Novel Enhanced Mammalian Cell Transient Expression Vector via Promoter Combination
Abstract
1. Introduction
2. Results
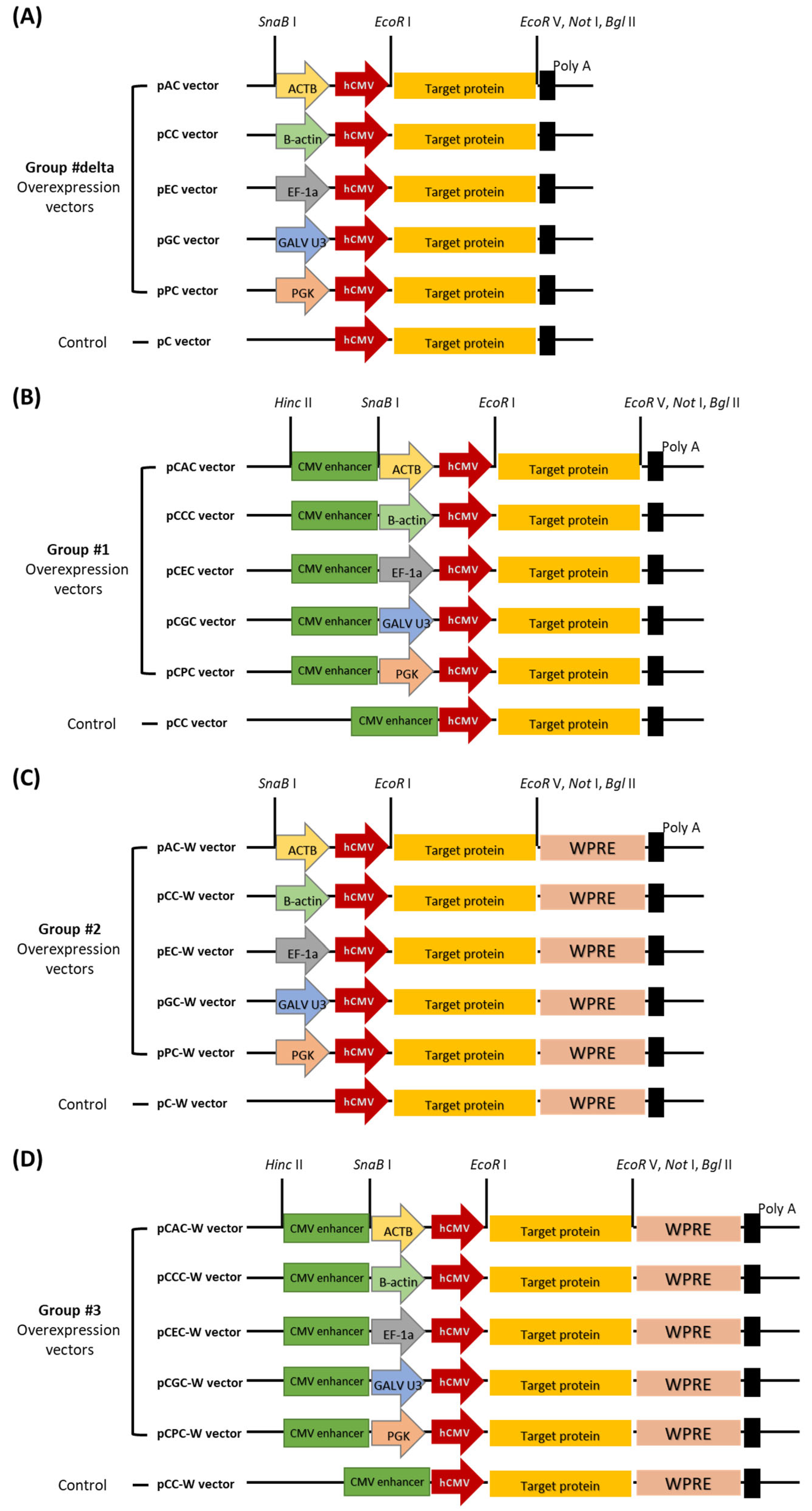
2.1. Construction of Mammalian Overexpression Vectors
2.2. Determination of the Optimal Expression Time Point
2.3. Comparative Evaluation of Overexpression Vectors through EGFP Expression
2.4. Application of Antigenic Protein Expression of Overexpression Vectors
3. Discussion
4. Materials and Methods
4.1. Cells
4.2. Plasmid Transfection
4.3. Fluorescence Observation and Intensity Measurement
4.4. Selection of Expression Elements and Establishment of a Vector Construction Plan
4.5. Construction of Expression Vectors
4.6. Western Blotting Analysis
4.7. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gifre, L.; Arís, A.; Bach, À.; Garcia-Fruitós, E. Trends in recombinant protein use in animal production. Microb. Cell Factories 2017, 16, 40. [Google Scholar] [CrossRef] [PubMed]
- Excler, J.L.; Saville, M.; Berkley, S.; Kim, J.H. Vaccine development for emerging infectious diseases. Nat. Med. 2021, 27, 591–600. [Google Scholar] [CrossRef] [PubMed]
- Ball, P. The lightning-fast quest for COVID vaccines—And what it means for other diseases. Nature 2021, 589, 16–18. [Google Scholar] [CrossRef] [PubMed]
- Balawejder, F.; Sampson, S.; Stratton, T. Industrial Strategy Council Home Page. Available online: https://industrialstrategycouncil.org/lessons-industrial-policy-development-oxfordastrazeneca-covid-19-vaccine (accessed on 12 February 2024).
- Cheng, N.; Liu, M.; Li, W.; Sun, B.; Liu, D.; Wang, G.; Shi, J.; Li, L. Protein post-translational modification in SARS-CoV-2 and host interaction. Front. Immunol. 2022, 13, 1068449. [Google Scholar] [CrossRef] [PubMed]
- Jefferis, R. Posttranslational modifications and the immunogenicity of biotherapeutics. J. Immunol. Res. 2016, 2016, 5358272. [Google Scholar] [CrossRef] [PubMed]
- Carpenter, E.P.; Beis, K.; Cameron, A.D.; Iwata, S. Overcoming the challenges of membrane protein crystallography. Curr. Opin. Struct. Biol. 2008, 18, 581–586. [Google Scholar] [CrossRef] [PubMed]
- Gubellini, F.; Verdon, G.; Karpowich, N.K.; Luff, J.D.; Boël, G.; Gauthier, N.; Handelman, S.K.; Ades, S.E.; Hunt, J.F. Physiological response to membrane protein overexpression in E. coli. Mol. Cell. Proteom. 2011, 10, M111.007930. [Google Scholar] [CrossRef]
- Klepsch, M.M.; Persson, J.O.; De Gier, J.W.L. Consequences of the overexpression of a eukaryotic membrane protein, the human KDEL receptor, in Escherichia coli. J. Mol. Biol. 2011, 407, 532–542. [Google Scholar] [CrossRef]
- Wagner, S.; Baars, L.; Ytterberg, A.J.; Klussmeier, A.; Wagner, C.S.; Nord, O.; Nygren, P.A.; van Wijk, K.J.; de Gier, J.W. Consequences of membrane protein overexpression in Escherichia coli. Mol. Cell. Proteom. 2007, 6, 1527–1550. [Google Scholar] [CrossRef]
- Hartley, J.L. Why proteins in mammalian cells. Methods Mol. Biol. 2012, 801, 1–12. [Google Scholar]
- Contreras-Gómez, A.; Sánchez-Mirón, A.; García-Camacho, F.; Molina-Grima, E.; Chisti, Y. Protein production using the baculovirus-insect cell expression system. Biotechnol. Prog. 2014, 30, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Palomares, L.A.; Srivastava, I.K.; Ramírez, O.T.; Cox, M.M.J. Glycobiotechnology of the insect cell-baculovirus expression system technology. Adv. Biochem. Eng. Biotechnol. 2021, 175, 71–92. [Google Scholar]
- Schuster, S.M. Biotechnology: Applying the genetic revolution by David P. Clark and Nanette J. Pazdernik. Biochem. Mol. Biol. Educ. 2009, 37, 262–263. [Google Scholar] [CrossRef]
- Nettleship, J.E.; Assenberg, R.; Diprose, J.M.; Rahman-Huq, N.; Owens, R.J. Recent advances in the production of proteins in insect and mammalian cells for structural biology. J. Struct. Biol. 2010, 172, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Li, G.; Ren, X.; Herrler, G. Select what you need: A comparative evaluation of the advantages and limitations of frequently used expression systems for foreign genes. J. Biotechnol. 2007, 127, 335–347. [Google Scholar] [CrossRef] [PubMed]
- Khan, K.H. Gene expression in mammalian cells and its applications. Adv. Pharm. Bull. 2013, 3, 257–263. [Google Scholar]
- Carter, J.; Zhang, J.; Dang, T.L.; Hasegawa, H.; Cheng, J.D.; Gianan, I.; O’Neill, J.W.; Wolfson, M.; Siu, S.; Qu, S.; et al. Fusion partners can increase the expression of recombinant interleukins via transient transfection in 2936E cells. Protein Sci. 2010, 19, 357–362. [Google Scholar] [CrossRef]
- Freudl, R. Signal peptides for recombinant protein secretion in bacterial expression systems. Microb. Cell Fact. 2018, 17, 52. [Google Scholar] [CrossRef]
- Li, Y. Self-cleaving fusion tags for recombinant protein production. Biotechnol. Lett. 2011, 33, 869–881. [Google Scholar] [CrossRef]
- Lee, J.H.; Gwak, W.; Bae, S.; Choi, J.; Han, B.; Woo, S. Increased productivity of the baculovirus expression vector system by combining enhancing factors. J. Asia Pac. Entomol. 2018, 21, 1079–1084. [Google Scholar] [CrossRef]
- Rao, Y.; Cai, D.; Wang, H.; Xu, Y.; Xiong, S.; Gao, L.; Xiong, M.; Wang, Z.; Chen, S.; Ma, X. Construction and application of a dual promoter system for efficient protein production and metabolic pathway enhancement in Bacillus licheniformis. J. Biotechnol. 2020, 312, 1–10. [Google Scholar] [CrossRef]
- Öztürk, S.; Ergün, B.G.; Çalık, P. Double promoter expression systems for recombinant protein production by industrial microorganisms. Appl. Microbiol. Biotechnol. 2017, 101, 7459–7475. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Guo, X.; Li, Y.M.; Wang, X.Y.; Yang, X.J.; Wang, Y.F.; Wang, T.Y. Enhanced transgene expression using cis-acting elements combined with the EF1 promoter in a mammalian expression system. Eur. J. Pharm. Sci. 2018, 123, 539–545. [Google Scholar] [CrossRef] [PubMed]
- Zufferey, R.; Donello, J.E.; Trono, D.; Hope, T.J. Woodchuck hepatitis virus posttranscriptional regulatory element enhances expression of transgenes delivered by retroviral vectors. J. Virol. 1999, 73, 2886–2892. [Google Scholar] [CrossRef] [PubMed]
- Loeb, J.E.; Cordier, W.S.; Harris, M.E.; Weitzman, M.D.; Hope, T.J. Enhanced expression of transgenes from adeno-associated virus vectors with the woodchuck hepatitis virus posttranscriptional regulatory element: Implications for gene therapy. Hum. Gene Ther. 1999, 10, 2295–2305. [Google Scholar] [CrossRef]
- Mastroyiannopoulos, N.P.; Feldman, M.L.; Uney, J.B.; Mahadevan, M.S.; Phylactou, L.A. Woodchuck post-transcriptional element induces nuclear export of myotonic dystrophy 3′ untranslated region transcripts. EMBO Rep. 2005, 6, 458–463. [Google Scholar] [CrossRef] [PubMed]
- Klein, R.; Ruttkowski, B.; Knapp, E.; Salmons, B.; Günzburg, W.H.; Hohenadl, C. WPRE-mediated enhancement of gene expression is promoter and cell line specific. Gene 2006, 372, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Kraunus, J.; Schaumann, D.H.; Meyer, J.; Modlich, U.; Fehse, B.; Brandenburg, G.; von Laer, D.; Klump, H.; Schambach, A.; Bohne, J.; et al. Self-inactivating retroviral vectors with improved RNA processing. Gene Ther. 2004, 11, 1568–1578. [Google Scholar] [CrossRef]
- Bosch, B.J.; van der Zee, R.; de Haan, C.A.; Rottier, P.J. The coronavirus spike protein is a class I virus fusion protein: Structural and functional characterization of the fusion core complex. J. Virol. 2003, 77, 8801–8811. [Google Scholar] [CrossRef]
- Huang, Y.; Yang, C.; Xu, X.F.; Xu, W.; Liu, S.W. Structural and functional properties of SARS-CoV-2 spike protein: Potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020, 41, 1141–1149. [Google Scholar] [CrossRef]
- Errasti-Murugarren, E.; Bartoccioni, P.; Palacín, M. Membrane protein stabilization strategies for structural and functional studies. Membranes 2021, 11, 155. [Google Scholar] [CrossRef] [PubMed]
- Jiang, P.; Ren, L.; Zhi, L.; Hu, X.; Xiao, R.P. Protocol for cell preparation and gene delivery in HEK293T and C2C12 cells. Star Protoc. 2021, 2, 100497. [Google Scholar] [CrossRef] [PubMed]
- Xia, W.; Bringmann, P.; McClary, J.; Jones, P.P.; Manzana, W.; Zhu, Y.; Wang, S.; Liu, Y.; Harvey, S.; Madlansacay, M.R.; et al. High levels of protein expression using different mammalian CMV promoters in several cell lines. Protein Expr. Purif. 2006, 45, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Alexopoulou, A.N.; Couchman, J.R.; Whiteford, J.R. The CMV early enhancer/chicken β actin (CAG) promoter can be used to drive transgene expression during the differentiation of murine embryonic stem cells into vascular progenitors. BMC Cell Biol. 2008, 9, 2. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xu, Z.; Tian, Z.; Zhang, X.; Xu, D.; Li, Q.; Zhang, J.; Wang, T. The EF-1α promoter maintains high-level transgene expression from episomal vectors in transfected CHO-K1 cells. J. Cell. Mol. Med. 2017, 21, 3044–3054. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Jones, K.L.; Sumer, H.; Verma, P.J. Stable transgene expression in human embryonic stem cells after simple chemical transfection. Mol. Reprod. Dev. 2009, 76, 580–586. [Google Scholar] [CrossRef] [PubMed]
- Ramezani, A.; Hawley, T.S.; Hawley, R.G. Lentiviral vectors for enhanced gene expression in human hematopoietic cells. Mol. Ther. 2000, 2, 458–469. [Google Scholar] [CrossRef] [PubMed]
- Norrman, K.; Fischer, Y.; Bonnamy, B.; Wolfhagen, S.F.; Ravassard, P.; Semb, H. Quantitative comparison of constitutive promoters in human ES cells. PLoS ONE 2010, 5, e12413. [Google Scholar] [CrossRef]
- Isomura, H.; Stinski, M.F. The human cytomegalovirus major immediate-early enhancer determines the efficiency of immediate-early gene transcription and viral replication in permissive cells at low multiplicity of infection. J. Virol. 2003, 77, 3602–3614. [Google Scholar] [CrossRef]



| Expression Elements | Origin | Size (bp) | Application |
|---|---|---|---|
| CMV promoter | Human cytomegalovirus | 204 | The most widely used promoter: very strong gene expression promoter in most cellular systems. |
| Chicken B-actin promoter | Chicken | 276 | Strong expression promoter; highly efficient in stem cells. |
| Human EF1-a promoter | Human | 1163 | Strong expression promoter; highly efficient in stem cells. |
| PGK promoter | Mouse | 500 | Mouse phosphoglycerate kinase one promoter. Medium expression promoter. |
| GALV U3 promoter | Gibbon ape leukemia virus | 322 | GaLV envelope protein: enables gene transfer to various host cell domains of GALV and helps increase the cell infection rate. |
| ACTB promoter | Human | 1505 | Housekeeping genes: promoter with a CpG island that extends the proximal transcription start site (TSS). |
| CMV Enhancer | Human cytomegalovirus | 380 | Cis-regulatory acting element (CRE); immediate early gene transcription enhancer. |
| WPRE | Woodchuck hepatitis virus | 688 | Post-transcriptional regulatory element |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yoon, S.; Park, S.; Lee, J.; Kim, B.; Gwak, W. Novel Enhanced Mammalian Cell Transient Expression Vector via Promoter Combination. Int. J. Mol. Sci. 2024, 25, 2330. https://doi.org/10.3390/ijms25042330
Yoon S, Park S, Lee J, Kim B, Gwak W. Novel Enhanced Mammalian Cell Transient Expression Vector via Promoter Combination. International Journal of Molecular Sciences. 2024; 25(4):2330. https://doi.org/10.3390/ijms25042330
Chicago/Turabian StyleYoon, SunKyung, SeJin Park, JuneWoo Lee, Byoungguk Kim, and WonSeok Gwak. 2024. "Novel Enhanced Mammalian Cell Transient Expression Vector via Promoter Combination" International Journal of Molecular Sciences 25, no. 4: 2330. https://doi.org/10.3390/ijms25042330
APA StyleYoon, S., Park, S., Lee, J., Kim, B., & Gwak, W. (2024). Novel Enhanced Mammalian Cell Transient Expression Vector via Promoter Combination. International Journal of Molecular Sciences, 25(4), 2330. https://doi.org/10.3390/ijms25042330






