Characterization of MYBL1 Gene in Triple-Negative Breast Cancers and the Genes’ Relationship to Alterations Identified at the Chromosome 8q Loci
Abstract
1. Introduction
2. Results
2.1. Analyses of MYBL1 in TNBC
2.2. Analyses of the Transcript Variants Deposited at NCBI
2.3. Analyses of the Protein Isoforms and Characterization of Sequences Demonstrating Differential Expression in MDA-M-231 Cells
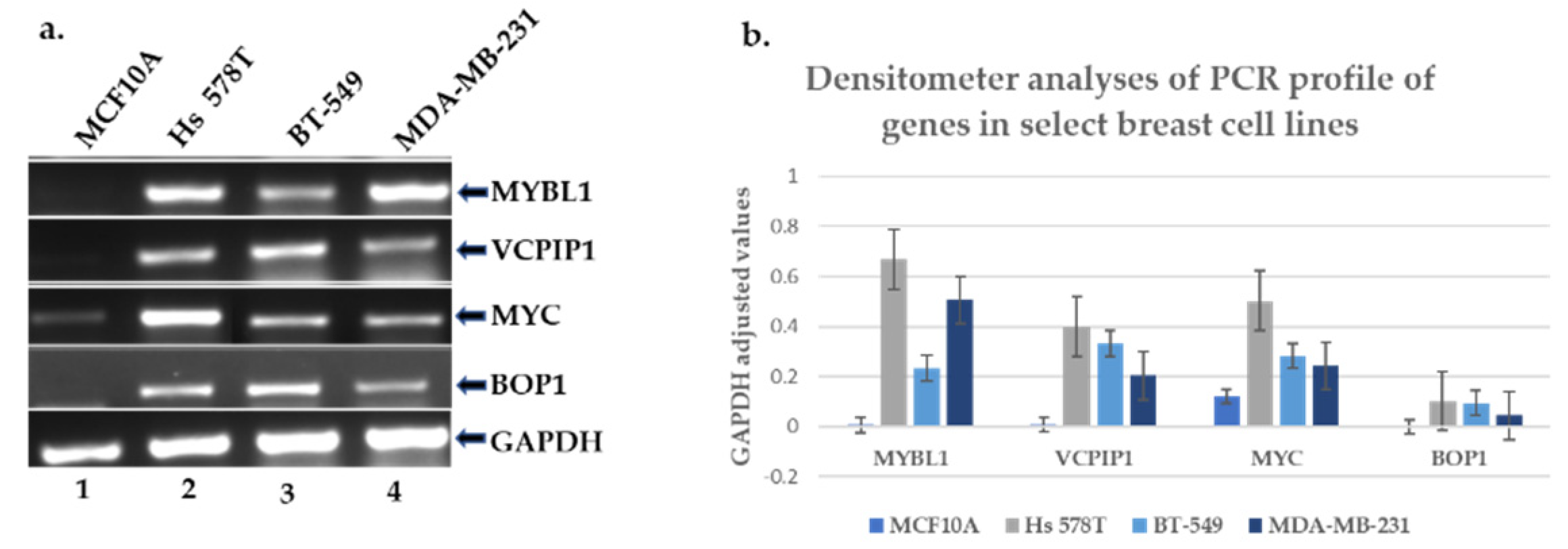
2.4. Experimental Validation of the Transcript and Protein Expression Levels of MYBL1, VCPIP1, MYC and BOP1 Genes Located at Chromosome 8q loci
| GENE | FUNCTION |
|---|---|
| MYBL1 | Proto-oncogene, a transcription factor regulating cell proliferation [24] |
| VCPIP1 | Protein deubiquitination; Golgi mitosis |
| repair; regulates protein localization to chromatin [25] | |
| MYC | Proto-oncogene, transcription factor, cell cycle apoptosis [26,27] |
| BOP1 | Regulation of cell cycle, p53 signal transduction class mediator [28,29] |
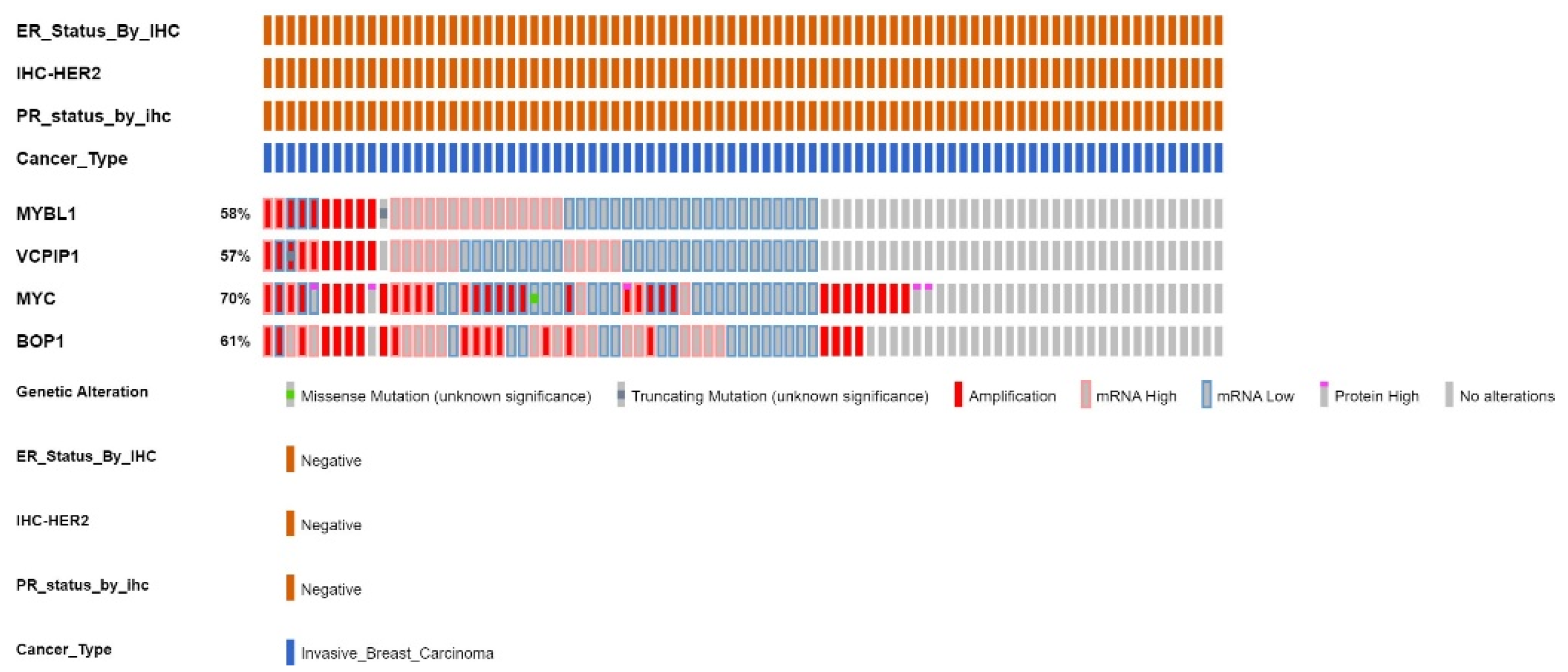
2.5. Analyses of the The-Cancer-Genome-Atlas Patient Sample Dataset for Gene Expression and Genetic Alterations of MYBL1, VCPIP1, MYC and BOP1 Genes

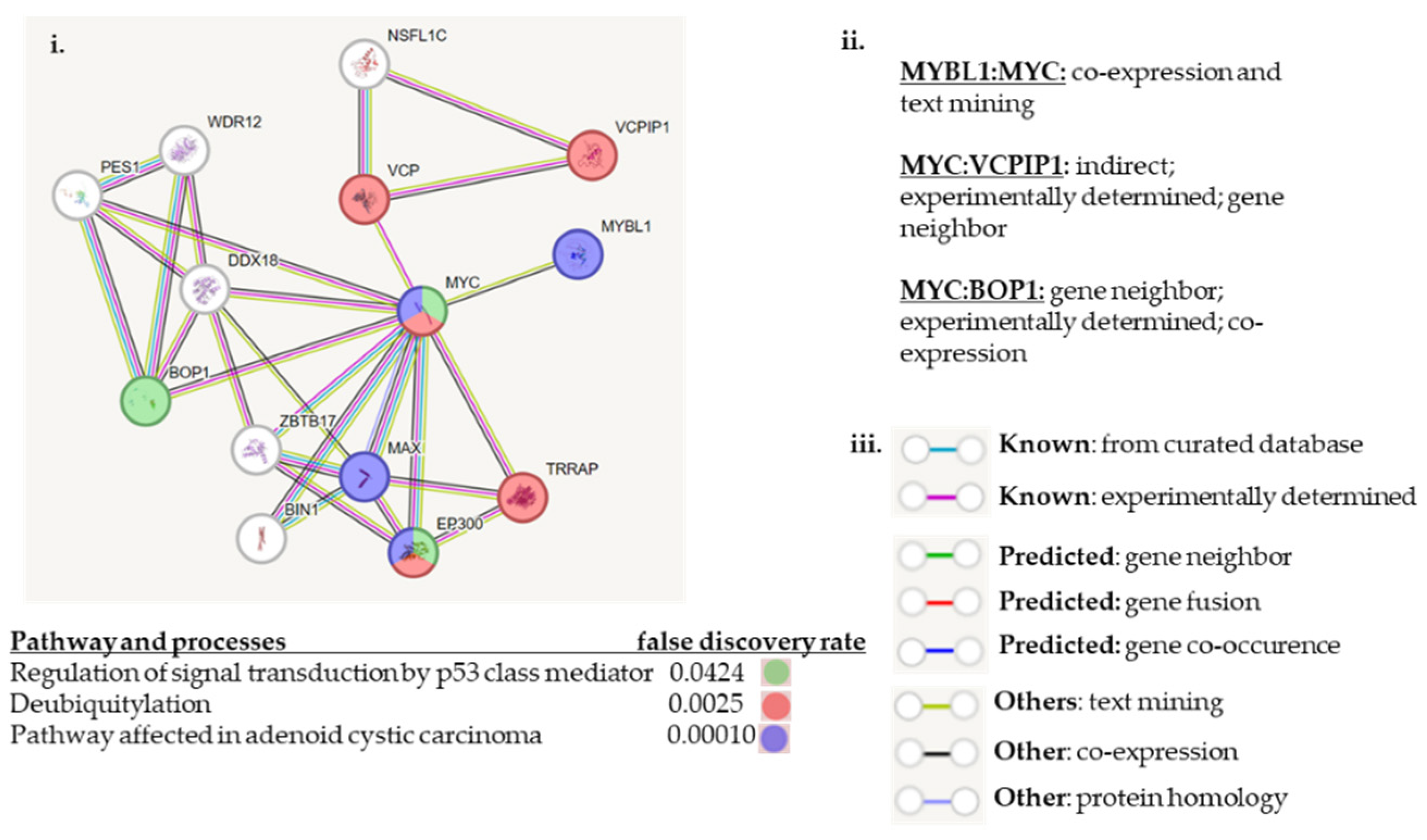
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Patient Sample Datasets
4.2. Ribonucleic Acid (RNA) Isolation, cDNA Generation, PCR and Gene Analyses
4.3. PCR Gene Primer Sets Generation
4.4. Data Analyses, Including Sequence Alignment Comparison and the cBioPortal Open Source Database
4.5. Data Analyses of Protein-Protein Interaction Using STRINGTM Open Source Database and PhosphoSitePlusTM
4.6. Western Blotting
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cicirò, Y.; Sala, A.A.-O. MYB oncoproteins: Emerging players and potential therapeutic targets in human cancer. Oncogenesis 2021, 10, 19. [Google Scholar] [CrossRef]
- Soza-Ried, C.; Hess, I.; Netuschil, N.; Schorpp MBoehm, T. Essential Role c-Myb in definitive hematopoiesis is evolutionarily conserved. Proc. Natl. Acad. Sci. USA 2010, 107, 17304–17308. [Google Scholar] [CrossRef]
- Lieu, Y.K.; Reddy, E.P. Conditional c-myb knockout in adult hematopoietic stem cells leads to loss of self-renewal due to impaired proliferation and accelerated differentiation. Proc. Natl. Acad. Sci. USA 2009, 106, 21689–21694. [Google Scholar] [CrossRef]
- Frerich, C.A.; Sedam, H.N.; Kang, H.; Mitani, Y.A.-O.; El-Naggar, A.K.; Ness, S.A.-O. N-Terminal Truncated Myb with New Transcriptional Activity Produced Through Use of an Alternative MYB Promoter in Salivary Gland Adenoid Cystic Carcinoma. Cancers 2019, 12, 45. [Google Scholar] [CrossRef] [PubMed]
- Musa, J.; Aynaud, M.M.; Mirabeau, O.; Delattre, O.; Grünewald, T.G. MYBL2 (B-Myb): A central regulator of cell proliferation, cell survival and differentiation involved in tumorigenesis. Cell Death Dis. 2017, 8, e2895. [Google Scholar] [CrossRef] [PubMed]
- Sleeman, J.P. Xenopus A-myb is expressed during early spermatogenesis. Oncogene 1993, 7, 1931–1941. [Google Scholar]
- Zhu, J.; Wu, Y.; Yu, Y.; Li, Y.; Shen, J.; Zhang, R.A.-O.X. MYBL1 induces transcriptional activation of ANGPT2 to promote tumor angiogenesis and confer sorafenib resistance in human hepatocellular carcinoma. Cell Death Dis. 2022, 13, 727. [Google Scholar] [CrossRef] [PubMed]
- Kalelioglu, T.A.-O.; Rama, B.A.-O.; Cho, B.B.; Lopes, B.M.; Patel, S.H. Pediatric-type diffuse low-grade glioma with MYB/MYBL1 alteration: Report of 2 cases. Neuroradiol. J. 2023, 36, 232–235. [Google Scholar] [CrossRef] [PubMed]
- Togashi, Y.; Dobashi, A.; Sakata, S.; Sato, Y.A.-O.; Baba, S.; Seto, A.; Mitani, H.; Kawabata, K.; Takeuchi, K.A.-O. MYB and MYBL1 in adenoid cystic carcinoma: Diversity in the mode of genomic rearrangement and transcripts. Mod. Pathol. 2018, 31, 934–946. [Google Scholar] [CrossRef]
- Pei, J.; Flieder, D.B.; Patchefsky, A.; Talarchek, J.N.; Cooper, H.S.; Testa, J.A.-O.; Wei, S. Detecting MYB and MYBL1 fusion genes in tracheobronchial adenoid cystic carcinoma by targeted RNA-sequencing. Mod. Pathol. 2019, 32, 1416–1420. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Jian, W.; Xue, W.; Meng, Y.; Xia, Z.; Li, Q.; Xu, S.; Dong, Y.; Mao, A.; Zhang, C. Integration analysis identifies MYBL1 as a novel immunotherapy biomarker affecting the immune microenvironment in clear cell renal cell carcinoma: Evidence based on machine learning and experiments. Front. Immunol. 2022, 14, 1080403. [Google Scholar] [CrossRef]
- Player, A.; Abraham, N.; Burrell, K.; Bengone, I.O.; Harris, A.; Nunez, L.; Willaims, T.; Kwende, S.; Walls, W. Identification of candidate genes associated with triple negative breast cancer. Genes. Cancer 2017, 8, 659–672. [Google Scholar] [CrossRef]
- Fujii, K.; Murase, T.; Beppu, S.; Saida, K.; Takino, H.; Masaki, A.; Ijichi, K.; Kusafuka, K.; Iida, Y.; Onitsuka, T.; et al. MYB, MYBL1, MYBL2 and NFIB gene alterations and MYC overexpression in salivary gland adenoid cystic carcinoma. Histopathology 2017, 71, 823–834. [Google Scholar] [CrossRef]
- Clough, E.; Barrett, T. The Gene Expression Omnibus Database. Methods Mol. Biol. 2016, 1418, 93–110. [Google Scholar]
- Borri, F.; Granaglia, A. Pathology of triple negative breast cancer. Semin. Cancer Biol. 2021, 72, 136–145. [Google Scholar] [CrossRef]
- Sayers, E.W.; Beck, J.; Bolton, E.E.; Bourexis, D.; Brister, J.R.; Canese, K.; Comeau, D.C.; Funk, K.; Kim, S.; Klimke, W.; et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2021, 49, D10–D17. [Google Scholar] [CrossRef]
- Takahashi, T.; Nakagoshi, H.; Sarai, A.; Nomura, N.; Yamamoto, T.; Ishii, S. Human A-myb gene encodes a transcriptional activator containing the negative regulatory domains. FEBS Lett. 1995, 358, 89–96. [Google Scholar] [CrossRef]
- Player, A.A.N.; Abdulrahman, N.; Nsende, E.; Cunningham, S.; Rogers, S. MYBL1 Knockdown in a Triple Negative Breast Cancer Line: Evidence of Down-Regulation of MYBL2, TCF19 and KIF18b Expression. Am. J. Cancer Clin. Res. 2021, 8, 1–11. [Google Scholar]
- Hornbeck, P.V.; Zhang, B.; Murray, B.; Kornhauser, J.M.; Latham, V.; Skrzypek, E. PhosphoSitePlus, 2014: Mutations, PTMs and recalibrations. Nucleic Acids Res. 2015, 43, D512–D520. [Google Scholar] [CrossRef]
- Facchinetti, V.; Loffarelli, L.; Schreek, S.; Oelgeschläger, M.; Lüscher, B.; Introna, M.; Golay, J. Regulatory domains of the A-Myb transcription factor and its interaction with the CBP/p300 adaptor molecules. Biochem. J. 1997, 324, 729–736. [Google Scholar] [CrossRef]
- Milioli, H.H.; Tishchenko, I.; Riveros, C.; Berretta, R.; Moscato, P.; Aine, M.; Boyaci, C.; Hartman, J.; Häkkinen, J.; Mitra, S.; et al. Basal-like breast cancer: Molecular profiles, clinical features and survival outcomes Molecular analyses of triple-negative breast cancer in the young and elderly. BMC Med. Genom. 2017, 10, 19. [Google Scholar] [CrossRef]
- Parris, T.Z.; Kovács, A.; Hajizadeh, S.; Nemes, S.; Semaan, M.; Levin, M.; Karlsson, P.; Helou, K. Frequent MYC coamplification and DNA hypomethylation of multiple genes on 8q in 8p11-p12-amplified breast carcinomas. Oncogenesis 2014, 3, e95. [Google Scholar] [CrossRef]
- Blancato, J.; Singh, B.; Liu, A.; Liao, D.J.; Dickson, R.B. Correlation of amplification and overexpression of the c-myc oncogene in high-grade breast cancer: FISH, in situ hybridisation and immunohistochemical analyses. Oncogenesis 2021, 10, 19. [Google Scholar] [CrossRef]
- Rushton, J.J.; Davis, L.M.; Lei, W.; Mo, X.; Leutz, A.; Ness, S.A. Distinct changes in gene expression induced by A-Myb, B-Myb and c-Myb proteins. Oncogene 2003, 22, 308–313. [Google Scholar] [CrossRef]
- Chan, W.A.-O.; Liu, X.; Magin, R.S.; Girardi, N.M.; Ficarro, S.B.; Hu, W.; Tarazona Guzman, M.I.; Starnbach, C.A.; Felix, A.; Adelmant, G.A.-O.; et al. Accelerating inhibitor discovery for deubiquitinating enzymes. Nat. Commun. 2023, 14, 686. [Google Scholar] [CrossRef]
- Liao, D.J.; Dickson, R.B. c-Myc in breast cancer. Endocr. Relat. Cancer 2000, 7, 143–164. [Google Scholar] [CrossRef]
- Llombart, V.; Mansour, M.R. Therapeutic targeting of “undruggable” MYC. eBioMedicine 2022, 75, 103756. [Google Scholar] [CrossRef]
- Li, W.; Song, P.; Zhao, M.; Gao, L.; Xie, J.; You, C. BOP1 Used as a Novel Prognostic Marker and Correlated with Tumor Microenvironment in Pan-Cancer. J. Oncol. 2021, 2021, 3603030. [Google Scholar] [CrossRef]
- Li, Y.Z.; Zhang, C.; Pei, J.P.; Zhang, W.C.; Zhang, C.D.; Dai, D.Q. The functional role of Pescadillo ribosomal biogenesis factor 1 in cancer. J. Cancer 2022, 13, 268–277. [Google Scholar] [CrossRef]
- Ciriello, G.; Gatza, M.L.; Beck, A.H.; Wilkerson, M.D.; Rhie, S.K.; Pastore, A.; Zhang, H.; McLellan, M.; Yau, C.; Kandoth, C.; et al. Comprehensive Molecular Portraits of Invasive Lobular Breast Cancer. Cell 2015, 163, 506–519. [Google Scholar] [CrossRef]
- Doherty, L.A.-O.; Mills, C.A.-O.; Boswell, S.A.-O.; Liu, X.; Hoyt, C.A.-O.; Gyori, B.A.-O.; Buhrlage, S.A.-O.; Sorger, P.A.-O. Integrating multi-omics data reveals function and therapeutic potential of deubiquitinating enzymes. eLife 2022, 11, e72879. [Google Scholar] [CrossRef]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- The ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium. Pan-cancer analysis of whole genomes. Nature 2020, 578, 82–93. [Google Scholar] [CrossRef] [PubMed]
- Bubola, J.; MacMillan, C.M.; Demicco, E.G.; Chami, R.A.; Chung, C.T.; Leong, I.; Marrano, P.; Onkal, Z.; Swanson, D.; Veremis, B.M.; et al. Targeted RNA sequencing in the routine clinical detection of fusion genes in salivary gland tumors. Genes. Chromosomes Cancer 2021, 60, 695–708. [Google Scholar] [CrossRef]
- Chung, K.Y.; Cheng, I.K.; Ching, A.K.; Chu, J.-H.; Lai, P.B.S.; Wong, N. Block of proliferation 1 (BOP1) plays an oncogenic role in hepatocellular carcinoma by promoting epithelial-to-mesenchymal transition. Hepatology 2011, 54, 307–318. [Google Scholar] [CrossRef]
- Heidelberger, J.B.; Voigt, A.; Borisova, M.E.; Petrosino, G.; Ruf, S.; Wagner, S.A.-O.; Beli, P.A.-O. Proteomic profiling of VCP substrates links VCP to K6-linked ubiquitylation and c-Myc function. EMBO Rep. 2018, 19, e44754. [Google Scholar] [CrossRef]
- UniProt Consortium. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2018, 16, 2699. [Google Scholar]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.A.-O.; Nastou, K.A.-O.; Mehryary, F.A.-O.; Hachilif, R.; Gable, A.L.; Fang, T.A.-O.; Doncheva, N.A.-O.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef] [PubMed]
- Saini, K.A.-O.; Punie, K.; Twelves, C.; Bortini, S.; de Azambuja, E.A.-O.; Anderson, S.; Criscitiello, C.; Awada, A.A.-O.; Loi, S. Antibody-drug conjugates, immune-checkpoint inhibitors, and their combination in breast cancer therapeutics. Expert. Opin. Biol. Ther. 2021, 21, 945–962. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, B.D.; Pietenpol, J.A.; Tan, A.R. Triple-Negative Breast Cancer: Molecular Subtypes and New Targets for Therapy. Am. Soc. Clin. Oncol. Educ. Book 2015, 35, e31–e39. [Google Scholar] [CrossRef]
- Rummukainen, J.; Kytölä, S.; Karhu, R.; Farnebo, F.; Larsson, C.; Isola, J.J. Aberrations of chromosome 8 in 16 breast cancer cell lines by comparative genomic hybridization, fluorescence in situ hybridization, and spectral karyotyping. Cancer Genet. Cytogenet. 2001, 126, 1–7. [Google Scholar] [CrossRef]
- Mahmood, S.F.; Gruel, N.; Chapeaublanc, E.; Lescure, A.; Jones, T.; Reyal, F.; Vincent-Salomon, A.; Raynal, V.; Pierron, G.; Perez, F.; et al. A siRNA screen identifies RAD21, EIF3H, CHRAC1 and TANC2 as driver genes within the 8q23, 8q24.3 and 17q23 amplicons in breast cancer with effects on cell growth, survival and transformation. Carcinogenesis 2014, 35, 670–682. [Google Scholar] [CrossRef]
- Buerger, H.; Otterbach, F.; Simon, R.; Poremba, C.; Diallo, R.; Decker, T.; Riethdorf, L.; Brinkschmidt, C.; Dockhorn-Dworniczak, B.; Boecker, W. Comparative genomic hybridization of ductal carcinoma in situ of the breast-evidence of multiple genetic pathways. J. Pathol. 1999, 187, 396–402. [Google Scholar] [CrossRef]
- Cingoz, S.; Altungoz, O.; Canda, T.; Saydam, S.; Aksakoglu, G.; Sakizli, M. DNA copy number changes detected by comparative genomic hybridization and their association with clinicopathologic parameters in breast tumors. Cancer Genet. Cytogenet. 2003, 145, 108–114. [Google Scholar] [CrossRef]
- Marhamati, D.J.; Bellas, R.E.; Arsura, M.; Kypreos, K.E.; Sonenshein, G.E. A-myb is expressed in bovine vascular smooth muscle cells during the late G1-to-S phase transition and cooperates with c-myc to mediate progression to S phase. Mol. Cell Biol. 1997, 17, 2448–2457. [Google Scholar] [CrossRef]
- Uchiyama, K.; Jokitalo, E.; Kano, F.; Murata, M.; Zhang, X.; Canas, B.; Newman, R.; Rabouille, C.; Pappin, D.; Freemont, P.; et al. VCIP135, a novel essential factor for p97/p47-mediated membrane fusion, is required for Golgi and ER assembly in vivo. J. Cell Biol. 2002, 159, 855–866. [Google Scholar] [CrossRef]
- Huang, S.; Wang, Y.A.-O. Golgi structure formation, function, and post-translational modifications in mammalian cells. F1000Research 2017, 6, 2050. [Google Scholar] [CrossRef]
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Stein, T.I.; Nudel, R.; Lieder, I.; Mazor, Y.; et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr. Protoc. Bioinform. 2016, 54, 1–30. [Google Scholar] [CrossRef]
- Fallah, Y.; Brundage, J.; Allegakoen, P.; Shajahan-Haq, A.N. MYC-Driven Pathways in Breast Cancer Subtypes. Biomolecules 2017, 7, 53. [Google Scholar] [CrossRef]
- Duffy, M.J.; O’Grady, S.; Tang, M.; Crown, J. MYC as a target for cancer treatment. Cancer Treat. Rev. 2021, 94, 102154. [Google Scholar] [CrossRef]
- Doha, Z.O.; Wang, X.; Calistri, N.A.-O.; Eng, J.A.-O.; Daniel, C.J.; Ternes, L.; Kim, E.N.; Pelz, C.; Munks, M.; Betts, C.; et al. MYC Deregulation and PTEN Loss Model Tumor and Stromal Heterogeneity of Aggressive Triple-Negative Breast Cancer. Nat. Commun. 2023, 14, 5665. [Google Scholar] [CrossRef]
- Zimmerli, D.; Brambillasca, C.S.; Talens, F.; Bhin, J.A.-O.; Linstra, R.A.-O.; Romanens, L.; Bhattacharya, A.A.-O.; Joosten, S.E.P.; Da Silva, A.M.; Padrao, N.; et al. MYC promotes immune-suppression in triple-negative breast cancer via inhibition of interferon signaling. Nat. Commun. 2022, 13, 6579. [Google Scholar] [CrossRef]
- AlSultan, D.; Kavanagh, E.; O’Grady, S.A.-O.; Eustace, A.J.; Castell, A.; Larsson, L.G.; Crown, J.; Madden, S.F.; Duffy, M.J. The novel low molecular weight MYC antagonist MYCMI-6 inhibits proliferation and induces apoptosis in breast cancer cells. Investig. New Drugs 2021, 39, 587–594. [Google Scholar] [CrossRef]
- Garrido-Castro, A.A.-O.; Lin, N.U.; Polyak, K. Insights into Molecular Classifications of Triple-Negative Breast Cancer: Improving Patient Selection for Treatment. Cancer Discov. 2019, 9, 176–198. [Google Scholar] [CrossRef]
- Wu, Z.; Wu, C.; Shi, J.; Huang, Z.; Lu, S.; Tan, Y.; You, R.; Hai, L.; Huang, J.; Guo, S.; et al. Elucidating the pharmacological effects of Compound Kushen injection on MYC-P15-CCND1 signaling pathway in nasopharyngeal carcinoma—An in vitro study. J. Ethnopharmacol. 2023, 315, 116702. [Google Scholar] [CrossRef]
- Mitani, Y.; Liu, B.; Rao, P.H.; Borra, V.J.; Zafereo, M.; Weber, R.S.; Kies, M.; Lozano, G.; Futreal, P.A.; Caulin, C.; et al. Novel MYBL1 Gene Rearrangements with Recurrent MYBL1-NFIB Fusions in Salivary Adenoid Cystic Carcinomas Lacking t(6;9) Translocations. Clin. Cancer Res. 2016, 22, 725–733. [Google Scholar] [CrossRef]
- Endo, Y.; Kuwamoto, S.; Ohira, T.; Matsushita, M.; Matsushige, T.; Fukuhara, T.; Nakamoto, S.; Hayashi, K.; Kugoh, H.; Hirooka, Y. Possible Relationship Between MYBL1 Alterations and Specific Primary Sites in Adenoid Cystic Carcinoma: A Clinicopathological and Molecular Study of 36 Cases. Yonago Acta Med. 2019, 62, 67–76. [Google Scholar] [CrossRef]
- Chae, Y.K.; Chung, S.Y.; Davis, A.A.; Carneiro, B.A.; Chandra, S.; Kaplan, J.; Kalyan, A.; Giles, F.J. Adenoid cystic carcinoma: Current therapy and potential therapeutic advances based on genomic profiling. Oncotarget 2015, 6, 37117–37134. [Google Scholar] [CrossRef]
- Deming, S.L.; Nass, S.j.; Dickson, R.B.; Trock, B.J. C-myc amplification in breast cancer: A meta-analysis of its occurrence and prognostic relevance. Br. J. Cancer 2000, 3, 1688–1695. [Google Scholar] [CrossRef]
- Chrzan, P.; Skokowski, J.; Karmolinski, A.; Pawelczyk, T. Amplification of c-myc gene and overexpression of c-Myc protein in breast cancer and adjacent non-neoplastic tissue. Clin. Biochem. 2001, 34, 557–562. [Google Scholar] [CrossRef]
- Hölzel, M.; Rohrmoser, M.; Schlee, M.; Grimm, T.; Harasim, T.; Malamoussi, A.; Gruber-Eber, A.; Kremmer, E.; Hiddemann, W.; Bornkamm, G.W.; et al. Mammalian WDR12 is a novel member of the Pes1-Bop1 complex and is required for ribosome biogenesis and cell proliferation. J. Cell Biol. 2005, 170, 367–378. [Google Scholar] [CrossRef]
- Pestov, D.G.; Strezoska, Z.; Lau, L.F. Evidence of p53-dependent cross-talk between ribosome biogenesis and the cell cycle: Effects of nucleolar protein Bop1 on G(1)/S transition. Mol. Cell Biol. 2001, 21, 4246–4255. [Google Scholar] [CrossRef]
- Li, S.; Wu, H.; Huang, X.; Jian, Y.; Kong, L.; Xu, H.; Ouyang, Y.; Chen, X.; Wu, G.; Yu, L.; et al. BOP1 confers chemoresistance of triple-negative breast cancer by promoting CBP-mediated β-catenin acetylation. Proc. Natl. Acad. Sci. USA 2009, 106, 21689–21694. [Google Scholar] [CrossRef]
- Gupta, R.; Bugide, S.; Wang, B.; Green, M.A.-O.; Johnson, D.B.; Wajapeyee, N. Loss of BOP1 confers resistance to BRAF kinase inhibitors in melanoma by activating MAP kinase pathway. Proc. Natl. Acad. Sci. USA 2019, 116, 4583–4591. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3--new capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef]
- Kent, W.J.; Sugnet, C.W.; Furey, T.S.; Roskin, K.M.; Pringle, T.H.; Zahler, A.M.; Haussler, D.; Haussler, D. The human genome browser at UCSC. Genome Res. 2002, 12, 996–1006. [Google Scholar] [CrossRef]
- Corpet, F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1998, 16, 10881–10890. [Google Scholar] [CrossRef]
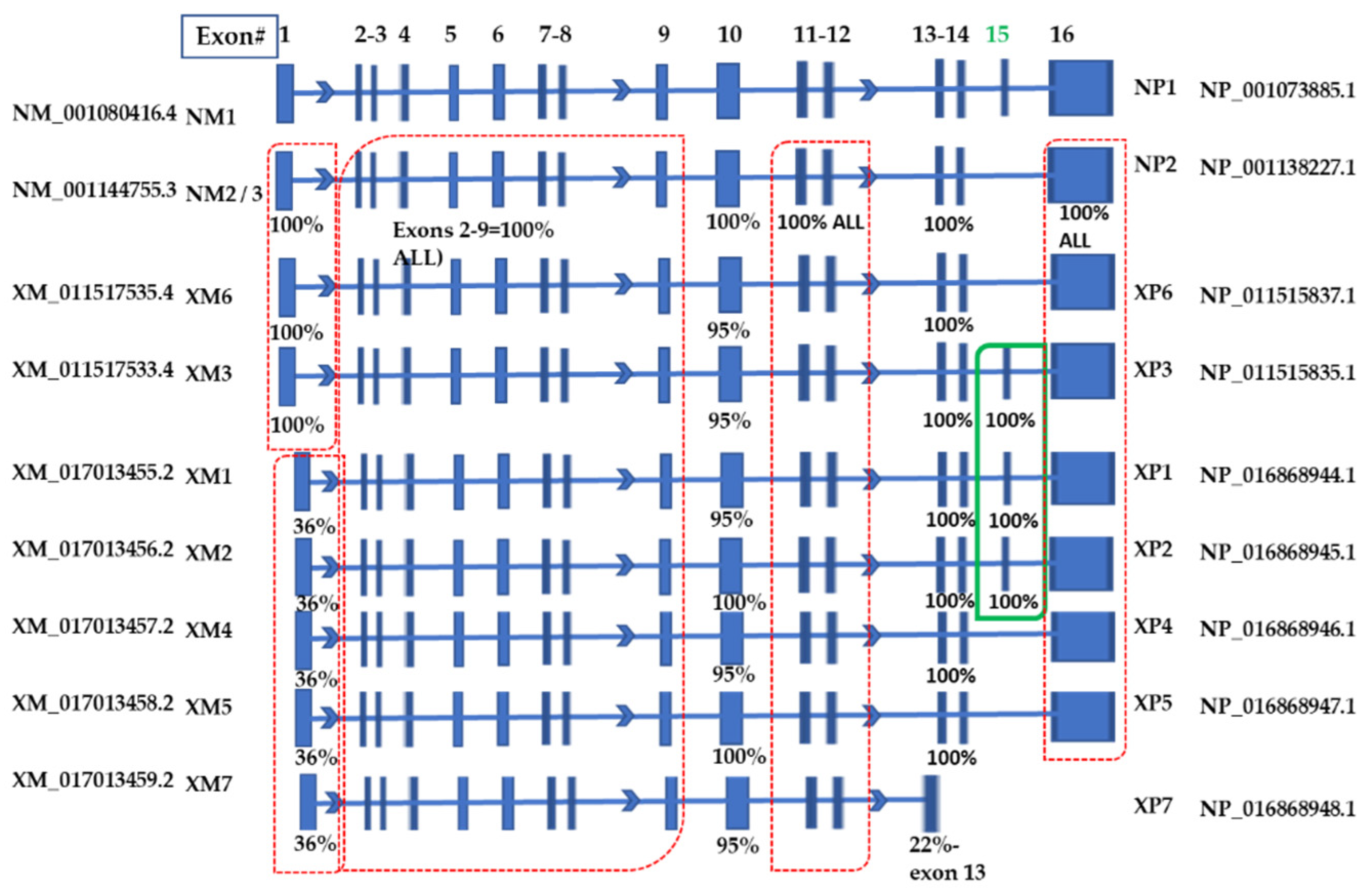


| NUCLEOTIDE | Abbreviation | SIZE (Base Pairs) | Start Site (bp) | PROTEIN | Abbreviation | SIZE (Amino Acids) |
|---|---|---|---|---|---|---|
| NM_001294282.1 | NM3 | 4878 | 381 | NP_001281211.1 | NP3 | 691 |
| NM_001144755.3 | NM2 | 4981 | 381 | NP_001138227.1 | NP2 | 692 |
| NM_001080416.4 | NM1 | 5161 | 381 | NP_001073885.1 | NP1 | 752 |
| XM_011517535.4 | XM6 | 4996 | 381 | NP_011515837.1 | XP6 | 697 |
| XM_011517533.4 | XM3 | 5176 | 381 | NP_011515835.1 | XP3 | 757 |
| XM_017013455.2 | XM1 | 5054 | 220 | NP_016868944.1 | XP1 | 770 |
| XM_017013456.2 | XM2 | 5036 | 217 | NP_016868945.1 | XP2 | 765 |
| XM_017013457.2 | XM4 | 4871 | 217 | NP_016868946.1 | XP4 | 710 |
| XM_017013458.2 | XM5 | 4853 | 241 | NP_016868947.1 | XP5 | 705 |
| XM_017013459.2 | XM7 | 2270 | 222 | NP_016868948.1 | XP7 | 597 |
| MYBL1: PROT>=2 PROT<=-2 AMP HOMDEL EXP>2 EXP<2 MUT FUSION HETLOSS; |
| VCPIP1: PROT>=2 PROT<=-2 AMP HOMDEL EXP>2 EXP<2 MUT FUSION HETLOSS; |
| MYC: PROT>=2 PROT<=-2 AMP HOMDEL EXP>2 EXP<2 MUT FUSION HETLOSS; |
| BOP1: PROT>=2 PROT<=-2 AMP HOMDEL EXP>2 EXP<2 MUT FUSION HETLOSS; |
| %ALTERATIONS /Profiled | # Patients/ALTERATIONS | AMPLIFICATIONS | HIGH mRNA | LOW mRNA | NONE | |
|---|---|---|---|---|---|---|
| MYBL1 | 58% of 83 | 26 patients | 10/83 | 17/83 | 25/83 | 36/83 |
| VCPIP1 | 57% of 83 | 21 patients (16 same) | 9/83 (9 same) | 14/83 (8 same) | 28/83 (18 same) | 37/83 (36 same) |
| MYC | 70% of 83 | 40 patients (22 same) | 32/83 (8 same) | 11/83 (7 same) | 28 (15 same) | 25/83 (25 same) |
| BOP1 | 61% of 83 | 36 patients (22 same) | 20/83 (7 same) | 26/83 (14 same) | 16/83 (12 same) | 32/83 (31 same) |
| A: GENE | LEFT PRIMER | RIGHT PRIMER | Size of Amplicon (Bp = Base Pair) |
|---|---|---|---|
| MYBL1 | TGGATAAGTCTGGGCTTATTGG | CCATGCAAGTATGGCTGCTA | 210 BP |
| MYC | TGGCTGCTTGTGAGTACAGG | TGAACTGGCTTCTTCCCAGG | 229 BP |
| BOP1 | CACCCCCAGCTTCTATGACC | CTGGATGAAGCGTCCGTAGG | 256 BP |
| VCPIP1 | AGGACATTAAGCGGGCCAAT | GGGAACACCCTCAGGTGGTA | 266 BP |
| GAPDH | TCCCTGAGCTGAACGGGAAG | GGAGGAGTGGGTGTCGCTGT | 217 BP |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Player, A.; Cunningham, S.; Philio, D.; Roy, R.; Haynes, C.; Dixon, C.; Thirston, L.; Ibikunle, F.; Boswell, T.A.; Alnakhalah, A.; et al. Characterization of MYBL1 Gene in Triple-Negative Breast Cancers and the Genes’ Relationship to Alterations Identified at the Chromosome 8q Loci. Int. J. Mol. Sci. 2024, 25, 2539. https://doi.org/10.3390/ijms25052539
Player A, Cunningham S, Philio D, Roy R, Haynes C, Dixon C, Thirston L, Ibikunle F, Boswell TA, Alnakhalah A, et al. Characterization of MYBL1 Gene in Triple-Negative Breast Cancers and the Genes’ Relationship to Alterations Identified at the Chromosome 8q Loci. International Journal of Molecular Sciences. 2024; 25(5):2539. https://doi.org/10.3390/ijms25052539
Chicago/Turabian StylePlayer, Audrey, Sierra Cunningham, Deshai Philio, Renata Roy, Cydney Haynes, Christopher Dixon, Lataja Thirston, Fawaz Ibikunle, Taylor Allen Boswell, Ayah Alnakhalah, and et al. 2024. "Characterization of MYBL1 Gene in Triple-Negative Breast Cancers and the Genes’ Relationship to Alterations Identified at the Chromosome 8q Loci" International Journal of Molecular Sciences 25, no. 5: 2539. https://doi.org/10.3390/ijms25052539
APA StylePlayer, A., Cunningham, S., Philio, D., Roy, R., Haynes, C., Dixon, C., Thirston, L., Ibikunle, F., Boswell, T. A., Alnakhalah, A., Contreras, J., Bell, M., McGuffery, T., Bryant, S., Nganya, C., & Kanu, S. (2024). Characterization of MYBL1 Gene in Triple-Negative Breast Cancers and the Genes’ Relationship to Alterations Identified at the Chromosome 8q Loci. International Journal of Molecular Sciences, 25(5), 2539. https://doi.org/10.3390/ijms25052539





