Cellular and Molecular Determinants of Biologic Drugs Resistance and Therapeutic Failure in Inflammatory Bowel Disease
Abstract
:1. Introduction
2. Materials and Methods
3. Genetic Polymorphisms
- Human Leukocyte Antigens variants
- Other Polymorphisms
- -
- Polymorphisms of the TNFα gene (leading to increased cytokine secretion) and TNFα receptors genes (TNFR1/2, resulting in increased response after interaction with TNFα).
- -
- Polymorphisms of innate immunity-related genes (TLR4, CD14, IL-6, and IL-1β).
- -
- Polymorphisms of apoptosis- and autophagy-related genes (FASL, CASP9 and ATG16L1), probably through an inhibitory effect on the apoptosis of immune cells.
4. Transcriptional Profiles
5. Epigenetic Modifications
- -
- There were 26 associated with specific immune compartment cells (B cells, T cells, granulocytes, and monocytes).
- -
- A further 125 had been previously associated by epigenome-wide association studies (EWAS) with alcohol consumption, body mass index, smoking, C reactive protein, and IBD type [72].
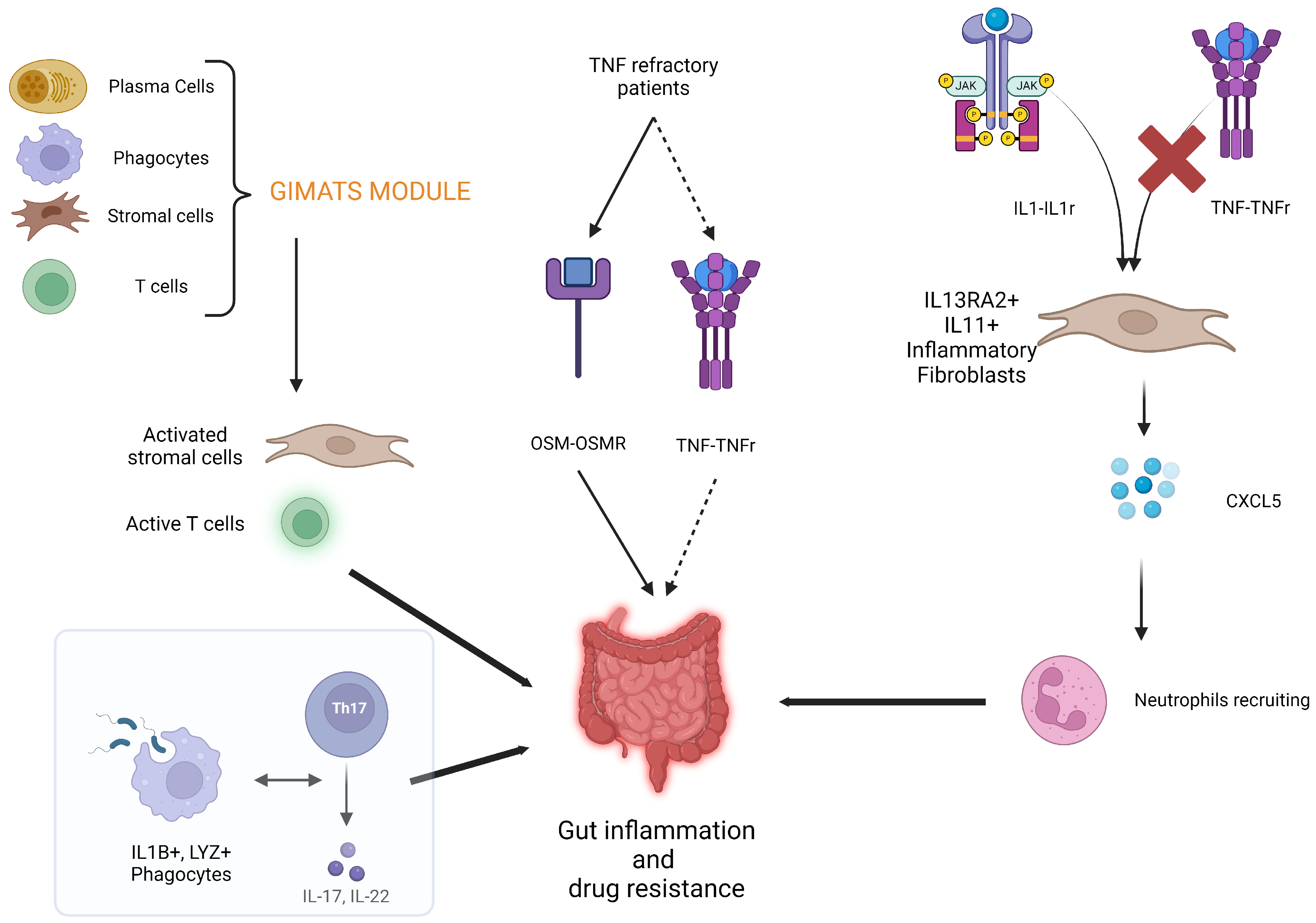
6. Other Mechanisms
7. Conclusions, Limitations and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Kaplan, G.G. The global burden of IBD: From 2015 to 2025. Nat. Rev. Gastroenterol. Hepatol. 2015, 12, 720–727. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Di, B.; Xu, L.-L. Recent advances in the treatment of IBD: Targets, mechanisms and related therapies. Cytokine Growth Factor Rev. 2023, 71–72, 1–12. [Google Scholar] [CrossRef]
- Jarmakiewicz-Czaja, S.; Zielińska, M.; Sokal, A.; Filip, R. Genetic and Epigenetic Etiology of Inflammatory Bowel Disease: An Update. Genes 2022, 13, 2388. [Google Scholar] [CrossRef] [PubMed]
- Link, J.; Ryner, M.L.; Fink, K.; Hermanrud, C.; Lima, I.; Brynedal, B.; Kockum, I.; Hillert, J.; Fogdell-Hahn, A. Human leukocyte antigen genes and interferon beta preparations influence risk of developing neutralizing anti-drug antibodies in multiple sclerosis. PLoS ONE 2014, 9, e90479. [Google Scholar] [CrossRef] [PubMed]
- Hayney, M.S.; Poland, G.A.; Jacobson, R.M.; Rabe, D.; Schaid, D.J.; Jacobsen, S.J.; Lipsky, J.J. Relationship of HLA-DQA1 alleles and humoral antibody following measles vaccination. Int. J. Infect. Dis. 1998, 2, 143–146. [Google Scholar] [CrossRef]
- Wilson, A.; Peel, C.; Wang, Q.; Pananos, A.D.; Kim, R.B. HLADQA1*05 genotype predicts anti-drug antibody formation and loss of response during infliximab therapy for inflammatory bowel disease. Aliment. Pharmacol. Ther. 2020, 51, 356–363. [Google Scholar] [CrossRef]
- Sazonovs, A.; Kennedy, N.A.; Moutsianas, L.; Heap, G.A.; Rice, D.L.; Reppell, M.; Bewshea, C.M.; Chanchlani, N.; Walker, G.J.; Perry, M.H.; et al. HLA-DQA1*05 Carriage Associated With Development of Anti-Drug Antibodies to Infliximab and Adalimumab in Patients With Crohn’s Disease. Gastroenterology 2020, 158, 189–199. [Google Scholar] [CrossRef]
- DelBaugh, R.M.; Cook, L.J.; Siegel, C.A.; Tsongalis, G.J.; Khan, W.A. Validation of a rapid HLA-DQA1*05 pharmacogenomics assay to identify at-risk resistance to anti–tumor necrosis factor therapy among patients with inflammatory bowel disease. Am. J. Clin. Pathol. 2023, 160, 194–199. [Google Scholar] [CrossRef]
- Pascual-Oliver, A.; Casas-Deza, D.; Cuarán, C.; García-López, S.; Corsino-Roche, P.; Sierra-Moros, E.; Olier-Martínez, P.; González-Tarancón, R.; Vicente-Lidón, R. HLA-DQA1*05 Was Not Associated With Primary Nonresponse or Loss of Response to First Anti-TNF in Real-World Inflammatory Bowel Disease Patients. Inflamm. Bowel Dis. 2023, izad130. [Google Scholar] [CrossRef]
- González, M.R.D.; Ballester, M.P.; Romero-González, E.; Sánchez-Pardo, A.M.; Marti-Aguado, D.; Tosca, J.; Suria, C.; Ausejo, R.A.; Moreno, I.P.; Silvestre, M.D.P.; et al. Biological treatment interruption in inflammatory bowel disease: Motivation and predictive factors. Gastroenterol. Hepatol. 2023, 46, 671–681. [Google Scholar] [CrossRef]
- Hernández, P.N.; Bellido, P.d.P.; González, L.L.; Rodríguez, C.G.; Pérez, A.P.; Arias, F.A. The HLA-DQA1*05 genotype does not influence the clinical response to ustekinumab and vedolizumab. Rev. Esp. Enfermedades Dig. 2023, 46, 671–681. [Google Scholar] [CrossRef]
- Colombel, J.-F.; Martín-Arranz, M.D.; Brinkman, B.; Guan, M.; Hart, A.; Gasink, C. HLA-DQA1*05 Not Associated With Ustekinumab Loss of Response and Antidrug Antibodies in Ulcerative Colitis and Crohn’s Disease Patients. Inflamm. Bowel Dis. 2023, izad273. [Google Scholar] [CrossRef] [PubMed]
- Dubinsky, M.C.; Mei, L.; Friedman, M.; Dhere, T.; Haritunians, T.; Hakonarson, H.; Kim, C.; Glessner, J.; Targan, S.R.; McGovern, D.P.; et al. Genome wide association (GWA) predictors of anti-TNFα therapeutic responsiveness in pediatric inflammatory bowel disease. Inflamm. Bowel Dis. 2010, 16, 1357–1366. [Google Scholar] [CrossRef] [PubMed]
- Salvador-Martín, S.; Zapata-Cobo, P.; Velasco, M.; Palomino, L.M.; Clemente, S.; Segarra, O.; Sánchez, C.; Tolín, M.; Moreno-Álvarez, A.; Fernández-Lorenzo, A.; et al. Association between HLA DNA Variants and Long-Term Response to Anti-TNF Drugs in a Spanish Pediatric Inflammatory Bowel Disease Cohort. Int. J. Mol. Sci. 2023, 24, 1797. [Google Scholar] [CrossRef] [PubMed]
- Billiet, T.; Casteele, N.V.; Van Stappen, T.; Princen, F.; Singh, S.; Gils, A.; Ferrante, M.; Van Assche, G.; Cleynen, I.; Vermeire, S. Immunogenicity to infliximab is associated with HLA-DRB1. Gut 2015, 64, 1344–1345. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Degner, J.; Davis, J.W.; Idler, K.B.; Nader, A.; Mostafa, N.M.; Waring, J.F. Identification of HLA-DRB1 association to adalimumab immunogenicity. PLoS ONE 2018, 13, e0195325. [Google Scholar] [CrossRef]
- Bek, S.; Nielsen, J.V.; Bojesen, A.B.; Franke, A.; Bank, S.; Vogel, U.; Andersen, V. Systematic review: Genetic biomarkers associated with anti-TNF treatment response in inflammatory bowel diseases. Aliment. Pharmacol. Ther. 2016, 44, 554–567. [Google Scholar] [CrossRef]
- Lauro, R.; Mannino, F.; Irrera, N.; Squadrito, F.; Altavilla, D.; Squadrito, G.; Pallio, G.; Bitto, A. Pharmacogenetics of Biological Agents Used in Inflammatory Bowel Disease: A Systematic Review. Biomedicines 2021, 9, 1748. [Google Scholar] [CrossRef]
- Lykowska-Szuber, L.; Walczak, M.; Stawczyk-Eder, K.; Krela-Kazmierczak, I.; Eder, P.; Zakerska-Banaszak, O.; Dobrowolska, A.; Skrzypczak-Zielinska, M. Variants of the CASP9 gene as candidate markers for primary response to anti-TNF therapy in Crohn’s disease patients. J. Appl. Genet. 2023, 64, 759–768. [Google Scholar] [CrossRef]
- Anderson, C.A.; Boucher, G.; Lees, C.W.; Franke, A.; D’Amato, M.; Taylor, K.D.; Lee, J.C.; Goyette, P.; Imielinski, M.; Latiano, A.; et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat. Genet. 2011, 43, 246–252. [Google Scholar] [CrossRef]
- Vermeire, S.; Louis, E.; Rutgeerts, P.; De Vos, M.; Van Gossum, A.; Belaiche, J.; Pescatore, P.; Fiasse, R.; Pelckmans, P.; Vlietinck, R.; et al. NOD2/CARD15 does not influence response to infliximab in Crohn’s disease. Gastroenterology 2002, 123, 106–111. [Google Scholar] [CrossRef] [PubMed]
- Schäffler, H.; Geiss, D.; Gittel, N.; Rohde, S.; Huth, A.; Glass, Ä.; Brandhorst, G.; Jaster, R.; Lamprecht, G. Mutations in theNOD2gene are associated with a specific phenotype and lower anti-tumor necrosis factor trough levels in Crohn’s disease. J. Dig. Dis. 2018, 19, 678–684. [Google Scholar] [CrossRef] [PubMed]
- Brinar, M.; Cukovic-Cavka, S.; Bozina, N.; Ravic, K.G.; Markos, P.; Ladic, A.; Cota, M.; Krznaric, Z.; Vucelic, B. MDR1polymorphisms are associated with inflammatory bowel disease in a cohort of Croatian IBD patients. BMC Gastroenterol. 2013, 13, 57. [Google Scholar] [CrossRef] [PubMed]
- Potočnik, U.; Ferkolj, I.; Glavač, D.; Dean, M. Polymorphisms in multidrug resistance 1 (MDR1) gene are associated with refractory Crohn disease and ulcerative colitis. Genes Immun. 2004, 5, 530–539. [Google Scholar] [CrossRef] [PubMed]
- Fischer, S.; Lakatos, P.L.; The Hungarian Ibd Study Group; Lakatos, L.; Kovacs, A.; Molnar, T.; Altorjay, I.; Papp, M.; Szilvasi, A.; Tulassay, Z.; et al. ATP-binding cassette transporter ABCG2 (BCRP) and ABCB1 (MDR1) variants are not associated with disease susceptibility, disease phenotype response to medical therapy or need for surgeryin Hungarian patients with inflammatory bowel diseases. Scand. J. Gastroenterol. 2007, 42, 726–733. [Google Scholar] [CrossRef] [PubMed]
- Fung, K.L.; Gottesman, M.M. A synonymous polymorphism in a common MDR1 (ABCB1) haplotype shapes protein function. Biochim. Biophys. Acta (BBA)—Proteins Proteom. 2009, 1794, 860–871. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Zhang, C.-B.; Lyu, K.-S.; Jin, Z.-M.; Guan, S.-X.; You, N.; Huang, M.; Wang, X.-D.; Gao, X. Association of polymorphisms in C1orf106, IL1RN, and IL10 with post-induction infliximab trough level in Crohn’s disease patients. Gastroenterol. Rep. 2019, 8, 367–373. [Google Scholar] [CrossRef]
- Salvador-Martín, S.; Pujol-Muncunill, G.; Bossacoma, F.; Navas-López, V.M.; Gallego-Fernández, C.; Segarra, O.; Clemente, S.; Muñoz-Codoceo, R.; Viada, J.; Magallares, L.; et al. Pharmacogenetics of trough serum anti-TNF levels in paediatric inflammatory bowel disease. Br. J. Clin. Pharmacol. 2021, 87, 447–457. [Google Scholar] [CrossRef]
- Salvador-Martín, S.; López-Cauce, B.; Nuñez, O.; Laserna-Mendieta, E.J.; García, M.I.; Lobato, E.; Abarca-Zabalía, J.; Sanjurjo-Saez, M.; Lucendo, A.J.; Marín-Jiménez, I.; et al. Genetic predictors of long-term response and trough levels of infliximab in crohn’s disease. Pharmacol. Res. 2019, 149, 104478. [Google Scholar] [CrossRef]
- Sewell, G.W.; Kaser, A. Interleukin-23 in the Pathogenesis of Inflammatory Bowel Disease and Implications for Therapeutic Intervention. J. Crohns Colitis 2022, 16 (Suppl. 2), ii3–ii19. [Google Scholar] [CrossRef] [PubMed]
- Cummings, F.J.; Ahmad, T.; Geremia, A.; Beckly, J.; Cooney, R.; Hancock, L.; Pathan, S.; Guo, C.; Cardon, L.R.; Jewell, D.P. Contribution of the novel inflammatory bowel disease gene IL23R to disease susceptibility and phenotype. Inflamm. Bowel Dis. 2007, 13, 1063–1068. [Google Scholar] [CrossRef]
- Kim, S.W.; Kim, E.S.; Moon, C.M.; Park, J.J.; Kim, T.I.; Kim, W.H.; Cheon, J.H. Genetic polymorphisms of IL-23R and IL-17A and novel insights into their associations with inflammatory bowel disease. Gut 2011, 60, 1527–1536. [Google Scholar] [CrossRef]
- Jürgens, M.; Laubender, R.P.; Hartl, F.; Weidinger, M.; Seiderer, J.; Wagner, J.; Wetzke, M.; Beigel, F.; Pfennig, S.; Stallhofer, J.; et al. Disease activity, ANCA, and IL23R genotype status determine early response to infliximab in patients with ulcerative colitis. Am. J. Gastroenterol. 2010, 105, 1811–1819. [Google Scholar] [CrossRef]
- Laserna-Mendieta, E.; Salvador-Martín, S.; Arias, A.; López-Cauce, B.; Marín-Jiménez, I.; Menchén, L.; Marín-Rubio, L.; Rodríguez, J.O.; López-Fernández, L.; Lucendo, A. Single nucleotide polymorphisms in ADAM17, IL23R and SLCO1C1 genes protect against infliximab failure in adults with Crohn’s disease. Biomed. Pharmacother. 2023, 159, 114225. [Google Scholar] [CrossRef]
- Sherlock, M.E.; Walters, T.; Tabbers, M.M.; Frost, K.; Zachos, M.; Muise, A.; Pope, E.; Griffiths, A.M. Infliximab-induced psoriasis and psoriasiform skin lesions in pediatric crohn disease and a potential association with IL-23 receptor polymorphisms. J. Pediatr. Gastroenterol. Nutr. 2013, 56, 512–518. [Google Scholar] [CrossRef] [PubMed]
- Cravo, M.L.; Ferreira, P.A.; Sousa, P.; Moura-Santos, P.; Velho, S.; Tavares, L.; de Deus, J.R.; Ministro, P.; Peixe, P.; Correia, L.A.; et al. IL23R polymorphisms influence phenotype and response to therapy in patients with ulcerative colitis. Eur. J. Gastroenterol. Hepatol. 2014, 26, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Tabatabaei-Panah, P.-S.; Moravvej, H.; Delpasand, S.; Jafari, M.; Sepehri, S.; Abgoon, R.; Ludwig, R.J.; Akbarzadeh, R. IL12B and IL23R polymorphisms are associated with alopecia areata. Genes Immun. 2020, 21, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Tillack, C.; Ehmann, L.M.; Friedrich, M.; Laubender, R.P.; Papay, P.; Vogelsang, H.; Stallhofer, J.; Beigel, F.; Bedynek, A.; Wetzke, M.; et al. Anti-TNF antibody-induced psoriasiform skin lesions in patients with inflammatory bowel disease are characterised by interferon-γ-expressing Th1 cells and IL-17A/IL-22-expressing Th17 cells and respond to anti-IL-12/IL-23 antibody treatment. Gut 2014, 63, 567–577. [Google Scholar] [CrossRef] [PubMed]
- Cabaleiro, T.; Prieto-Pérez, R.; Navarro, R.; Solano, G.; Román, M.; Ochoa, D.; Abad-Santos, F.; Daudén, E. Paradoxical psoriasiform reactions to anti-TNFα drugs are associated with genetic polymorphisms in patients with psoriasis. Pharmacogenomics J. 2016, 16, 336–340. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, P.; Lamerz, D.; Hill, P.; Kirchner, M.; Gauss, A. Gene Polymorphisms of NOD2, IL23R, PTPN2 and ATG16L1 in Patients with Crohn’s Disease: On the Way to Personalized Medicine? Genes 2021, 12, 866. [Google Scholar] [CrossRef]
- West, N.R.; Owens, B.M.J.; Hegazy, A.N. The oncostatin M-stromal cell axis in health and disease. Scand. J. Immunol. 2018, 88, e12694. [Google Scholar] [CrossRef] [PubMed]
- Lantieri, F.; Bachetti, T. OSM/OSMR and Interleukin 6 Family Cytokines in Physiological and Pathological Condition. Int. J. Mol. Sci. 2022, 23, 11096. [Google Scholar] [CrossRef] [PubMed]
- West, N.R.; Oxford IBD Cohort Investigators; Hegazy, A.N.; Owens, B.M.J.; Bullers, S.J.; Linggi, B.; Buonocore, S.; Coccia, M.; Görtz, D.; This, S.; et al. Erratum: Oncostatin M drives intestinal inflammation and predicts response to tumor necrosis factor–neutralizing therapy in patients with inflammatory bowel disease. Nat. Med. 2017, 23, 788. [Google Scholar] [CrossRef] [PubMed]
- Bertani, L.; Fornai, M.; Fornili, M.; Antonioli, L.; Benvenuti, L.; Tapete, G.; Svizzero, G.B.; Ceccarelli, L.; Mumolo, M.G.; Baglietto, L.; et al. Serum oncostatin M at baseline predicts mucosal healing in Crohn’s disease patients treated with infliximab. Aliment. Pharmacol. Ther. 2020, 52, 284–291. [Google Scholar] [CrossRef] [PubMed]
- Guo, A.; Ross, C.; Chande, N.; Gregor, J.; Ponich, T.; Khanna, R.; Sey, M.; Beaton, M.; Yan, B.; Kim, R.B.; et al. High oncostatin M predicts lack of clinical remission for patients with inflammatory bowel disease on tumor necrosis factor α antagonists. Sci. Rep. 2022, 12, 1185. [Google Scholar] [CrossRef]
- Cao, Y.; Dai, Y.; Zhang, L.; Wang, D.; Hu, W.; Yu, Q.; Wang, X.; Yu, P.; Liu, W.; Ping, Y.; et al. Combined Use of Fecal Biomarkers in Inflammatory Bowel Diseases: Oncostatin M and Calprotectin. J. Inflamm. Res. 2021, 14, 6409–6419. [Google Scholar] [CrossRef]
- Bertani, L.; Barberio, B.; Fornili, M.; Antonioli, L.; Zanzi, F.; Casadei, C.; Benvenuti, L.; Facchin, S.; D’Antongiovanni, V.; Lorenzon, G.; et al. Serum oncostatin M predicts mucosal healing in patients with inflammatory bowel diseases treated with anti-TNF, but not vedolizumab. Dig. Liver Dis. 2022, 54, 1367–1373. [Google Scholar] [CrossRef]
- Nishioka, K.; Ogino, H.; Chinen, T.; Ihara, E.; Tanaka, Y.; Nakamura, K.; Ogawa, Y. Mucosal IL23A expression predicts the response to Ustekinumab in inflammatory bowel disease. J. Gastroenterol. 2021, 56, 976–987. [Google Scholar] [CrossRef]
- Gaujoux, R.; Starosvetsky, E.; Maimon, N.; Vallania, F.; Bar-Yoseph, H.; Pressman, S.; Weisshof, R.; Goren, I.; Rabinowitz, K.; Waterman, M.; et al. Cell-centred meta-analysis reveals baseline predictors of anti-TNFα non-response in biopsy and blood of patients with IBD. Gut 2019, 68, 604–614. [Google Scholar] [CrossRef]
- Verstockt, B.; Verstockt, S.; Dehairs, J.; Ballet, V.; Blevi, H.; Wollants, W.-J.; Breynaert, C.; Van Assche, G.; Vermeire, S.; Ferrante, M. Low TREM1 expression in whole blood predicts anti-TNF response in inflammatory bowel disease. EBioMedicine 2019, 40, 733–742. [Google Scholar] [CrossRef]
- Arijs, I.; Quintens, R.; Van Lommel, L.; Van Steen, K.; De Hertogh, G.; Lemaire, K.; Schraenen, A.; Perrier, C.; Van Assche, G.; Vermeire, S.; et al. Predictive value of epithelial gene expression profiles for response to infliximab in Crohn’s disease. Inflamm. Bowel Dis. 2010, 16, 2090–2098. [Google Scholar] [CrossRef]
- Arijs, I.; Li, K.; Toedter, G.; Quintens, R.; Van Lommel, L.; Van Steen, K.; Leemans, P.; De Hertogh, G.; Lemaire, K.; Ferrante, M.; et al. Mucosal gene signatures to predict response to infliximab in patients with ulcerative colitis. Gut 2009, 58, 1612–1619. [Google Scholar] [CrossRef]
- Verstockt, B.; Verstockt, S.; Creyns, B.; Tops, S.; Van Assche, G.; Gils, A.; Ceuppens, J.L.; Vermeire, S.; Ferrante, M.; Breynaert, C. Mucosal IL13RA2 expression predicts nonresponse to anti-TNF therapy in Crohn’s disease. Aliment. Pharmacol. Ther. 2019, 49, 572–581. [Google Scholar] [CrossRef]
- Clarkston, K.; Karns, R.; Jegga, A.G.; Sharma, M.; Fox, S.; Ojo, B.A.; Minar, P.; Walters, T.D.; Griffiths, A.M.; Mack, D.R.; et al. Targeted Assessment of Mucosal Immune Gene Expression Predicts Clinical Outcomes in Children with Ulcerative Colitis. J. Crohns Colitis 2022, 16, 1735–1750. [Google Scholar] [CrossRef]
- Nazari, M.H.D.; Shahrokh, S.; Ghanbari-Maman, L.; Maleknia, S.; Ghorbaninejad, M.; Meyfour, A. Prediction of anti-TNF therapy failure in ulcerative colitis patients by ensemble machine learning: A prospective study. Heliyon 2023, 9, e21154. [Google Scholar] [CrossRef] [PubMed]
- Karmele, E.P.; Pasricha, T.S.; Ramalingam, T.R.; Thompson, R.W.; Gieseck, R.L.; Knilans, K.J.; Hegen, M.; Farmer, M.; Jin, F.; Kleinman, A.; et al. Anti-IL-13Rα2 therapy promotes recovery in a murine model of inflammatory bowel disease. Mucosal Immunol. 2019, 12, 1174–1186. [Google Scholar] [CrossRef] [PubMed]
- Verstockt, B.; Perrier, C.; De Hertogh, G.; Cremer, J.; Creyns, B.; Van Assche, G.; Ferrante, M.; Ceuppens, J.L.; Vermeire, S.; Breynaert, C. Effects of Epithelial IL-13Rα2 Expression in Inflammatory Bowel Disease. Front. Immunol. 2018, 9, 2983. [Google Scholar] [CrossRef] [PubMed]
- Verstockt, B.; Verstockt, S.; Veny, M.; Dehairs, J.; Arnauts, K.; Van Assche, G.; De Hertogh, G.; Vermeire, S.; Salas, A.; Ferrante, M. Expression Levels of 4 Genes in Colon Tissue Might Be Used to Predict Which Patients Will Enter Endoscopic Remission After Vedolizumab Therapy for Inflammatory Bowel Diseases. Clin. Gastroenterol. Hepatol. 2020, 18, 1142–1151.e10. [Google Scholar] [CrossRef]
- Haglund, S.; Söderman, J.; Almer, S. Differences in Whole-Blood Transcriptional Profiles in Inflammatory Bowel Disease Patients Responding to Vedolizumab Compared with Non-Responders. Int. J. Mol. Sci. 2023, 24, 5820. [Google Scholar] [CrossRef]
- Peña-Cearra, A.; Castelo, J.; Lavín, J.L.; Gonzalez-Lopez, M.; Pascual-Itoiz, M.A.; Fuertes, M.; de Juan, V.G.; Bárcena, L.; Martín-Ruiz, I.; Pellón, A.; et al. Mitochondrial dysfunction-associated microbiota establishes a transmissible refractory response to anti-TNF therapy during ulcerative colitis. Gut Microbes 2023, 15, 2266626. [Google Scholar] [CrossRef]
- Smillie, C.S.; Biton, M.; Ordovas-Montanes, J.; Sullivan, K.M.; Burgin, G.; Graham, D.B.; Herbst, R.H.; Rogel, N.; Slyper, M.; Waldman, J.; et al. Intra- and Inter-cellular Rewiring of the Human Colon during Ulcerative Colitis. Cell 2019, 178, 714–730.e22. [Google Scholar] [CrossRef]
- Martin, J.C.; Chang, C.; Boschetti, G.; Ungaro, R.; Giri, M.; Grout, J.A.; Gettler, K.; Chuang, L.-S.; Nayar, S.; Greenstein, A.J.; et al. Single-Cell Analysis of Crohn’s Disease Lesions Identifies a Pathogenic Cellular Module Associated with Resistance to Anti-TNF Therapy. Cell 2019, 178, 1493–1508.e20. [Google Scholar] [CrossRef]
- Friedrich, M.; Pohin, M.; Jackson, M.A.; Korsunsky, I.; Bullers, S.J.; Rue-Albrecht, K.; Christoforidou, Z.; Sathananthan, D.; Thomas, T.; Ravindran, R.; et al. IL-1-driven stromal–neutrophil interactions define a subset of patients with inflammatory bowel disease that does not respond to therapies. Nat. Med. 2021, 27, 1970–1981. [Google Scholar] [CrossRef]
- Mayer, A.T.; Holman, D.R.; Sood, A.; Tandon, U.; Bhate, S.S.; Bodapati, S.; Barlow, G.L.; Chang, J.; Black, S.; Crenshaw, E.C.; et al. A tissue atlas of ulcerative colitis revealing evidence of sex-dependent differences in disease-driving inflammatory cell types and resistance to TNF inhibitor therapy. Sci. Adv. 2023, 9, eadd1166. [Google Scholar] [CrossRef]
- Devlin, J.C.; Axelrad, J.; Hine, A.M.; Chang, S.; Sarkar, S.; Lin, J.-D.; Ruggles, K.V.; Hudesman, D.; Cadwell, K.; Loke, P. Single-Cell Transcriptional Survey of Ileal-Anal Pouch Immune Cells From Ulcerative Colitis Patients. Gastroenterology 2021, 160, 1679–1693. [Google Scholar] [CrossRef] [PubMed]
- Fiocchi, C. Omics and Multi-Omics in IBD: No Integration, No Breakthroughs. Int. J. Mol. Sci. 2023, 24, 14912. [Google Scholar] [CrossRef] [PubMed]
- Dor, Y.; Cedar, H. Principles of DNA methylation and their implications for biology and medicine. Lancet 2018, 392, 777–786. [Google Scholar] [CrossRef] [PubMed]
- McDermott, E.; Ryan, E.J.; Tosetto, M.; Gibson, D.; Burrage, J.; Keegan, D.; Byrne, K.; Crowe, E.; Sexton, G.; Malone, K.; et al. DNA Methylation Profiling in Inflammatory Bowel Disease Provides New Insights into Disease Pathogenesis. J. Crohns Colitis 2016, 10, 77–86. [Google Scholar] [CrossRef]
- Joustra, V.; Hageman, I.L.; Satsangi, J.; Adams, A.; Ventham, N.T.; de Jonge, W.J.; Henneman, P.; D’haens, G.R.; Yim, A.Y.F.L. Systematic Review and Meta-analysis of Peripheral Blood DNA Methylation Studies in Inflammatory Bowel Disease. J. Crohns Colitis 2023, 17, 185–198. [Google Scholar] [CrossRef]
- Birney, E.; Smith, G.D.; Greally, J.M. Epigenome-wide Association Studies and the Interpretation of Disease -Omics. PLoS Genet. 2016, 12, e1006105. [Google Scholar] [CrossRef]
- Kennedy, N.A.; Heap, G.A.; Green, H.D.; Hamilton, B.; Bewshea, C.; Walker, G.J.; Thomas, A.; Nice, R.; Perry, M.H.; Bouri, S.; et al. Predictors of anti-TNF treatment failure in anti-TNF-naive patients with active luminal Crohn’s disease: A prospective, multicentre, cohort study. Lancet Gastroenterol. Hepatol. 2019, 4, 341–353. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Hannon, E.; Reppell, M.; Waring, J.F.; Smaoui, N.; Pivorunas, V.; Guay, H.; Chanchlani, N.; Bewshea, C.; Bai, B.Y.H.; et al. Whole blood DNA methylation changes are associated with anti-TNF drug concentration in patients with crohn’s disease. J. Crohns Colitis 2023, jjad133. [Google Scholar] [CrossRef] [PubMed]
- Bai, B.Y.H.; Reppell, M.; Smaoui, N.; Waring, J.F.; Pivorunas, V.; Guay, H.; Lin, S.; Chanchlani, N.; Bewshea, C.; Goodhand, J.R.; et al. Baseline expression of immune gene modules in blood is associated with primary response to anti-TNF therapy in crohn’s disease patients. J. Crohns Colitis 2023, jjad166. [Google Scholar] [CrossRef] [PubMed]
- Julià, A.; Gómez, A.; López-Lasanta, M.; Blanco, F.; Erra, A.; Fernández-Nebro, A.; Mas, A.J.; Pérez-García, C.; Vivar, M.L.G.; Sánchez-Fernández, S.; et al. Longitudinal analysis of blood DNA methylation identifies mechanisms of response to tumor necrosis factor inhibitor therapy in rheumatoid arthritis. EBioMedicine 2022, 80, 104053. [Google Scholar] [CrossRef] [PubMed]
- Mishra, N.; Aden, K.; Blase, J.I.; Baran, N.; Bordoni, D.; Tran, F.; Conrad, C.; Avalos, D.; Jaeckel, C.; Scherer, M.; et al. Longitudinal multi-omics analysis identifies early blood-based predictors of anti-TNF therapy response in inflammatory bowel disease. Genome Med. 2022, 14, 110. [Google Scholar] [CrossRef] [PubMed]
- Dhuppar, S.; Murugaiyan, G. miRNA effects on gut homeostasis: Therapeutic implications for inflammatory bowel disease. Trends Immunol. 2022, 43, 917–931. [Google Scholar] [CrossRef] [PubMed]
- Nag, S.; Mitra, O.; Tripathi, G.; Samanta, S.; Bhattacharya, B.; Chandane, P.; Mohanto, S.; Sundararajan, V.; Malik, S.; Rustagi, S.; et al. Exploring the theranostic potentials of miRNA and epigenetic networks in autoimmune diseases: A comprehensive review. Immun. Inflamm. Dis. 2023, 11, e1121. [Google Scholar] [CrossRef]
- Batra, S.K.; Heier, C.R.; Diaz-Calderon, L.; Tully, C.B.; Fiorillo, A.A.; Anker, J.v.D.; Conklin, L.S. Serum miRNAs Are Pharmacodynamic Biomarkers Associated With Therapeutic Response in Pediatric Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2020, 26, 1597–1606. [Google Scholar] [CrossRef]
- Sáez-González, E.; Moret-Tatay, I.; Bastida, G.; Aguas, M.; Iborra, M.; Nos, P.; Beltrán, B. MicroRNA and granulocyte-monocyte adsorption apheresis combotherapy after inadequate response to anti-TNF agents in ulcerative colitis. J. Clin. Apher. 2023, 39, e22101. [Google Scholar] [CrossRef]
- Papaconstantinou, I.; Kapizioni, C.; Legaki, E.; Xourgia, E.; Karamanolis, G.; Gklavas, A.; Gazouli, M. Association of miR-146 rs2910164, miR-196a rs11614913, miR-221 rs113054794 and miR-224 rs188519172 polymorphisms with anti-TNF treatment response in a Greek population with Crohn’s disease. World J. Gastrointest. Pharmacol. Ther. 2017, 8, 193–200. [Google Scholar] [CrossRef]
- Casertano, M.; Trotta, M.C.; Cenni, S.; Creoli, M.; Miele, E.; Martinelli, M.; Lepre, C.C.; Russo, M.; Alfano, R.; D’Amico, M.; et al. Infliximab therapy decreases the expression of serum and faecal miR-126 and miR-20a in paediatric Crohn’s disease: A pilot study. Acta Paediatr. 2023, 113, 590–597. [Google Scholar] [CrossRef]
- Dahlén, R.; Magnusson, M.K.; Bajor, A.; Lasson, A.; Ung, K.-A.; Strid, H.; Öhman, L. Global mucosal and serum cytokine profile in patients with ulcerative colitis undergoing anti-TNF therapy. Scand. J. Gastroenterol. 2015, 50, 1118–1126. [Google Scholar] [CrossRef]
- Magnusson, M.K.; Strid, H.; Sapnara, M.; Lasson, A.; Bajor, A.; Ung, K.-A.; Öhman, L. Anti-TNF Therapy Response in Patients with Ulcerative Colitis Is Associated with Colonic Antimicrobial Peptide Expression and Microbiota Composition. J. Crohns Colitis 2016, 10, 943–952. [Google Scholar] [CrossRef]
- Aden, K.; Rehman, A.; Waschina, S.; Pan, W.-H.; Walker, A.; Lucio, M.; Nunez, A.M.; Bharti, R.; Zimmerman, J.; Bethge, J.; et al. Metabolic Functions of Gut Microbes Associate With Efficacy of Tumor Necrosis Factor Antagonists in Patients With Inflammatory Bowel Diseases. Gastroenterology 2019, 157, 1279–1292.e11. [Google Scholar] [CrossRef]
- Ananthakrishnan, A.N.; Luo, C.; Yajnik, V.; Khalili, H.; Garber, J.J.; Stevens, B.W.; Cleland, T.; Xavier, R.J. Gut Microbiome Function Predicts Response to Anti-integrin Biologic Therapy in Inflammatory Bowel Diseases. Cell Host Microbe 2017, 21, 603–610.e3. [Google Scholar] [CrossRef]
- Doherty, M.K.; Ding, T.; Koumpouras, C.; Telesco, S.E.; Monast, C.; Das, A.; Brodmerkel, C.; Schloss, P.D. Fecal Microbiota Signatures Are Associated with Response to Ustekinumab Therapy among Crohn’s Disease Patients. mBio 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Wyant, T.; Fedyk, E.; Abhyankar, B. An Overview of the Mechanism of Action of the Monoclonal Antibody Vedolizumab. J. Crohns Colitis 2016, 10, 1437–1444. [Google Scholar] [CrossRef] [PubMed]
- Deveuve, Q.; Lajoie, L.; Barrault, B.; Thibault, G. The Proteolytic Cleavage of Therapeutic Monoclonal Antibody Hinge Region: More Than a Matter of Subclass. Front. Immunol. 2020, 11, 168. [Google Scholar] [CrossRef] [PubMed]
- Derijks, L.J.J.; Wong, D.R.; Hommes, D.W.; van Bodegraven, A.A. Clinical Pharmacokinetic and Pharmacodynamic Considerations in the Treatment of Inflammatory Bowel Disease. Clin. Pharmacokinet. 2018, 57, 1075–1106. [Google Scholar] [CrossRef] [PubMed]
- Rizzello, F.; Saracino, I.M.; Gionchetti, P.; Valerii, M.C.; Ricci, C.; Imbesi, V.; Filippone, E.; Bellocchio, I.; Dussias, N.K.; Dervieux, T.; et al. Nutritional Biomarkers for the Prediction of Response to Anti-TNF-α Therapy in Crohn’s Disease: New Tools for New Approaches. Nutrients 2024, 16, 280. [Google Scholar] [CrossRef]
- Abraham, B.P.; Fan, C.; Thurston, T.; Moskow, J.; Malaty, H.M. The Role of Vitamin D in Patients with Inflammatory Bowel Disease Treated with Vedolizumab. Nutrients 2023, 15, 4847. [Google Scholar] [CrossRef] [PubMed]
- Hizarcioglu-Gulsen, H.; Kaplan, J.L.; Moran, C.J.; Israel, E.J.; Lee, H.; Winter, H. The Impact of Vitamin D on Response to Anti-tumor Necrosis Factor-α Therapy in Children With Inflammatory Bowel Disease. J. Pediatr. Gastroenterol. Nutr. 2021, 72, e125–e131. [Google Scholar] [CrossRef] [PubMed]
- Chanchlani, N.; Lin, S.; Smith, R.; Roberts, C.; Nice, R.; McDonald, T.J.; Hamilton, B.; Bishara, M.; Bewshea, C.; Kennedy, N.A.; et al. Pretreatment Vitamin D Concentrations Do Not Predict Therapeutic Outcome to Anti-TNF Therapies in Biologic-Naïve Patients With Active Luminal Crohn’s Disease. Crohns Colitis 360 2023, 5, otad026. [Google Scholar] [CrossRef] [PubMed]
- Kaazan, P.; Seow, W.; Yong, S.; Heilbronn, L.K.; Segal, J.P. The Impact of Obesity on Inflammatory Bowel Disease. Biomedicines 2023, 11, 3256. [Google Scholar] [CrossRef]
- Levine, L. Effect of obesity on vedolizumab response in inflammatory bowel disease. Ann. Gastroenterol. 2022, 35, 275–280. [Google Scholar] [CrossRef]
- Plaza-Diaz, J.; Álvarez-Mercado, A.I.; Yu, B.; Sungthong, R. Editorial: Molecular mechanisms underlying obesity and their links with other comorbidities. Front. Mol. Biosci. 2024, 10, 1334024. [Google Scholar] [CrossRef]
- Wetwittayakhlang, P.; Lakatos, P.L. Current Evidence for Combined Targeted Therapy for the Treatment of Inflammatory Bowel Disease. J. Can. Assoc. Gastroenterol. 2023, 7, 22–29. [Google Scholar] [CrossRef]
| SNPs | Molecule | Disease | Drug | Effect | References |
|---|---|---|---|---|---|
| rs2097432 | HLA-DQA1*05 | P-UC P-CD UC CD | IFX ADA | Decreased response | [10,14] |
| rs2395185 | HLA-DRB9 | P-UC P-CD | IFX | Decreased response | [13,14] |
| rs1052571 rs4645978 | CASP9 | CD | IFX ADA | Increased response | [19] |
| rs7234029 | PTPN2 | CD | UST | Decreased response | [20] |
| rs442905 | C1orf106 | CD | IFX | Decreased IFX levels | [27] |
| rs7587051 | ATG16L1 | CD | IFX | Decreased IFX levels | [27] |
| rs3213448 | IL1RN | CD | IFX | Increased IFX levels | [27] |
| rs5030728 | TLR4 | P-UC P-CD | IFX | Decreased IFX levels | [28] |
| rs11465996 | LY96 | P-UC P-CD | IFX | Decreased IFX levels | [28] |
| rs1816702 | TLR2 | P-UC P-CD | ADA | Decreased ADA levels | [28] |
| rs3397 | TNFRSF1B | P-UC P-CD | ADA | Decreased ADA levels | [28] |
| rs3024505 | IL-10 | CD | IFX | Decreased IFX levels | [29] |
| SNPs | Molecule | Disease | Drug | Effect | References |
|---|---|---|---|---|---|
| rs1004819 rs2201841 rs10889677 rs11209032 rs1495965 | IL-23R | UC | IFX | Increased response | [33] |
| rs7517847 rs10489629 rs11465804 rs1343151 | IL-23R | UC | IFX | Decreased response | [33] |
| rs10489629 | IL-23R | CD | IFX | Increased response | [34] |
| rs10489628 rs10789229 rs1343151 | IL-23R | P-CD | IFX | Increased IFX-induced psoriasis rates | [35] |
| rs1004819 rs10889677 | IL-23R | UC | IFX | Increased EIMs rates | [36] |
| rs11209026 | IL-23R | UC, CD, Psoriasis | IFX, ADA | Increased Anti-TNF-induced psoriasiform skin lesion rates; Increased risk of CD-related surgery | [38,39,40] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Puca, P.; Capobianco, I.; Coppola, G.; Di Vincenzo, F.; Trapani, V.; Petito, V.; Laterza, L.; Pugliese, D.; Lopetuso, L.R.; Scaldaferri, F. Cellular and Molecular Determinants of Biologic Drugs Resistance and Therapeutic Failure in Inflammatory Bowel Disease. Int. J. Mol. Sci. 2024, 25, 2789. https://doi.org/10.3390/ijms25052789
Puca P, Capobianco I, Coppola G, Di Vincenzo F, Trapani V, Petito V, Laterza L, Pugliese D, Lopetuso LR, Scaldaferri F. Cellular and Molecular Determinants of Biologic Drugs Resistance and Therapeutic Failure in Inflammatory Bowel Disease. International Journal of Molecular Sciences. 2024; 25(5):2789. https://doi.org/10.3390/ijms25052789
Chicago/Turabian StylePuca, Pierluigi, Ivan Capobianco, Gaetano Coppola, Federica Di Vincenzo, Valentina Trapani, Valentina Petito, Lucrezia Laterza, Daniela Pugliese, Loris Riccardo Lopetuso, and Franco Scaldaferri. 2024. "Cellular and Molecular Determinants of Biologic Drugs Resistance and Therapeutic Failure in Inflammatory Bowel Disease" International Journal of Molecular Sciences 25, no. 5: 2789. https://doi.org/10.3390/ijms25052789






