A Direct Comparison of rAAV5 Variants Derived from the Baculovirus Expression System Using LC-MS Workflows Demonstrates Key Differences in Overall Production Yield, Product Quality and Vector Efficiency
Abstract
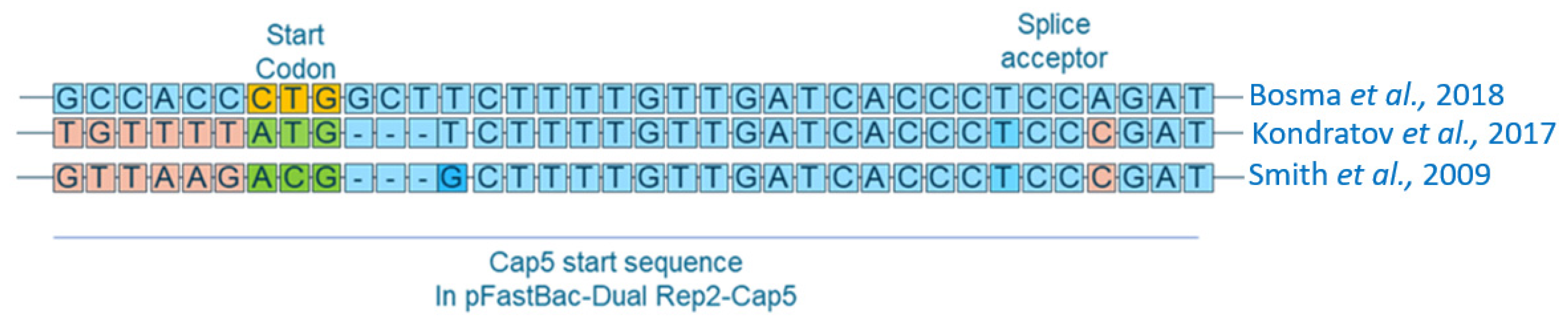
1. Introduction
2. Results and Discussion
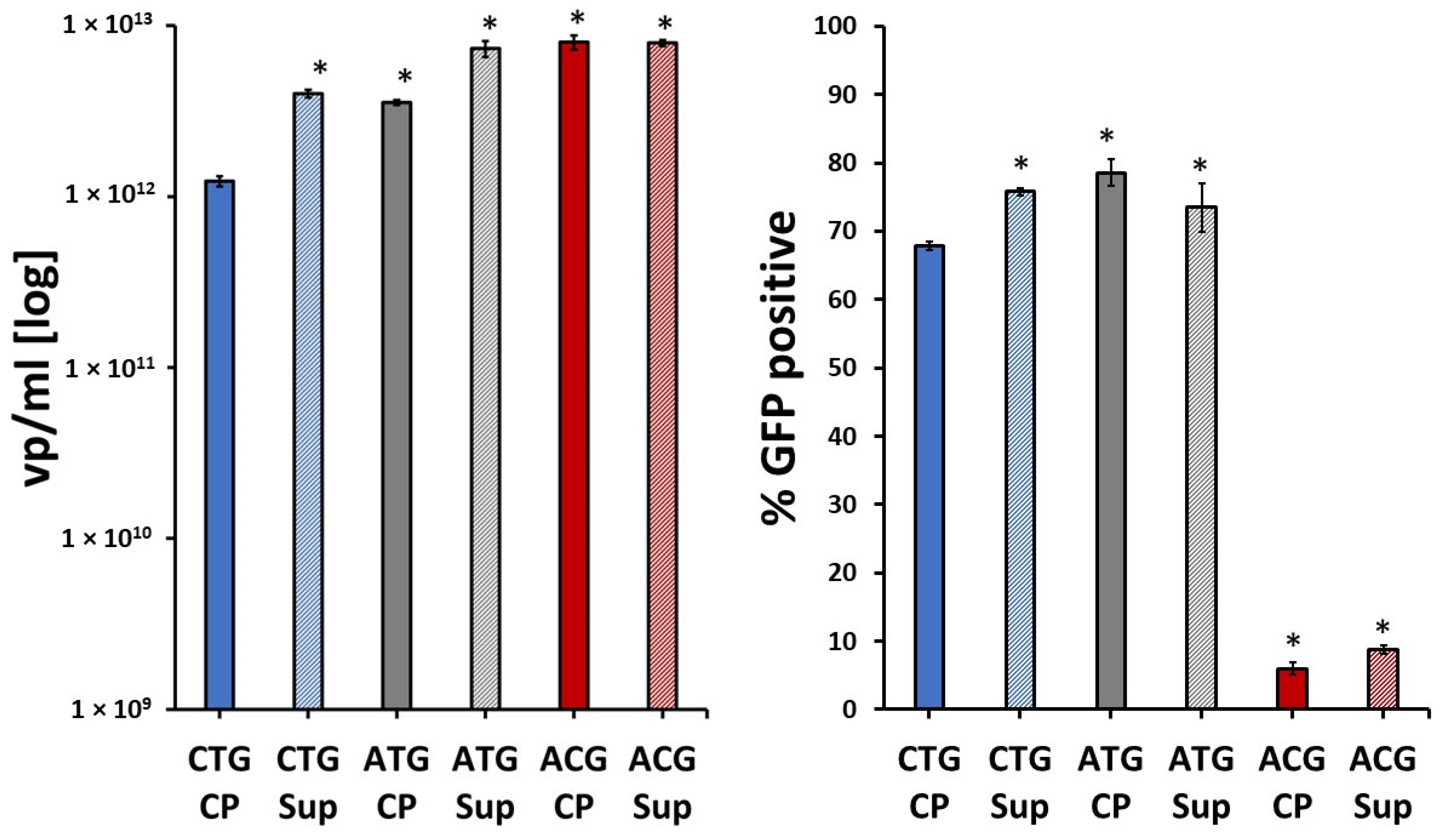
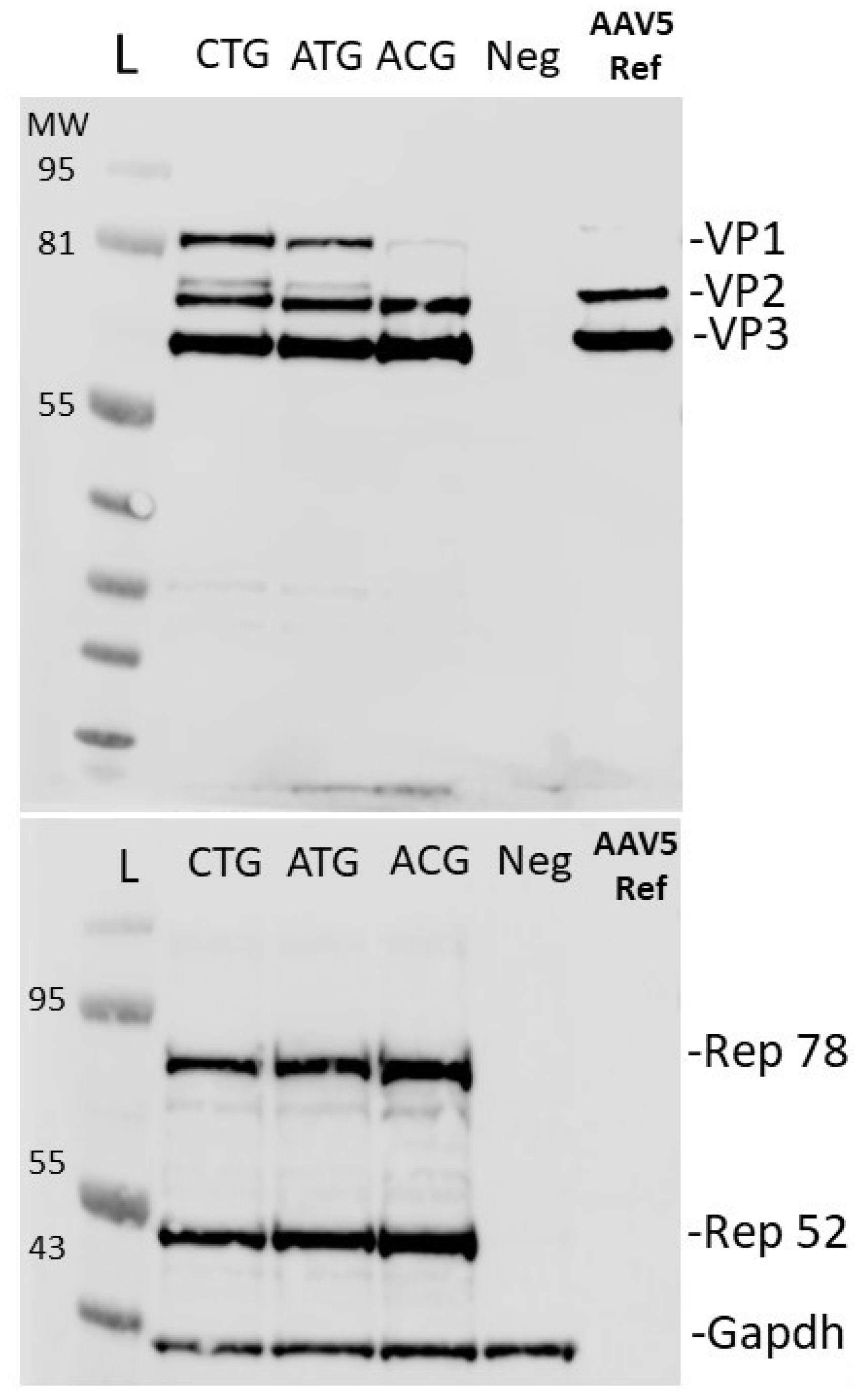
2.1. Yield and Transduction Efficiency
2.2. VP Ratio Analysis
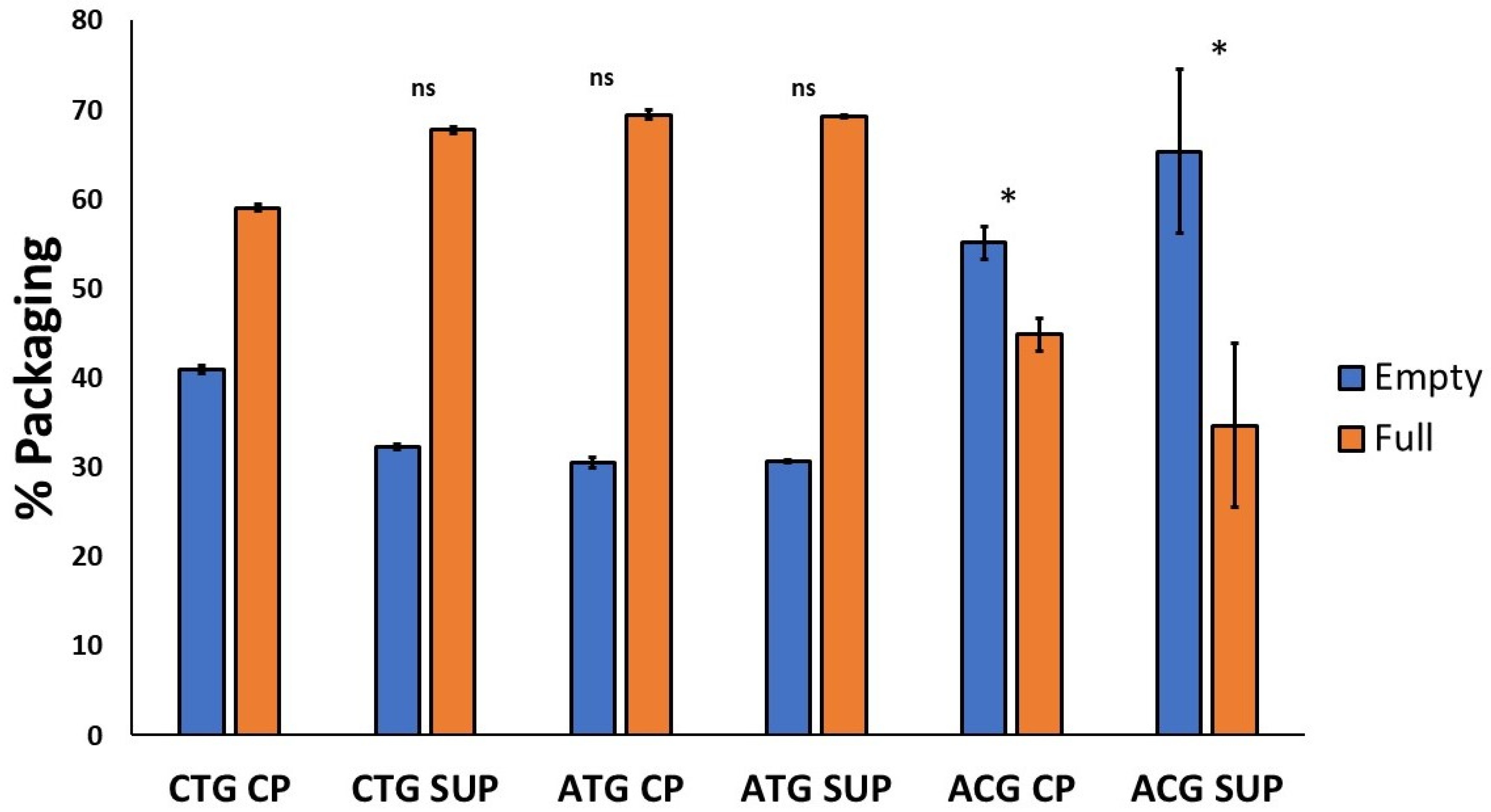
2.3. Packaging Efficiency
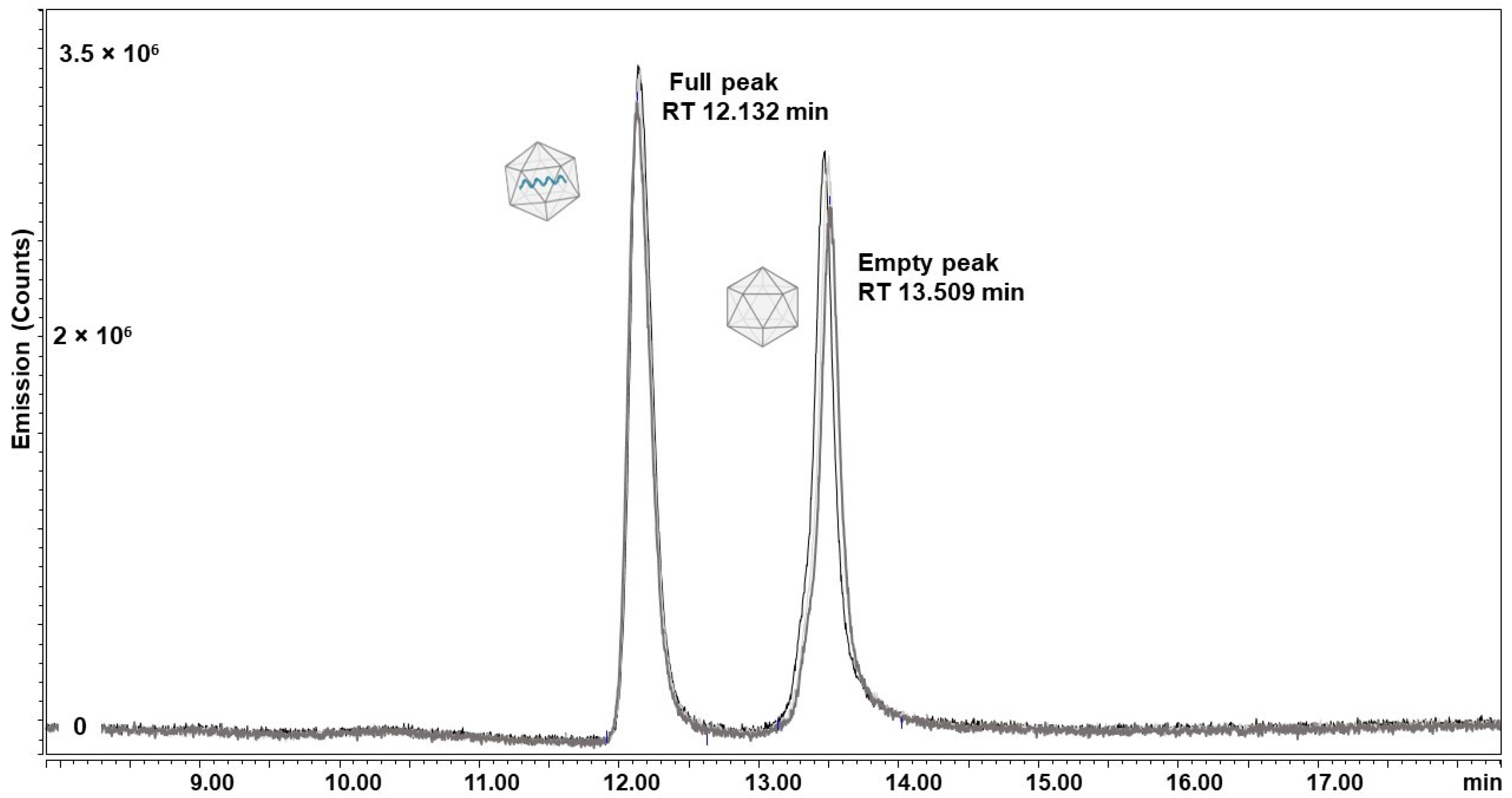
2.3.1. AEX-LC
2.3.2. Native MS
2.3.3. Packaging Assessment by Orbitrap-Based Charge Detection Mass Spectrometry (CDMS)
3. Materials and Methods
3.1. Plasmid and BV Construction
3.2. Sf9 Cell Culture and Transductions
Sf9 Transduction
3.3. AAV Harvest
3.4. Viral Vector Purification
3.5. Western Blot and Silver Stain
3.6. Titre Determination by qPCR and Capsid ELISA
3.6.1. qPCR
3.6.2. ELISA
3.7. Transduction Efficiency Assay
3.8. Hydrophilic Interaction Liquid Chromatography (HILIC) Intact Separation of VP Proteins
3.9. Anion Exchange Liquid Chromatography (AEX-LC) Analysis of Empty and Full Capsids
3.10. Native and Charge Detection Mass Spectrometry of Empty and Full Capsid Ratios
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kessler, P.D.; Podsakoff, G.M.; Chen, X.; McQuiston, S.A.; Colosi, P.C.; Matelis, L.A.; Kurtzman, G.J.; Byrne, B.J. Gene delivery to skeletal muscle results in sustained expression and systemic delivery of a therapeutic protein. Proc. Natl. Acad. Sci. USA 1996, 93, 14082–14087. [Google Scholar] [CrossRef]
- Shirley, J.L.; de Jong, Y.P.; Terhorst, C.; Herzog, R.W. Immune Responses to Viral Gene Therapy Vectors. Mol. Ther. 2020, 28, 709–722. [Google Scholar] [CrossRef]
- Samulski, R.J.; Muzyczka, N. AAV-mediated gene therapy for research and therapeutic purposes. Annu. Rev. Virol. 2014, 1, 427–451. [Google Scholar] [CrossRef]
- Keeler, A.M.; Flotte, T.R. Recombinant adeno-associated virus gene therapy in light of Luxturna (and Zolgensma and Glybera): Where are we, and how did we get here? Annu. Rev. Virol. 2019, 6, 601–621. [Google Scholar] [CrossRef]
- van Oers, M.M. Opportunities and challenges for the baculovirus expression system. J. Invertebr. Pathol. 2011, 107, S3–S15. [Google Scholar] [CrossRef]
- Urabe, M.; Ding, C.; Kotin, R.M. Insect Cells as a Factory to Produce Adeno-Associated Virus Type 2 Vectors. Hum. Gene Ther. 2002, 13, 1935–1943. [Google Scholar] [CrossRef]
- Urabe, M.; Nakakura, T.; Xin, K.-Q.; Obara, Y.; Mizukami, H.; Kume, A.; Kotin Robert, M.; Ozawa, K. Scalable Generation of High-Titer Recombinant Adeno-Associated Virus Type 5 in Insect Cells. J. Virol. 2006, 80, 1874–1885. [Google Scholar] [CrossRef]
- Bosma, B.; du Plessis, F.; Ehlert, E.; Nijmeijer, B.; de Haan, M.; Petry, H.; Lubelski, J. Optimization of viral protein ratios for production of rAAV serotype 5 in the baculovirus system. Gene Ther. 2018, 25, 415–424. [Google Scholar] [CrossRef]
- Gimpel, A.L.; Katsikis, G.; Sha, S.; Maloney, A.J.; Hong, M.S.; Nguyen, T.N.T.; Wolfrum, J.; Springs, S.L.; Sinskey, A.J.; Manalis, S.R.; et al. Analytical methods for process and product characterization of recombinant adeno-associated virus-based gene therapies. Mol. Ther.-Methods Clin. Dev. 2021, 20, 740–754. [Google Scholar] [CrossRef]
- Smith, R.H.; Levy, J.R.; Kotin, R.M. A Simplified Baculovirus-AAV Expression Vector System Coupled With One-step Affinity Purification Yields High-titer rAAV Stocks From Insect Cells. Mol. Ther. 2009, 17, 1888–1896. [Google Scholar] [CrossRef]
- Wu, Y.; Jiang, L.; Geng, H.; Yang, T.; Han, Z.; He, X.; Lin, K.; Xu, F. A Recombinant Baculovirus Efficiently Generates Recombinant Adeno-Associated Virus Vectors in Cultured Insect Cells and Larvae. Mol. Ther. Methods Clin. Dev. 2018, 10, 38–47. [Google Scholar] [CrossRef] [PubMed]
- Kondratov, O.; Marsic, D.; Crosson, S.M.; Mendez-Gomez, H.R.; Moskalenko, O.; Mietzsch, M.; Heilbronn, R.; Allison, J.R.; Green, K.B.; Agbandje-McKenna, M. Direct head-to-head evaluation of recombinant adeno-associated viral vectors manufactured in human versus insect cells. Mol. Ther. 2017, 25, 2661–2675. [Google Scholar] [CrossRef] [PubMed]
- Xiao, P.-J.; Samulski, R.J. Cytoplasmic Trafficking, Endosomal Escape, and Perinuclear Accumulation of Adeno-Associated Virus Type 2 Particles Are Facilitated by Microtubule Network. J. Virol. 2012, 86, 10462–10473. [Google Scholar] [CrossRef] [PubMed]
- Sonntag, F.; Bleker, S.; Leuchs, B.; Fischer, R.; Kleinschmidt, J.A. Adeno-Associated Virus Type 2 Capsids with Externalized VP1/VP2 Trafficking Domains Are Generated prior to Passage through the Cytoplasm and Are Maintained until Uncoating Occurs in the Nucleus. J. Virol. 2006, 80, 11040–11054. [Google Scholar] [CrossRef] [PubMed]
- Popa-Wagner, R.; Porwal, M.; Kann, M.; Reuss, M.; Weimer, M.; Florin, L.; Kleinschmidt, J.A. Impact of VP1-Specific Protein Sequence Motifs on Adeno-Associated Virus Type 2 Intracellular Trafficking and Nuclear Entry. J. Virol. 2012, 86, 9163. [Google Scholar] [CrossRef]
- Ellis, B.L.; Hirsch, M.L.; Barker, J.C.; Connelly, J.P.; Steininger, R.J.; Porteus, M.H. A survey of ex vivo/in vitro transduction efficiency of mammalian primary cells and cell lines with Nine natural adeno-associated virus (AAV1-9) and one engineered adeno-associated virus serotype. Virol. J. 2013, 10, 74. [Google Scholar] [CrossRef] [PubMed]
- Girod, A.; Wobus, C.E.; Zádori, Z.; Ried, M.; Leike, K.; Tijssen, P.; Kleinschmidt, J.A.; Hallek, M. The VP1 capsid protein of adeno-associated virus type 2 is carrying a phospholipase A2 domain required for virus infectivity. J. Gen. Virol. 2002, 83, 973–978. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.P.; Patel, S.K.; Xing, T.; Yan, Y.; Wang, S.; Li, N. Characterization of Adeno-Associated Virus Capsid Proteins Using Hydrophilic Interaction Chromatography Coupled with Mass Spectrometry. J. Pharm. Biomed. Anal. 2020, 189, 113481. [Google Scholar] [CrossRef]
- Smith, J.; Guapo, F.; Strasser, L.; Millan-Martin, S.; Milian, S.; Snyder, R.; Bones, J. Development of a Rapid Adeno-Associated Virus (AAV) Identity Testing Platform through Comprehensive Mass Analysis of Full-length AAV Capsid Proteins. J. Proteome Res. 2023, 23, 161–174. [Google Scholar] [CrossRef]
- Bleker, S.; Sonntag, F.; Kleinschmidt, J.A. Mutational Analysis of Narrow Pores at the Fivefold Symmetry Axes of Adeno-Associated Virus Type 2 Capsids Reveals a Dual Role in Genome Packaging and Activation of Phospholipase A2 Activity. J. Virol. 2005, 79, 2528–2540. [Google Scholar] [CrossRef]
- Grieger, J.C.; Soltys, S.M.; Samulski, R.J. Production of recombinant adeno-associated virus vectors using suspension HEK293 cells and continuous harvest of vector from the culture media for GMP FIX and FLT1 clinical vector. Mol. Ther. 2016, 24, 287–297. [Google Scholar] [CrossRef] [PubMed]
- Stahnke, S.; Lux, K.; Uhrig, S.; Kreppel, F.; Hösel, M.; Coutelle, O.; Ogris, M.; Hallek, M.; Büning, H. Intrinsic phospholipase A2 activity of adeno-associated virus is involved in endosomal escape of incoming particles. Virology 2011, 409, 77–83. [Google Scholar] [CrossRef]
- Strasser, L.; Morgan, T.E.; Guapo, F.; Füssl, F.; Forsey, D.; Anderson, I.; Bones, J. A Native Mass Spectrometry-Based Assay for Rapid Assessment of the Empty:Full Capsid Ratio in Adeno-Associated Virus Gene Therapy Products. Anal. Chem. 2021, 93, 12817–12821. [Google Scholar] [CrossRef] [PubMed]
- Keifer, D.Z.; Pierson, E.E.; Jarrold, M.F. Charge detection mass spectrometry: Weighing heavier things. Analyst 2017, 142, 1654–1671. [Google Scholar] [CrossRef]
- Pierson, E.E.; Keifer, D.Z.; Asokan, A.; Jarrold, M.F. Resolving adeno-associated viral particle diversity with charge detection mass spectrometry. Anal. Chem. 2016, 88, 6718–6725. [Google Scholar] [CrossRef] [PubMed]
- Barnes, L.F.; Draper, B.E.; Chen, Y.-T.; Powers, T.W.; Jarrold, M.F. Quantitative analysis of genome packaging in recombinant AAV vectors by charge detection mass spectrometry. Mol. Ther.-Methods Clin. Dev. 2021, 23, 87–97. [Google Scholar] [CrossRef]
- Wörner, T.P.; Snijder, J.; Friese, O.; Powers, T.; Heck, A.J. Assessment of genome packaging in AAVs using Orbitrap-based charge-detection mass spectrometry. Mol. Ther.-Methods Clin. Dev. 2022, 24, 40–47. [Google Scholar] [CrossRef]
- Wörner, T.P.; Snijder, J.; Bennett, A.; Agbandje-McKenna, M.; Makarov, A.A.; Heck, A.J. Resolving heterogeneous macromolecular assemblies by Orbitrap-based single-particle charge detection mass spectrometry. Nat. Methods 2020, 17, 395–398. [Google Scholar] [CrossRef]
- Venkatakrishnan, B.; Yarbrough, J.; Domsic, J.; Bennett, A.; Bothner, B.; Kozyreva, O.G.; Samulski, R.J.; Muzyczka, N.; McKenna, R.; Agbandje-McKenna, M. Structure and dynamics of adeno-associated virus serotype 1 VP1-unique N-terminal domain and its role in capsid trafficking. J. Virol. 2013, 87, 4974–4984. [Google Scholar] [CrossRef]
- Li, C.; He, Y.; Nicolson, S.; Hirsch, M.; Weinberg, M.S.; Zhang, P.; Kafri, T.; Samulski, R.J. Adeno-associated virus capsid antigen presentation is dependent on endosomal escape. J. Clin. Investig. 2013, 123, 1390–1401. [Google Scholar] [CrossRef]
- Gao, K.; Li, M.; Zhong, L.; Su, Q.; Li, J.; Li, S.; He, R.; Zhang, Y.; Hendricks, G.; Wang, J.; et al. Empty virions in AAV8 vector preparations reduce transduction efficiency and may cause total viral particle dose-limiting side effects. Mol. Ther.-Methods Clin. Dev. 2014, 1, 9. [Google Scholar] [CrossRef] [PubMed]
- Gao, G.; Vandenberghe, L.H.; Alvira, M.R.; Lu, Y.; Calcedo, R.; Zhou, X.; Wilson, J.M. Clades of Adeno-associated viruses are widely disseminated in human tissues. J. Virol. 2004, 78, 6381–6388. [Google Scholar] [CrossRef] [PubMed]
- Kafader, J.O.; Melani, R.D.; Durbin, K.R.; Ikwuagwu, B.; Early, B.P.; Fellers, R.T.; Beu, S.C.; Zabrouskov, V.; Makarov, A.A.; Maze, J.T.; et al. Multiplexed mass spectrometry of individual ions improves measurement of proteoforms and their complexes. Nat. Methods 2020, 17, 391–394. [Google Scholar] [CrossRef] [PubMed]
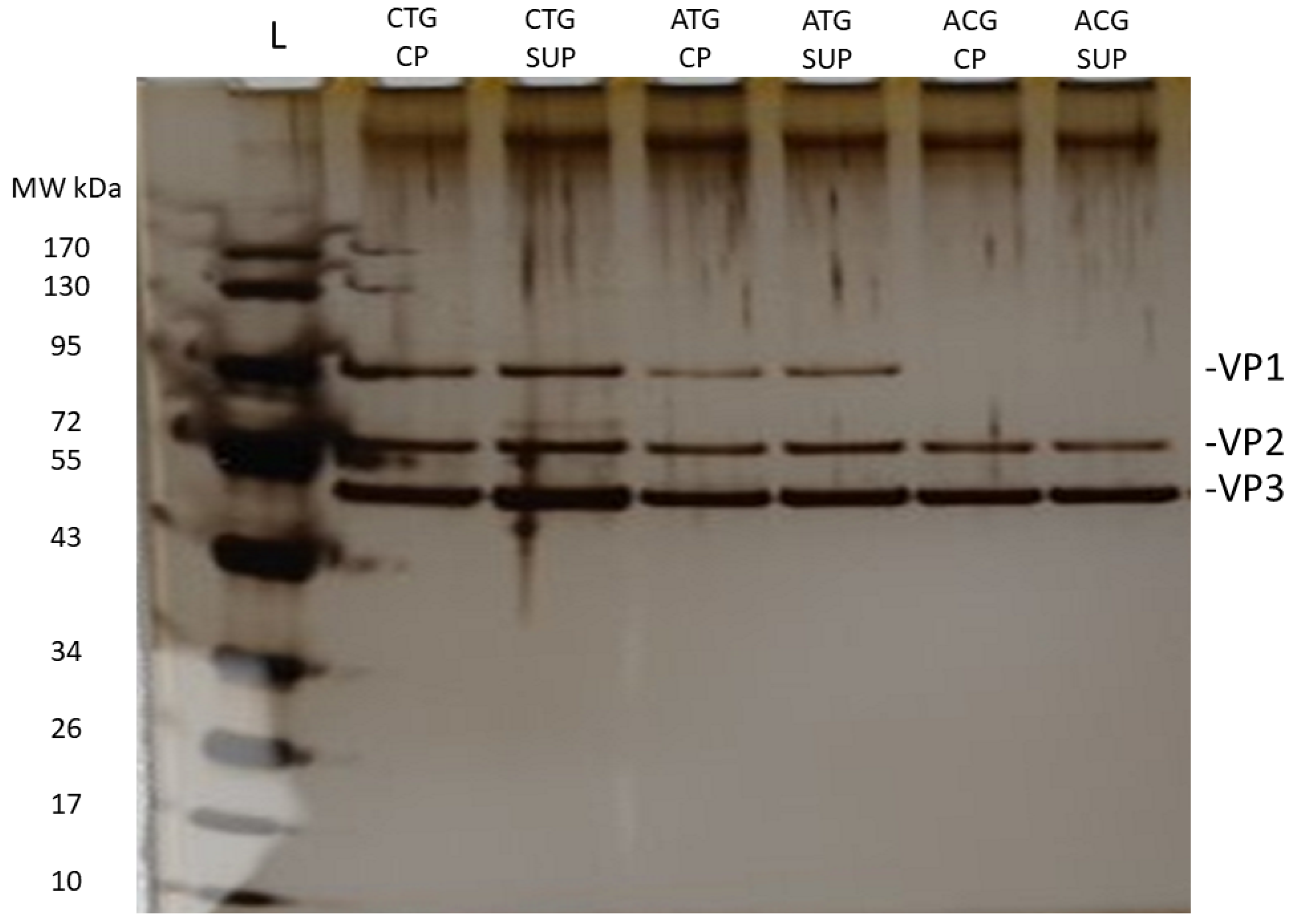
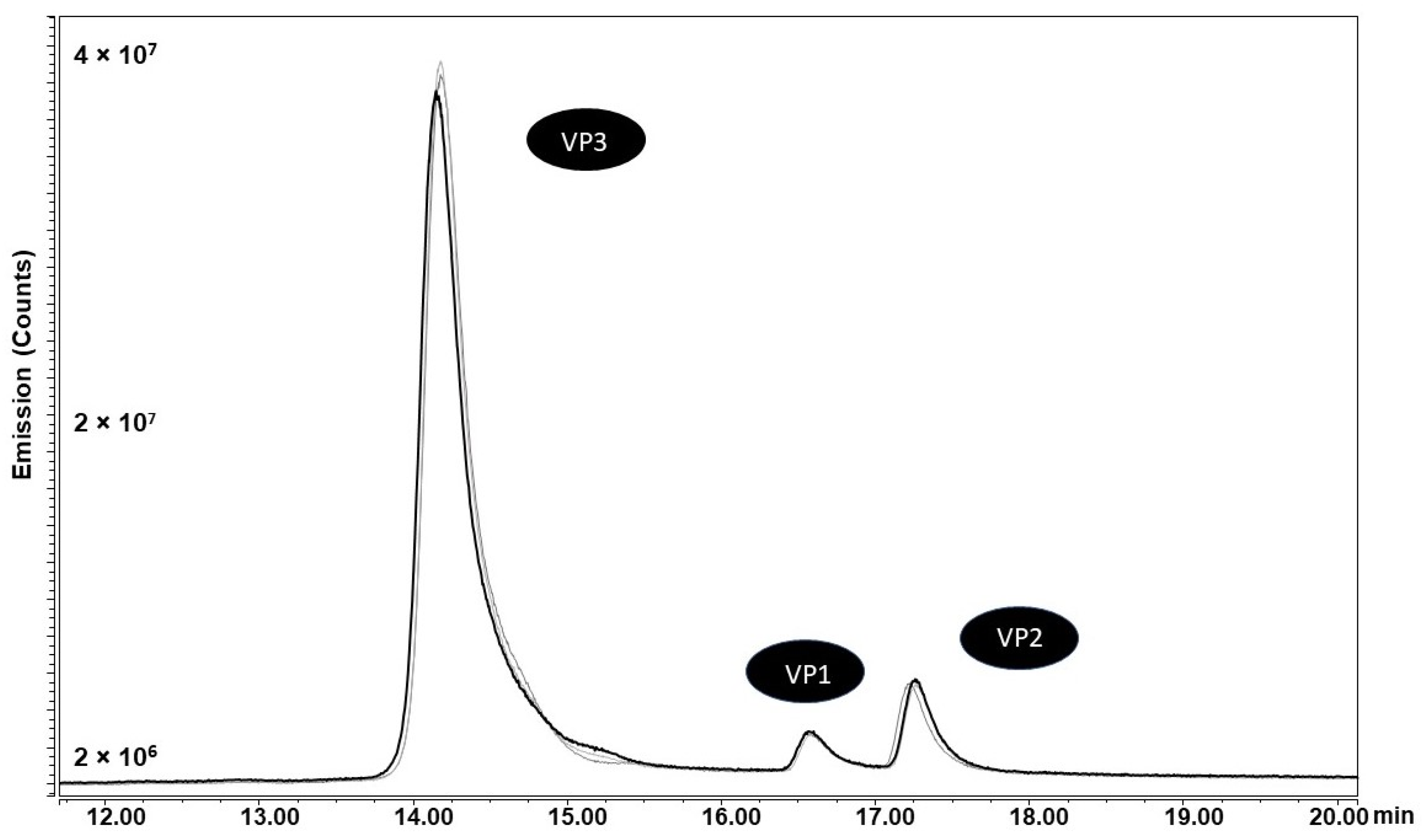
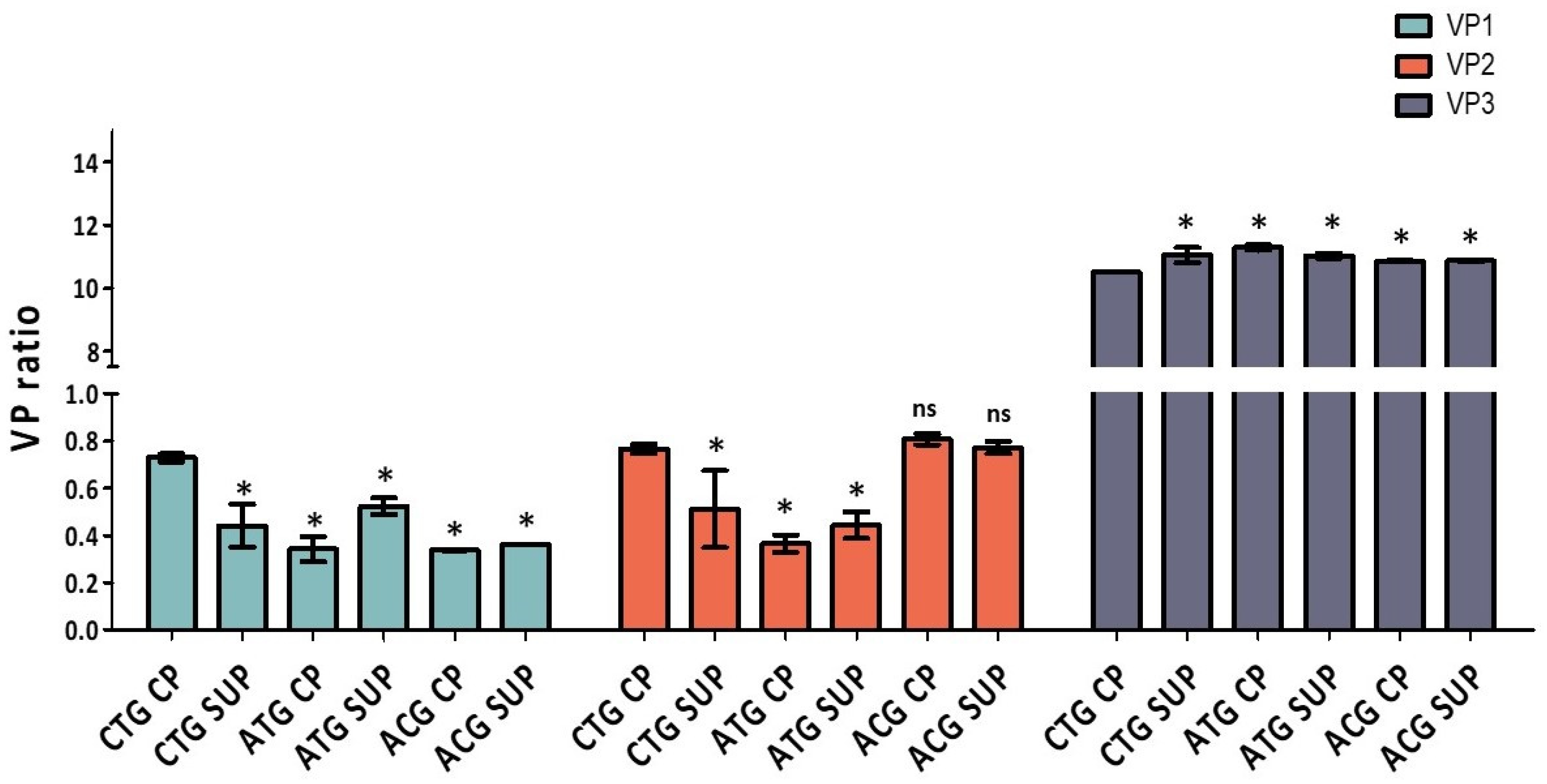

| VP1 | VP2 | VP3 | VP1/VP2 | VP1/VP3 | VP2/VP3 | |
|---|---|---|---|---|---|---|
| CTG CP | 0.7 | 0.8 | 10.5 | 0.95 | 0.07 | 0.07 |
| CTG SUP | 0.4 | 0.5 | 11.0 | 0.86 | 0.04 | 0.05 |
| ATG CP | 0.3 | 0.4 | 11.3 | 0.94 | 0.03 | 0.03 |
| ATG SUP | 0.5 | 0.4 | 11.0 | 1.18 | 0.05 | 0.04 |
| ACG CP | 0.3 | 0.8 | 10.9 | 0.42 | 0.03 | 0.07 |
| ACG SUP | 0.4 | 0.8 | 10.9 | 0.47 | 0.03 | 0.07 |
| Sample | Mass kDa Empty | Mass kDa Full | Δm E/F kDa | Mass Deviation % * |
|---|---|---|---|---|
| CTG CP | 3742.44 | 4884.41 | 1141.97 | 12.3 |
| CTG SUP | 3713.26 | 4906.12 | 1192.85 | 17.3 |
| ATG CP | 3654.10 | 4755.79 | 1101.70 | 8.3 |
| ATG SUP | 3653.32 | 4714.71 | 1061.39 | 4.4 |
| ACG CP | 3583.57 | 4737.56 | 1153.98 | 13.5 |
| ACG SUP | 3594.25 | 4525.09 | 930.839 | −8.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guapo, F.; Donohue, N.; Strasser, L.; Boi, S.; Füssl, F.; Rainbow-Fletcher, A.; Getty, P.; Anderson, I.; Barron, N.; Bones, J. A Direct Comparison of rAAV5 Variants Derived from the Baculovirus Expression System Using LC-MS Workflows Demonstrates Key Differences in Overall Production Yield, Product Quality and Vector Efficiency. Int. J. Mol. Sci. 2024, 25, 2785. https://doi.org/10.3390/ijms25052785
Guapo F, Donohue N, Strasser L, Boi S, Füssl F, Rainbow-Fletcher A, Getty P, Anderson I, Barron N, Bones J. A Direct Comparison of rAAV5 Variants Derived from the Baculovirus Expression System Using LC-MS Workflows Demonstrates Key Differences in Overall Production Yield, Product Quality and Vector Efficiency. International Journal of Molecular Sciences. 2024; 25(5):2785. https://doi.org/10.3390/ijms25052785
Chicago/Turabian StyleGuapo, Felipe, Nicholas Donohue, Lisa Strasser, Stefano Boi, Florian Füssl, Alana Rainbow-Fletcher, Paul Getty, Ian Anderson, Niall Barron, and Jonathan Bones. 2024. "A Direct Comparison of rAAV5 Variants Derived from the Baculovirus Expression System Using LC-MS Workflows Demonstrates Key Differences in Overall Production Yield, Product Quality and Vector Efficiency" International Journal of Molecular Sciences 25, no. 5: 2785. https://doi.org/10.3390/ijms25052785
APA StyleGuapo, F., Donohue, N., Strasser, L., Boi, S., Füssl, F., Rainbow-Fletcher, A., Getty, P., Anderson, I., Barron, N., & Bones, J. (2024). A Direct Comparison of rAAV5 Variants Derived from the Baculovirus Expression System Using LC-MS Workflows Demonstrates Key Differences in Overall Production Yield, Product Quality and Vector Efficiency. International Journal of Molecular Sciences, 25(5), 2785. https://doi.org/10.3390/ijms25052785







