Circulating microRNA Panel for Prediction of Recurrence and Survival in Early-Stage Lung Adenocarcinoma
Abstract
:1. Introduction
2. Results
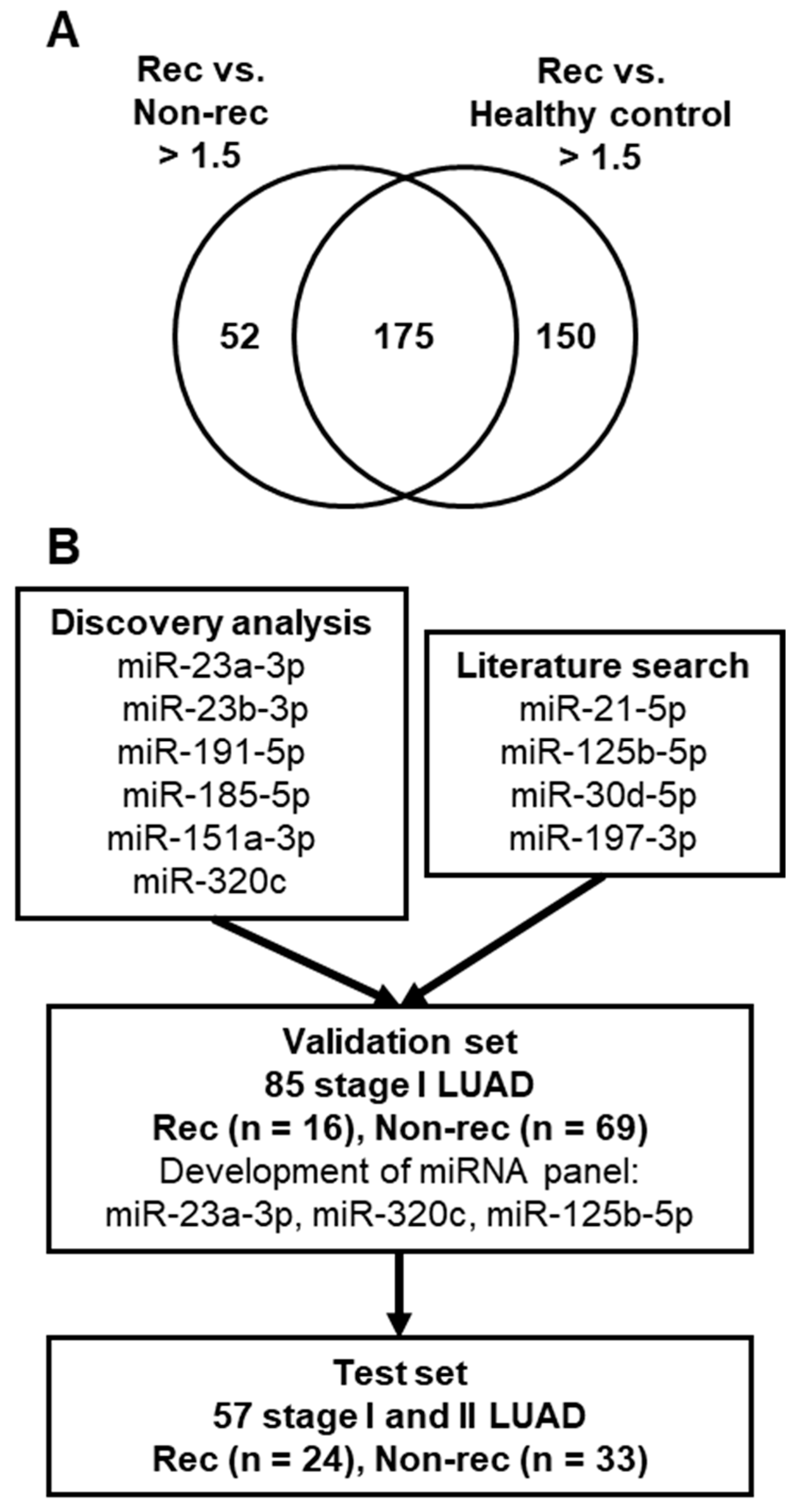
2.1. Comparison of Plasma miRNA Profiles of Stage I LUAD Patients with or without Recurrence
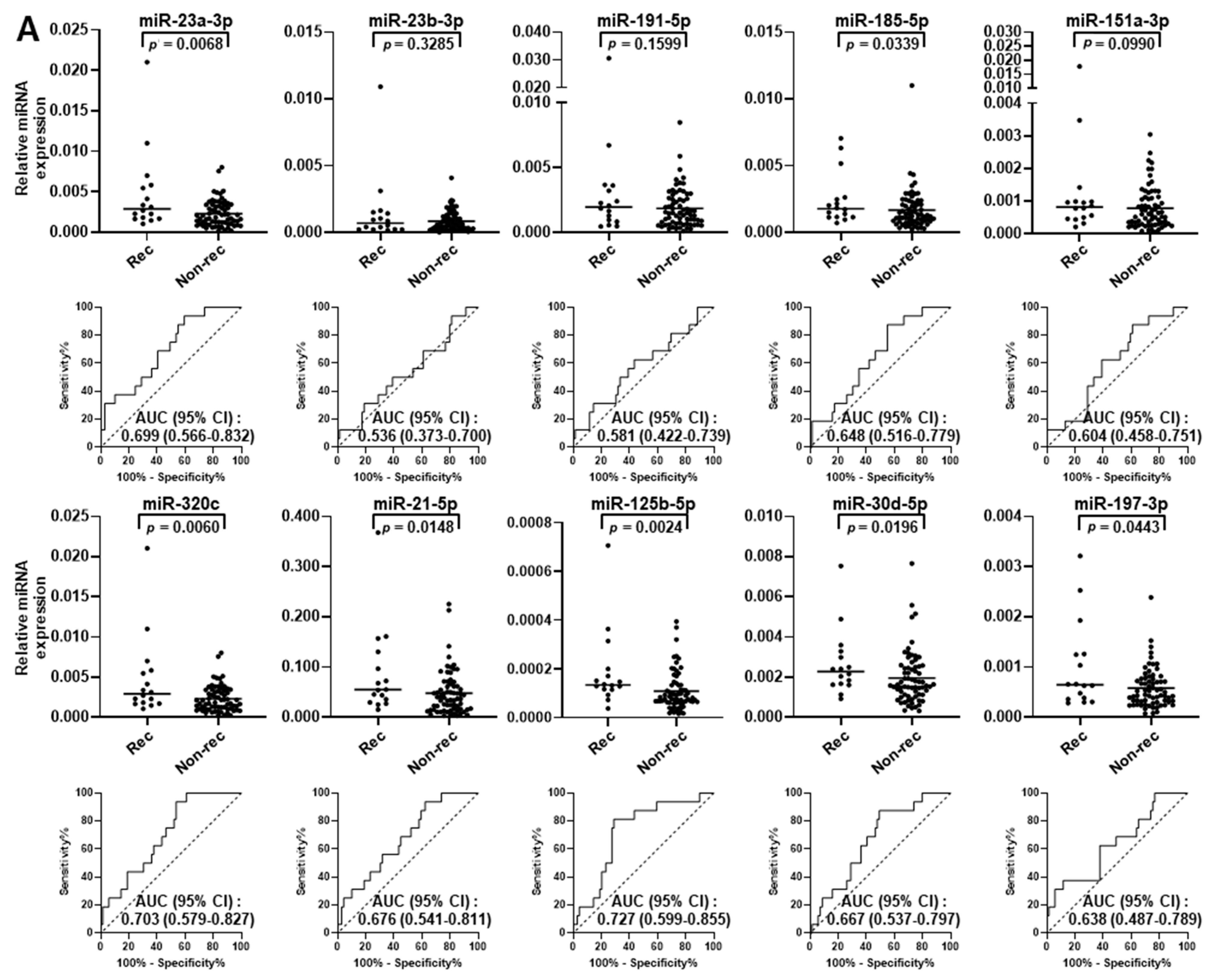
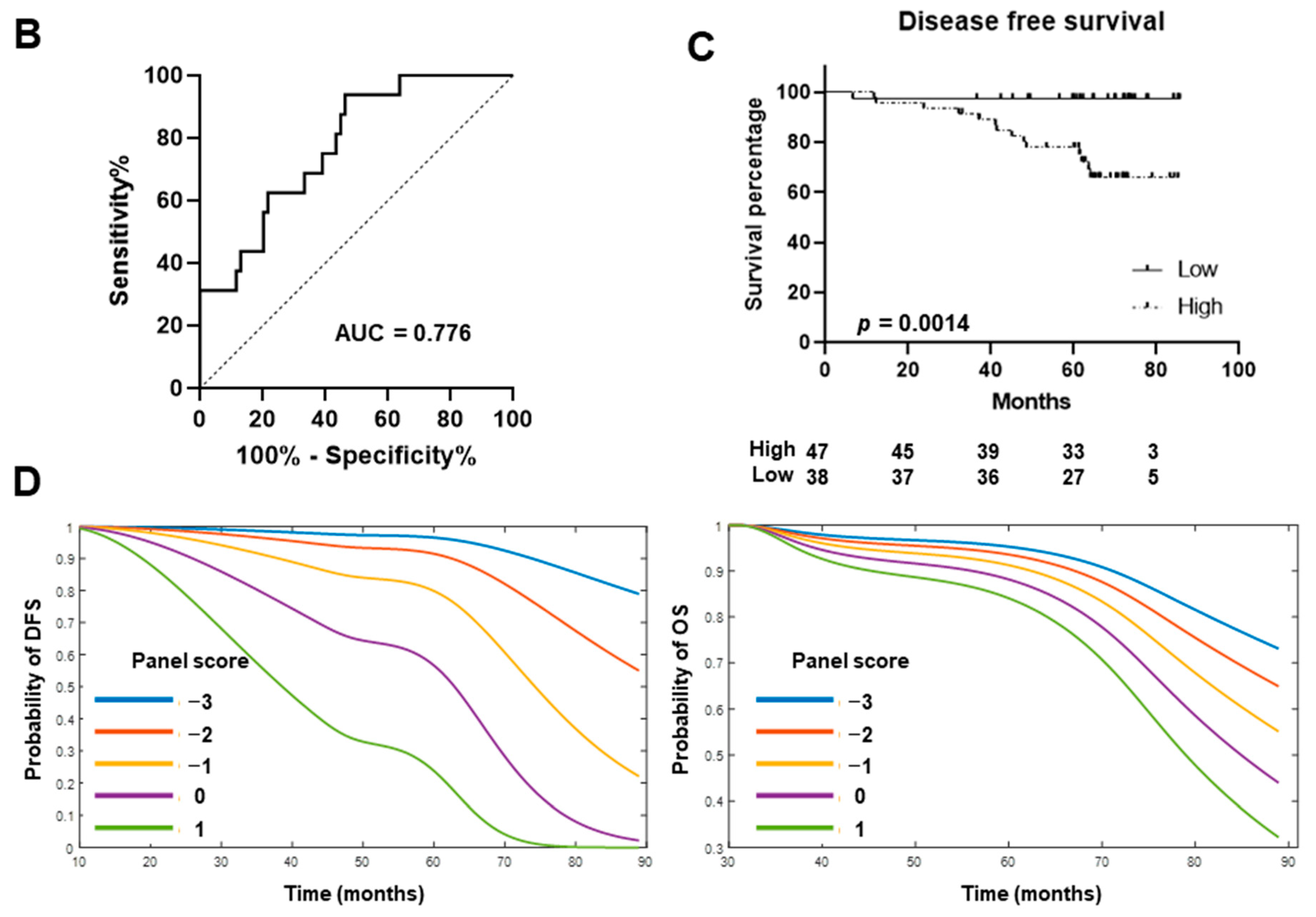
2.2. Validation of miRNA Biomarker Candidates and Development of Circulating miRNA Panel Using Plasma Samples from Stage I LUAD Patients
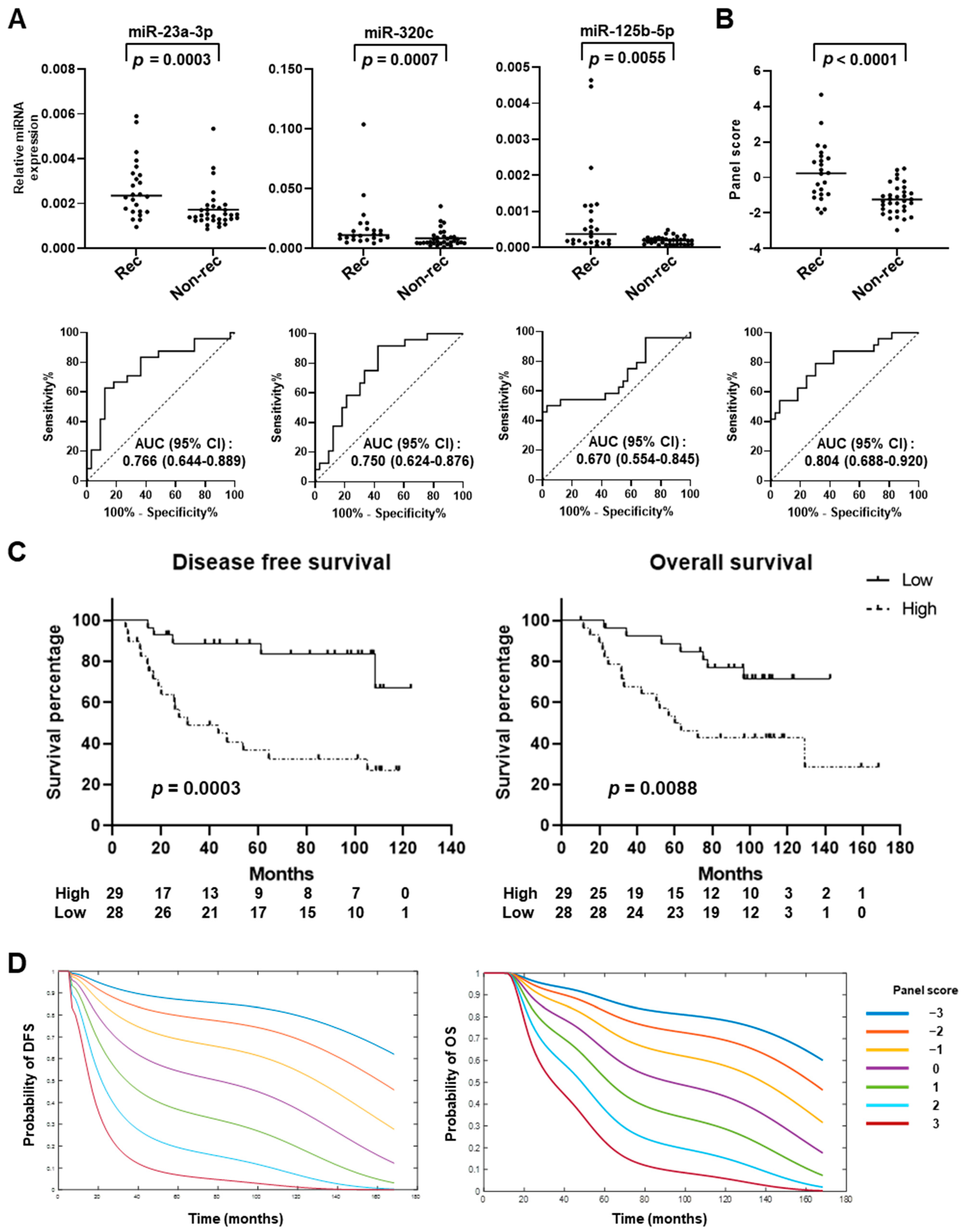
2.3. Testing of miRNA Panel in an Independent Set of Early-Stage LUAD Plasma Samples
3. Discussion
4. Materials and Methods
4.1. Human Plasma Samples
4.2. Plasma RNA Isolation
4.3. microRNA Microarray
4.4. Quantitative Real-Time PCR Analysis of LUAD Cell Lines
4.5. Quantitative Real-Time PCR Analysis of Plasma Samples
4.6. CEA ELISA
4.7. Development of miRNA Panel
4.8. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Goldstraw, P.; Chansky, K.; Crowley, J.; Rami-Porta, R.; Asamura, H.; Eberhardt, W.E.; Nicholson, A.G.; Groome, P.; Mitchell, A.; Bolejack, V.; et al. The IASLC Lung Cancer Staging Project: Proposals for Revision of the TNM Stage Groupings in the Forthcoming (Eighth) Edition of the TNM Classification for Lung Cancer. J. Thorac. Oncol. 2016, 11, 39–51. [Google Scholar] [CrossRef]
- Kris, M.G.; Gaspar, L.E.; Chaft, J.E.; Kennedy, E.B.; Azzoli, C.G.; Ellis, P.M.; Lin, S.H.; Pass, H.I.; Seth, R.; Shepherd, F.A.; et al. Adjuvant Systemic Therapy and Adjuvant Radiation Therapy for Stage I to IIIA Completely Resected Non-Small-Cell Lung Cancers: American Society of Clinical Oncology/Cancer Care Ontario Clinical Practice Guideline Update. J. Clin. Oncol. 2017, 35, 2960–2974. [Google Scholar] [CrossRef]
- Pignon, J.P.; Tribodet, H.; Scagliotti, G.V.; Douillard, J.Y.; Shepherd, F.A.; Stephens, R.J.; Dunant, A.; Torri, V.; Rosell, R.; Seymour, L.; et al. Lung adjuvant cisplatin evaluation: A pooled analysis by the LACE Collaborative Group. J. Clin. Oncol. 2008, 26, 3552–3559. [Google Scholar] [CrossRef]
- McDonald, F.; De Waele, M.; Hendriks, L.E.; Faivre-Finn, C.; Dingemans, A.C.; Van Schil, P.E. Management of stage I and II nonsmall cell lung cancer. Eur. Respir. J. 2017, 49, 1600764. [Google Scholar] [CrossRef]
- Chaft, J.E.; Rimner, A.; Weder, W.; Azzoli, C.G.; Kris, M.G.; Cascone, T. Evolution of systemic therapy for stages I-III non-metastatic non-small-cell lung cancer. Nat. Rev. Clin. Oncol. 2021, 18, 547–557. [Google Scholar] [CrossRef]
- Petrelli, F.; Barni, S. Non-cancer-related mortality after cisplatin-based adjuvant chemotherapy for non-small cell lung cancer: A study-level meta-analysis of 16 randomized trials. Med. Oncol. 2013, 30, 641. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.Y.; Zhang, C.; Wang, X.; Zhai, L.; Ma, Y.; Mao, Y.; Qian, K.; Sun, C.; Liu, Z.; Jiang, S.; et al. Integrative Proteomic Characterization of Human Lung Adenocarcinoma. Cell 2020, 182, 245–261.e17. [Google Scholar] [CrossRef]
- Tomida, S.; Takeuchi, T.; Shimada, Y.; Arima, C.; Matsuo, K.; Mitsudomi, T.; Yatabe, Y.; Takahashi, T. Relapse-related molecular signature in lung adenocarcinomas identifies patients with dismal prognosis. J. Clin. Oncol. 2009, 27, 2793–2799. [Google Scholar] [CrossRef] [PubMed]
- Beer, D.G.; Kardia, S.L.; Huang, C.C.; Giordano, T.J.; Levin, A.M.; Misek, D.E.; Lin, L.; Chen, G.; Gharib, T.G.; Thomas, D.G.; et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat. Med. 2002, 8, 816–824. [Google Scholar] [CrossRef]
- McGranahan, N.; Swanton, C. Clonal Heterogeneity and Tumor Evolution: Past, Present, and the Future. Cell 2017, 168, 613–628. [Google Scholar] [CrossRef] [PubMed]
- Pellini, B.; Chaudhuri, A.A. Circulating Tumor DNA Minimal Residual Disease Detection of Non-Small-Cell Lung Cancer Treated With Curative Intent. J. Clin. Oncol. 2022, 40, 567–575. [Google Scholar] [CrossRef] [PubMed]
- Duffy, M.J.; O’Byrne, K. Tissue and Blood Biomarkers in Lung Cancer: A Review. In Advances in Clinical Chemistry; Elsevier Ltd.: Amsterdam, The Netherlands, 2018; Volume 86, pp. 1–21. [Google Scholar] [CrossRef]
- Sexauer, D.; Gray, E.; Zaenker, P. Tumour- associated autoantibodies as prognostic cancer biomarkers- a review. Autoimmun. Rev. 2022, 21, 103041. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Gregory, R.I. MicroRNA biogenesis pathways in cancer. Nat. Rev. Cancer 2015, 15, 321–333. [Google Scholar] [CrossRef] [PubMed]
- Anfossi, S.; Babayan, A.; Pantel, K.; Calin, G.A. Clinical utility of circulating non-coding RNAs—An update. Nat. Rev. Clin. Oncol. 2018, 15, 541–563. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Chen, X.; Zhao, Y.; Tian, T.; Jin, G.; Shu, Y.; Chen, Y.; Xu, L.; Zen, K.; Zhang, C.; et al. Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J. Clin. Oncol. 2010, 28, 1721–1726. [Google Scholar] [CrossRef] [PubMed]
- Sozzi, G.; Boeri, M.; Rossi, M.; Verri, C.; Suatoni, P.; Bravi, F.; Roz, L.; Conte, D.; Grassi, M.; Sverzellati, N.; et al. Clinical utility of a plasma-based miRNA signature classifier within computed tomography lung cancer screening: A correlative MILD trial study. J. Clin. Oncol. 2014, 32, 768–773. [Google Scholar] [CrossRef]
- Cheng, T.Y.; Cramb, S.M.; Baade, P.D.; Youlden, D.R.; Nwogu, C.; Reid, M.E. The International Epidemiology of Lung Cancer: Latest Trends, Disparities, and Tumor Characteristics. J. Thorac. Oncol. 2016, 11, 1653–1671. [Google Scholar] [CrossRef]
- Pritchard, C.C.; Kroh, E.; Wood, B.; Arroyo, J.D.; Dougherty, K.J.; Miyaji, M.M.; Tait, J.F.; Tewari, M. Blood cell origin of circulating microRNAs: A cautionary note for cancer biomarker studies. Cancer Prev. Res. 2012, 5, 492–497. [Google Scholar] [CrossRef]
- Zhang, Y.; Hu, Y.; Fang, J.Y.; Xu, J. Gain-of-function miRNA signature by mutant p53 associates with poor cancer outcome. Oncotarget 2016, 7, 11056–11066. [Google Scholar] [CrossRef]
- Munoz-Culla, M.; Irizar, H.; Saenz-Cuesta, M.; Castillo-Trivino, T.; Osorio-Querejeta, I.; Sepulveda, L.; Lopez de Munain, A.; Olascoaga, J.; Otaegui, D. SncRNA (microRNA &snoRNA) opposite expression pattern found in multiple sclerosis relapse and remission is sex dependent. Sci. Rep. 2016, 6, 20126. [Google Scholar] [CrossRef]
- Schliekelman, M.J.; Taguchi, A.; Zhu, J.; Dai, X.; Rodriguez, J.; Celiktas, M.; Zhang, Q.; Chin, A.; Wong, C.H.; Wang, H.; et al. Molecular portraits of epithelial, mesenchymal, and hybrid States in lung adenocarcinoma and their relevance to survival. Cancer Res. 2015, 75, 1789–1800. [Google Scholar] [CrossRef]
- Wang, B.; Hsu, S.H.; Frankel, W.; Ghoshal, K.; Jacob, S.T. Stat3-mediated activation of microRNA-23a suppresses gluconeogenesis in hepatocellular carcinoma by down-regulating glucose-6-phosphatase and peroxisome proliferator-activated receptor gamma, coactivator 1 alpha. Hepatology 2012, 56, 186–197. [Google Scholar] [CrossRef]
- Jahid, S.; Sun, J.; Edwards, R.A.; Dizon, D.; Panarelli, N.C.; Milsom, J.W.; Sikandar, S.S.; Gumus, Z.H.; Lipkin, S.M. miR-23a promotes the transition from indolent to invasive colorectal cancer. Cancer Discov. 2012, 2, 540–553. [Google Scholar] [CrossRef]
- Hatzl, S.; Geiger, O.; Kuepper, M.K.; Caraffini, V.; Seime, T.; Furlan, T.; Nussbaumer, E.; Wieser, R.; Pichler, M.; Scheideler, M.; et al. Increased Expression of miR-23a Mediates a Loss of Expression in the RAF Kinase Inhibitor Protein RKIP. Cancer Res. 2016, 76, 3644–3654. [Google Scholar] [CrossRef]
- Fan, X.; Tao, S.; Li, Q.; Deng, B.; Tan, Q.Y.; Jin, H. The miR-23a/27a/24-2 cluster promotes postoperative progression of early-stage non-small cell lung cancer. Mol. Ther. Oncolytics 2022, 24, 205–217. [Google Scholar] [CrossRef]
- Wang, N.; Tan, H.Y.; Feng, Y.G.; Zhang, C.; Chen, F.; Feng, Y. microRNA-23a in Human Cancer: Its Roles, Mechanisms and Therapeutic Relevance. Cancers 2018, 11, 7. [Google Scholar] [CrossRef]
- Ostenfeld, M.S.; Jensen, S.G.; Jeppesen, D.K.; Christensen, L.L.; Thorsen, S.B.; Stenvang, J.; Hvam, M.L.; Thomsen, A.; Mouritzen, P.; Rasmussen, M.H.; et al. miRNA profiling of circulating EpCAM+ extracellular vesicles: Promising biomarkers of colorectal cancer. J. Extracell. Vesicles 2016, 5, 31488. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, L.; Fredsoe, J.; Kristensen, H.; Strand, S.H.; Rasmussen, A.; Hoyer, S.; Borre, M.; Mouritzen, P.; Orntoft, T.; Sorensen, K.D. Training and validation of a novel 4-miRNA ratio model (MiCaP) for prediction of postoperative outcome in prostate cancer patients. Ann. Oncol. 2018, 29, 2003–2009. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.X.; Yu, R.; Wu, X.; Wu, S.Y.; Pi, C.; Chen, Z.H.; Zhang, X.C.; Gao, C.Y.; Shao, Y.W.; Liu, L.; et al. Correlation of plasma exosomal microRNAs with the efficacy of immunotherapy in EGFR / ALK wild-type advanced non-small cell lung cancer. J Immunother. Cancer 2020, 8, e000376. [Google Scholar] [CrossRef] [PubMed]
- Pontis, F.; Roz, L.; Mensah, M.; Segale, M.; Moro, M.; Bertolini, G.; Petraroia, I.; Centonze, G.; Ferretti, A.M.; Suatoni, P.; et al. Circulating extracellular vesicles from individuals at high-risk of lung cancer induce pro-tumorigenic conversion of stromal cells through transfer of miR-126 and miR-320. J. Exp. Clin. Cancer Res. 2021, 40, 237. [Google Scholar] [CrossRef] [PubMed]
- Fortunato, O.; Borzi, C.; Milione, M.; Centonze, G.; Conte, D.; Boeri, M.; Verri, C.; Moro, M.; Facchinetti, F.; Andriani, F.; et al. Circulating mir-320a promotes immunosuppressive macrophages M2 phenotype associated with lung cancer risk. Int. J. Cancer 2019, 144, 2746–2761. [Google Scholar] [CrossRef]
- Lin, R.; Chen, L.; Chen, G.; Hu, C.; Jiang, S.; Sevilla, J.; Wan, Y.; Sampson, J.H.; Zhu, B.; Li, Q.J. Targeting miR-23a in CD8+ cytotoxic T lymphocytes prevents tumor-dependent immunosuppression. J. Clin. Investig. 2014, 124, 5352–5367. [Google Scholar] [CrossRef]
- Zhu, Y.; Zhang, S.; Li, Z.; Wang, H.; Li, Z.; Hu, Y.; Chen, H.; Zhang, X.; Cui, L.; Zhang, J.; et al. miR-125b-5p and miR-99a-5p downregulate human γδ T-cell activation and cytotoxicity. Cell. Mol. Immunol. 2019, 16, 112–125. [Google Scholar] [CrossRef] [PubMed]
- Boeri, M.; Milione, M.; Proto, C.; Signorelli, D.; Lo Russo, G.; Galeone, C.; Verri, C.; Mensah, M.; Centonze, G.; Martinetti, A.; et al. Circulating miRNAs and PD-L1 Tumor Expression Are Associated with Survival in Advanced NSCLC Patients Treated with Immunotherapy: A Prospective Study. Clin. Cancer Res. 2019, 25, 2166–2173. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Lin, J.; Ye, Y.; Oba, T.; Gentile, E.; Lian, J.; Wang, J.; Zhao, Y.; Gu, J.; Wistuba, I.I.; et al. Serum MicroRNA-150 Predicts Prognosis for Early-Stage Non-Small Cell Lung Cancer and Promotes Tumor Cell Proliferation by Targeting Tumor Suppressor Gene SRCIN1. Clin. Pharmacol. Ther. 2018, 103, 1061–1073. [Google Scholar] [CrossRef]
- Silva, J.; Garcia, V.; Zaballos, A.; Provencio, M.; Lombardia, L.; Almonacid, L.; Garcia, J.M.; Dominguez, G.; Pena, C.; Diaz, R.; et al. Vesicle-related microRNAs in plasma of nonsmall cell lung cancer patients and correlation with survival. Eur. Respir. J. 2011, 37, 617–623. [Google Scholar] [CrossRef] [PubMed]
- Andreu, Z.; Rivas, E.; Sanguino-Pascual, A.; Lamana, A.; Marazuela, M.; Gonzalez-Alvaro, I.; Sanchez-Madrid, F.; de la Fuente, H.; Yanez-Mo, M. Comparative analysis of EV isolation procedures for miRNAs detection in serum samples. J. Extracell. Vesicles 2016, 5, 31655. [Google Scholar] [CrossRef] [PubMed]
- Katayama, H.; Kobayashi, M.; Irajizad, E.; Sevillarno, A.; Patel, N.; Mao, X.; Rusling, L.; Vykoukal, J.; Cai, Y.; Hsiao, F.; et al. Protein citrullination as a source of cancer neoantigens. J. Immunother. Cancer 2021, 9, e002549. [Google Scholar] [CrossRef]
- Bantis, L.E.; Tsimikas, J.V.; Georgiou, S.D. Survival estimation through the cumulative hazard function with monotone natural cubic splines. Lifetime Data Anal. 2012, 18, 364–396. [Google Scholar] [CrossRef]
- Bantis, L.E.; Tsimikas, J.V.; Georgiou, S.D. Survival estimation through the cumulative hazard with monotone natural cubic splines using convex optimization-the HCNS approach. Comput. Methods Programs Biomed. 2020, 190, 105357. [Google Scholar] [CrossRef]

| miRNA | Ratio of Rec vs. Non Rec | Ratio of Rec vs. Healthy Control | Ratio of H1299 vs. PBMC | Average in 34 LUAD Cell Lines (Log2 Intensity) | Average Ct Values in 10 LUAD Cell Lines |
|---|---|---|---|---|---|
| miR-23a-3p | 3.48 | 5.86 | 2.11 | 5.60 | - |
| miR-23b-3p | 1.67 | 3.36 | 1.88 | 5.31 | - |
| miR-191-5p | 1.60 | 5.26 | 1.87 | 1.72 | - |
| miR-185-5p | 1.76 | 15.90 | 1.58 | 1.01 | - |
| miR-151a-3p | 2.13 | 2.70 | 3.45 | 0.98 | - |
| miR-130b-3p | 1.70 | 2.05 | 3.16 | 2.06 | - |
| miR-23a-5p | 1.98 | 1.55 | 1.51 | −5.81 | - |
| miR-361-5p | 3.63 | 4.23 | 1.89 | −6.01 | - |
| miR-320c | 2.56 | 1.62 | 1.60 | NA | 26.9 |
| miR-4532 | 2.84 | 51.08 | 1.61 | NA | >35 |
| miR-6511a-3p | 1.83 | 1.88 | 1.55 | NA | >35 |
| miR-4745-5p | 2.39 | 3.41 | 1.93 | NA | Not detected |
| miR-4459 | 8.29 | 12.61 | 1.80 | NA | Not detected |
| miR-3935 | 2.44 | 1.73 | 2.12 | NA | Not detected |
| miR-574-5p | 14.70 | 37.16 | 1.59 | NA | Not available |
| Characteristics | Total | Recurrence | Non-Recurrence | p Value |
|---|---|---|---|---|
| No. (%) | No. (%) | |||
| Validation set | ||||
| Total | 85 | 16 (18.8) | 69 (81.2) | |
| Gender | ||||
| Male | 43 | 8 (18.6) | 35 (81.4) | >0.9999 |
| Female | 42 | 8 (19.0) | 34 (81.0) | |
| Age | ||||
| Range, years (mean ± SD) | 49–77 (68.9 ± 7.9) | 47–84 (68.5 ± 6.7) | 0.8258 | |
| >65 years | 63 | 13 (20.6) | 50 (79.4) | 0.5458 |
| <65 years | 22 | 3 (13.6) | 19 (86.4) | |
| Smoking status | ||||
| Smoker (current, former) | 42 | 7 (16.7) | 35 (83.3) | 0.7825 |
| Non-smoker | 43 | 9 (20.9) | 34 (79.1) | |
| CEA | ||||
| Mean ± SD, ng/mL | 2.78 ± 3.88 | 2.75 ± 3.68 | 0.9753 | |
| Test set | ||||
| Total | 57 | 24 (42.1) | 33 (57.9) | |
| Gender | ||||
| Male | 25 | 11 (44.0) | 14 (56.0) | >0.9999 |
| Female | 32 | 13 (40.6) | 19 (59.4) | |
| Age | ||||
| Range, years (mean ± SD) | 32–79 (64.5 ± 10.5) | 50–87 (66.9 ± 9.9) | 0.3910 | |
| >65 years | 31 | 15 (48.4) | 16 (51.6) | 0.4197 |
| <65 years | 26 | 9 (34.6) | 17 (65.4) | |
| Smoking status | ||||
| Smoker (current, former) | 47 | 22 (46.8) | 25 (53.2) | 0.1661 |
| Non-smoker | 10 | 2 (20.0) | 8 (80.0) | |
| Stage (AJCC 7th Edition) | ||||
| I | 36 | 10 (27.8) | 26 (72.2) | 0.0058 |
| II | 21 | 14 (66.7) | 7 (33.3) | |
| CEA | ||||
| Mean ± SD, ng/mL | 1.93 ± 2.03 | 1.40 ± 2.01 | 0.3313 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tai, M.-C.; Bantis, L.E.; Parhy, G.; Kato, T.; Tanaka, I.; Chow, C.-W.; Fujimoto, J.; Behrens, C.; Hase, T.; Kawaguchi, K.; et al. Circulating microRNA Panel for Prediction of Recurrence and Survival in Early-Stage Lung Adenocarcinoma. Int. J. Mol. Sci. 2024, 25, 2331. https://doi.org/10.3390/ijms25042331
Tai M-C, Bantis LE, Parhy G, Kato T, Tanaka I, Chow C-W, Fujimoto J, Behrens C, Hase T, Kawaguchi K, et al. Circulating microRNA Panel for Prediction of Recurrence and Survival in Early-Stage Lung Adenocarcinoma. International Journal of Molecular Sciences. 2024; 25(4):2331. https://doi.org/10.3390/ijms25042331
Chicago/Turabian StyleTai, Mei-Chee, Leonidas E. Bantis, Gargy Parhy, Taketo Kato, Ichidai Tanaka, Chi-Wan Chow, Junya Fujimoto, Carmen Behrens, Tetsunari Hase, Koji Kawaguchi, and et al. 2024. "Circulating microRNA Panel for Prediction of Recurrence and Survival in Early-Stage Lung Adenocarcinoma" International Journal of Molecular Sciences 25, no. 4: 2331. https://doi.org/10.3390/ijms25042331
APA StyleTai, M.-C., Bantis, L. E., Parhy, G., Kato, T., Tanaka, I., Chow, C.-W., Fujimoto, J., Behrens, C., Hase, T., Kawaguchi, K., Fahrmann, J. F., Ostrin, E. J., Yokoi, K., Chen-Yoshikawa, T. F., Hasegawa, Y., Hanash, S. M., Wistuba, I. I., & Taguchi, A. (2024). Circulating microRNA Panel for Prediction of Recurrence and Survival in Early-Stage Lung Adenocarcinoma. International Journal of Molecular Sciences, 25(4), 2331. https://doi.org/10.3390/ijms25042331








