Inhibition of Multifunctional Protein p32/C1QBP Promotes Cytostatic Effects in Colon Cancer Cells by Altering Mitogenic Signaling Pathways and Promoting Mitochondrial Damage
Abstract
1. Introduction
2. Results
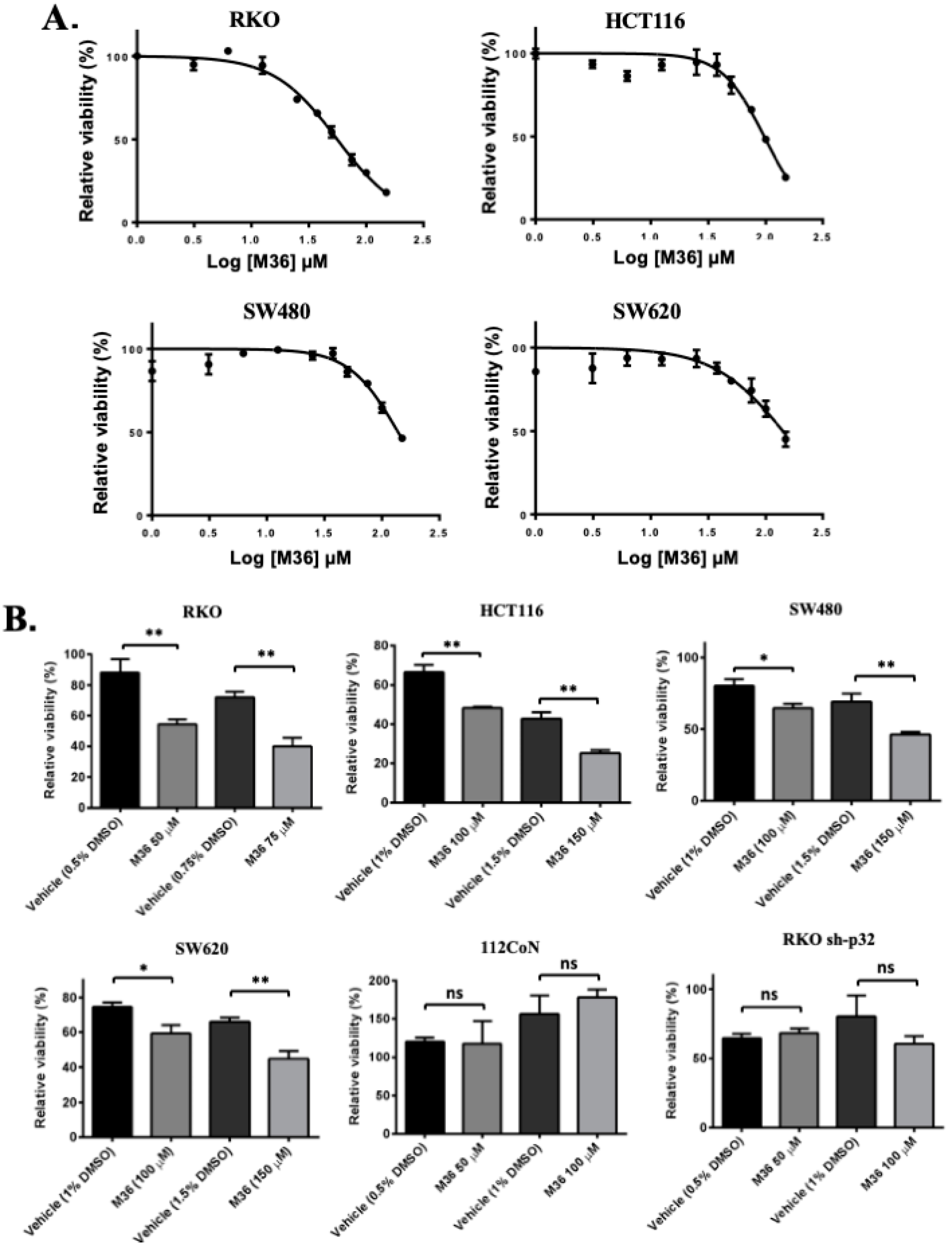
2.1. Pharmacological Inhibition of the p32/C1QBP Protein Negatively Affects the Viability of Colon Cancer Cells
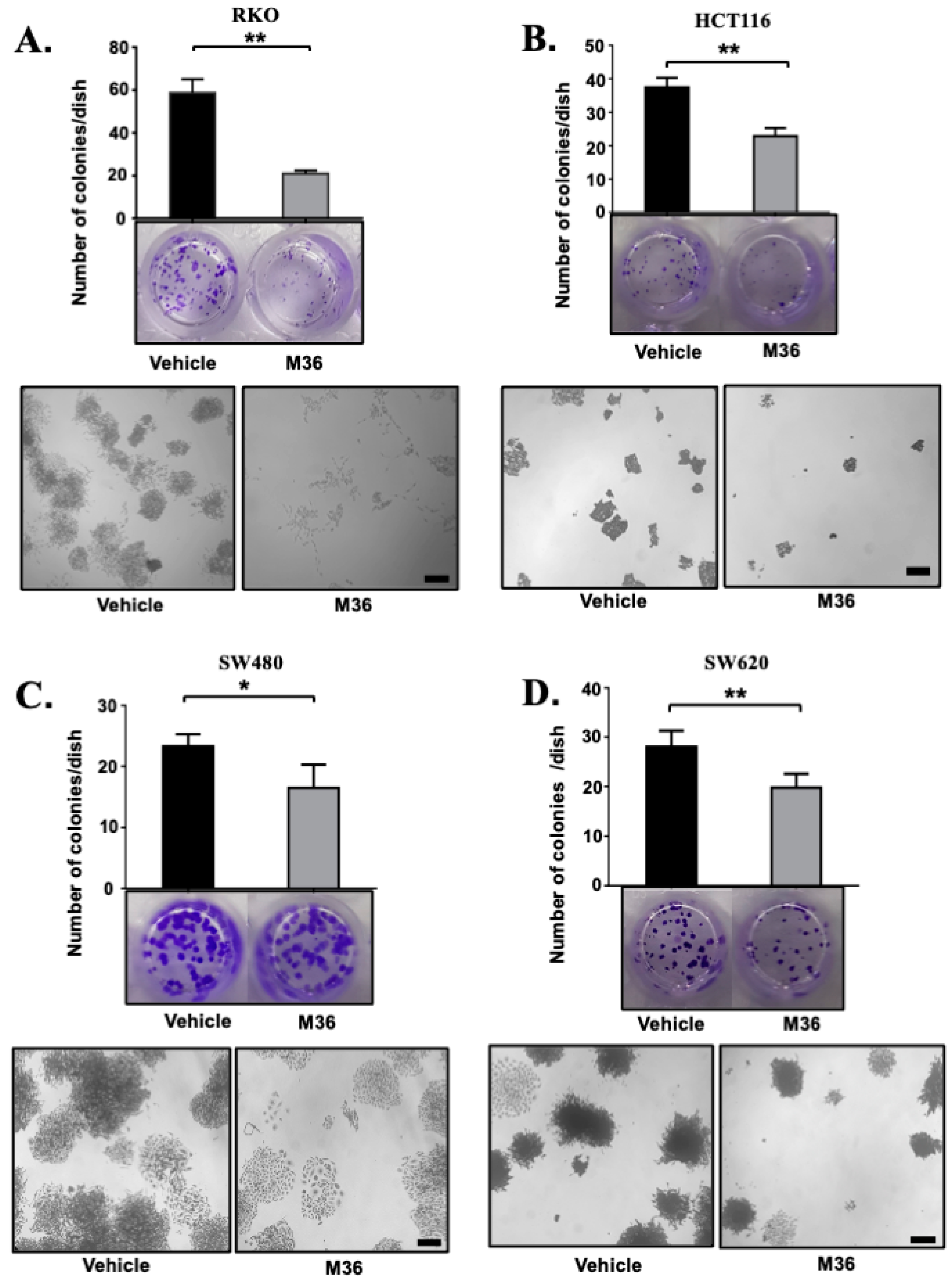
2.2. Pharmacological Inhibition of p32 Protein Significantly Decreased the Clonogenic Capacity of Colon Cancer Cells
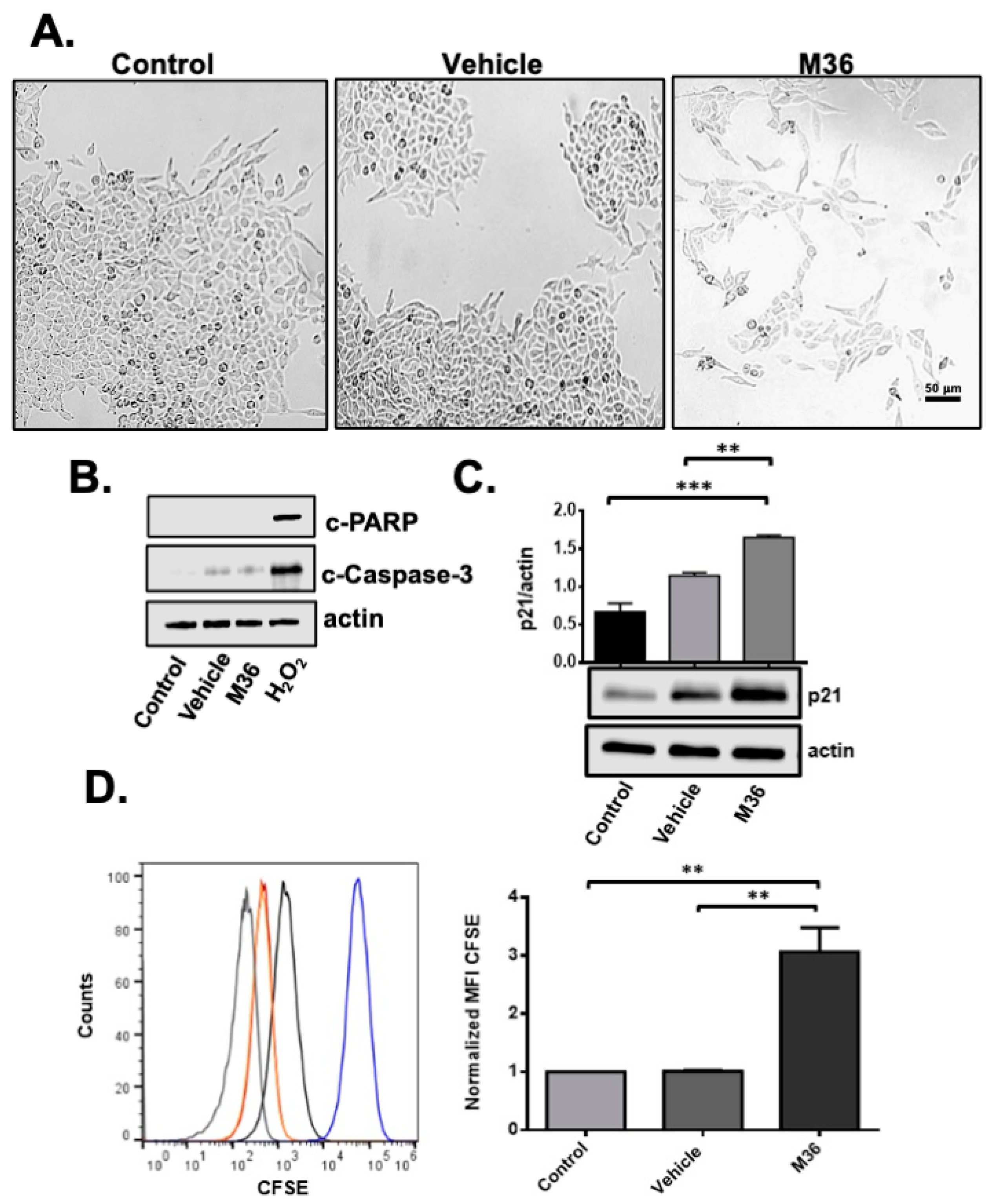
2.3. Pharmacological Inhibition of p32 Protein Induces a Cytostaic but Not Cytotoxic Effect on RKO Colon Cancer Cells
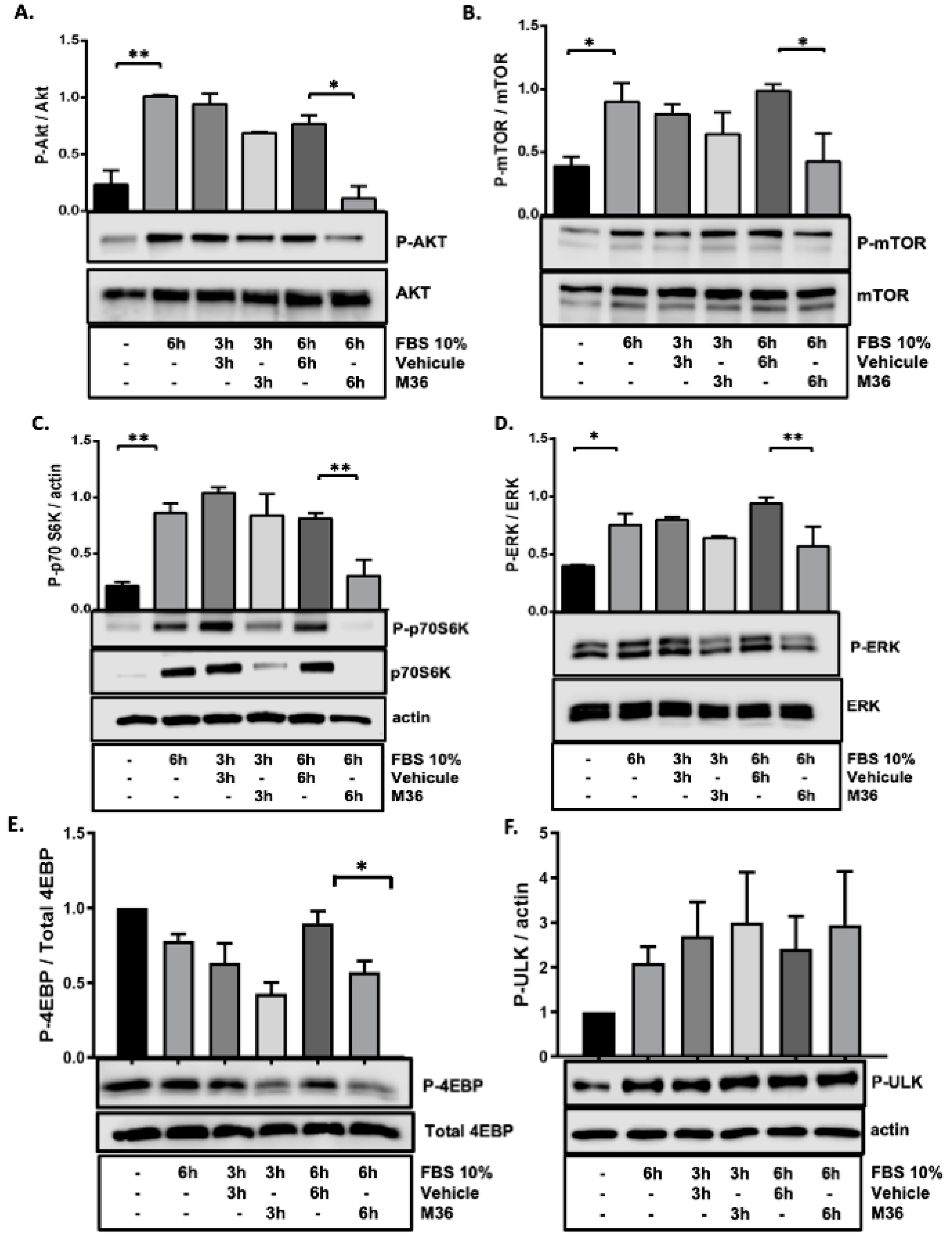
2.4. Pharmacological Inhibition of the p32/C1QBP Protein Negatively Affects the Rate of the Activation of Mitogenic Signaling Pathways
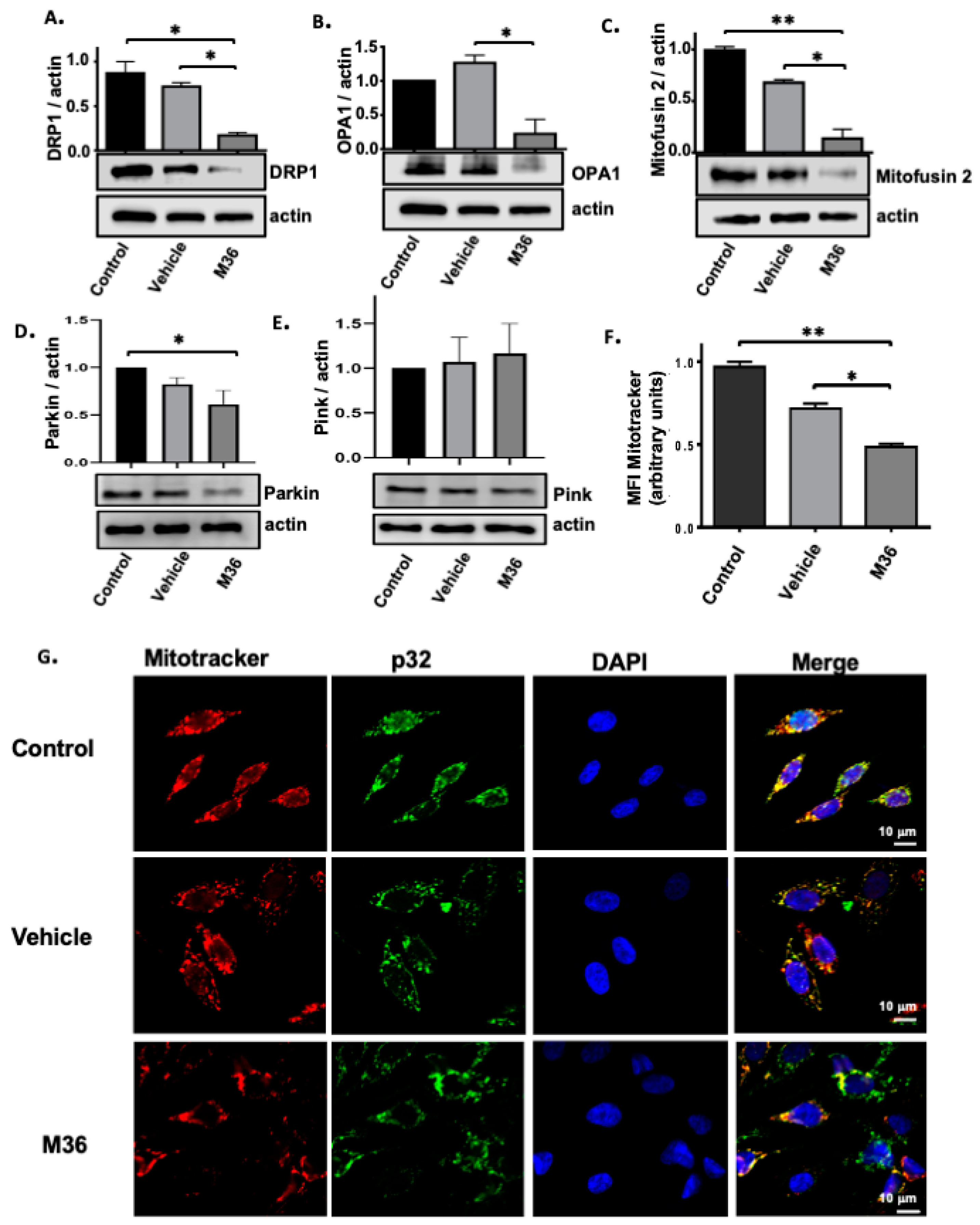
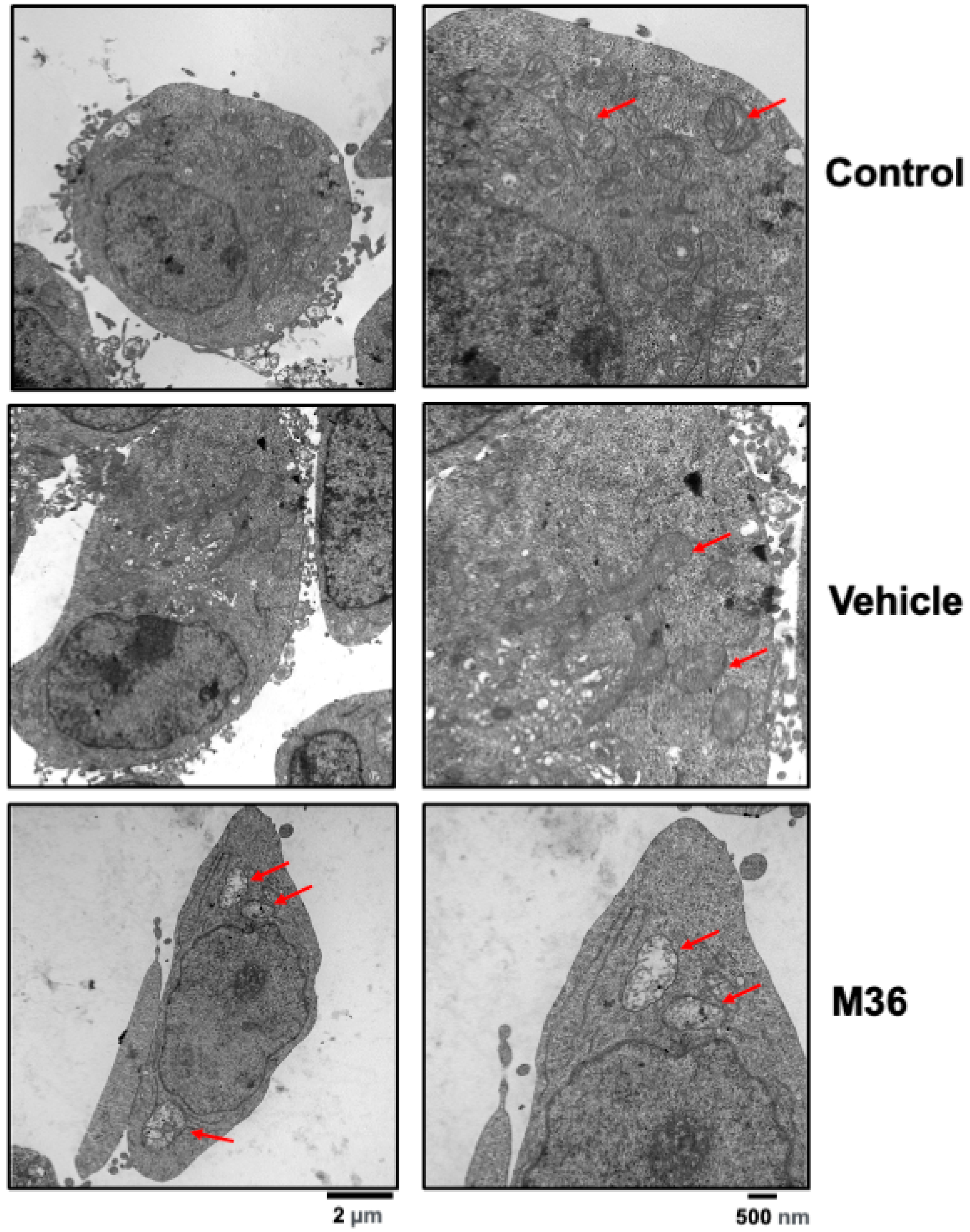
2.5. Pharmacological Inhibition of the p32/C1QBP Protein Negatively Affects the Level of Expression Levels of Proteins Involved in Mitochondrial Dynamics and Induces Mitochondrial Damage
3. Discussion
4. Materials and Methods
4.1. Antibodies and Reagents
4.2. Cell Culture
4.3. Viability Assays
4.4. MitoTracker Assay
4.5. Confocal Immunofluorescence Microscopy
4.6. Colony Formation Assay
4.7. Proliferation Assay
4.8. Western Blot
4.9. Evaluation of the Akt/mTOR and MAPK Cell Signaling Pathways
4.10. Transmission Electron Microscopy (TEM)
4.11. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- GLOBOCAN. GLOBOCAN 2020: New Global Cancer Data. 2020. Available online: https://www.uicc.org/news/globocan-2020-new-global-cancer-data (accessed on 14 December 2023).
- Morris, V.K.; Kennedy, E.B.; Baxter, N.N.; Benson, A.B.; Cercek, A.; Cho, M.; Ciombor, K.K.; Cremolini, C.; Davis, A.; Deming, D.A.; et al. Treatment of Metastatic Colorectal Cancer: ASCO Guideline. J. Clin. Oncol. 2022, 41, 678–700. [Google Scholar] [CrossRef] [PubMed]
- Egusquiza, C.A.; Robles, M. An approach to p32/gC1qR/HABP1: A multifunctional protein with an essential role in cancer. J. Cancer Res. Clin. Oncol. 2022, 148, 1831–1854. [Google Scholar] [CrossRef]
- Ghebrehiwet, B.; Geisbrecht, B.V.; Xu, X.; Savitt, A.G.; Peerschke, E.I.B. The C1q Receptors: Focus on gC1qR/p33 (C1qBP, p32, HABP-1). Semin. Inmunol. 2019, 45, 101338. [Google Scholar] [CrossRef]
- Peerschke, E.I.B.; Ghebrehiwet, B. cC1qR/CR and gC1qR/p33: Observations in cancer. Mol. Immunol. 2014, 61, 100–109. [Google Scholar] [CrossRef]
- Jiang, J.; Zhang, Y.; Krainer, A.R.; Xu, R.M. Crystal structure of human p32, a doughnut-shaped acidic mitochondrial matrix protein. Proc. Natl. Acad. Sci. USA 1999, 96, 3572–3577. [Google Scholar] [CrossRef]
- Dedio, J.; Jahnen-Dechent, W.; Bachman, M.; Müller-Esterl, W. The Multiligand-Binding Protein gC1qR, Putative C1q Receptor, Is a Mitochondrial Protein. J. Inmunol. 1998, 160, 3534–3542. [Google Scholar] [CrossRef]
- Muta, T.; Kang, D.; Kitajima, S.; Fujiwara, T.; Hamasaki, N. p32 Protein, a Splicing Factor 2-associated Protein, Is Localized in Mitochondrial Matrix and Is Functionally Important in Maintaining Oxidative Phosphorylation. J. Biol. Chem. 1997, 272, 24363–24370. [Google Scholar] [CrossRef] [PubMed]
- Ghebrehiwet, B.; Lim, B.; Kumar, R.; Feng, X.; Peerschke, E.I.B. gC1q-R/p33, a member of a new class of multifunctional and multicompartmental cellular proteins, is involved in inflammation and infection. Immunol. Rev. 2001, 180, 65–77. [Google Scholar] [CrossRef] [PubMed]
- Jha, B.K.; Salunke, D.M.; Datta, K. Structural Flexibility of Multifunctional HABP1 May Be Important for Regulating Its Binding to Different Ligands. J. Biol. Chem. 2003, 278, 27464–27472. [Google Scholar] [CrossRef]
- Storz, P.; Hausser, A.; Link, G.; Dedio, J.; Ghebrehiwet, B.; Pfizenmaier, K.; Johannes, F.J. Protein kinase C μ is regulated by the multifunctional chaperon protein p32. J. Biol. Chem. 2000, 275, 24601–24607. [Google Scholar] [CrossRef]
- Robles, M.; Rendón, E.; González, H.; Mendoza, G.; Islas, S.; Mendoza, V.; Ponce-Castañeda, M.V.; González-Mariscal, L.; López-Casillas, F. p32 (gC1qBP) is a general protein kinase C (PKC)-binding protein. Interaction and cellular localization of p32-PKC complexes in rat hepatocytes. J. Biol. Chem. 2002, 277, 5247–5255. [Google Scholar] [CrossRef]
- Krainer, A.R.; Mayeda, A.; Kozak, D.; Binns, G. Functional Expression of Cloned Human Splicing Factor SF2: Homology to RNA-Binding Proteins, Ul 70K, and Drosophila Splicing Regulators. Cell 1991, 66, 383–394. [Google Scholar] [CrossRef]
- Petersen, S.K.; Estmer, C.; Öhrmalm, C.; Matthews, D.A.; Russell, W.C.; Akusjärvi, G. The splicing factor-associated protein, p32, regulates RNA splicing by inhibiting ASF/SF2 RNA binding and phosphorylation. EMBO J. 1999, 18, 1014–1024. [Google Scholar] [CrossRef] [PubMed]
- Ghate, N.B.; Kim, J.; Shin, Y.; Situ, A.; Ulmer, T.S. p32 is a negative regulator of p53 tetramerization and transactivation. Mol. Oncol. 2019, 13, 1976–1992. [Google Scholar] [CrossRef]
- Seytter, T.; Lottspeich, F.; Neupert, W.; Schwarz, E. Mam33p, an Oligomeric, Acidic Protein in the Mitochondrial Matrix of Saccharomyces cerevisiae is Related to the Human Complement Receptor gC1q-R. Yeast 1998, 14, 303–310. [Google Scholar] [CrossRef]
- Yagi, M.; Uchiumi, T.; Takazaki, S.; Okuno, B.; Nomura, M.; Yoshida, S.I.; Kanki, T.; Kang, D. P32/gC1qR is indispensable for fetal development and mitochondrial translation: Importance of its RNA-binding ability. Nucleic Acids Res. 2012, 40, 9717–9737. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.J.; Crawford, S.A.; Henstridge, D.C.; Ng, I.H.W.; Boey, E.J.H.; Xu, Y.; Febbraio, M.A.; Jans, D.A.; Bogoyevitch, M.A. P32 Protein Levels Are Integral To Mitochondrial and Endoplasmic Reticulum Morphology, Cell Metabolism and Survival. Biochem. J. 2013, 453, 381–391. [Google Scholar] [CrossRef] [PubMed]
- Noh, S.; Phorl, S.; Naskar, R.; Oeum, K.; Seo, Y.; Kim, E.; Kweon, H.S.; Lee, J.Y. p32/C1QBP regulates OMA1-dependent proteolytic processing of OPA1 to maintain mitochondrial connectivity related to mitochondrial dysfunction and apoptosis. Sci. Rep. 2020, 10, 10618. [Google Scholar] [CrossRef] [PubMed]
- McGee, A.M.; Baines, C.P. Complement 1q-binding protein inhibits the mitochondrial permeability transition pore and protects against oxidative stress-induced death. Biochem. J. 2011, 433, 119–125. [Google Scholar] [CrossRef] [PubMed]
- McGee, A.M.; Douglas, D.L.; Liang, Y.; Hyder, S.M.; Baines, C.P. The mitochondrial protein C1qbp promotes cell proliferation, migration and resistance to cell death. Cell Cycle 2011, 10, 4119–4127. [Google Scholar] [CrossRef]
- Sunayama, J.; Ando, Y.; Itoh, N.; Tomiyama, A.; Sakurada, K.; Sugiyama, A.; Kang, D.; Tashiro, F.; Gotoh, Y.; Kuchino, Y.; et al. Physical and functional interaction between BH3-only protein Hrk and mitochondrial pore-forming protein p32. Cell Death Differ. 2004, 11, 771–781. [Google Scholar] [CrossRef] [PubMed]
- Reef, S.; Shifman, O.; Oren, M.; Kimchi, A. The autophagic inducer smARF interacts with and is stabilized by the mitochondrial p32 protein. Oncogene 2007, 26, 6677–6683. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Loewenstein, P.M.; Zhang, Z.; Green, M. In vitro interaction of the human immunodeficiency virus type 1 Tat transactivator and the general transcription factor TFIIB with the cellular protein TAP. J. Virol. 1995, 69, 3017–3023. [Google Scholar] [CrossRef]
- Chattopadhyay, C.; Hawke, D.; Kobayashi, R.; Maity, S.N. Human p32, interacts with B subunit of the CCAAT-binding factor, CBF/NF-Y, and inhibits CBF-mediated transcription activation in vitro. Nucleic Acids Res. 2004, 32, 3632–3641. [Google Scholar] [CrossRef][Green Version]
- Waggoner, S.N.; Cruise, M.W.; Kassel, R.; Hahn, Y.S. gC1q Receptor Ligation Selectively Down-Regulates Human IL-12 Production through Activation of the Phosphoinositide 3-Kinase Pathway. J. Immunol. 2005, 175, 4706–4714. [Google Scholar] [CrossRef] [PubMed]
- Dembitzer, F.R.; Kinoshita, Y.; Burstein, D.; Phelps, R.G.; Beasley, M.B.; Garcia, R.; Harpaz, N.; Jaffer, S.; Thung, S.N.; Unger, P.D.; et al. gC1qR Expression in Normal and Pathologic Human Tissues: Differential Expression in Tissues of Epithelial and Mesenchymal Origin. J. Histochem. Cytochem. 2012, 60, 467–474. [Google Scholar] [CrossRef]
- Egusquiza, C.A.; Castañeda, M.C.; Albarran, S.; Gonzalez, H.; Moreno, A.P.; Maldonado, V.; Melendez-Zajgla, J.; Robles-Flores, M. Overexpression of Multifunctional Protein p32 Promotes a Malignant Phenotype in Colorectal Cancer Cells. Front. Oncol. 2021, 11, 642940. [Google Scholar] [CrossRef]
- Fogal, V.; Zhang, L.; Krajewski, S.; Ruoslahti, E. Mitochondrial/Cell-Surface Protein p32/gC1qR as a Molecular Target in Tumor Cells and Tumor Stroma. Cancer Res. 2008, 68, 7210–7219. [Google Scholar] [CrossRef]
- Fogal, V.; Babic, I.; Chao, Y.; Pastorino, S.; Mukthavaram, R.; Jiang, P.; Cho, Y.J.; Pingle, S.C.; Crawford, J.R.; Piccioni, D.E.; et al. Mitochondrial p32 is upregulated in Myc expressing brain cancers and mediates glutamine addiction. Oncotarget 2015, 6, 1157–1170. [Google Scholar] [CrossRef]
- Rubinstein, D.B.; Stortchevoi, A.; Boosalis, M.; Ashfaq, R.; Ghebrehiwet, B.; Peerschke, E.I.B.; Calvo, F.; Guillaume, T. Receptor for the globular heads of C1q (gC1q-R, p33, hyaluronan-binding protein) is preferentially expressed by adenocarcinoma cells. Int. J. Cancer 2004, 110, 741–750. [Google Scholar] [CrossRef]
- Saha, S.; Kim, K.; Islam, S.M.; Cho, S.G.; Gil, M. Systematic Multiomics Analysis of Alterations in C1QBP mRNA Expression and Relevance for Clinical Outcomes in Cancers. J. Clin. Med. 2019, 8, 513. [Google Scholar] [CrossRef]
- Fogal, V.; Richardson, A.D.; Karmali, P.P.; Scheffler, I.E.; Smith, J.W.; Ruoslahti, E. Mitochondrial p32 Protein Is a Critical Regulator of Tumor Metabolism via Maintenance of Oxidative Phosphorylation. Mol. Cell. Biol. 2010, 30, 1303–1318. [Google Scholar] [CrossRef]
- Kim, K.B.; Yi, J.S.; Nguyen, N.; Lee, J.H.; Kwon, Y.C.; Ahn, B.Y.; Cho, H.; Kim, Y.K.; Yoo, H.J.; Lee, J.S.; et al. Cell-surface receptor for complement component C1q (gC1qR) is a key regulator for lamellipodia formation and cancer metastasis. J. Biol. Chem. 2011, 286, 23093–23101. [Google Scholar] [CrossRef]
- Peerschke, E.; Stier, K.; Li, X.; Kandov, E.; de Stanchina, E.; Chang, Q.; Xiong, Y.; Manova-Todorova, K.; Fan, N.; Barlas, A.; et al. gC1qR/HABP1/p32 Is a Potential New Therapeutic Target Against Mesothelioma. Front. Oncol. 2020, 10, 1413. [Google Scholar] [CrossRef]
- Kim, B.C.; Hwang, H.J.; An, H.T.; Lee, H.; Park, J.S.; Hong, J.; Ko, J.; Kim, C.; Lee, J.S.; Ko, Y.G. Antibody neutralization of cell-surface gC1qR/HABP1/SF2-p32 prevents lamellipodia formation and tumorigenesis. Oncotarget 2016, 7, 49972–49985. [Google Scholar] [CrossRef] [PubMed]
- Yenugonda, V.; Nomura, N.; Kouznetsova, V.; Tsigelny, I.; Fogal, V.; Nurmemmedov, E.; Kesari, S.; Babic, I. A novel small molecule inhibitor of p32 mitochondrial protein overexpressed in glioma. J. Transl. Med. 2017, 15, 210. [Google Scholar] [CrossRef] [PubMed]
- Shamloo, B.; Usluer, S. P21 in cancer research. Cancers 2019, 11, 1178. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Fang, W.; Liu, M.; Fu, D. Complement component 1, q subcomponent binding protein (C1QBP) in lipid rafts mediates hepatic metastasis of pancreatic cancer by regulating IGF-1/IGF-1R signaling. Int. J. Cancer 2017, 141, 1389–1401. [Google Scholar] [CrossRef] [PubMed]
- Ravikumar, B.; Vacher, C.; Berger, Z.; Davies, J.E.; Luo, S.; Oroz, L.G.; Scaravilli, F.; Easton, D.F.; Duden, R.; O’Kane, C.J.; et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat. Genet. 2004, 36, 585–595. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; White, E.P.; Mehnert, J.M. Coordinate autophagy and mTOR pathway inhibition enhances cell death in melanoma. PLoS ONE 2013, 8, e55096. [Google Scholar] [CrossRef]
- Jiao, H.; Su, G.Q.; Dong, W.; Zhang, L.; Xie, W.; Yao, L.M.; Chen, P.; Wang, Z.X.; Liou, Y.C.; You, H. Cell Chaperone-like protein p32 regulates ULK1 stability and autophagy. Death Differ. 2015, 22, 1812–1823. [Google Scholar] [CrossRef]
- Jiao, H.; You, H. p32: A new player in autophagy. Mol. Cell. Oncol. 2016, 3, e1061097. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Kundu, M.; Viollet, B.; Guan, K.L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 2011, 13, 132–141. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; She, H.; Zhang, T.; Xu, H.; Cheng, L.; Yepes, M.; Zhao, Y.; Mao, Z. p38 MAPK inhibits autophagy and promotes microgial inflammatory responses by phosphorylating ULK1. J. Cell Biol. 2018, 217, 315–328. [Google Scholar] [CrossRef]
- Wang, Q.; Chai, D.; Sobhani, N.; Sun, N.; Neeli, P.; Zheng, J.; Tian, H. C1QBP regulates mitochondrial plasticity to impact tumor progression and antitumor immune response. Front. Physiol. 2022, 13, 1012112. [Google Scholar] [CrossRef] [PubMed]
- Yapa, N.M.B.; Lisnyak, V.; Reljic, B.; Ryan, M.T. Mitochondrial dynamics in health and disease. FEBS Lett. 2021, 595, 1184–1204. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Ashraf, R.; Aparna, C.K. Mitochondrial dynamics regulators: Implications for therapeutic intervention in cancer. Cell Biol. Toxicol. 2022, 38, 377–406. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Hong, Y.; Shu, Y.; Wu, C.; Ye, G.; Chen, H.; Zhou, H.; Gao, R.; Zhang, J. The involvement of Parkin-dependent mitophagy in the anti-cancer activity of Ginsenoside. J. Ginseng Res. 2022, 46, 266–274. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Zhang, D.; Wang, J.; Wang, Y.; Hu, J.; Zhou, Y.; Zhou, X.; Nie, S.; Xie, M. Aloe gel glucomannan induced colon cancer cell death via mitochondrial damage-driven PINK1/Parkin mitophagy pathway. Carbohydr. Polym. 2022, 295, 119841. [Google Scholar] [CrossRef]
- Li, Y.; Wan, O.W.; Xie, W.; Chung, K.K.K. P32 regulates mitochondrial morphology and dynamics through parkin. Neuroscience 2011, 199, 346–358. [Google Scholar] [CrossRef]
- Desai, S.; Grefte, S.; Westerlo, E.; Lauwen, S.; Paters, A.; Prehn, J.H.M.; Gan, Z.; Keijer, J.; Adjobo-Hermans, M.J.W.; Koopman, W.J.H. Performance of TMRM and Mitotrackers in mitochondrial morphofunctional analysis of primary human skin fibroblasts. Biochim. Biophys. Acta Bioenerg. 2023, 1865, 149027. [Google Scholar] [CrossRef] [PubMed]
- Santoyo-Ramos, P.; Likhatcheva, M.; Castañeda-Patlán, C.; García-Zepeda, E.; Robles-Flores, M. Hypoxia-inducible factors participate in the modulation of stemness and malignancy of colon cancer cells playing opposite roles in canonical Wnt signaling. PLoS ONE 2014, 9, e112580. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Egusquiza-Alvarez, C.A.; Moreno-Londoño, A.P.; Alvarado-Ortiz, E.; Ramos-Godínez, M.d.P.; Sarabia-Sánchez, M.A.; Castañeda-Patlán, M.C.; Robles-Flores, M. Inhibition of Multifunctional Protein p32/C1QBP Promotes Cytostatic Effects in Colon Cancer Cells by Altering Mitogenic Signaling Pathways and Promoting Mitochondrial Damage. Int. J. Mol. Sci. 2024, 25, 2712. https://doi.org/10.3390/ijms25052712
Egusquiza-Alvarez CA, Moreno-Londoño AP, Alvarado-Ortiz E, Ramos-Godínez MdP, Sarabia-Sánchez MA, Castañeda-Patlán MC, Robles-Flores M. Inhibition of Multifunctional Protein p32/C1QBP Promotes Cytostatic Effects in Colon Cancer Cells by Altering Mitogenic Signaling Pathways and Promoting Mitochondrial Damage. International Journal of Molecular Sciences. 2024; 25(5):2712. https://doi.org/10.3390/ijms25052712
Chicago/Turabian StyleEgusquiza-Alvarez, Carlos Alejandro, Angela Patricia Moreno-Londoño, Eduardo Alvarado-Ortiz, María del Pilar Ramos-Godínez, Miguel Angel Sarabia-Sánchez, María Cristina Castañeda-Patlán, and Martha Robles-Flores. 2024. "Inhibition of Multifunctional Protein p32/C1QBP Promotes Cytostatic Effects in Colon Cancer Cells by Altering Mitogenic Signaling Pathways and Promoting Mitochondrial Damage" International Journal of Molecular Sciences 25, no. 5: 2712. https://doi.org/10.3390/ijms25052712
APA StyleEgusquiza-Alvarez, C. A., Moreno-Londoño, A. P., Alvarado-Ortiz, E., Ramos-Godínez, M. d. P., Sarabia-Sánchez, M. A., Castañeda-Patlán, M. C., & Robles-Flores, M. (2024). Inhibition of Multifunctional Protein p32/C1QBP Promotes Cytostatic Effects in Colon Cancer Cells by Altering Mitogenic Signaling Pathways and Promoting Mitochondrial Damage. International Journal of Molecular Sciences, 25(5), 2712. https://doi.org/10.3390/ijms25052712





