In Silico Screening of Bacteriocin Gene Clusters within a Set of Marine Bacillota Genomes
Abstract
1. Introduction
2. Results
2.1. Distribution of Putative Bacteriocin Gene Clusters in the Selected Marine Bacillota Genomes
2.2. Abundance of Different Classes of Bacteriocins Gene Clusters Identified by In Silico Analysis
2.2.1. Class I: The RiPPs (Ribosomally Synthesized and Post-Translationally Modified Peptides)
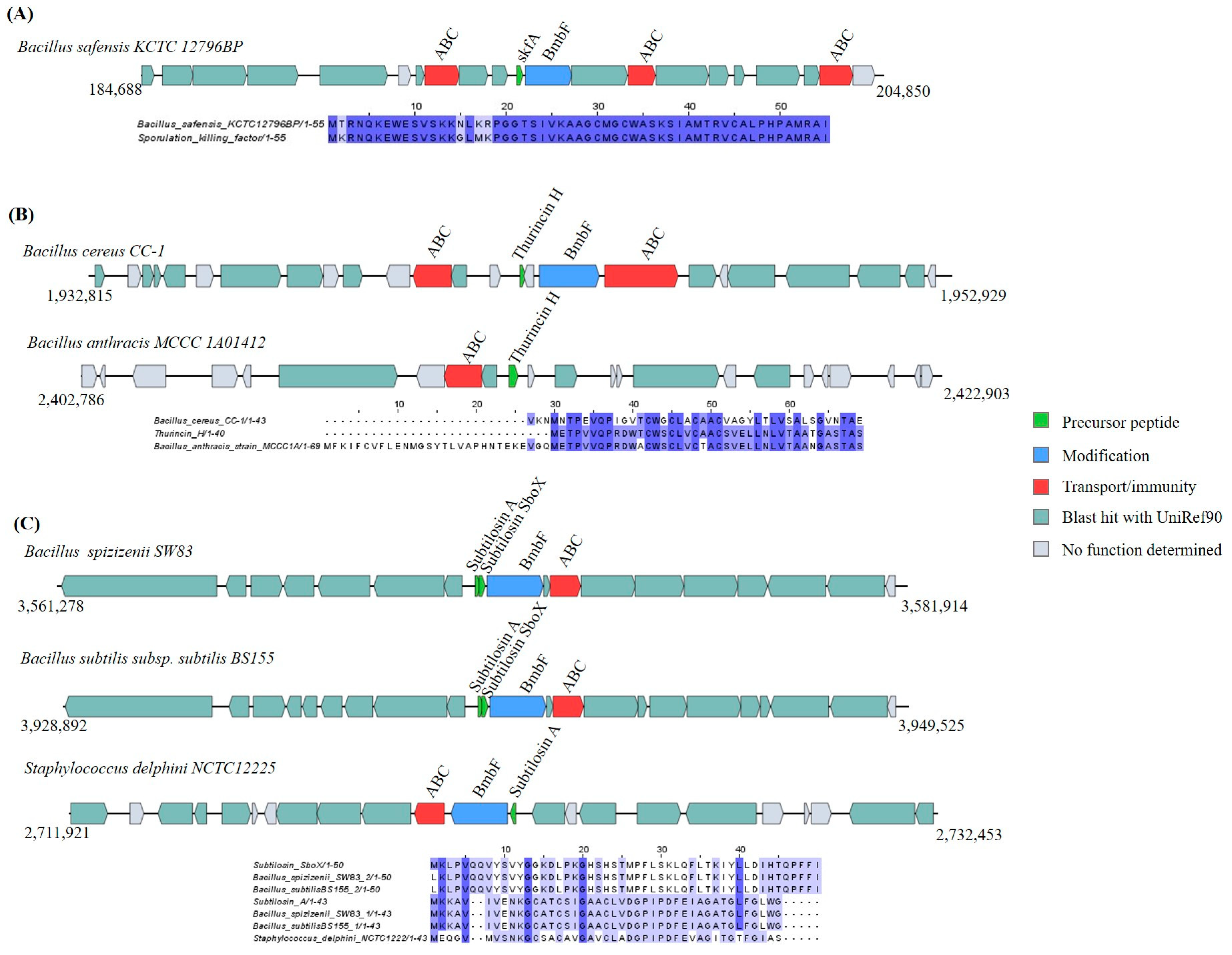
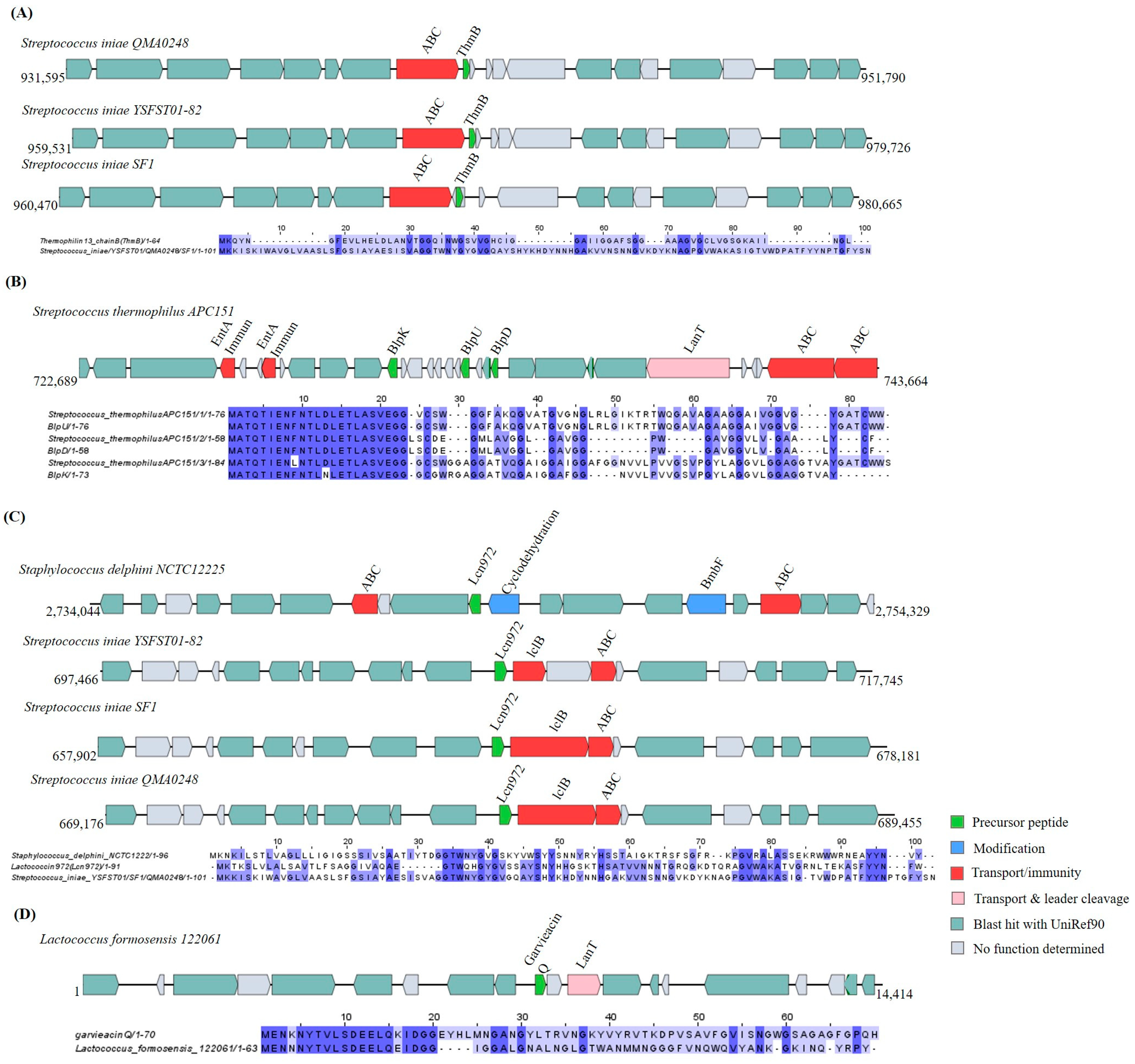
Sactipeptides
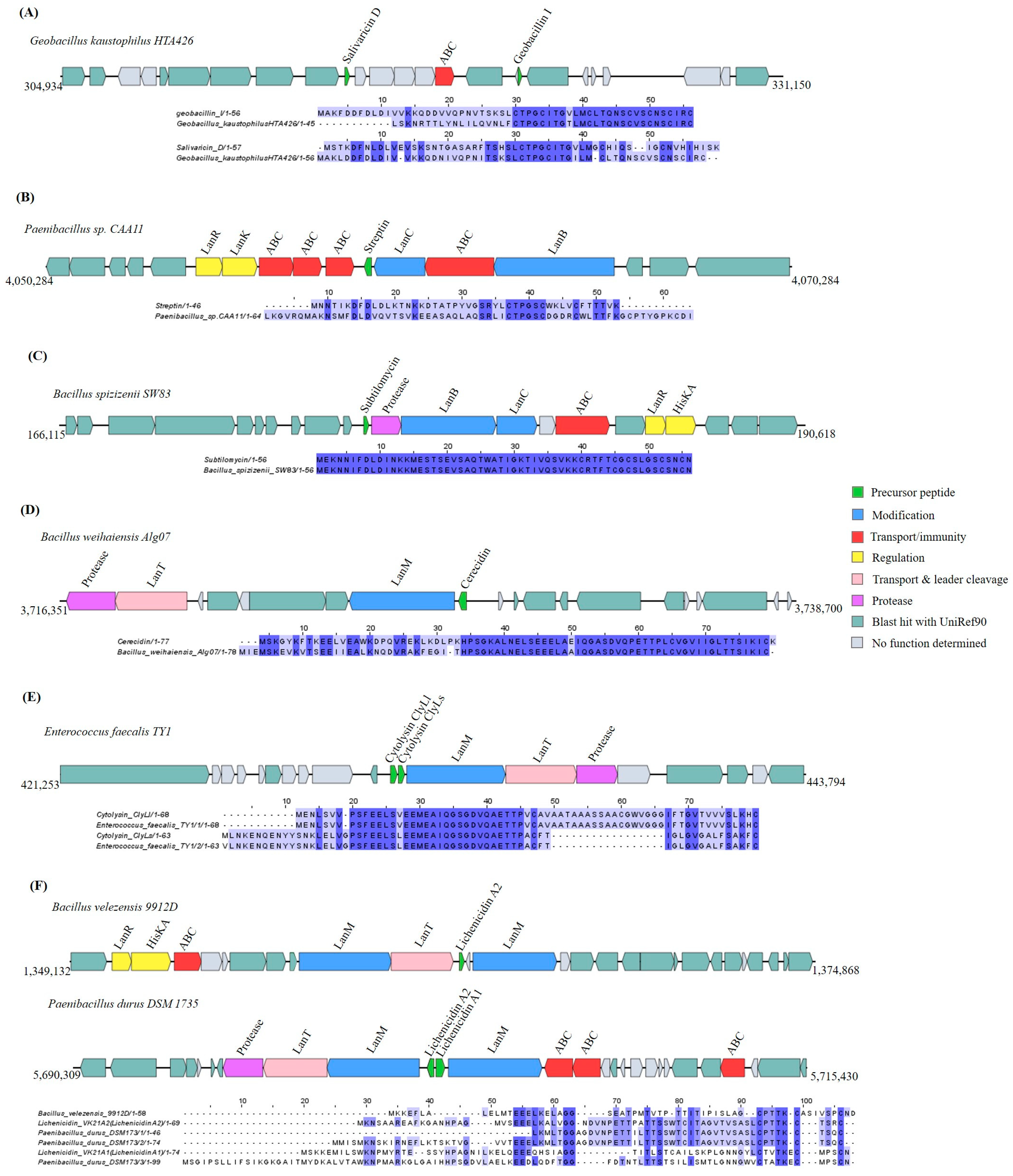
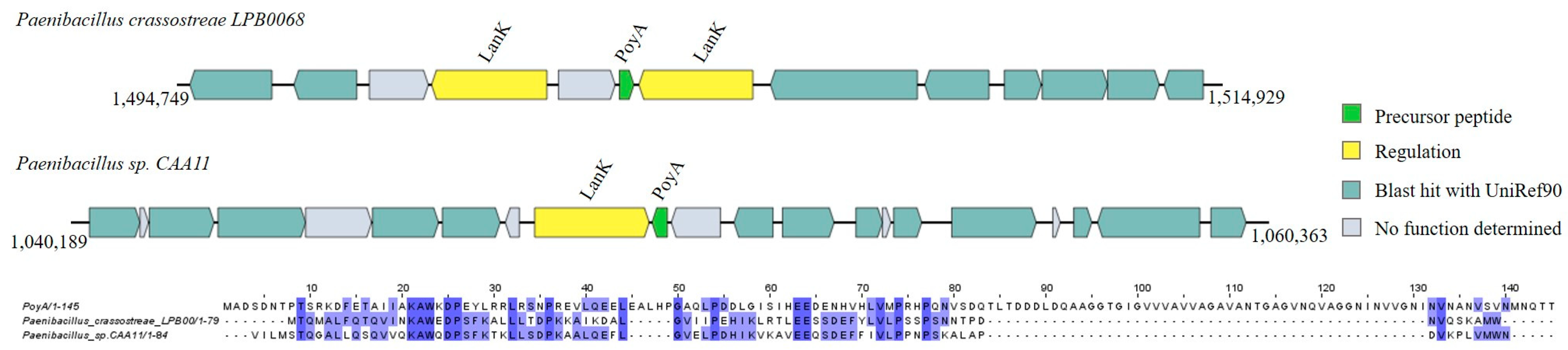
Lanthipeptides
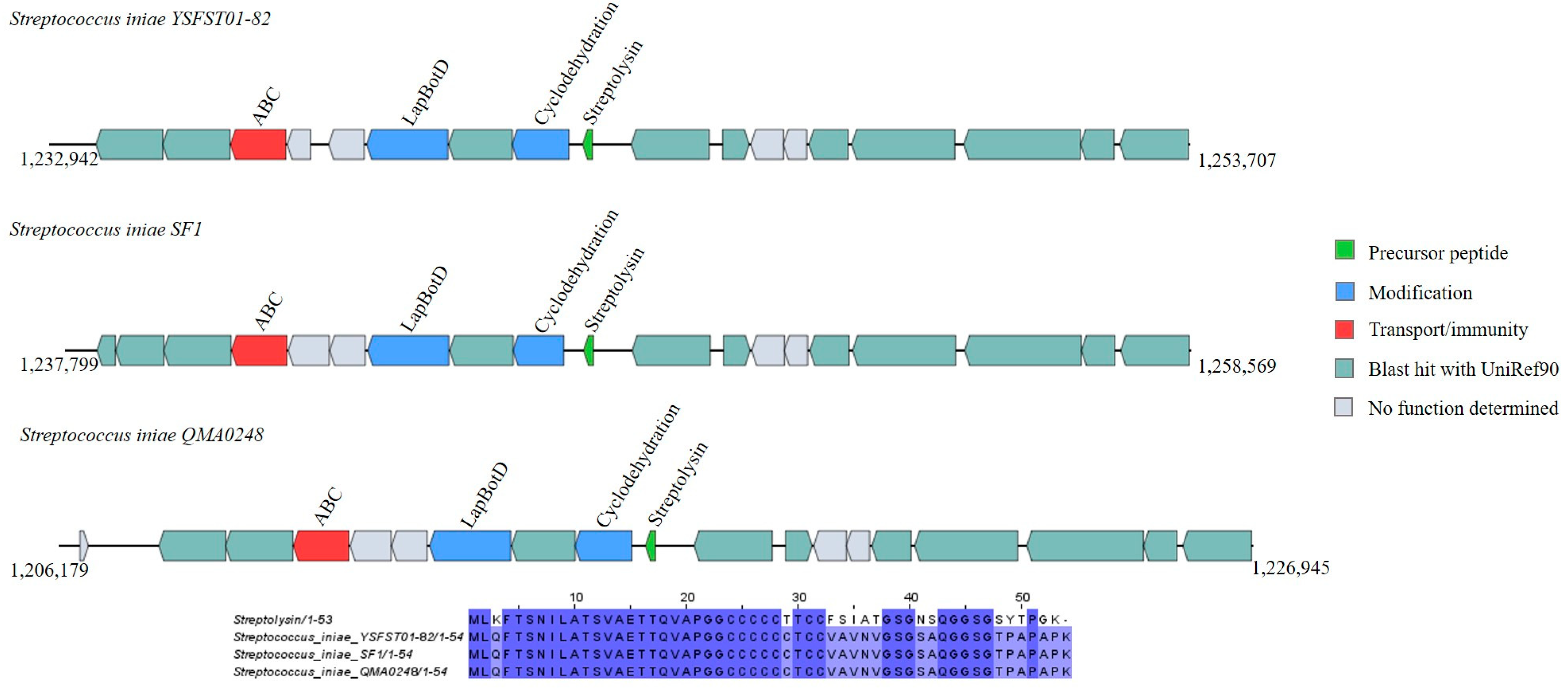
Linear Azole-Containing Peptides (LAPs)
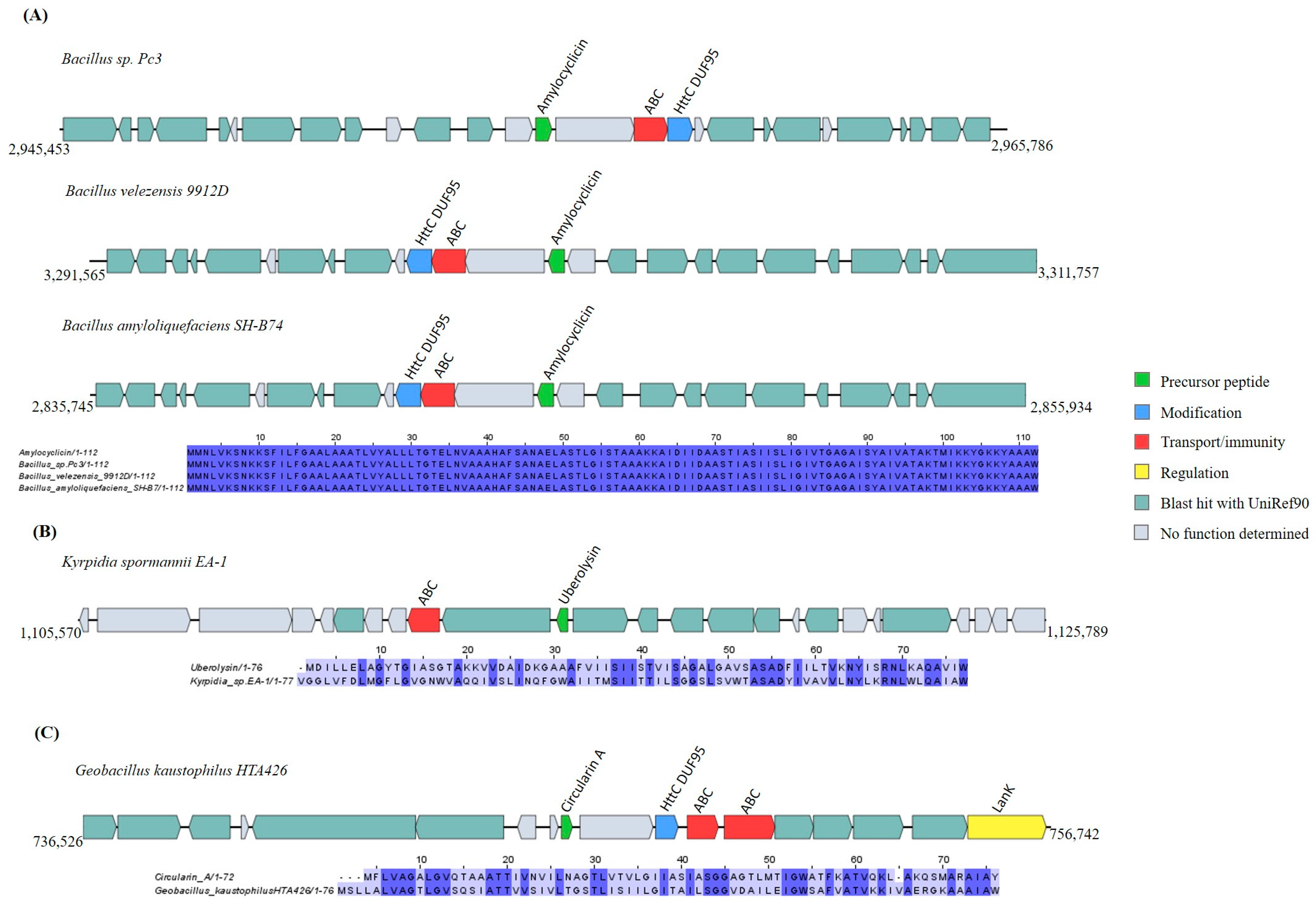
Head-to-Tail Cyclized Peptides
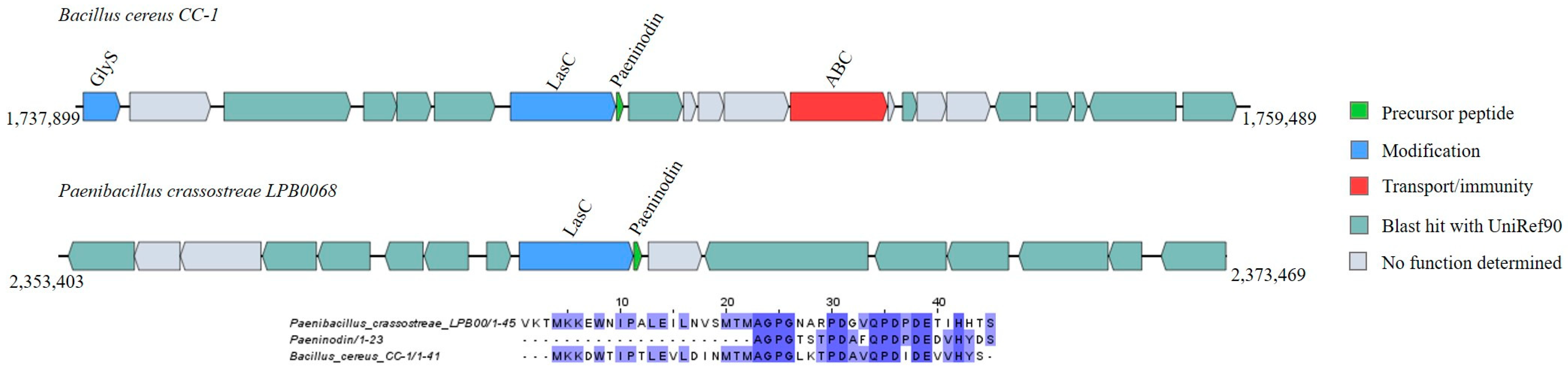
Lasso Peptides
Proteusins
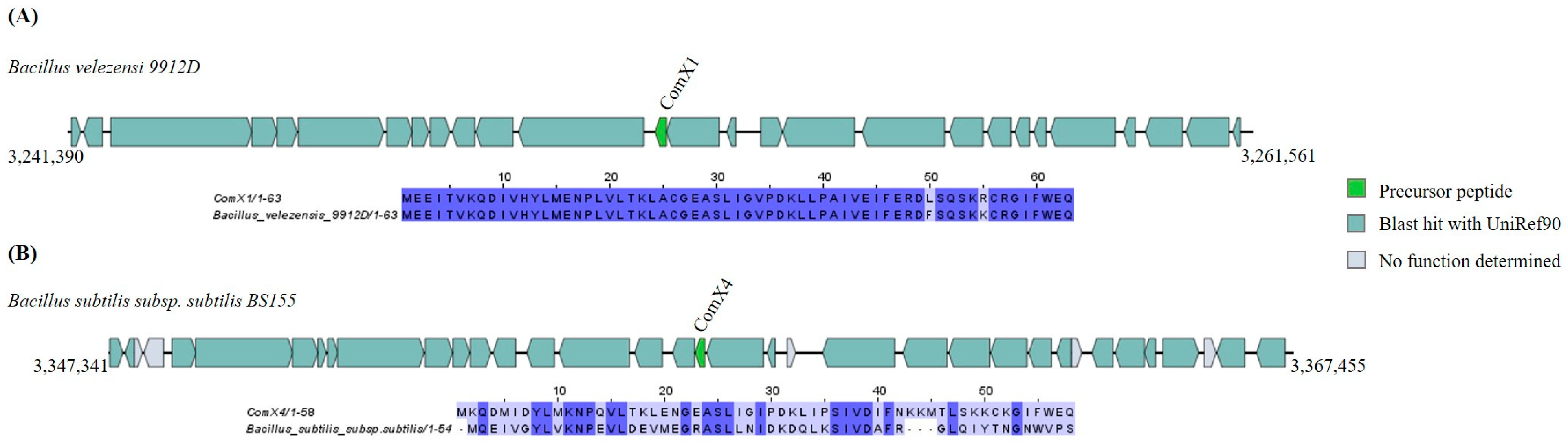
ComX
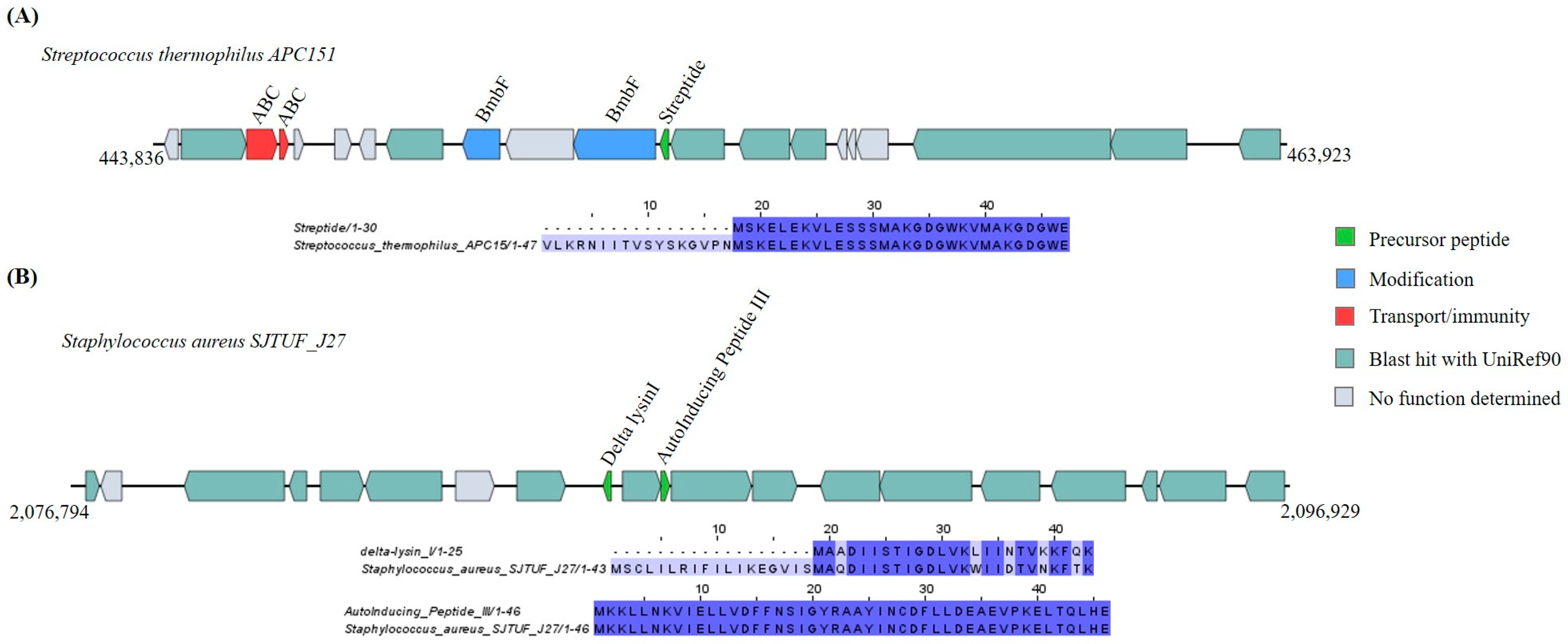
Lysine-to-Tryptophan Crosslink and Autoinducing Peptides
Class II
Class III
3. Discussion
4. Methods
4.1. Data Collection
4.2. Identification of Putative Bacteriocin Gene Clusters
4.3. Construction of Phylogenetic Tree of Bacillota Strains
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Antimicrobial Resistance. Available online: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance (accessed on 21 December 2023).
- Acedo, J.Z.; Chiorean, S.; Vederas, J.C.; van Belkum, M.J. The expanding structural variety among bacteriocins from Gram-positive bacteria. FEMS Microbiol. Rev. 2018, 42, 805–828. [Google Scholar] [CrossRef]
- Egan, K.; Field, D.; Ross, R.P.; Cotter, P.D.; Hill, C. In silico prediction and exploration of potential bacteriocin gene clusters within the bacterial genus Geobacillus. Front. Microbiol. 2018, 9, 2116. [Google Scholar] [CrossRef]
- Walsh, C.J.; Guinane, C.M.; Hill, C.; Ross, R.P.; O’Toole, P.W.; Cotter, P.D.; Ross, R. In silico identification of bacteriocin gene clusters in the gastrointestinal tract, based on the Human Microbiome Project’s reference genome database. BMC Microbiol. 2015, 15, 183. [Google Scholar] [CrossRef]
- Van Heel, A.J.; De Jong, A.; Song, C.; Viel, J.H.; Kok, J.; Kuipers, O.P. BAGEL4: A user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res. 2018, 46, W278–W281. [Google Scholar] [CrossRef]
- Morton, J.T.; Freed, S.D.; Lee, S.W.; Friedberg, I. A large scale prediction of bacteriocin gene blocks suggests a wide functional spectrum for bacteriocins. BMC Bioinform. 2015, 16, 381. [Google Scholar] [CrossRef]
- Sabino, Y.N.V.; de Araújo, K.C.; Assis, F.G.d.V.d.; Moreira, S.M.; Lopes, T.d.S.; Mendes, T.A.d.O.; Huws, S.A.; Mantovani, H.C. In silico screening unveil the great potential of ruminal bacteria synthesizing lasso peptides. Front. Microbiol. 2020, 11, 576738. [Google Scholar] [CrossRef]
- Azevedo, A.C.; Bento, C.B.P.; Ruiz, J.C.; Queiroz, M.V.; Mantovani, H.C. Distribution and genetic diversity of bacteriocin gene clusters in rumen microbial genomes. Appl. Environ. Microbiol. 2015, 81, 7290–7304. [Google Scholar] [CrossRef] [PubMed]
- Gontijo, M.T.P.; Silva, J.d.S.; Vidigal, P.M.P.; Martin, J.G.P. Phylogenetic distribution of the bacteriocin repertoire of lactic acid bacteria species associated with artisanal cheese. Food Res. Int. 2020, 128, 108783. [Google Scholar] [CrossRef] [PubMed]
- Bakkal, S.; Robinson, S.M.; Riley, M.A. Bacteriocins of aquatic microorganisms and their potential applications in the seafood industry. In Health and Environment in Aquaculture; Carvalho, D., David, G.S., Reinaldo, J.S., Eds.; Intech Open: London, UK, 2012; pp. 303–328. [Google Scholar]
- Uniacke-Lowe, S.; Collins, F.W.J.; Hill, C.; Ross, R.P. Bioactivity screening and genomic analysis reveals deep-sea fish microbiome isolates as sources of novel antimicrobials. Mar. Drugs 2023, 21, 444. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Ye, T.; Li, C.; Praveen, P.; Hu, Z.; Li, W.; Shang, C. Embracing the era of antimicrobial peptides with marine organisms. Nat. Prod. Rep. 2024; Advance Article. [Google Scholar] [CrossRef] [PubMed]
- Saidumohamed, B.E.; Johny, T.K.; Raveendran, A.T.; Sheela, U.B.; Sreeraganathan, M.; Sasidharan, R.S.; Bhat, S.G. 3D structure elucidation and appraisal of mode of action of a bacteriocin BaCf3 with anticancer potential produced by marine Bacillus amyloliquefaciens BTSS3. Re GEN Open 2022, 2, 45–56. [Google Scholar] [CrossRef]
- Voser, T.M.; Campbell, M.D.; Carroll, A.R. How different are marine microbial natural products compared to their terrestrial counterparts? Nat. Prod. Rep. 2022, 39, 7–19. [Google Scholar] [CrossRef]
- Srinivasan, R.; Kannappan, A.; Shi, C.; Lin, X. Marine bacterial secondary metabolites: A treasure house for structurally unique and effective antimicrobial compounds. Mar. Drugs 2021, 19, 530. [Google Scholar] [CrossRef]
- Luo, S.; Dong, S.-H. Recent advances in the discovery and biosynthetic study of eukaryotic RiPP natural products. Molecules 2019, 24, 1541. [Google Scholar] [CrossRef]
- Russell, A.H.; Truman, A.W. Genome mining strategies for ribosomally synthesised and post-translationally modified peptides. Comput. Struct. Biotechnol. J. 2020, 18, 1838–1851. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, J.; Li, G.; Yang, Y.; Ding, W. Current advancements in sactipeptide natural products. Front. Chem. 2021, 9, 595991. [Google Scholar] [CrossRef] [PubMed]
- Alkhalili, R.N.; Canbäck, B. Identification of putative novel class-I lanthipeptides in Firmicutes: A combinatorial in silico analysis approach performed on genome sequenced bacteria and a close inspection of Z-geobacillin lanthipeptide biosynthesis gene cluster of the thermophilic Geobacillus sp. strain ZGt-1. Int. J. Mol. Sci. 2018, 19, 2650. [Google Scholar] [CrossRef] [PubMed]
- Basi-Chipalu, S. A review: Lantibiotics, a promising antimicrobial agent. J. Inst. Sci. Technol. 2016, 21, 119–128. [Google Scholar] [CrossRef][Green Version]
- Yu, Y.; Zhang, Q.; van der Donk, W.A. Insights into the evolution of lanthipeptide biosynthesis. Protein Sci. 2013, 22, 1478–1489. [Google Scholar] [CrossRef]
- Bartholomae, M.; Buivydas, A.; Viel, J.H.; Montalbán-López, M.; Kuipers, O.P. Major gene-regulatory mechanisms operating in ribosomally synthesized and post-translationally modified peptide (RiPP) biosynthesis. Mol. Microbiol. 2017, 106, 186–206. [Google Scholar] [CrossRef]
- Arnison, P.G.; Bibb, M.J.; Bierbaum, G.; Bowers, A.A.; Bugni, T.S.; Bulaj, G.; Camarero, J.A.; Campopiano, D.J.; Challis, G.L.; Clardy, J.; et al. Ribosomally synthesized and post-translationally modified peptide natural products: Overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 2013, 30, 108–160. [Google Scholar] [CrossRef]
- Van Heel, A.J.; Montalbán-López, M.; Quentin, O.; Kuipers, O.P. Genome-guided identification of novel head-to-tail cyclized antimicrobial peptides, exemplified by the discovery of pumilarin. Microb. Genom. 2017, 3, e000134. [Google Scholar] [CrossRef]
- Freeman, M.F.; Gurgui, C.; Helf, M.J.; Morinaka, B.I.; Uria, A.R.; Oldham, N.J.; Sahl, H.-G.; Matsunaga, S.; Piel, J. Metagenome mining reveals polytheonamides as posttranslationally modified ribosomal peptides. Science 2012, 338, 387–390. [Google Scholar] [CrossRef]
- Schramma, K.R.; Bushin, L.B.; Seyedsayamdost, M.R. Structure and biosynthesis of a macrocyclic peptide containing an unprecedented lysine-to-tryptophan crosslink. Nat. Chem. 2015, 7, 431–437. [Google Scholar] [CrossRef]
- Teng, K.; Huang, F.; Liu, Y.; Wang, Y.; Xia, T.; Yun, F.; Zhong, J. Food and gut originated bacteriocins involved in gut microbe-host interactions. Crit. Rev. Microbiol. 2022, 49, 515–527. [Google Scholar] [CrossRef]
- Zimina, M.; Babich, O.; Prosekov, A.; Sukhikh, S.; Ivanova, S.; Shevchenko, M.; Noskova, S. Overview of global trends in classification, methods of preparation and application of bacteriocins. Antibiotics 2020, 9, 553. [Google Scholar] [CrossRef]
- Bédard, F.; Biron, E. Recent progress in the chemical synthesis of class II and S-glycosylated bacteriocins. Front. Microbiol. 2018, 9, 1048. [Google Scholar] [CrossRef] [PubMed]
- Fields, F.R.; Lee, S.W.; McConnell, M.J. Using bacterial genomes and essential genes for the development of new antibiotics. Biochem. Pharmacol. 2017, 134, 74–86. [Google Scholar] [CrossRef] [PubMed]
- Gradisteanu Pircalabioru, G.; Popa, L.I.; Marutescu, L.; Gheorghe, I.; Popa, M.; Czobor Barbu, I.; Cristescu, R.; Chifiriuc, M.-C. Bacteriocins in the era of antibiotic resistance: Rising to the challenge. Pharmaceutics 2021, 13, 196. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Kuipers, O.P. Identification and classification of known and putative antimicrobial compounds produced by a wide variety of Bacillales species. BMC Genom. 2016, 17, 882. [Google Scholar] [CrossRef]
- Bindiya, E.S.; Bhat, S.G. Marine bacteriocins: A review. J. Bacteriol. Mycol. Open Access 2016, 2, 140–147. [Google Scholar] [CrossRef]
- Rather, I.A.; Galope, R.; Bajpai, V.K.; Lim, J.; Paek, W.K.; Park, Y.H. Diversity of marine bacteria and their bacteriocins: Applications in aquaculture. Rev. Fish. Sci. Aquac. 2017, 25, 257–269. [Google Scholar] [CrossRef]
- Santos, J.D.; Vitorino, I.; Reyes, F.; Vicente, F.; Lage, O.M. From ocean to medicine: Pharmaceutical applications of metabolites from marine bacteria. Antibiotics 2020, 9, 455. [Google Scholar] [CrossRef]
- Alam, K.; Islam, M.M.; Gong, K.; Abbasi, M.N.; Li, R.; Zhang, Y.; Li, A. In silico genome mining of potential novel biosynthetic gene clusters for drug discovery from Burkholderia bacteria. Comput. Biol. Med. 2022, 140, 105046. [Google Scholar] [CrossRef]
- Choi, S.; Baek, M.-g.; Chung, M.-J.; Lim, S.; Yi, H. Distribution of bacteriocin genes in the lineages of Lactiplantibacillus plantarum. Sci. Rep. 2021, 11, 20063. [Google Scholar] [CrossRef] [PubMed]
- Gardner, A.; West, S.A.; Buckling, A. Bacteriocins, spite and virulence. Proc. R. Soc. B Biol. Sci. 2004, 271, 1529–1535. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Gutiérrez, R.-A.; López-Ramírez, V.; Islas, Á.; Alcaraz, L.D.; Hernández-González, I.; Olivera, B.C.L.; Santillán, M.; Eguiarte, L.E.; Souza, V.; Travisano, M.; et al. Antagonism influences assembly of a Bacillus guild in a local community and is depicted as a food-chain network. ISME J. 2013, 7, 487–497. [Google Scholar] [CrossRef] [PubMed]
- Hawlena, H.; Bashey, F.; Lively, C.M. Bacteriocin-mediated interactions within and between coexisting species. Ecol. Evol. 2012, 2, 2516–2521. [Google Scholar] [CrossRef] [PubMed]
- Heilbronner, S.; Krismer, B.; Brötz-Oesterhelt, H.; Peschel, A. The microbiome-shaping roles of bacteriocins. Nat. Rev. Microbiol. 2021, 19, 726–739. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.G.; Becattini, S.; Moody, T.U.; Shliaha, P.V.; Littmann, E.R.; Seok, R.; Gjonbalaj, M.; Eaton, V.; Fontana, E.; Amoretti, L.; et al. Microbiota-derived lantibiotic restores resistance against vancomycin-resistant Enterococcus. Nature 2019, 572, 665–669. [Google Scholar] [CrossRef] [PubMed]
- Rossi, F.; Rizzotti, L.; Felis, G.E.; Torriani, S. Horizontal gene transfer among microorganisms in food: Current knowledge and future perspectives. Food Microbiol. 2014, 42, 232–243. [Google Scholar] [CrossRef] [PubMed]
- Xia, L.; Miao, Y.; Cao, A.; Liu, Y.; Liu, Z.; Sun, X.; Xue, Y.; Xu, Z.; Xun, W.; Shen, Q.; et al. Biosynthetic gene cluster profiling predicts the positive association between antagonism and phylogeny in Bacillus. Nat. Commun. 2022, 13, 1023. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Chen, Y.; Imre, K.; Arslan-Acaroz, D.; Istanbullugil, F.R.; Fang, Y.; Ros, G.; Zhu, K.; Acaroz, U. Mechanisms of probiotic Bacillus against enteric bacterial infections. One Health Adv. 2023, 1, 21. [Google Scholar] [CrossRef]
- Gonzalez, D.; Mavridou, D.A.I. Making the best of aggression: The many dimensions of bacterial toxin regulation. Trends Microbiol. 2019, 27, 897–905. [Google Scholar] [CrossRef] [PubMed]
- Skinnider, M.A.; Johnston, C.W.; Edgar, R.E.; Dejong, C.A.; Merwin, N.J.; Rees, P.N.; Magarvey, N.A. Genomic charting of ribosomally synthesized natural product chemical space facilitates targeted mining. Proc. Natl. Acad. Sci. USA 2016, 113, E6343–E6351. [Google Scholar] [CrossRef]
- Fobofou, S.A.; Savidge, T. Microbial metabolites: Cause or consequence in gastrointestinal disease? Am. J. Physiol. Liver Physiol. 2022, 322, G535–G552. [Google Scholar] [CrossRef]
- Hudson, G.A.; Burkhart, B.J.; DiCaprio, A.J.; Schwalen, C.J.; Kille, B.; Pogorelov, T.V.; Mitchell, D.A. Bioinformatic mapping of radical S-adenosylmethionine-dependent ribosomally synthesized and post-translationally modified peptides identifies new Cα, Cβ, and Cγ-linked thioether-containing peptides. J. Am. Chem. Soc. 2019, 141, 8228–8238. [Google Scholar] [CrossRef]
- Hudson, G.A.; Mitchell, D.A. RiPP antibiotics: Biosynthesis and engineering potential. Curr. Opin. Microbiol. 2018, 45, 61–69. [Google Scholar] [CrossRef]
- Akhter, S.; Miller, J.H. BaPreS: A software tool for predicting bacteriocins using an optimal set of features. BMC Bioinform. 2023, 24, 313. [Google Scholar] [CrossRef]
- Cui, Y.; Luo, L.; Wang, X.; Lu, Y.; Yi, Y.; Shan, Y.; Liu, B.; Zhou, Y.; Lü, X. Mining, heterologous expression, purification, antibactericidal mechanism, and application of bacteriocins: A review. Compr. Rev. Food Sci. Food Saf. 2021, 20, 863–899. [Google Scholar] [CrossRef]
- Lee, H.; Churey, J.J.; Worobo, R.W. Biosynthesis and transcriptional analysis of thurincin H, a tandem repeated bacteriocin genetic locus, produced by Bacillus thuringiensis SF361. FEMS Microbiol. Lett. 2009, 299, 205–213. [Google Scholar] [CrossRef]
- Ortiz-Rodríguez, T.; Mendoza-Acosta, F.; Martínez-Zavala, S.A.; Salcedo-Hernández, R.; Casados-Vázquez, L.E.; Bideshi, D.K.; Barboza-Corona, J.E. Thurincin H is a nonhemolytic bacteriocin of Bacillus thuringiensis with potential for applied use. Probiotics Antimicrob. Proteins 2023, 15, 955–966. [Google Scholar] [CrossRef]
- Wang, G.; Manns, D.C.; Churey, J.J.; Worobo, R.W. Development of a homologous expression system for and systematic site-directed mutagenesis analysis of thurincin H, a bacteriocin produced by Bacillus thuringiensis SF361. Appl. Environ. Microbiol. 2014, 80, 3576–3584. [Google Scholar] [CrossRef]
- Himes, P.M.; Allen, S.E.; Hwang, S.; Bowers, A.A. Production of sactipeptides in Escherichia coli: Probing the substrate promiscuity of subtilosin A biosynthesis. ACS Chem. Biol. 2016, 11, 1737–1744. [Google Scholar] [CrossRef] [PubMed]
- Phelan, R.W.; Barret, M.; Cotter, P.D.; O’Connor, P.M.; Chen, R.; Morrissey, J.P.; Dobson, A.D.W.; O’Gara, F.; Barbosa, T.M. Subtilomycin: A new lantibiotic from Bacillus subtilis strain MMA7 isolated from the marine sponge Haliclona simulans. Mar. Drugs 2013, 11, 1878–1898. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Hegemann, J.D.; Fage, C.D.; Zimmermann, M.; Xie, X.; Linne, U.; Marahiel, M.A. Insights into the unique phosphorylation of the lasso peptide paeninodin. J. Biol. Chem. 2016, 291, 13662–13678. [Google Scholar] [CrossRef] [PubMed]
- Zakrzewski, A.J.; Kurpas, M.; Zadernowska, A.; Chajęcka-Wierzchowska, W.; Fraqueza, M.J. A comprehensive virulence and resistance characteristics of Listeria monocytogenes isolated from fish and the fish industry environment. Int. J. Mol. Sci. 2023, 24, 3581. [Google Scholar] [CrossRef] [PubMed]
- Begley, M.; Cotter, P.D.; Hill, C.; Ross, R.P. Identification of a novel two-peptide lantibiotic, lichenicidin, following rational genome mining for LanM proteins. Appl. Environ. Microbiol. 2009, 75, 5451–5460. [Google Scholar] [CrossRef]
- Tymoszewska, A.; DIep, D.B.; Wirtek, P.; Aleksandrzak-Piekarczyk, T. The non-lantibiotic bacteriocin garvicin Q targets man-pts in a broad spectrum of sensitive bacterial genera. Sci. Rep. 2017, 7, 8359. [Google Scholar] [CrossRef]
- Soltani, M.; Baldisserotto, B.; Hosseini Shekarabi, S.P.; Shafiei, S.; Bashiri, M. Lactococcosis a re-emerging disease in aquaculture: Disease significant and phytotherapy. Vet. Sci. 2021, 8, 181. [Google Scholar] [CrossRef] [PubMed]
- Dobson, A.; Cotter, P.D.; Ross, R.P.; Hill, C. Bacteriocin production: A probiotic trait? Appl. Environ. Microbiol. 2012, 78, 1–6. [Google Scholar] [CrossRef]
- Kloosterman, A.M.; Medema, M.H.; van Wezel, G.P. Omics-based strategies to discover novel classes of RiPP natural products. Curr. Opin. Biotechnol. 2021, 69, 60–67. [Google Scholar] [CrossRef]
- Zheng, J.; Gänzle, M.G.; Lin, X.B.; Ruan, L.; Sun, M. Diversity and dynamics of bacteriocins from human microbiome. Environ. Microbiol. 2015, 17, 2133–2143. [Google Scholar] [CrossRef]
- Klemetsen, T.; Raknes, I.A.; Fu, J.; Agafonov, A.; Balasundaram, S.V.; Tartari, G.; Robertsen, E.; Willassen, N.P. The MAR databases: Development and implementation of databases specific for marine metagenomics. Nucleic Acids Res. 2018, 46, D693–D699. [Google Scholar] [CrossRef]
- Sievers, F.; Higgins, D.G. Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 2018, 27, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef]
- Asnicar, F.; Thomas, A.M.; Beghini, F.; Mengoni, C.; Manara, S.; Manghi, P.; Zhu, Q.; Bolzan, M.; Cumbo, F.; May, U.; et al. Precise phylogenetic analysis of microbial isolates and genomes from metagenomes using PhyloPhlAn 3.0. Nat. Commun. 2020, 11, 2500. [Google Scholar] [CrossRef] [PubMed]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2014, 12, 59–60. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Teber, R.; Asakawa, S. In Silico Screening of Bacteriocin Gene Clusters within a Set of Marine Bacillota Genomes. Int. J. Mol. Sci. 2024, 25, 2566. https://doi.org/10.3390/ijms25052566
Teber R, Asakawa S. In Silico Screening of Bacteriocin Gene Clusters within a Set of Marine Bacillota Genomes. International Journal of Molecular Sciences. 2024; 25(5):2566. https://doi.org/10.3390/ijms25052566
Chicago/Turabian StyleTeber, Rabeb, and Shuichi Asakawa. 2024. "In Silico Screening of Bacteriocin Gene Clusters within a Set of Marine Bacillota Genomes" International Journal of Molecular Sciences 25, no. 5: 2566. https://doi.org/10.3390/ijms25052566
APA StyleTeber, R., & Asakawa, S. (2024). In Silico Screening of Bacteriocin Gene Clusters within a Set of Marine Bacillota Genomes. International Journal of Molecular Sciences, 25(5), 2566. https://doi.org/10.3390/ijms25052566





