Integrative Metabolomic Analysis of Serum and Selected Serum Exosomal microRNA in Metastatic Castration-Resistant Prostate Cancer
Abstract
:1. Introduction
2. Results
2.1. Relative Changes in Blood Metabolites
2.2. Exosomal miRNA Expression in the Serum Samples of mCRPC
2.3. Correlations between miRs and Metabolites Levels
3. Discussion
4. Materials and Methods
4.1. Study Population
4.2. 1H-NMR Metabolomics
4.3. Exosomal miRs Expression
4.4. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bergengren, O.; Pekala, K.R.; Matsoukas, K.; Fainberg, J.; Mungovan, S.F.; Bratt, O.; Bray, F.; Brawley, O.; Luckenbaugh, A.N.; Mucci, L.; et al. 2022 Update on Prostate Cancer Epidemiology and Risk Factors—A Systematic Review. Eur. Urol. 2023, 84, 191–206. [Google Scholar] [CrossRef] [PubMed]
- Descotes, J.-L. Diagnosis of prostate cancer. Asian J. Urol. 2019, 6, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Bruno, S.M.; Falagario, U.G.; D’altilia, N.; Recchia, M.; Mancini, V.; Selvaggio, O.; Sanguedolce, F.; Del Giudice, F.; Maggi, M.; Ferro, M.; et al. PSA Density Help to Identify Patients With Elevated PSA Due to Prostate Cancer Rather Than Intraprostatic Inflammation: A Prospective Single Center Study. Front. Oncol. 2021, 11, 693684. [Google Scholar] [CrossRef] [PubMed]
- Fujita, K.; Nonomura, N. Urinary biomarkers of prostate cancer. Int. J. Urol. 2018, 25, 770–779. [Google Scholar] [CrossRef] [PubMed]
- Das, C.J.; Kubihal, V.; Sharma, S.; Kumar, R.; Seth, A.; Kumar, R.; Kaushal, S.; Sarangi, J.; Gupta, R. Multiparametric magnetic resonance imaging, 68Ga prostate-specific membrane antigen positron emission tomography–Computed tomography, and respective quantitative parameters in detection and localization of clinically significant prostate cancer in intermediate- and high-risk group patients: An Indian demographic study. Indian J. Nucl. Med. 2021, 36, 362–370. [Google Scholar] [CrossRef]
- Masoom, S.N.; Sundaram, K.M.; Ghanouni, P.; Fütterer, J.; Oto, A.; Ayyagari, R.; Sprenkle, P.; Weinreb, J.; Arora, S. Real-Time MRI-Guided Prostate Interventions. Cancers 2022, 14, 1860. [Google Scholar] [CrossRef]
- Ferretti, S.; Mercinelli, C.; Marandino, L.; Litterio, G.; Marchioni, M.; Schips, L. Metastatic Castration-Resistant Prostate Cancer: Insights on Current Therapy and Promising Experimental Drugs. Res. Rep. Urol. 2023, 15, 243–259. [Google Scholar] [CrossRef]
- Myint, Z.W.; Allison, D.B.; Ellis, C.S. A Case Report of Metastatic Castration-Resistant Prostate Cancer Harboring a PTEN Loss. Front. Oncol. 2021, 11, 731002. [Google Scholar] [CrossRef]
- Mottet, N.; van den Bergh, R.C.N.; Briers, E.; van den Broeck, T.; Cumberbatch, M.G.; De Santis, M.; Fanti, S.; Fossati, N.; Gandaglia, G.; Gillessen, S.; et al. EAU-EANM-ESTRO-ESUR-SIOG Guidelines on Prostate Cancer—2020 Update. Part 1: Screening, Diagnosis, and Local Treatment with Curative Intent. Eur. Urol. 2021, 79, 243–262. [Google Scholar] [CrossRef]
- Warburg, O. On the Origin of Cancer Cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef]
- Cardoso, H.J.; Carvalho, T.M.A.; Fonseca, L.R.S.; Figueira, M.I.; Vaz, C.V.; Socorro, S. Revisiting prostate cancer metabolism: From metabolites to disease and therapy. Med. Res. Rev. 2021, 41, 1499–1538. [Google Scholar] [CrossRef] [PubMed]
- Strmiska, V.; Michalek, P.; Eckschlager, T.; Stiborova, M.; Adam, V.; Krizkova, S.; Heger, Z. Prostate cancer-specific hallmarks of amino acids metabolism: Towards a paradigm of precision medicine. Biochim. Biophys. Acta (BBA)-Rev. Cancer 2019, 1871, 248–258. [Google Scholar] [CrossRef] [PubMed]
- Costello, L.C.; Feng, P.; Milon, B.; Tan, M.; Franklin, R.B. Role of zinc in the pathogenesis and treatment of prostate cancer: Critical issues to resolve. Prostate Cancer Prostatic Dis. 2004, 7, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Twum-Ampofo, J.; Fu, D.-X.; Passaniti, A.; Hussain, A.; Siddiqui, M.M. Metabolic targets for potential prostate cancer therapeutics. Curr. Opin. Oncol. 2016, 28, 241–247. [Google Scholar] [CrossRef] [PubMed]
- Beier, A.-M.K.; Puhr, M.; Stope, M.B.; Thomas, C.; Erb, H.H.H. Metabolic changes during prostate cancer development and progression. J. Cancer Res. Clin. Oncol. 2023, 149, 2259–2270. [Google Scholar] [CrossRef] [PubMed]
- Halliday, K.R.; Fenoglio-Preiser, C.; Sillerud, L.O. Differentiation of human tumors from nonmalignant tissue by natural-abundance 13C NMR spectroscopy. Magn. Reson. Med. 1988, 7, 384–411. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, H. Reprogramming of glucose, fatty acid and amino acid metabolism for cancer progression. Cell. Mol. Life Sci. 2016, 73, 377–392. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Zidan, H.E.; Abdul-Maksoud, R.S.; Elsayed, W.S.H.; Desoky, E.A.M. Diagnostic and prognostic value of serum miR-15a and miR-16-1 expression among egyptian patients with prostate cancer. IUBMB Life 2018, 70, 437–444. [Google Scholar] [CrossRef] [PubMed]
- Cutruzzolà, F.; Giardina, G.; Marani, M.; Macone, A.; Paiardini, A.; Rinaldo, S.; Paone, A. Glucose Metabolism in the Progression of Prostate Cancer. Front. Physiol. 2017, 8, 97. [Google Scholar] [CrossRef] [PubMed]
- Santotoribio, J.D.; Jimenez-Romero, M.E. Serum biomarkers of inflammation for diagnosis of prostate cancer in patients with nonspecific elevations of serum prostate specific antigen levels. Transl. Cancer Res. 2019, 8, 273–278. [Google Scholar] [CrossRef]
- DE LA Cruz-López, K.G.; Castro-Muñoz, L.J.; Reyes-Hernández, D.O.; García-Carrancá, A.; Manzo-Merino, J. Lactate in the Regulation of Tumor Microenvironment and Therapeutic Approaches. Front. Oncol. 2019, 9, 1143. [Google Scholar] [CrossRef]
- Chetta, P.; Sriram, R.; Zadra, G. Lactate as Key Metabolite in Prostate Cancer Progression: What Are the Clinical Implications? Cancers 2023, 15, 3473. [Google Scholar] [CrossRef]
- Lima, A.R.; Pinto, J.; Bastos, M.d.L.; Carvalho, M.; de Pinho, P.G. NMR-based metabolomics studies of human prostate cancer tissue. Metabolomics 2018, 14, 88. [Google Scholar] [CrossRef]
- Tessem, M.; Swanson, M.G.; Keshari, K.R.; Albers, M.J.; Joun, D.; Tabatabai, Z.L.; Simko, J.P.; Shinohara, K.; Nelson, S.J.; Vigneron, D.B.; et al. Evaluation of lactate and alanine as metabolic biomarkers of prostate cancer using 1H HR-MAS spectroscopy of biopsy tissues. Magn. Reson. Med. 2008, 60, 510–516. [Google Scholar] [CrossRef]
- van Asten, J.J.A.; Cuijpers, V.; de Kaa, C.H.-V.; Soede-Huijbregts, C.; Witjes, J.A.; Verhofstad, A.; Heerschap, A. High resolution magic angle spinning NMR spectroscopy for metabolic assessment of cancer presence and Gleason score in human prostate needle biopsies. Magn. Reson. Mater. Phys. Biol. Med. 2008, 21, 435–442. [Google Scholar] [CrossRef]
- Costello, L.C.; Franklin, R.B. The clinical relevance of the metabolism of prostate cancer; zinc and tumor suppression: Connecting the dots. Mol. Cancer 2006, 5, 17. [Google Scholar] [CrossRef] [PubMed]
- Costello, L.C.; Franklin, R.B. Concepts of citrate production and secretion by prostate: 2. Hormonal relationships in normal and neoplastic prostate. Prostate 1991, 19, 181–205. [Google Scholar] [CrossRef] [PubMed]
- Costello, L.C.; Franklin, R.B. Plasma Citrate Homeostasis: How It Is Regulated; and Its Physiological and Clinical Implications. An Important, but Neglected, Relationship in Medicine. J. Hum. Endocrinol. 2016, 1, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Yuan, X.-H.; Wang, J.-J.; Wang, Y.-C.; Li, S.-L. The diagnostic value of miRNA-141 in prostate cancer: A Systematic Review and PRISMA-Compliant Meta-Analysis. Medicine 2020, 99, e19993. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Zhu, Y.; Shao, X.; Cai, A.; Dong, B.; Xue, W.; Gao, H. Distinct Metabolic Signatures of Hormone-Sensitive and Castration-Resistant Prostate Cancer Revealed by a 1H NMR-Based Metabolomics of Biopsy Tissue. J. Proteome Res. 2020, 19, 3741–3749. [Google Scholar] [CrossRef]
- Kumar, D.; Gupta, A.; Mandhani, A.; Sankhwar, S.N. NMR spectroscopy of filtered serum of prostate cancer: A new frontier in metabolomics. Prostate 2016, 76, 1106–1119. [Google Scholar] [CrossRef]
- Costello, L.; Franklin, R.; Feng, P. Mitochondrial function, zinc, and intermediary metabolism relationships in normal prostate and prostate cancer. Mitochondrion 2005, 5, 143–153. [Google Scholar] [CrossRef]
- Haferkamp, S.; Drexler, K.; Federlin, M.; Schlitt, H.J.; Berneburg, M.; Adamski, J.; Gaumann, A.; Geissler, E.K.; Ganapathy, V.; Parkinson, E.K.; et al. Extracellular Citrate Fuels Cancer Cell Metabolism and Growth. Front. Cell Dev. Biol. 2020, 8, 602476. [Google Scholar] [CrossRef]
- Mycielska, M.E.; Dettmer, K.; Rümmele, P.; Schmidt, K.M.; Prehn, C.; Milenkovic, V.M.; Jagla, W.; Madej, G.M.; Lantow, M.; Schladt, M.T.; et al. Extracellular Citrate Affects Critical Elements of Cancer Cell Metabolism and Supports Cancer Development In Vivo. Cancer Res 2018, 78, 2513–2523. [Google Scholar] [CrossRef] [PubMed]
- Icard, P.; Simula, L.; Zahn, G.; Alifano, M.; Mycielska, M.E. The dual role of citrate in cancer. Biochim. Biophys. Acta (BBA)-Rev. Cancer 2023, 1878, 188987. [Google Scholar] [CrossRef] [PubMed]
- Buszewska-Forajta, M.; Monedeiro, F.; Gołębiowski, A.; Adamczyk, P.; Buszewski, B. Citric Acid as a Potential Prostate Cancer Biomarker Determined in Various Biological Samples. Metabolites 2022, 12, 268. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Mondul, A.M.; Weinstein, S.J.; Karoly, E.D.; Sampson, J.N.; Albanes, D. Prospective serum metabolomic profile of prostate cancer by size and extent of primary tumor. Oncotarget 2017, 8, 45190–45199. [Google Scholar] [CrossRef] [PubMed]
- Whitburn, J.; Edwards, C.M. Metabolism in the Tumour-Bone Microenvironment. Curr. Osteoporos. Rep. 2021, 19, 494–499. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.J.; Pollock, C.B.; Kelly, K. Mechanisms of cancer metastasis to the bone. Cell Res. 2005, 15, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Outschoorn, U.E.; Peiris-Pagés, M.; Pestell, R.G.; Sotgia, F.; Lisanti, M.P. Cancer metabolism: A therapeutic perspective. Nat. Rev. Clin. Oncol. 2017, 14, 11–31. [Google Scholar] [CrossRef]
- McGarrah, R.W.; White, P.J. Branched-chain amino acids in cardiovascular disease. Nat. Rev. Cardiol. 2023, 20, 77–89. [Google Scholar] [CrossRef]
- Jung, M.K.; Okekunle, A.P.; Lee, J.E.; Sung, M.K.; Lim, Y.J. Role of Branched-chain Amino Acid Metabolism in Tumor Development and Progression. J. Cancer Prev. 2021, 26, 237–243. [Google Scholar] [CrossRef]
- Ericksen, R.E.; Lim, S.L.; McDonnell, E.; Shuen, W.H.; Vadiveloo, M.; White, P.J.; Ding, Z.; Kwok, R.; Lee, P.; Radda, G.K.; et al. Loss of BCAA Catabolism during Carcinogenesis Enhances mTORC1 Activity and Promotes Tumor Development and Progression. Cell Metab. 2019, 29, 1151–1165.e6. [Google Scholar] [CrossRef]
- Hattori, A.; Tsunoda, M.; Konuma, T.; Kobayashi, M.; Nagy, T.; Glushka, J.; Tayyari, F.; McSkimming, D.; Kannan, N.; Tojo, A.; et al. Cancer progression by reprogrammed BCAA metabolism in myeloid leukaemia. Nature 2017, 545, 500–504. [Google Scholar] [CrossRef]
- Mayers, J.R.; Wu, C.; Clish, C.B.; Kraft, P.; Torrence, M.E.; Fiske, B.P.; Yuan, C.; Bao, Y.; Townsend, M.K.; Tworoger, S.S.; et al. Elevation of circulating branched-chain amino acids is an early event in human pancreatic adenocarcinoma development. Nat. Med. 2014, 20, 1193–1198. [Google Scholar] [CrossRef] [PubMed]
- Giskeødegård, G.F.; Hansen, A.F.; Bertilsson, H.; Gonzalez, S.V.; Kristiansen, K.A.; Bruheim, P.; Mjøs, S.A.; Angelsen, A.; Bathen, T.F.; Tessem, M.-B. Metabolic markers in blood can separate prostate cancer from benign prostatic hyperplasia. Br. J. Cancer 2015, 113, 1712–1719. [Google Scholar] [CrossRef] [PubMed]
- Dereziński, P.; Klupczynska, A.; Sawicki, W.; Pałka, J.A.; Kokot, Z.J. Amino Acid Profiles of Serum and Urine in Search for Prostate Cancer Biomarkers: A Pilot Study. Int. J. Med. Sci. 2017, 14, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Miyagi, Y.; Higashiyama, M.; Gochi, A.; Akaike, M.; Ishikawa, T.; Miura, T.; Saruki, N.; Bando, E.; Kimura, H.; Imamura, F.; et al. Plasma Free Amino Acid Profiling of Five Types of Cancer Patients and Its Application for Early Detection. PLoS ONE 2011, 6, e24143. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Xia, B.; Zheng, H.; Ning, J.; Zhu, Y.; Shao, X.; Liu, B.; Dong, B.; Gao, H. Identification of characteristic metabolic panels for different stages of prostate cancer by 1H NMR-based metabolomics analysis. J. Transl. Med. 2022, 20, 275. [Google Scholar] [CrossRef] [PubMed]
- Sakata, T.; Ferdous, G.; Tsuruta, T.; Satoh, T.; Baba, S.; Muto, T.; Ueno, A.; Kanai, Y.; Endou, H.; Okayasu, I. L-type amino-acid transporter 1 as a novel biomarker for high-grade malignancy in prostate cancer. Pathol. Int. 2009, 59, 7–18. [Google Scholar] [CrossRef]
- Peng, H.; Wang, Y.; Luo, W. Multifaceted role of branched-chain amino acid metabolism in cancer. Oncogene 2020, 39, 6747–6756. [Google Scholar] [CrossRef]
- Seyfried, T.N.; Flores, R.E.; Poff, A.M.; D’Agostino, D.P. Cancer as a metabolic disease: Implications for novel therapeutics. Carcinogenesis 2014, 35, 515–527. [Google Scholar] [CrossRef]
- Puchalska, P.; Crawford, P.A. Multi-dimensional Roles of Ketone Bodies in Fuel Metabolism, Signaling, and Therapeutics. Cell Metab. 2017, 25, 262–284. [Google Scholar] [CrossRef]
- Hwang, C.Y.; Choe, W.; Yoon, K.-S.; Ha, J.; Kim, S.S.; Yeo, E.-J.; Kang, I. Molecular Mechanisms for Ketone Body Metabolism, Signaling Functions, and Therapeutic Potential in Cancer. Nutrients 2022, 14, 4932. [Google Scholar] [CrossRef]
- Rodrigues, L.M.; Uribe-Lewis, S.; Madhu, B.; Honess, D.J.; Stubbs, M.; Griffiths, J.R. The action of β-hydroxybutyrate on the growth, metabolism and global histone H3 acetylation of spontaneous mouse mammary tumours: Evidence of a β-hydroxybutyrate paradox. Cancer Metab. 2017, 5, 4. [Google Scholar] [CrossRef] [PubMed]
- Ferrere, G.; Alou, M.T.; Liu, P.; Goubet, A.-G.; Fidelle, M.; Kepp, O.; Durand, S.; Iebba, V.; Fluckiger, A.; Daillère, R.; et al. Ketogenic diet and ketone bodies enhance the anticancer effects of PD-1 blockade. J. Clin. Investig. 2021, 6, e145207. [Google Scholar] [CrossRef] [PubMed]
- Saraon, P.; Cretu, D.; Musrap, N.; Karagiannis, G.S.; Batruch, I.; Drabovich, A.P.; van der Kwast, T.; Mizokami, A.; Morrissey, C.; Jarvi, K.; et al. Quantitative Proteomics Reveals That Enzymes of the Ketogenic Pathway Are Associated with Prostate Cancer Progression. Mol. Cell. Proteom. 2013, 12, 1589–1601. [Google Scholar] [CrossRef] [PubMed]
- Labanca, E.; Bizzotto, J.; Sanchis, P.; Anselmino, N.; Yang, J.; Shepherd, P.D.A.; Paez, A.; Antico-Arciuch, V.; Lage-Vickers, S.; Hoang, A.G.; et al. Prostate cancer castrate resistant progression usage of non-canonical androgen receptor signaling and ketone body fuel. Oncogene 2021, 40, 6284–6298. [Google Scholar] [CrossRef]
- Yoshii, Y.; Furukawa, T.; Saga, T.; Fujibayashi, Y. Acetate/acetyl-CoA metabolism associated with cancer fatty acid synthesis: Overview and application. Cancer Lett. 2015, 356, 211–216. [Google Scholar] [CrossRef] [PubMed]
- Zadra, G.; Loda, M. When fat goes down, prostate cancer is on the ropes. Mol. Cell. Oncol. 2019, 6, 1595308. [Google Scholar] [CrossRef]
- Gupta, A.; Gupta, S.; Mahdi, A.A. 1H NMR-derived serum metabolomics of leukoplakia and squamous cell carcinoma. Clin. Chim. Acta 2015, 441, 47–55. [Google Scholar] [CrossRef]
- Singh, A.; Prakash, V.; Gupta, N.; Kumar, A.; Kant, R.; Kumar, D. Serum Metabolic Disturbances in Lung Cancer Investigated through an Elaborative NMR-Based Serum Metabolomics Approach. ACS Omega 2022, 7, 5510–5520. [Google Scholar] [CrossRef]
- Gu, J.; Xiao, Y.; Shu, D.; Liang, X.; Hu, X.; Xie, Y.; Lin, D.; Li, H. Metabolomics Analysis in Serum from Patients with Colorectal Polyp and Colorectal Cancer by 1H-NMR Spectrometry. Dis. Markers 2019, 2019, 3491852. [Google Scholar] [CrossRef]
- Hosios, A.M.; Vander Heiden, M.G. Acetate metabolism in cancer cells. Cancer Metab. 2014, 2, 27. [Google Scholar] [CrossRef] [PubMed]
- Aqeilan, R.I.; Calin, G.A.; Croce, C.M. miR-15a and miR-16-1 in cancer: Discovery, function and future perspectives. Cell Death Differ. 2010, 17, 215–220. [Google Scholar] [CrossRef]
- Feng, Y.; Mao, Z.G.; Wang, X.; Du, Q.; Jian, M.; Zhu, D.; Xiao, Z.; Wang, H.J.; Zhu, Y.H. MicroRNAs and Target Genes in Pituitary Adenomas. Horm. Metab. Res. 2018, 50, 179–192. [Google Scholar] [CrossRef]
- Kim, J.; Cho, S.; Park, Y.; Lee, J.; Park, J. Evaluation of micro-RNA in extracellular vesicles from blood of patients with prostate cancer. PLoS ONE 2021, 16, e0262017. [Google Scholar] [CrossRef] [PubMed]
- Jin, W.; Chen, F.; Wang, K.; Song, Y.; Fei, X.; Wu, B. miR-15a/miR-16 cluster inhibits invasion of prostate cancer cells by suppressing TGF-β signaling pathway. Biomed. Pharmacother. 2018, 104, 637–644. [Google Scholar] [CrossRef] [PubMed]
- Musumeci, M.; Coppola, V.; Addario, A.; Patrizii, M.; Maugeri-Saccà, M.; Memeo, L.; Colarossi, C.; Francescangeli, F.; Biffoni, M.; Collura, D.; et al. Control of tumor and microenvironment cross-talk by miR-15a and miR-16 in prostate cancer. Oncogene 2011, 30, 4231–4242. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, X.; Shi, J.; Cao, P.; Wan, M.; Zhang, Q.; Wang, Y.; Kridel, S.J.; Liu, W.; Xu, J.; et al. Fatty acid synthase is a primary target of MiR-15a and MiR-16-1 in breast cancer. Oncotarget 2016, 7, 78566–78576. [Google Scholar] [CrossRef]
- Arora, S.; Singh, P.; Tabassum, G.; Dohare, R.; Syed, M.A. miR-16-5p regulates aerobic glycolysis and tumorigenesis of NSCLC cells via LDH-A/lactate/NF-κB signaling. Life Sci. 2022, 304, 120722. [Google Scholar] [CrossRef]
- Feng, Y.-G.; Zhao, J.-F.; Xiao, L.; Rao, W.-Y.; Ran, C.; Xiao, Y.-H. MicroRNA-19a-3p suppresses invasion and metastasis of prostate cancer via inhibiting SOX4. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 6245–6251. [Google Scholar] [PubMed]
- Wa, Q.; Li, L.; Lin, H.; Peng, X.; Ren, N.; Huang, Y.; He, P.; Huang, S. Downregulation of miR-19a-3p promotes invasion, migration and bone metastasis via activating TGF-β signaling in prostate cancer. Oncol. Rep. 2018, 39, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Pinto, S.K.; Lamon, S.; Stephenson, E.J.; Kalanon, M.; Mikovic, J.; Koch, L.G.; Britton, S.L.; Hawley, J.A.; Camera, D.M. Expression of microRNAs and target proteins in skeletal muscle of rats selectively bred for high and low running capacity. Am. J. Physiol. Endocrinol. Metab. 2017, 313, E335–E343. [Google Scholar] [CrossRef] [PubMed]
- Schramedei, K.; Mörbt, N.; Pfeifer, G.; Läuter, J.; Rosolowski, M.; Tomm, J.M.; von Bergen, M.; Horn, F.; Brocke-Heidrich, K. MicroRNA-21 targets tumor suppressor genes ANP32A and SMARCA4. Oncogene 2011, 30, 2975–2985. [Google Scholar] [CrossRef] [PubMed]
- Bautista-Sánchez, D.; Arriaga-Canon, C.; Pedroza-Torres, A.; De La Rosa-Velázquez, I.A.; González-Barrios, R.; Contreras-Espinosa, L.; Montiel-Manríquez, R.; Castro-Hernández, C.; Fragoso-Ontiveros, V.; Álvarez-Gómez, R.M.; et al. The Promising Role of MiR-21 as a Cancer Biomarker and Its Importance in RNA-Based Therapeutics. Mol. Ther. Nucleic Acids 2020, 20, 409–420. [Google Scholar] [CrossRef] [PubMed]
- Dai, Q.; Li, N.; Zhou, X. Increased miR-21a provides metabolic advantages through suppression of FBP1 expression in non-small cell lung cancer cells. Am. J. Cancer Res. 2017, 7, 2121–2130. [Google Scholar] [PubMed]
- Chen, S.; Chen, X.; Shan, T.; Ma, J.; Lin, W.; Li, W.; Kang, Y. MiR-21-mediated Metabolic Alteration of Cancer-associated Fibroblasts and Its Effect on Pancreatic Cancer Cell Behavior. Int. J. Biol. Sci. 2018, 14, 100–110. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Cappello, T.; Wang, L. Emerging role of microRNAs in lipid metabolism. Acta Pharm. Sin. B 2015, 5, 145–150. [Google Scholar] [CrossRef]
- Ni, K.; Ni, K.; Wang, D.; Wang, D.; Xu, H.; Xu, H.; Mei, F.; Mei, F.; Wu, C.; Wu, C.; et al. miR-21 promotes non-small cell lung cancer cells growth by regulating fatty acid metabolism. Cancer Cell Int. 2019, 19, 219. [Google Scholar] [CrossRef] [PubMed]
- Kanagasabai, T.; Li, G.; Shen, T.H.; Gladoun, N.; Castillo-Martin, M.; Celada, S.I.; Xie, Y.; Brown, L.K.; Mark, Z.A.; Ochieng, J.; et al. MicroRNA-21 deficiency suppresses prostate cancer progression through downregulation of the IRS1-SREBP-1 signaling pathway. Cancer Lett. 2022, 525, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Hashemi, M.; Mirdamadi, M.S.A.; Talebi, Y.; Khaniabad, N.; Banaei, G.; Daneii, P.; Gholami, S.; Ghorbani, A.; Tavakolpournegari, A.; Farsani, Z.M.; et al. Pre-clinical and clinical importance of miR-21 in human cancers: Tumorigenesis, therapy response, delivery approaches and targeting agents. Pharmacol. Res. 2023, 187, 106568. [Google Scholar] [CrossRef] [PubMed]
- Damodaran, M.; Dandapani, M.C.; Raj, S.D.; Sundaram, S.; VenkatRamanan, S.; Ramachandran, I.; Venkatesan, V. Differentially expressed miR-20, miR-21, miR-100, miR-125a and miR-146a as a potential biomarker for prostate cancer. Mol. Biol. Rep. 2021, 48, 3349–3356. [Google Scholar] [CrossRef]
- Joković, S.M.; Dobrijević, Z.; Kotarac, N.; Filipović, L.; Popović, M.; Korać, A.; Vuković, I.; Savić-Pavićević, D.; Brajušković, G. MiR-375 and miR-21 as Potential Biomarkers of Prostate Cancer: Comparison of Matching Samples of Plasma and Exosomes. Genes 2022, 13, 2320. [Google Scholar] [CrossRef]
- Canturk, K.M.; Ozdemir, M.; Can, C.; Öner, S.; Emre, R.; Aslan, H.; Cilingir, O.; Ciftci, E.; Celayir, F.M.; Aldemir, O.; et al. Investigation of key miRNAs and target genes in bladder cancer using miRNA profiling and bioinformatic tools. Mol. Biol. Rep. 2014, 41, 8127–8135. [Google Scholar] [CrossRef]
- Scarpati, G.D.V.; Calura, E.; Di Marino, M.; Romualdi, C.; Beltrame, L.; Malapelle, U.; Troncone, G.; De Stefano, A.; Pepe, S.; De Placido, S.; et al. Analysis of Differential miRNA Expression in Primary Tumor and Stroma of Colorectal Cancer Patients. BioMed Res. Int. 2014, 2014, 840921. [Google Scholar] [CrossRef]
- Huang, S.; Wa, Q.; Pan, J.; Peng, X.; Ren, D.; Huang, Y.; Chen, X.; Tang, Y. Downregulation of miR-141-3p promotes bone metastasis via activating NF-κB signaling in prostate cancer. J. Exp. Clin. Cancer Res. 2017, 36, 173. [Google Scholar] [CrossRef]
- Ye, Y.; Li, S.-L.; Ma, Y.-Y.; Diao, Y.-J.; Yang, L.; Su, M.-Q.; Li, Z.; Ji, Y.; Wang, J.; Lei, L.; et al. Exosomal miR-141-3p regulates osteoblast activity to promote the osteoblastic metastasis of prostate cancer. Oncotarget 2017, 8, 94834–94849. [Google Scholar] [CrossRef] [PubMed]
- Akalin, I.; Erol, B.; Aslan, E.; Ozkanli, S.S.; Efiloglu, O.; Yildirim, S.; Caskurlu, T.; Yildirim, A.; Karaman, M.I. A New Promising Pathway in Aggressive Prostate Cancer: Treg/mir-let8c/lin28b. Arch. Esp. Urol. 2022, 75, 459–466. [Google Scholar] [CrossRef] [PubMed]
- Cornford, P.; van den Bergh, R.C.N.; Briers, E.; van den Broeck, T.; Cumberbatch, M.G.; De Santis, M.; Fanti, S.; Fossati, N.; Gandaglia, G.; Gillessen, S.; et al. EAU-EANM-ESTRO-ESUR-SIOG Guidelines on Prostate Cancer. Part II—2020 Update: Treatment of Relapsing and Metastatic Prostate Cancer. Eur. Urol. 2021, 79, 263–282. [Google Scholar] [CrossRef] [PubMed]
- Gowda, G.A.N.; Gowda, Y.N.; Raftery, D. Expanding the Limits of Human Blood Metabolite Quantitation Using NMR Spectroscopy. Anal. Chem. 2015, 87, 706–715. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Guo, A.; Oler, E.; Wang, F.; Anjum, A.; Peters, H.; Dizon, R.; Sayeeda, Z.; Tian, S.; Lee, B.L.; et al. HMDB 5.0: The Human Metabolome Database for 2022. Nucleic Acids Res. 2021, 50, D622–D631. [Google Scholar] [CrossRef] [PubMed]
- Sarlinova, M.; Halasa, M.; Mistuna, D.; Musak, L.; Iliev, R.; Slaby, O.; Mazuchova, J.; Valentova, V.; Plank, L.; Halasova, E. miR-21, miR-221 and miR-150 Are Deregulated in Peripheral Blood of Patients with Colorectal Cancer. Anticancer. Res. 2016, 36, 5449–5454. [Google Scholar] [CrossRef] [PubMed]
- Benko, J.; Sarlinova, M.; Mikusova, V.; Bolek, T.; Pec, M.J.; Halasova, E.; Galajda, P.; Samos, M.; Mokan, M. MiR-126 and miR-146a as markers of type 2 diabetes mellitus: A pilot study. Bratisl. Med. J. 2023, 124, 527–533. [Google Scholar] [CrossRef]
- Mishra, P.; Pandey, C.M.; Singh, U.; Gupta, A.; Sahu, C.; Keshri, A. Descriptive statistics and normality tests for sta-tistical data. Ann. Card. Anaesth. 2019, 22, 67–72. [Google Scholar] [CrossRef]
- Chughtai, B.; Forde, J.C.; Thomas, D.D.M.; Laor, L.; Hossack, T.; Woo, H.H.; Te, A.E.; Kaplan, S.A. Benign prostatic hyperplasia. Nat. Rev. Dis. Prim. 2016, 2, 16031. [Google Scholar] [CrossRef]
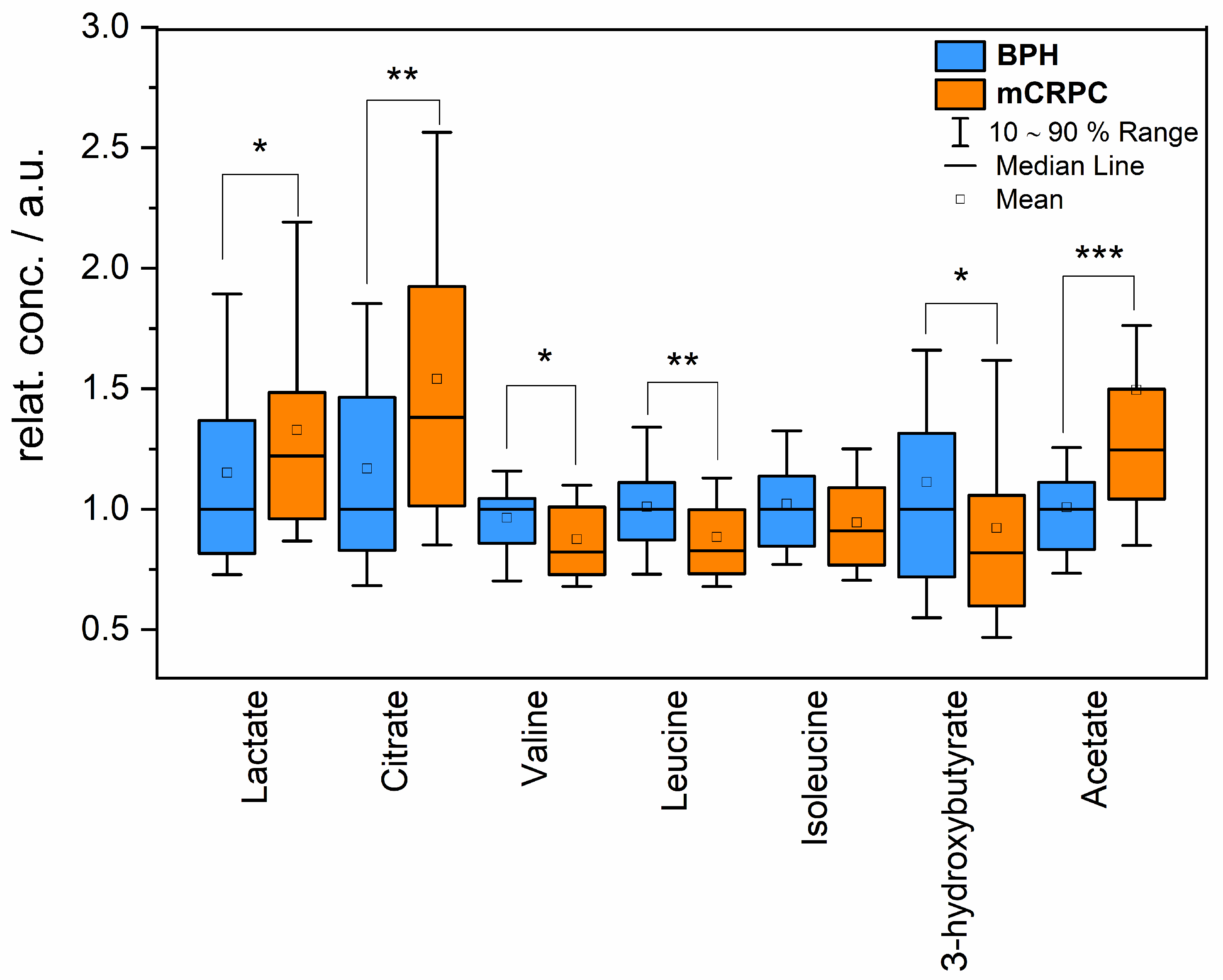
| Metabolite | Percentage Change mCRPC vs. BPH Group | p-Value |
|---|---|---|
| Lactate | 22% | 0.04 |
| Citrate | 38% | 0.003 |
| Valine | −18% | 0.02 |
| Leucine | −18% | 0.002 |
| Isoleucine | −11% | 0.06 |
| 3-hydroxybutyrate | −19% | 0.03 |
| Acetate | 24% | 0.00002 |
| miR | Fold Regulation +/− | p-Value |
|---|---|---|
| miR-15a | −2.00 | 0.006 |
| miR-16 | −3.24 | 4.33 × 10−6 |
| miR-19a-3p | −2.78 | 1.11 × 10−5 |
| miR-21 | −2.39 | 0.003 |
| miR-141a-3p | +1.23 | 0.41 |
| miR-15a | miR-16 | miR-19a-3p | miR-21 | miR-141a-3p | ||
|---|---|---|---|---|---|---|
| Lactate | Pearson Corr. | 0.35 | 0.31 | 0.33 | 0.27 | 0.01 |
| p-value | 0.01 | 0.03 | 0.03 | 0.06 | 0.92 | |
| Citrate | Pearson Corr. | −0.47 | −0.24 | −0.35 | −0.47 | −0.24 |
| p-value | 9.23 × 10−4 | 0.11 | 0.02 | 7.61 × 10−4 | 0.09 | |
| Valine | Pearson Corr. | 0.05 | 0.05 | 0.15 | 0.14 | −0.09 |
| p-value | 0.73 | 0.071 | 0.33 | 0.35 | 0.55 | |
| Leucine | Pearson Corr. | −0.04 | −0.02 | 0.05 | 0.08 | −0.09 |
| p-value | 0.78 | 0.89 | 0.72 | 0.55 | 0.55 | |
| Isoleucine | Pearson Corr. | −0.23 | −0.09 | −0.07 | −0.12 | −0.08 |
| p-value | 0.11 | 0.52 | 0.62 | 0.44 | 0.59 | |
| 3-hydroxybutyrate | Pearson Corr. | 0.28 | 0.09 | 0.17 | 0.33 | 0.05 |
| p-value | 0.05 | 0.51 | 0.24 | 0.03 | 0.72 | |
| Acetate | Pearson Corr. | 0.16 | 0.01 | 0.30 | 0.46 | 0.42 |
| p-value | 0.29 | 0.96 | 0.04 | 0.001 | 0.003 | |
| Glutamine | Pearson Corr. | 0.28 | 0.28 | 0.21 | 0.19 | −0.14 |
| p-value | 0.05 | 0.05 | 0.16 | 0.21 | 0.36 | |
| Lysine | Pearson Corr. | −0.44 | −0.22 | −0.29 | −0.39 | −0.04 |
| p-value | 0.002 | 0.14 | 0.05 | 0.006 | 0.81 |
| mCRPC Patients | BPH Patients | p-Value | |
|---|---|---|---|
| Number | 51 | 48 | |
| Age (years, mean ± SD) | 73.5 ± 7.65 | 67.5 ± 6.54 | <0.05 |
| Range | (58–85) | (55–84) | |
| PSA (ng/mL, mean ± SD) | 103.5 ± 1478.7 | 3.29 ± 9.77 | <0.05 |
| Range | (2.2–9506) | (0.2–66.5) | |
| T Staging | No. | N/A | – |
| T2 | 2 | ||
| T3 | 35 | ||
| T4 | 14 | ||
| N staging | No. | N/A | – |
| N0 | 40 | ||
| N1 | 11 | ||
| M staging * | No. | N/A | – |
| M1a | 8 | ||
| M1b | 51 | ||
| M1c | 4 | ||
| Gleason score | No. | N/A | – |
| 6 | 1 | ||
| 7 | 9 | ||
| 8 | 17 | ||
| 9 | 20 | ||
| 10 | 4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Evin, D.; Evinová, A.; Baranovičová, E.; Šarlinová, M.; Jurečeková, J.; Kaplán, P.; Poláček, H.; Halašová, E.; Dušenka, R.; Briš, L.; et al. Integrative Metabolomic Analysis of Serum and Selected Serum Exosomal microRNA in Metastatic Castration-Resistant Prostate Cancer. Int. J. Mol. Sci. 2024, 25, 2630. https://doi.org/10.3390/ijms25052630
Evin D, Evinová A, Baranovičová E, Šarlinová M, Jurečeková J, Kaplán P, Poláček H, Halašová E, Dušenka R, Briš L, et al. Integrative Metabolomic Analysis of Serum and Selected Serum Exosomal microRNA in Metastatic Castration-Resistant Prostate Cancer. International Journal of Molecular Sciences. 2024; 25(5):2630. https://doi.org/10.3390/ijms25052630
Chicago/Turabian StyleEvin, Daniel, Andrea Evinová, Eva Baranovičová, Miroslava Šarlinová, Jana Jurečeková, Peter Kaplán, Hubert Poláček, Erika Halašová, Róbert Dušenka, Lukáš Briš, and et al. 2024. "Integrative Metabolomic Analysis of Serum and Selected Serum Exosomal microRNA in Metastatic Castration-Resistant Prostate Cancer" International Journal of Molecular Sciences 25, no. 5: 2630. https://doi.org/10.3390/ijms25052630
APA StyleEvin, D., Evinová, A., Baranovičová, E., Šarlinová, M., Jurečeková, J., Kaplán, P., Poláček, H., Halašová, E., Dušenka, R., Briš, L., Brožová, M. K., & Sivoňová, M. K. (2024). Integrative Metabolomic Analysis of Serum and Selected Serum Exosomal microRNA in Metastatic Castration-Resistant Prostate Cancer. International Journal of Molecular Sciences, 25(5), 2630. https://doi.org/10.3390/ijms25052630






