectives. Cancers 2020, 12, 2699. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.-L.; Li, B.; Luo, Y.-X.; Lin, Q.; Liu, S.-R.; Zhang, X.-Q.; Zhou, H.; Yang, J.-H.; Qu, L.-H. Comprehensive Genomic Charac-terization of RNA-Binding Proteins across Human Cancers. Cell Rep. 2018, 22, 286–298. [Google Scholar] [CrossRef] [PubMed]
- Neelamraju, Y.; Gonzalez-Perez, A.; Bhat-Nakshatri, P.; Nakshatri, H.; Janga, S.C. Mutational landscape of RNA-binding pro-teins in human cancers. RNA Biol. 2018, 15, 115–129. [Google Scholar] [CrossRef] [PubMed]
- Gebauer, F.; Schwarzl, T.; Valcárcel, J.; Hentze, M.W. RNA-binding proteins in human genetic disease. Nat. Rev. Genet. 2021, 22, 185–198. [Google Scholar] [CrossRef] [PubMed]
- Chu, J.; Pelletier, J. Therapeutic Opportunities in Eukaryotic Translation. Cold Spring Harb. Perspect. Biol. 2018, 10, a032995. [Google Scholar] [CrossRef] [PubMed]
- Mohibi, S.; Chen, X.; Zhang, J. Cancer The’RBP’eutics—RNA-Binding Proteins as Therapeutic Targets for Cancer. Pharmacol. Ther. 2019, 203, 107390. [Google Scholar] [CrossRef]
- Kovalski, J.R.; Kuzuoglu-Ozturk, D.; Ruggero, D. Protein synthesis control in cancer: Selectivity and therapeutic targeting. EMBO J. 2022, 41, e109823. [Google Scholar] [CrossRef]
- Mir, C.; Garcia-Mayea, Y.; Lleonart, M.E. Targeting the “undruggable”: RNA-binding proteins in the spotlight in cancer ther-apy. Semin. Cancer Biol. 2022, 86, 69. [Google Scholar] [CrossRef]
- Wurth, L.; Papasaikas, P.; Olmeda, D.; Bley, N.; Calvo, G.T.; Guerrero, S.; Cerezo-Wallis, D.; Martinez-Useros, J.; García-Fernández, M.; Hüttelmaier, S.; et al. UNR/CSDE1 Drives a Post-transcriptional Program to Promote Melanoma Invasion and Metastasis. Cancer Cell 2016, 30, 694–707. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.; Zhou, Y.; Zhou, N.; Wang, Z.; Chen, J.; Chen, H.; Wang, D.; Zhou, L.; Wei, K.; Zhang, H.; et al. Epigenetic modification of CSDE1 locus dictates immune recognition of nascent tumorigenic cells. Sci. Transl. Med. 2023, 15, eabq6024. [Google Scholar] [CrossRef] [PubMed]
- Guo, A.-X.; Cui, J.-J.; Wang, L.-Y.; Yin, J.-Y. The role of CSDE1 in translational reprogramming and human diseases. Cell Commun. Signal. 2020, 18, 14. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Useros, J.; Georgiev-Hristov, T.; Fernández-Aceñero, M.J.; Borrero-Palacios, A.; Indacochea, A.; Guerrero, S.; Li, W.; Cebrián, A.; Gómez Del Pulgar, T.; Puime-Otin, A.; et al. UNR/CDSE1 ex-pression as prognosis biomarker in resectable pancreatic ductal adenocarcinoma patients: A proof-of-concept. PLoS ONE 2017, 12, e0182044. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Useros, J.; Garcia-Carbonero, N.; Li, W.; Fernandez-Aceñero, M.J.; Cristobal, I.; Rincon, R.; Rodriguez-Remirez, M.; Borrero-Palacios, A.; Garcia-Foncillas, J. UNR/CSDE1 Expression Is Critical to Maintain Invasive Phenotype of Colorectal Cancer through Regulation of c-MYC and Epithelial-to-Mesenchymal Transition. J. Clin. Med. 2019, 8, 560. [Google Scholar] [CrossRef]
- Liu, H.; Li, X.; Dun, M.D.; Faulkner, S.; Jiang, C.C.; Hondermarck, H. Cold Shock Domain Containing E1 (CSDE1) Protein is Overexpressed and Can be Targeted to Inhibit Invasiveness in Pancreatic Cancer Cells. Proteomics 2020, 20, e1900331. [Google Scholar] [CrossRef]
- Xie, P.; Guo, Y. LINC00205 promotes malignancy in lung cancer by recruiting FUS and stabilizing CSDE1. Biosci. Rep. 2020, 40, BSR20190701. [Google Scholar] [CrossRef]
- Fishbein, L.; Leshchiner, I.; Walter, V.; Danilova, L.; Robertson, A.G.; Johnson, A.R.; Lichtenberg, T.M.; Murray, B.A.; Ghayee, H.K.; Else, T.; et al. Comprehensive Molecular Characterization of Pheochromocytoma and Paraganglioma. Cancer Cell 2017, 31, 181. [Google Scholar] [CrossRef]
- Avolio, R.; Inglés-Ferrándiz, M.; Ciocia, A.; Coll, O.; Bonnin, S.; Guitart, T.; Ribó, A.; Gebauer, F. Coordinated post-transcriptional control of oncogene-induced senescence by UNR/CSDE1. Cell Rep. 2022, 38, 110211. [Google Scholar] [CrossRef] [PubMed]
- Andre, F.; Conforti, R.; Moeder, C.B.; Mauguen, A.; Arnedos, M.; Berrada, N.; Delaloge, S.; Tomasic, G.; Spielmann, M.; Esteva, F.J.; et al. Association between the nuclear to cytoplasmic ratio of p27 and the efficacy of adjuvant polychemotherapy in early breast cancer. Ann. Oncol. 2012, 23, 2059–2064. [Google Scholar] [CrossRef]
- Sung, W.-W.; Lin, Y.-M.; Wu, P.-R.; Yen, H.-H.; Lai, H.-W.; Su, T.-C.; Huang, R.-H.; Wen, C.-K.; Chen, C.-Y.; Chen, C.-J.; et al. High nuclear/cytoplasmic ratio of Cdk1 expression predicts poor prognosis in colorectal cancer patients. BMC Cancer 2014, 14, 951. [Google Scholar] [CrossRef]
- Xu, J.; Zhao, Z.; Ye, L.; Zhuge, W.; Han, Z.; Zhang, T.; Ye, S.; Chen, W.; Zhu, S.; Shi, L.; et al. Prognostic significance of Daxx NCR (Nuclear/Cytoplasmic Ratio) in gastric cancer. Cancer Med. 2017, 6, 2063–2075. [Google Scholar] [CrossRef]
- Mendaza, S.; Fernández-Irigoyen, J.; Santamaría, E.; Zudaire, T.; Guarch, R.; Guerrero-Setas, D.; Vidal, A.; Santos-Salas, J.; Ma-tias-Guiu, X.; Ausín, K.; et al. Absence of nuclear p16 is a diagnostic and in-dependent prognostic biomarker in squamous cell carcinoma of the cervix. Int. J. Mol. Sci. 2020, 21, 2125. [Google Scholar] [CrossRef]
- Grammatikakis, I.; Abdelmohsen, K.; Gorospe, M. Posttranslational control of HuR function. Wiley Interdiscip. Rev. RNA 2017, 8, e1372. [Google Scholar] [CrossRef]
- Schultz, C.W.; Preet, R.; Dhir, T.; Dixon, D.A.; Brody, J.R. Understanding and targeting the disease-related RNA binding protein human antigen R (HuR). Wiley Interdiscip. Rev. RNA 2020, 11, e1581. [Google Scholar] [CrossRef]
- Wu, X.; Xu, L. The RNA-binding protein HuR in human cancer: A friend or foe? Adv. Drug Deliv. Rev. 2022, 184, 114179. [Google Scholar] [CrossRef]
- Kim, Y.; Ko, J.Y.; Lee, S.B.; Oh, S.; Park, J.W.; Kang, H.G.; Kim, D.H.; Chung, D.; Lim, S.; Kong, H.; et al. Reduced miR-371b-5p expression drives tumor progression via CSDE1/RAC1 regulation in triple-negative breast cancer. Oncogene 2022, 41, 3151. [Google Scholar] [CrossRef] [PubMed]
- Militti, C.; Maenner, S.; Becker, P.B.; Gebauer, F. UNR facilitates the interaction of MLE with the lncRNA roX2 during Drosophila dosage compensation. Nat. Commun. 2014, 5, 4762. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Guijarro, E.; Karras, P.; Cifdaloz, M.; Martínez-Herranz, R.; Cañón, E.; Graña, O.; Horcajada-Reales, C.; Alonso-Curbelo, D.; Calvo, T.G.; Gómez-López, G.; et al. Lineage-specific roles of the cytoplasmic polyadenylation factor CPEB4 in the regulation of melanoma drivers. Nat. Commun. 2016, 7, 13418. [Google Scholar] [CrossRef] [PubMed]
- Cifdaloz, M.; Osterloh, L.; Graña, O.; Riveiro-Falkenbach, E.; Ximénez-Embún, P.; Muñoz, J.; Tejedo, C.; Calvo, T.G.; Karras, P.; Olmeda, D.; et al. Systems analysis identifies melanoma-enriched pro-oncogenic networks controlled by the RNA binding protein CELF1. Nat. Commun. 2017, 8, 2249. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Zheng, H.; Chan, M.T.; Wu, W.K. NOVA1 acts as an oncogene in melanoma via regulating FOXO3a expression. J. Cell. Mol. Med. 2018, 22, 2622–2630. [Google Scholar] [CrossRef] [PubMed]
- Phung, B.; Cieśla, M.; Sanna, A.; Guzzi, N.; Beneventi, G.; Ngoc, P.C.T.; Lauss, M.; Cabrita, R.; Cordero, E.; Bosch, A.; et al. The X-linked DDX3X RNA helicase dictates translation reprogramming and metastasis in melanoma. Cell Rep. 2019, 27, 3573. [Google Scholar] [CrossRef] [PubMed]
- Karras, P.; Riveiro-Falkenbach, E.; Cañón, E.; Tejedo, C.; Calvo, T.G.; Martínez-Herranz, R.; Alonso-Curbelo, D.; Cifdaloz, M.; Perez-Guijarro, E.; Gómez-López, G.; et al. p62/SQSTM1 fuels melanoma progression by opposing mRNA decay of a selective set of pro-metastatic factors. Cancer Cell 2019, 35, 46–63.e10. [Google Scholar] [CrossRef] [PubMed]
- Hanniford, D.; Ulloa-Morales, A.; Karz, A.; Berzoti-Coelho, M.G.; Moubarak, R.S.; Sánchez-Sendra, B.; Kloetgen, A.; Davalos, V.; Imig, J.; Wu, P.; et al. Epigenetic Silencing of CDR1as Drives IGF2BP3-Mediated Melanoma Invasion and Metastasis. Cancer Cell 2020, 37, 55–70. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Zhang, M.; Tian, N.; Li, D.; Wu, F.; Hu, P.; Wang, Z.; Wang, L.; Hao, W.; Kang, J.; et al. The antibiotic clofoctol suppresses glioma stem cell proliferation by activating KLF13. J. Clin. Investig. 2019, 129, 3072–3085. [Google Scholar] [CrossRef] [PubMed]
- Law, A.M.K.; Yin, J.X.M.; Castillo, L.; Young, A.I.J.; Piggin, C.; Rogers, S.; Caldon, C.E.; Burgess, A.; Millar, E.K.A.; O’toole, S.A.; et al. Andy’s Algorithms: New automated digital image analysis pipelines for FIJI. Sci. Rep. 2017, 7, 15171. [Google Scholar] [CrossRef]
- Drete, S.; Saccani, G.J.; Dowsset, M. A “quickscore” method for immunohistochemical semiquantitation: Validation for oes-trogen receptor in breast carcinomas. J. Clin. Pathol. 1995, 48, 876. [Google Scholar] [CrossRef]

| Feature | n (%) | |
|---|---|---|
| Gender | Male | 25 (50) |
| Female | 25 (50) | |
| Age (years) | <58 | 11 (22) |
| ≥58 | 39 (78) | |
| Location | Face, scalp or neck | 9 (18) |
| Trunk | 19 (38) | |
| Upper extremities | 6 (12) | |
| Lower extremities | 13 (26) | |
| NA * | 3 (6) | |
| Histopathological subtype | Superficial spreading | 25 (50) |
| Nodular | 14 (28) | |
| Other (in situ, acral, lentigo, mixed histology and nonclassifiable) | 11 (22) | |
| Breslow | In situ | 15 (30) |
| <1 mm | 13 (26) | |
| ≥1 mm | 20 (40) | |
| NA | 2 (4) | |
| Ulceration | No | 36 (72) |
| Yes | 7 (14) | |
| NA * | 7 (14) | |
| Mitotic index | <2 mitoses/mm2 | 20 (40) |
| ≥2 mitoses/mm2 | 16 (32) | |
| NA * | 14 (28) |
| Feature | n (%) | |
|---|---|---|
| Gender | Male | 32 (65) |
| Female | 17 (35) | |
| Age (years) | <58 | 13 (27) |
| ≥58 | 36 (73) | |
| Primary type | Cutaneous | 40 (82) |
| Noncutaneous | 3 (6) | |
| Unknown primary | 6 (12) | |
| Metastasis type | Cutaneous | 16 (33) |
| Lymph node | 21 (43) | |
| Visceral | 12 (24) | |
| Molecular subtype | BRAF | 20 (41) |
| NRAS | 7 (14) | |
| WT | 9 (18) | |
| NA * | 13 (27) | |
| Stage at diagnosis | Stage I | 4 (8.2) |
| Stage II | 12 (24.5) | |
| Stage III | 22 (44.9) | |
| Stage IV | 6 (12.2) | |
| NA * | 5 (10.2) | |
| LDH at diagnosis | Normal | 11 (22.5) |
| Increased | 5 (10.2) | |
| NA * | 33 (67.4) | |
| Treatment before sample collection | Naïve | 38 (78) |
| Interferon | 8 (16) | |
| Other | 3 (6) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Indacochea, A.; Guitart, T.; Boada, A.; Peg, V.; Quer, A.; Laayouni, H.; Condal, L.; Espinosa, P.; Manzano, J.L.; Gebauer, F. CSDE1 Intracellular Distribution as a Biomarker of Melanoma Prognosis. Int. J. Mol. Sci. 2024, 25, 2319. https://doi.org/10.3390/ijms25042319
Indacochea A, Guitart T, Boada A, Peg V, Quer A, Laayouni H, Condal L, Espinosa P, Manzano JL, Gebauer F. CSDE1 Intracellular Distribution as a Biomarker of Melanoma Prognosis. International Journal of Molecular Sciences. 2024; 25(4):2319. https://doi.org/10.3390/ijms25042319
Chicago/Turabian StyleIndacochea, Alberto, Tanit Guitart, Aram Boada, Vicente Peg, Ariadna Quer, Hafid Laayouni, Laura Condal, Pablo Espinosa, Jose Luis Manzano, and Fátima Gebauer. 2024. "CSDE1 Intracellular Distribution as a Biomarker of Melanoma Prognosis" International Journal of Molecular Sciences 25, no. 4: 2319. https://doi.org/10.3390/ijms25042319
APA StyleIndacochea, A., Guitart, T., Boada, A., Peg, V., Quer, A., Laayouni, H., Condal, L., Espinosa, P., Manzano, J. L., & Gebauer, F. (2024). CSDE1 Intracellular Distribution as a Biomarker of Melanoma Prognosis. International Journal of Molecular Sciences, 25(4), 2319. https://doi.org/10.3390/ijms25042319






