The Effect of 8,5′-Cyclo 2′-deoxyadenosine on the Activity of 10-23 DNAzyme: Experimental and Theoretical Study
Abstract
:1. Introduction
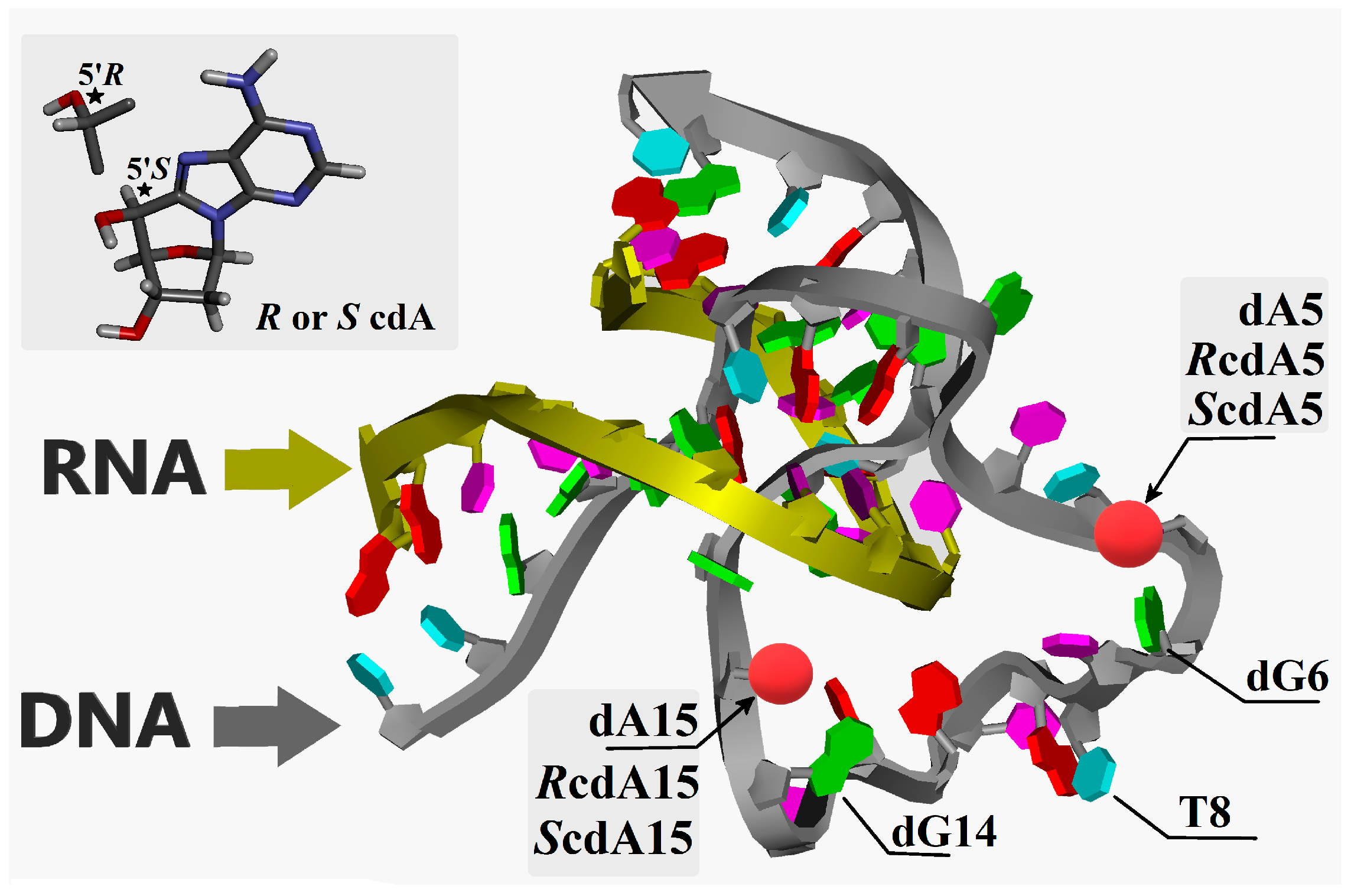
2. Results
2.1. 5′,8-Cyclo-2′deoxyadenosine (cdA) Doesn’t Alter the Stability of DNAzymes
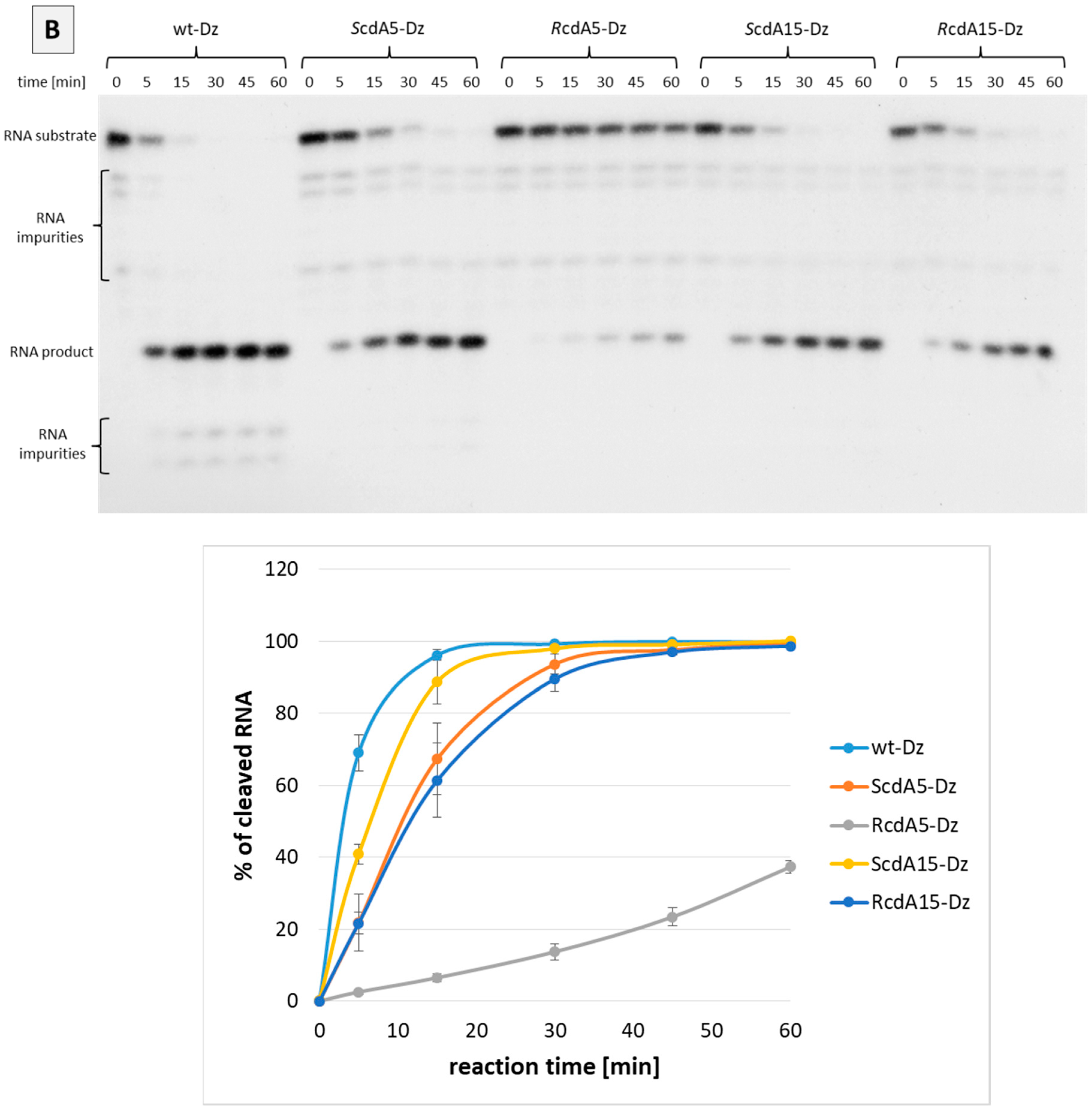
2.2. Effect of cdA on the Catalytic Activity of Dz10-23 in the Absence of Divalent Cations
2.3. Effect of cdA on Catalytic Activity of Dz10-23 in the Presence of Divalent Cations
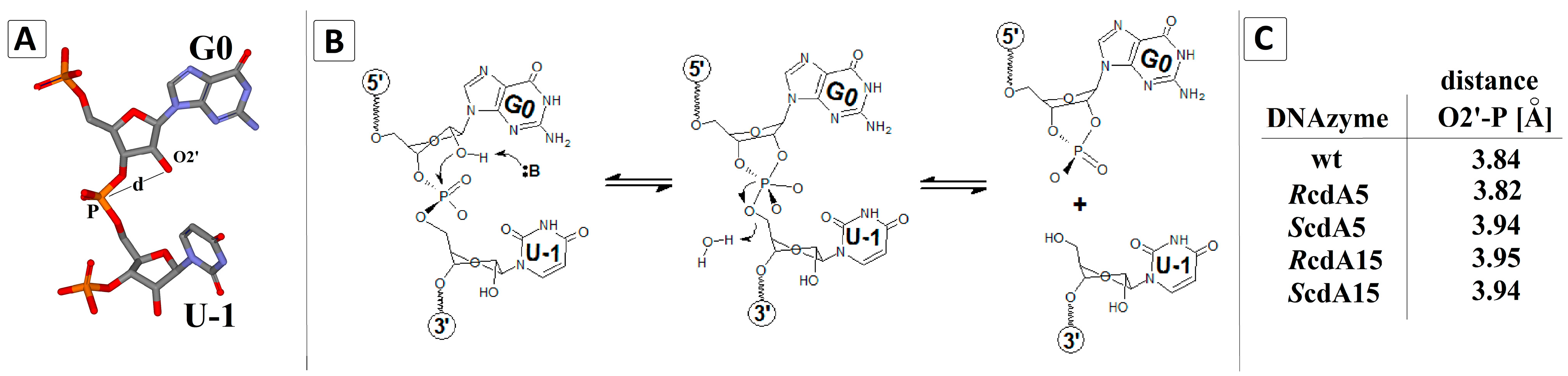
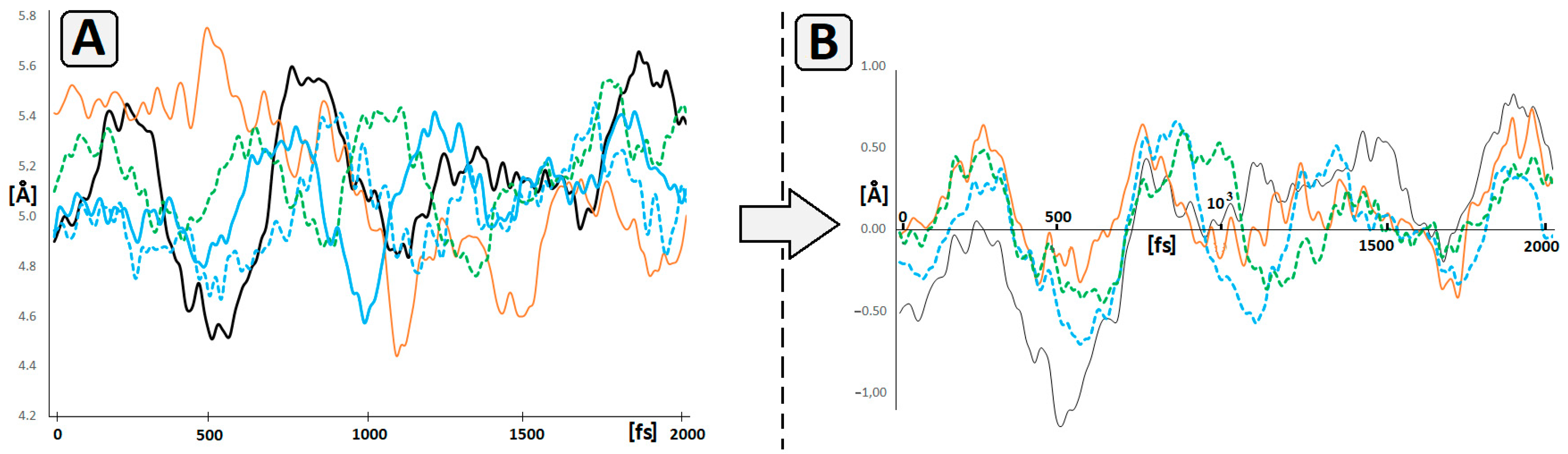
2.4. Theoretical Studies
3. Discussion
4. Materials and Methods
4.1. Synthesis of Oligonucleotides
- 5′ GAGTCCCATA GGCTAGCTACAACGA AAGACTTGAG 3′; wt-Dz (unmodified).
- 5′ GAGTCCCATA GGCT R-cdA GCTACAACGA AAGACTTGAG 3′; RcdA5-Dz.
- 5′ GAGTCCCATA GGCT S-cdA GCTACAACGA AAGACTTGAG 3′; ScdA5-Dz.
- 5′ GAGTCCCATA GGCTAGCTACAACG R-cdA AAGACTTGAG 3′; RcdA15-Dz.
- 5′ GAGTCCCATA GGCTAGCTACAACG S-cdA AAGACTTGAG 3′; ScdA15-Dz.
- 5′ *CUC AAG UCU UGU AUG GGA CUC 3′; RNA substrate.
4.2. RNA Phosphorylation with γ(32P)-ATP
4.3. Phosphorylation of the 5′ End of Dz10-23
4.4. Cleavage Assay
4.5. Polyacrylamide Gel Electrophoresis
4.6. Densitometry
4.7. Geometry Optimisation
4.8. Molecular Dynamics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Breaker, R.R.; Joyce, G.F. A DNA enzyme that cleaves RNA. Chem. Biol. 1994, 1, 223–299. [Google Scholar] [CrossRef]
- Chinnapen, D.J.; Sen, D. A deoxyribozyme that harnesses light to repair thymine dimers in DNA. Proc. Natl. Acad. Sci. USA 2004, 101, 65–69. [Google Scholar] [CrossRef] [PubMed]
- Cuenoud, B.; Szostak, J.W. A DNA metalloenzyme with DNA ligase activity. Nature 1995, 375, 611–614. [Google Scholar] [CrossRef] [PubMed]
- Hollenstein, M. DNA Catalysis: The Chemical Repertoire of DNAzymes. Molecules 2015, 20, 20777–20804. [Google Scholar] [CrossRef] [PubMed]
- Santoro, S.W.; Joyce, G.F. A general purpose RNA-cleaving DNA enzyme. Proc. Natl. Acad. Sci. USA 1997, 94, 4262–4266. [Google Scholar] [CrossRef] [PubMed]
- Cramer, E.R.; Starcovic, S.A.; Avey, R.M.; Kaya, A.I.; Robart, A.R. Structure of a 10–23 deoxyribozyme exhibiting a homodimer conformation. Commun. Chem. 2023, 6, 119. [Google Scholar] [CrossRef] [PubMed]
- Borggräfe, J.; Gertzen, C.G.W.; Viegas, A.; Gohlke, H.; Etzkorn, M. The architecture of the 10–23 DNAzyme and its implications for DNA-mediated catalysis. FEBS J. 2023, 290, 2011–2021. [Google Scholar] [CrossRef] [PubMed]
- Jakobsen, M.R.; Haasnoot, J.; Wengel, J.; Berkhout, B.; Kjems, J. Efficient inhibition of HIV-1 expression by LNA modified antisense oligonucleotides and DNAzymes targeted to functionally selected binding sites. Retrovirology 2007, 4, 29. [Google Scholar] [CrossRef] [PubMed]
- Fokina, A.A.; Meschaninova, M.I.; Durfort, T.; Venyaminova, A.G.; François, J.C. Targeting insulin-like growth factor I with 10–23 DNAzymes: 2′-O-methyl modifications in the catalytic core enhance mRNA cleavage. Biochemistry 2012, 51, 2181–2191. [Google Scholar] [CrossRef]
- Cieslak, M.; Niewiarowska, J.; Nawrot, M.; Koziolkiewicz, M.; Stec, W.J.; Cierniewski, C.S. DNAzymes to beta 1 and beta 3 mRNA down-regulate expression of the targeted integrins and inhibit endothelial cell capillary tube formation in fibrin and matrigel. J. Biol. Chem. 2002, 277, 6779–6787. [Google Scholar] [CrossRef]
- Zhong, X.; Yang, S.; Yang, P.; Du, H.; Hou, X.; Chen, J.; Zhou, R. Designing DNAzyme-Powered Nanomachines Simultaneously Responsive to Multiple MicroRNAs. Chemistry 2018, 24, 19024–19031. [Google Scholar] [CrossRef]
- Keck, K. Bildung von Cyclonucleotiden bei Bestrahlung wäßriger Lösungen von Purinnucleotiden. Z. Naturforsch. B Anorg. Chem. Org. Chem. Biochem. Biophys. Biol. 1968, 23, 1034–1043. [Google Scholar] [CrossRef]
- Dizdaroglu, M. Free-radical-induced formation of an 8,5′-cyclo-2′-deoxyguanosine moiety in deoxyribonucleic acid. Biochem. J. 1986, 238, 247–254. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Das, R.S.; Basu, A.K.; Stone, M.P. Structure of (5′S)-8,5′-cyclo-2′-deoxyguanosine in DNA. J. Am. Chem. Soc. 2011, 133, 20357–20368. [Google Scholar] [CrossRef] [PubMed]
- Zaliznyak, T.; Lukin, M.; de los Santos, C. Structure and stability of duplex DNA containing (5′S)-5′,8-cyclo-2′-deoxyadenosine: An oxidatively generated lesion repaired by NER. Chem. Res. Toxicol. 2012, 25, 2103–2111. [Google Scholar] [CrossRef] [PubMed]
- Dizdaroglu, M.; Coskun, E.; Jaruga, P. Measurement of oxidatively induced DNA damage and its repair, by mass spectrometric techniques. Free Radic. Res. 2015, 49, 525–548. [Google Scholar] [CrossRef]
- Kuraoka, I.; Bender, C.; Romieu, A.; Cadet, J.; Wood, R.D.; Lindahl, T. Removal of oxygen free-radical-induced 5′,8-purine cyclodeoxynucleosides from DNA by the nucleotide excision-repair pathway in human cells. Proc. Natl. Acad. Sci. USA 2000, 97, 3832–3837. [Google Scholar] [CrossRef] [PubMed]
- Sarmini, L.; Meabed, M.; Emmanouil, E.; Atsaves, G.; Robeska, E.; Karwowski, B.T.; Campalans, A.; Gimisis, T.; Khobta, A. Requirement of transcription-coupled nucleotide excision repair for the removal of a specific type of oxidatively induced DNA damage. Nucleic Acids Res. 2023, 51, 4982–4994. [Google Scholar] [CrossRef]
- Rosenbach, H.; Victor, J.; Etzkorn, M.; Steger, G.; Riesner, D.; Span, I. Molecular Features and Metal Ions That Influence 10–23 DNAzyme Activity. Molecules 2020, 25, 3100. [Google Scholar] [CrossRef]
- Santoro, S.W.; Joyce, G.F. Mechanism and utility of an RNA-cleaving DNA enzyme. Biochemistry 1998, 37, 13330–13342. [Google Scholar] [CrossRef]
- Karwowski, B.T. The role of (5′R) and (5′S) 5′,8-cyclo-2′-deoxyadenosine in ds-DNA structure: A comparative QM/MM theoretical study. Comput. Theor. Chem. 2013, 1010, 38–44. [Google Scholar] [CrossRef]
- Zaborowska, Z.; Fürste, J.P.; Erdmann, V.A.; Kurreck, J. Sequence requirements in the catalytic core of the “10–23” DNA enzyme. J. Biol. Chem. 2002, 277, 40617–40622. [Google Scholar] [CrossRef]
- Nawrot, B.; Widera, K.; Wojcik, M.; Rebowska, B.; Nowak, G.; Stec, W.J. Mapping of the functional phosphate groups in the catalytic core of deoxyribozyme 10–23. FEBS J. 2007, 274, 1062–1072. [Google Scholar] [CrossRef] [PubMed]
- Borggräfe, J.; Victor, J.; Rosenbach, H.; Viegas, A.; Gertzen, C.G.W.; Wuebben, C.; Kovacs, H.; Gopalswamy, M.; Riesner, D.; Steger, G.; et al. Time-resolved structural analysis of an RNA-cleaving DNA catalyst. Nature 2022, 601, 144–149. [Google Scholar] [CrossRef] [PubMed]
- Zaborowska, Z.; Schubert, S.; Kurreck, J.; Erdmann, V.A. Deletion analysis in the catalytic region of the 10–23 DNA enzyme. FEBS Lett. 2005, 579, 554–558. [Google Scholar] [CrossRef] [PubMed]
- Karwowski, B.T.; Gaillard, J.; Grand, A.; Cadet, J. Effect of (5′S)-5′,8-cyclo-2′-deoxyadenosine on the conformation of di and trinucleotides. A NMR and DFT study. Org. Biomol. Chem. 2008, 6, 3408–3413. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Cao, L.; Chiuman, W.; Li, Y.; Xi, Z. Probing the function of nucleotides in the catalytic cores of the 8–17 and 10–23 DNAzymes by abasic nucleotide and C3 spacer substitutions. Biochemistry 2010, 49, 7553–7562. [Google Scholar] [CrossRef]
- Karwowski, B.T. (5′R)-5′,8-Cyclo-2′-deoxyadenosine: NMR and DFT study of its influence on TPOcdA structure. Tetrahedron Asymmetry 2008, 19, 2390–2395. [Google Scholar] [CrossRef]
- Altona, C.; Sundaralingam, M. Conformational analysis of the sugar ring in nucleosides and nucleotides. A new description using the concept of pseudorotation. J. Am. Chem. Soc. 1972, 94, 8205–8212. [Google Scholar] [CrossRef] [PubMed]
- Karwowski, B.T.; Bellon, S.; O’Neill, P.; Lomax, M.E.; Cadet, J. Effects of (5′S)-5′,8-cyclo-2′-deoxyadenosine on the base excision repair of oxidatively generated clustered DNA damage. A biochemical and theoretical study. Org. Biomol. Chem. 2014, 12, 8671–8682. [Google Scholar] [CrossRef]
- Romieu, A.; Gasparutto, D.; Cadet, J. Synthesis and characterization of oligonucleotides containing 5′,8-cyclopurine 2′-deoxyribonucleosides: (5′R)-5′,8-cyclo-2′-deoxyadenosine, (5′S)-5′,8-cyclo-2′-deoxyguanosine, and (5′R)-5′,8-cyclo-2′-deoxyguanosine. Chem. Res. Toxicol. 1999, 12, 412–421. [Google Scholar] [CrossRef]
- de M Seabra, G.; Walker, R.C.; Elstner, M.; Case, D.A.; Roitberg, A.E. Implementation of the SCC-DFTB method for hybrid QM/MM simulations within the amber molecular dynamics package. J. Phys. Chem. A 2007, 111, 5655–5664. [Google Scholar] [CrossRef]
- Elstner, M.; Porezag, D.; Jungnickel, G.; Elsner, J.; Haugk, M.; Frauenheim, T.; Suhai, S.; Seifert, G. Self-consistent-charge density-functional tight-binding method for simulations of complex materials properties. Phys. Rev. B 1998, 58, 7260. [Google Scholar] [CrossRef]
- Gaus, M.; Goez, A.; Elstner, M. Parametrization and Benchmark of DFTB3 for Organic Molecules. J. Chem. Theory Comput. 2013, 9, 338–354. [Google Scholar] [CrossRef]
- Marenich, A.V.; Cramer, C.J.; Truhlar, D.G. Generalized Born Solvation Model SM12. J. Chem. Theory Comput. 2013, 9, 609–620. [Google Scholar] [CrossRef] [PubMed]
- Yong, X.; Zhang, L.T. Thermostats and thermostat strategies for molecular dynamics simulations of nanofluidics. J. Chem. Phys. 2013, 138, 084503. [Google Scholar] [CrossRef] [PubMed]
- te Velde, G.; Bickelhaupt, F.M.; Baerends, E.J.; Fonseca Guerra, C.; van Gisbergen, S.J.A.; Snijders, J.G.; Ziegler, T. Chemistry with ADF. J. Comput. Chem. 2001, 22, 931–967. [Google Scholar] [CrossRef]




| Dz10-23 | Spatial Geometry Comparison RMSD [Å2] | |
|---|---|---|
| DNA Part | RNA Part | |
| RcdA5 vs. wt | 0.238 | 0.261 |
| ScdA5 vs. wt | 0.796 | 0.533 |
| RcdA15 vs. wt | 0.818 | 0.525 |
| ScdA15 vs. wt | 0.833 | 0.476 |
| Dz10-23 | Distance (d) [Å] |  | |
| 5′P-dA5-3′P | 5′P-dA15-3′P | ||
| wt | 4.90 | 5.91 | |
| RcdA5 | 5.41 | 5.91 | |
| RcdA15 | 4.94 | 7.14 | |
| ScdA5 | 5.10 | 5.96 | |
| ScdA15 | 4.92 | 6.29 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cieślak, M.; Karwowski, B.T. The Effect of 8,5′-Cyclo 2′-deoxyadenosine on the Activity of 10-23 DNAzyme: Experimental and Theoretical Study. Int. J. Mol. Sci. 2024, 25, 2519. https://doi.org/10.3390/ijms25052519
Cieślak M, Karwowski BT. The Effect of 8,5′-Cyclo 2′-deoxyadenosine on the Activity of 10-23 DNAzyme: Experimental and Theoretical Study. International Journal of Molecular Sciences. 2024; 25(5):2519. https://doi.org/10.3390/ijms25052519
Chicago/Turabian StyleCieślak, Marcin, and Bolesław T. Karwowski. 2024. "The Effect of 8,5′-Cyclo 2′-deoxyadenosine on the Activity of 10-23 DNAzyme: Experimental and Theoretical Study" International Journal of Molecular Sciences 25, no. 5: 2519. https://doi.org/10.3390/ijms25052519





