TMEM119 (c.G143A, p.S48L) Mutation Is Involved in Primary Failure of Eruption by Attenuating Glycolysis-Mediated Osteogenesis
Abstract
:1. Introduction
2. Results
2.1. Clinical Characteristics of Primary Failure of Eruption (PFE) Individuals
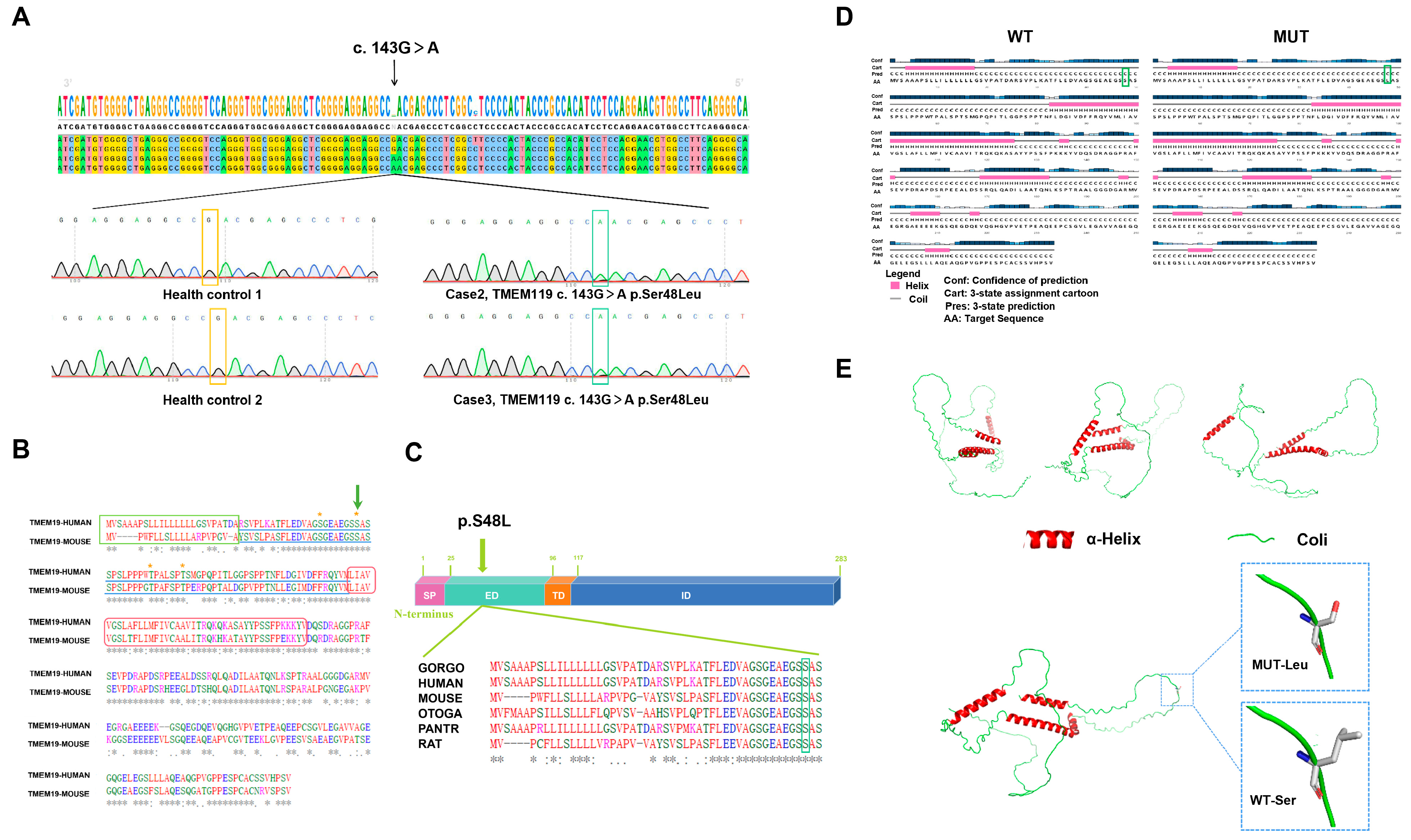
2.2. Identified Mutation and Pathogenic Analysis
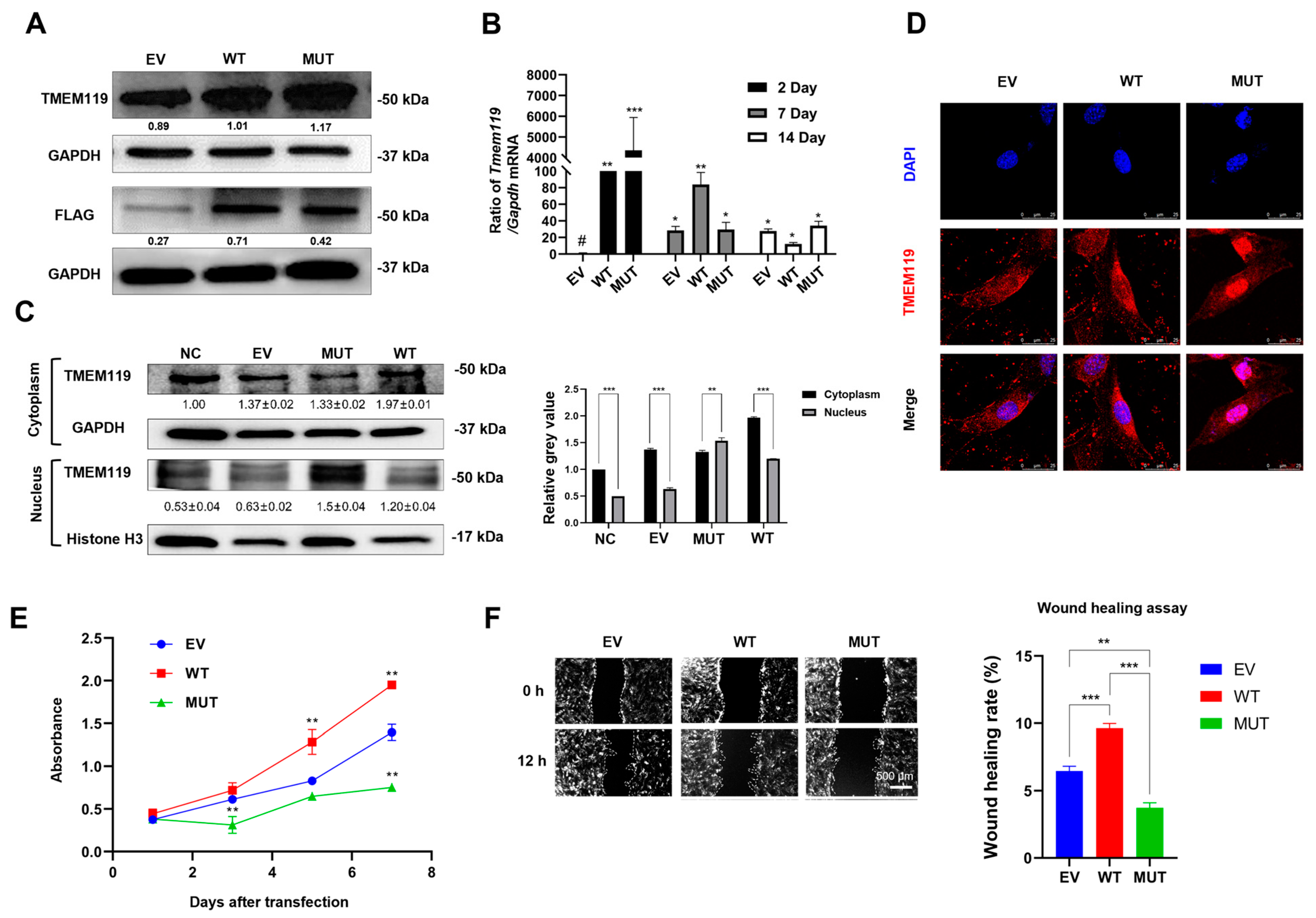
2.3. Overexpression, Subcellular Distribution, Proliferative Activity and Migration Ability Analysis of MC3T3-E1 Cells after TMEM119 Transfection
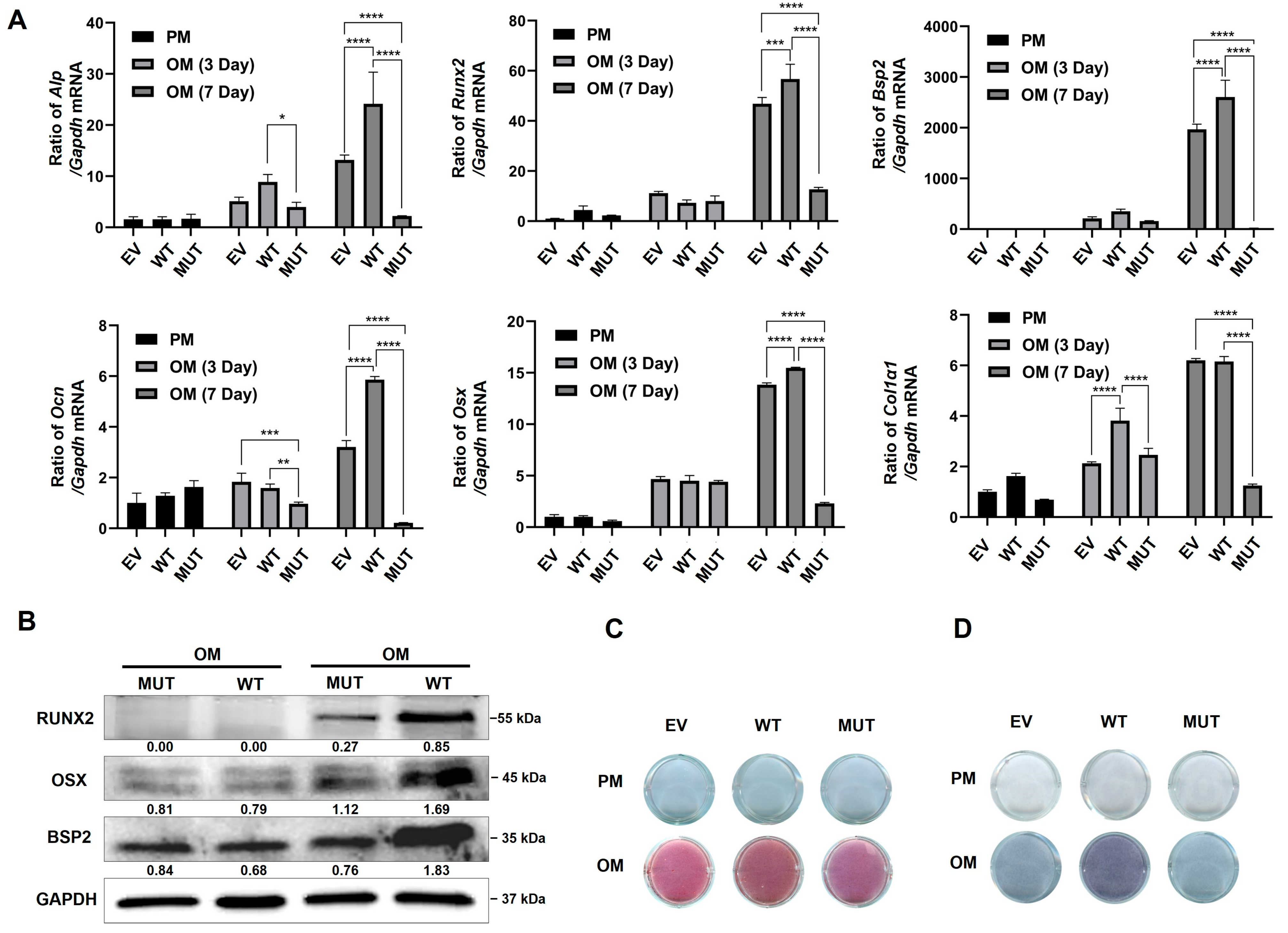
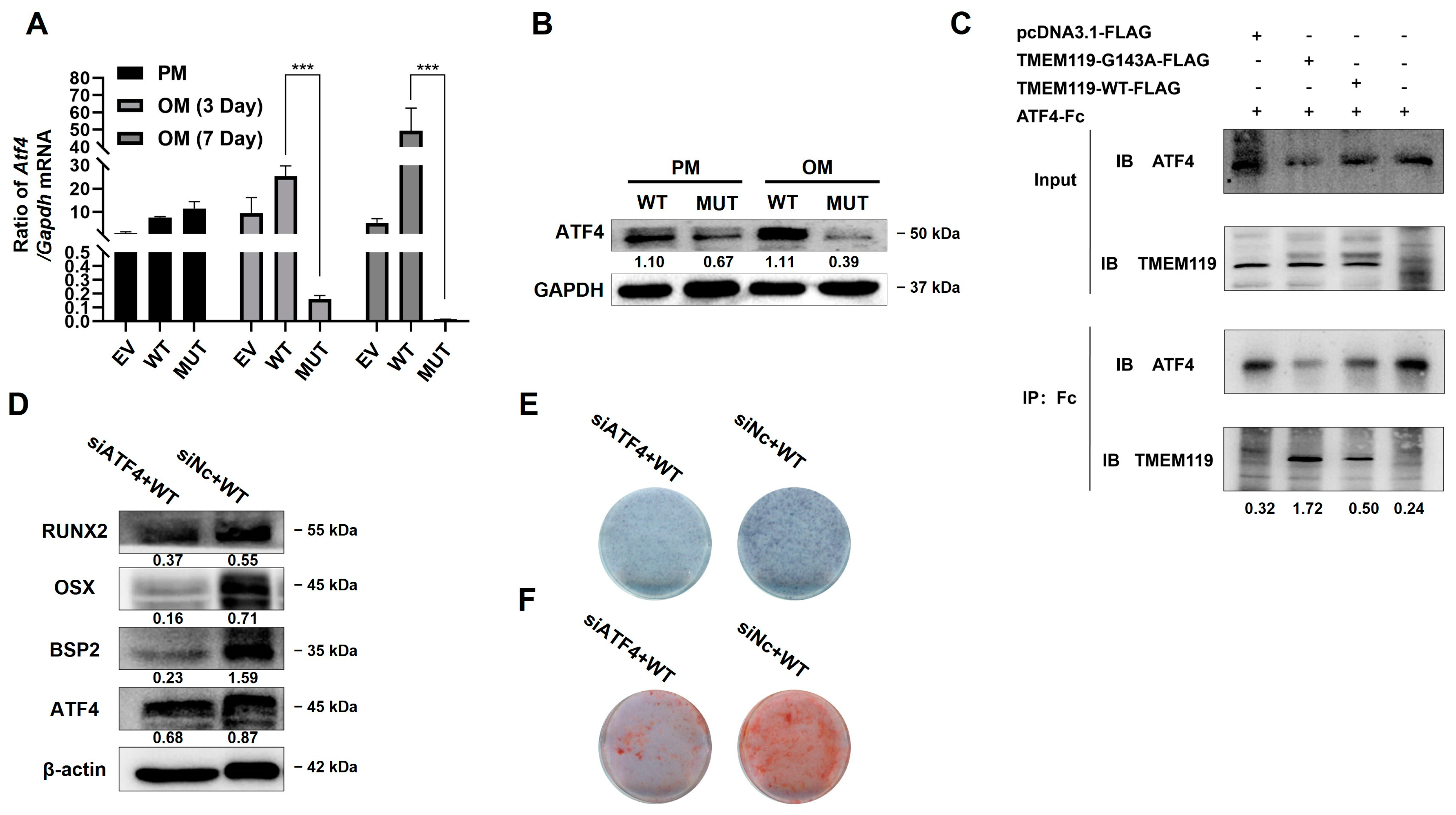
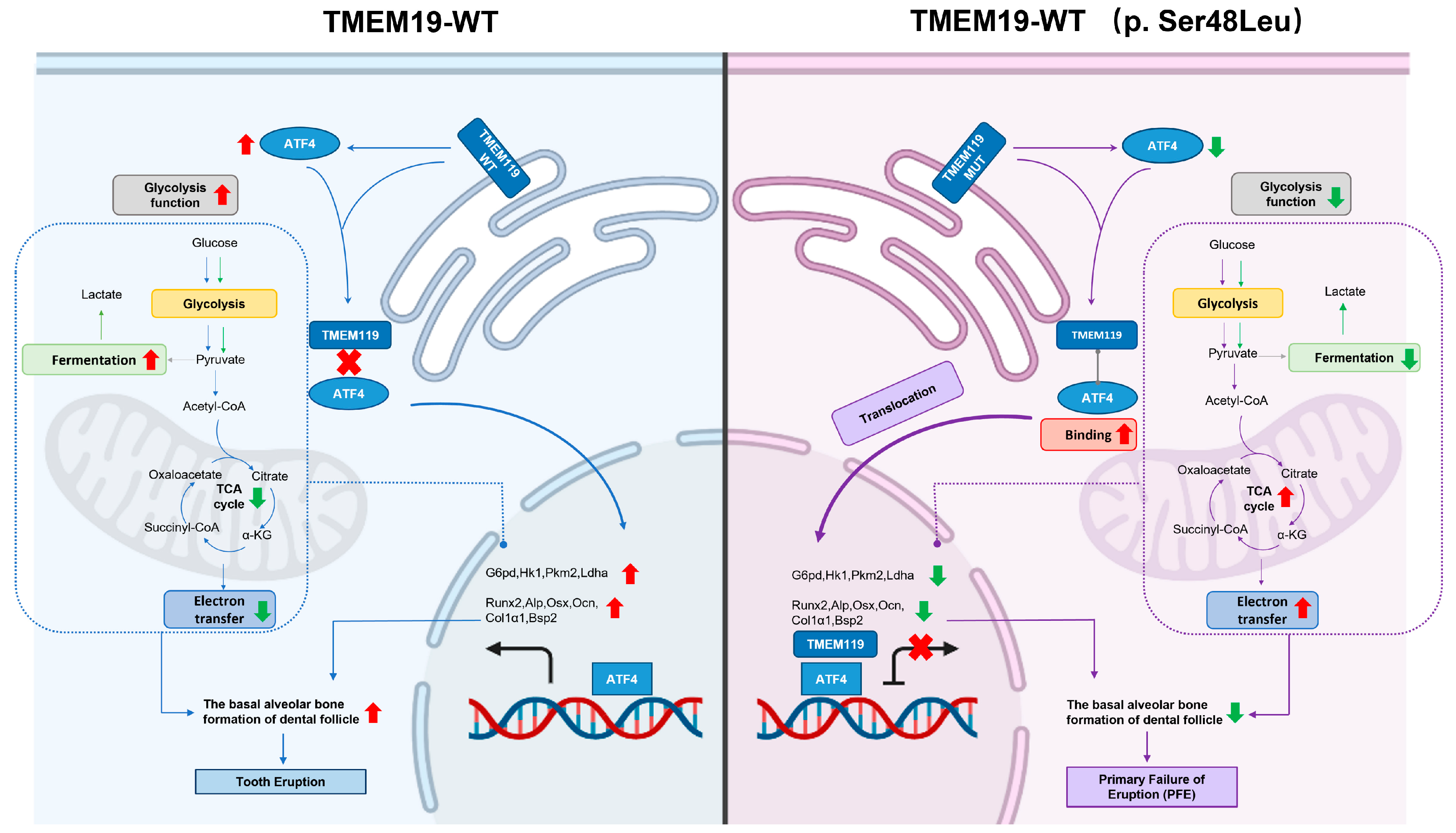
2.4. TMEM119 Affects Osteogenic Differentiation of MC3T3E1 Cells by Regulating ATF4
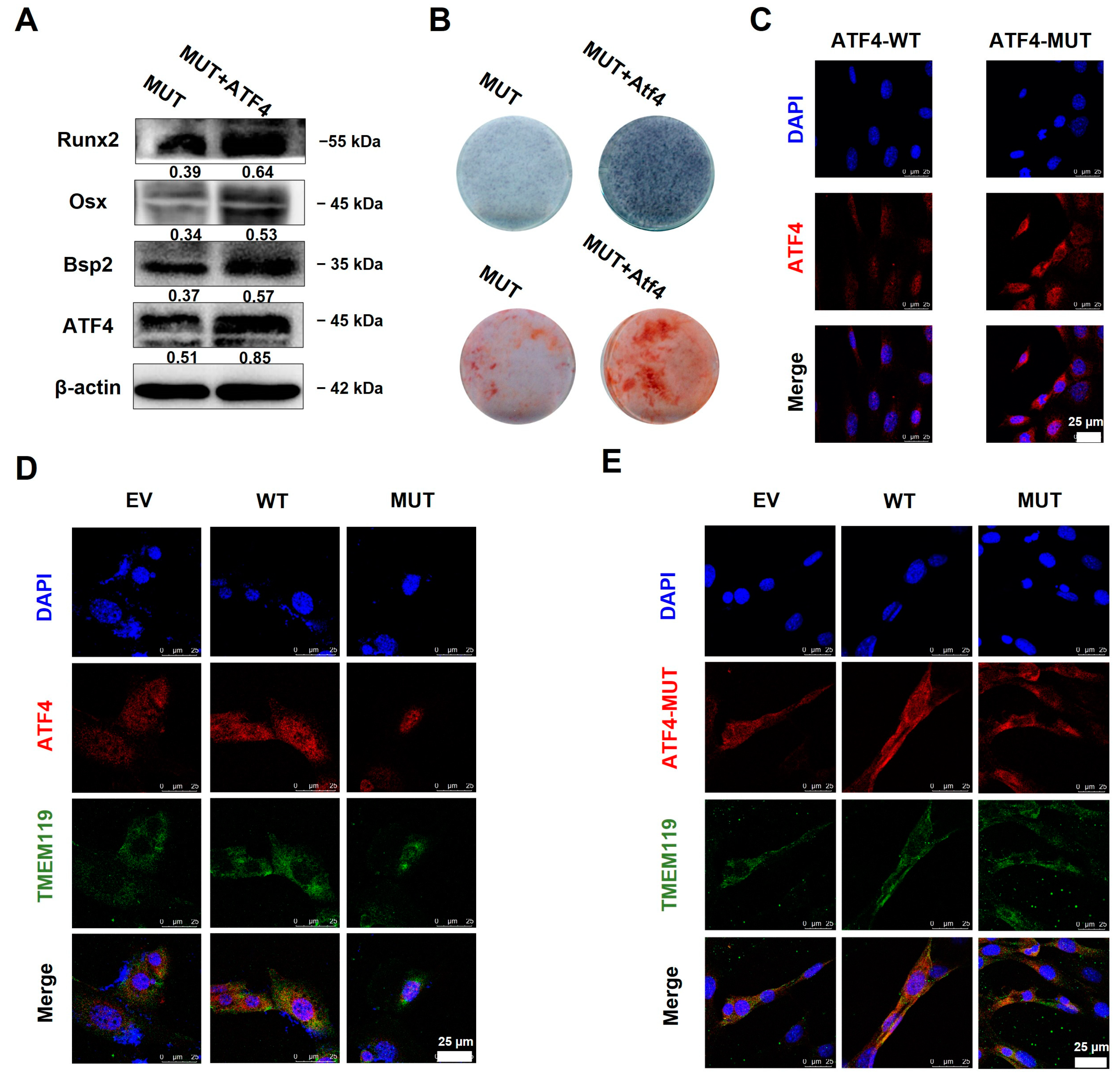
2.5. Overexpression of ATF4 Rescues TMEM119 Mutation-Caused Osteo-Differentiation Dysfunction and Nuclear Translocation Regulated by TMEM119-ATF4 Interaction in MC3T3-E1 Cells
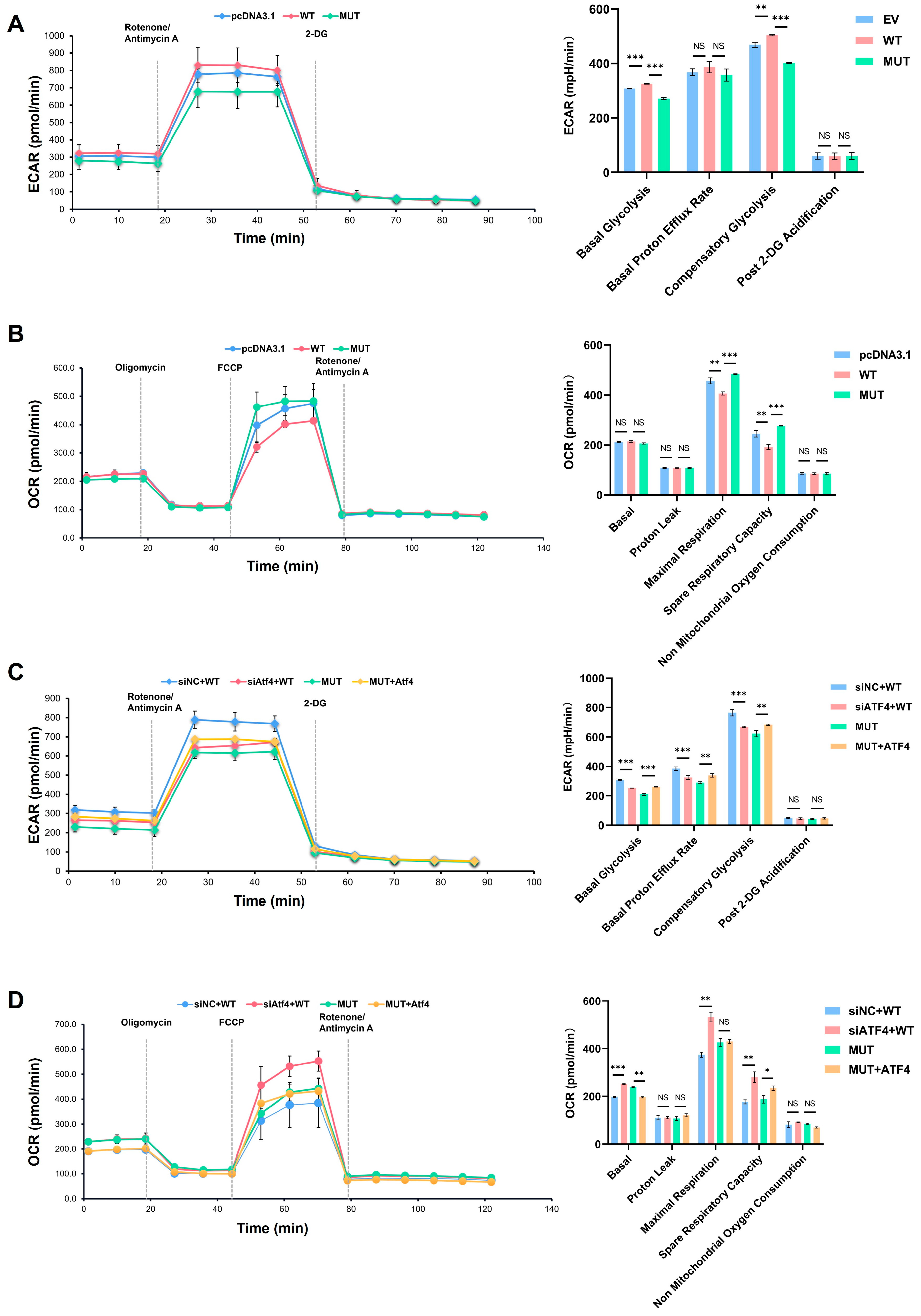
2.6. Decrease in Glycolysis Function Impacted by Reduced Expression of ATF4 Monitored by Seahorse Assay
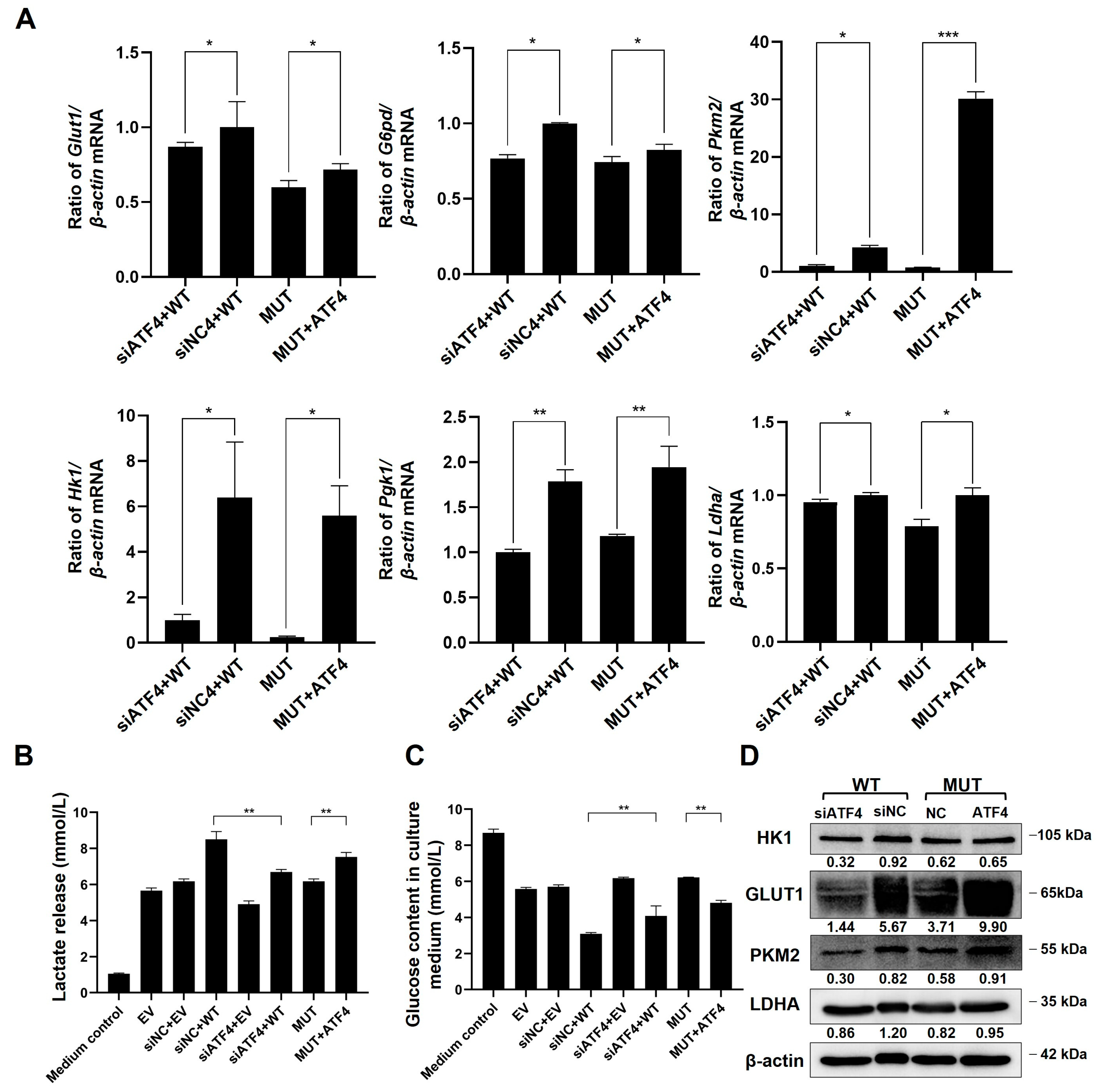
2.7. TMEM119 Mutation Inhibits Osteogenic Differentiation of MC3T3-E1 through ATF4-Mediated Glycolysis
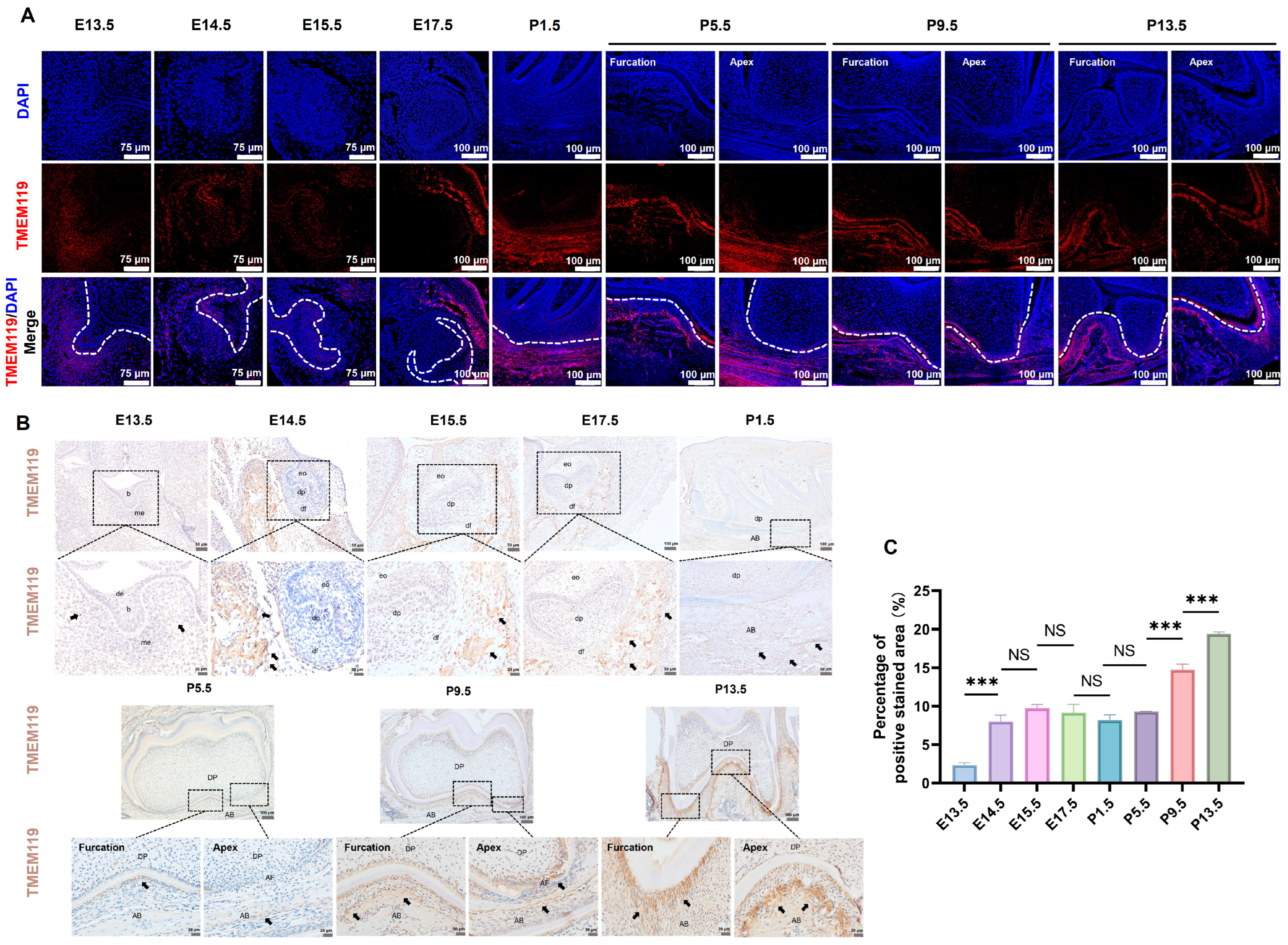
2.8. Expression of TMEM119 around Mouse Tooth Germ during Tooth Development
3. Discussion
4. Materials and Methods
4.1. Recruitment and Clinical Examination of Primary Failure of Eruption (PFE) Individuals
4.2. Whole-Exome Sequencing (WES) and Mutation Analysis
4.3. Conservation and Structural Analysis
4.4. Construction of Plasmids
4.5. Cell Culture and Liposome Transfection
4.6. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
4.7. Western Blot Analysis
4.8. In Vitro Mineralization Ability Detection
4.9. Cell Immunofluorescence
4.10. Co-Immunoprecipitation (Co-IP)
4.11. Cell Proliferation Assay
4.12. Wound-Healing Assay
4.13. Small Interfering RNA (siRNA) Construction and Transfection
4.14. Glycolytic Rate Assay and Mitochondrial Stress Test
4.15. Measurement of Glucose in Culture Medium
4.16. Measurement of Lactic Acid in Culture Medium
4.17. Mouse Treatment and Tissue Preparation
4.18. Tissue Immunohistochemistry and Immunofluorescence
4.19. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wise, G.E.; Frazier-Bowers, S.; D’Souza, R.N. Cellular, molecular, and genetic determinants of tooth eruption. Crit. Rev. Oral. Biol. Med. 2002, 13, 323–334. [Google Scholar] [CrossRef]
- Richman, J.M. Shedding new light on the mysteries of tooth eruption. Proc. Natl. Acad. Sci. USA 2019, 116, 353–355. [Google Scholar] [CrossRef]
- Kjaer, I. Mechanism of human tooth eruption: Review article including a new theory for future studies on the eruption process. Scientifica 2014, 2014, 341905. [Google Scholar] [CrossRef]
- Marks, S.J.; Cahill, D.R. Regional control by the dental follicle of alterations in alveolar bone metabolism during tooth eruption. J. Oral. Pathol. 1987, 16, 164–169. [Google Scholar]
- Mori, G.; Ballini, A.; Carbone, C.; Oranger, A.; Brunetti, G.; Di Benedetto, A.; Rapone, B.; Cantore, S.; Di Comite, M.; Colucci, S.; et al. Osteogenic differentiation of dental follicle stem cells. Int. J. Med. Sci. 2012, 9, 480–487. [Google Scholar] [CrossRef]
- Park, S.J.; Bae, H.S.; Cho, Y.S.; Lim, S.R.; Kang, S.A.; Park, J.C. Apoptosis of the reduced enamel epithelium and its implications for bone resorption during tooth eruption. J. Mol. Histol. 2013, 44, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Viale-Bouroncle, S.; Klingelhoffer, C.; Ettl, T.; Reichert, T.E.; Morsczeck, C. A protein kinase A (PKA)/beta-catenin pathway sustains the BMP2/DLX3-induced osteogenic differentiation in dental follicle cells (DFCs). Cell Signal 2015, 27, 598–605. [Google Scholar] [CrossRef] [PubMed]
- Wise, G.E.; Yao, S.; Henk, W.G. Bone formation as a potential motive force of tooth eruption in the rat molar. Clin. Anat. 2007, 20, 632–639. [Google Scholar] [CrossRef] [PubMed]
- Wise, G.E.; He, H.; Gutierrez, D.L.; Ring, S.; Yao, S. Requirement of alveolar bone formation for eruption of rat molars. Eur. J. Oral. Sci. 2011, 119, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Sharma, G.; Kneafsey, L.; Ashley, P.; Noar, J. Failure of eruption of permanent molars: A diagnostic dilemma. Int. J. Paediatr. Dent. 2016, 26, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Snider, T.N.; Wimer, H.F.; Yamada, S.; Yang, T.; Holmbeck, K.; Foster, B. Multiple essential MT1-MMP functions in tooth root formation, dentinogenesis, and tooth eruption. Matrix Biol. 2016, 52–54, 266–283. [Google Scholar] [CrossRef]
- Suri, L.; Gagari, E.; Vastardis, H. Delayed tooth eruption: Pathogenesis, diagnosis, and treatment. A literature review. Am. J. Orthod. Orthop. 2004, 126, 432–445. [Google Scholar] [CrossRef]
- Yamaguchi, T.; Hosomichi, K.; Shirota, T.; Miyamoto, Y.; Ono, W.; Ono, N. Primary failure of tooth eruption: Etiology and management. Jpn. Dent. Sci. Rev. 2022, 58, 258–267. [Google Scholar] [CrossRef]
- Hanisch, M.; Hanisch, L.; Kleinheinz, J.; Jung, S. Primary failure of eruption (PFE): A systematic review. Head. Face Med. 2018, 14, 5. [Google Scholar] [CrossRef]
- Proffit, W.R.; Vig, K.W. Primary failure of eruption: A possible cause of posterior open-bite. Am. J. Orthod. 1981, 80, 173–190. [Google Scholar] [CrossRef]
- Baccetti, T. Tooth anomalies associated with failure of eruption of first and second permanent molars. Am. J. Orthod. Orthop. 2000, 118, 608–610. [Google Scholar] [CrossRef] [PubMed]
- Frazier-Bowers, S.A.; Koehler, K.E.; Ackerman, J.L.; Proffit, W.R. Primary failure of eruption: Further characterization of a rare eruption disorder. Am. J. Orthod. Orthop. 2007, 131, 571–578. [Google Scholar] [CrossRef] [PubMed]
- Proffit, W.R.; Frazier-Bowers, S.A. Mechanism and control of tooth eruption: Overview and clinical implications. Orthod. Orthop. Res. 2009, 12, 59–66. [Google Scholar] [CrossRef]
- Dibiase, A.T.; Leggat, T.G. Primary failure of eruption in the permanent dentition of siblings. Int. J. Paediatr. Dent. 2000, 10, 153–157. [Google Scholar] [CrossRef]
- Ahmad, S.; Bister, D.; Cobourne, M.T. The clinical features and aetiological basis of primary eruption failure. Eur. J. Orthodont 2006, 28, 535–540. [Google Scholar] [CrossRef] [PubMed]
- Decker, E.; Stellzig-Eisenhauer, A.; Fiebig, B.S.; Rau, C.; Kress, W.; Saar, K.; Rüschendorf, F.; Hubner, N.; Grimm, T.; Weber, B.H. PTHR1 loss-of-function mutations in familial, nonsyndromic primary failure of tooth eruption. Am. J. Hum. Genet. 2008, 83, 781–786. [Google Scholar] [CrossRef]
- Frazier-Bowers, S.A.; Simmons, D.; Wright, J.T.; Proffit, W.R.; Ackerman, J.L. Primary failure of eruption and PTH1R: The importance of a genetic diagnosis for orthodontic treatment planning. Am. J. Orthod. Orthop. 2010, 137, 160–161. [Google Scholar] [CrossRef]
- Yamaguchi, T.; Hosomichi, K.; Narita, A.; Shirota, T.; Tomoyasu, Y.; Maki, K.; Inoue, I. Exome resequencing combined with linkage analysis identifies novel PTH1R variants in primary failure of tooth eruption in Japanese. J. Bone Miner. Res. 2011, 26, 1655–1661. [Google Scholar] [CrossRef] [PubMed]
- Stellzig-Eisenhauer, A.; Decker, E.; Meyer-Marcotty, P.; Rau, C.; Fiebig, B.S.; Kress, W.; Saar, K.; Rüschendorf, F.; Hubner, N.; Grimm, T.; et al. Primary failure of eruption (PFE). Clinical and molecular genetics analysis. Orthod. Fr. 2013, 84, 241–250. [Google Scholar] [CrossRef]
- Roth, H.; Fritsche, L.G.; Meier, C.; Pilz, P.; Eigenthaler, M.; Meyer-Marcotty, P.; Stellzig-Eisenhauer, A.; Proff, P.; Kanno, C.M.; Weber, B.H. Expanding the spectrum of PTH1R mutations in patients with primary failure of tooth eruption. Clin. Oral. Investig. 2014, 18, 377–384. [Google Scholar] [CrossRef] [PubMed]
- Jelani, M.; Kang, C.; Mohamoud, H.S.; Al-Rehaili, R.; Almramhi, M.M.; Serafi, R.; Yang, H.; Al-Aama, J.Y.; Naeem, M.; Alkhiary, Y.M. A novel homozygous PTH1R variant identified through whole-exome sequencing further expands the clinical spectrum of primary failure of tooth eruption in a consanguineous Saudi family. Arch. Oral. Biol. 2016, 67, 28–33. [Google Scholar] [CrossRef]
- Assiry, A.A.; Albalawi, A.M.; Zafar, M.S.; Khan, S.D.; Ullah, A.; Almatrafi, A.; Ramzan, K.; Basit, S. KMT2C, a histone methyltransferase, is mutated in a family segregating non-syndromic primary failure of tooth eruption. Sci. Rep.-UK 2019, 9, 16469. [Google Scholar] [CrossRef]
- Aziz, S.; Hermann, N.V.; Duno, M.; Risom, L.; Daugaard-Jensen, J.; Kreiborg, S. Primary failure of eruption of teeth in two siblings with a novel mutation in the PTH1R gene. Eur. Arch. Paediatr. 2019, 20, 295–300. [Google Scholar] [CrossRef]
- Risom, L.; Christoffersen, L.; Daugaard-Jensen, J.; Hove, H.D.; Andersen, H.S.; Andresen, B.S.; Kreiborg, S.; Duno, M. Identification of six novel PTH1R mutations in families with a history of primary failure of tooth eruption. PLoS ONE 2013, 8, e74601. [Google Scholar] [CrossRef]
- Wise, G.E.; Yao, S. Regional differences of expression of bone morphogenetic protein-2 and RANKL in the rat dental follicle. Eur. J. Oral. Sci. 2006, 114, 512–516. [Google Scholar] [CrossRef]
- Kanamoto, T.; Mizuhashi, K.; Terada, K.; Minami, T.; Yoshikawa, H.; Furukawa, T. Isolation and characterization of a novel plasma membrane protein, osteoblast induction factor (obif), associated with osteoblast differentiation. BMC Dev. Biol. 2009, 9, 70. [Google Scholar] [CrossRef]
- Kawao, N.; Matsumura, D.; Yamada, A.; Okumoto, K.; Ohira, T.; Mizukami, Y.; Hashimoto, D.; Kaji, H. Tmem119 is involved in bone anabolic effects of PTH through enhanced osteoblastic bone formation in mice. Bone 2024, 181, 117040. [Google Scholar] [CrossRef]
- Tanaka, K.; Kaji, H.; Yamaguchi, T.; Kanazawa, I.; Canaff, L.; Hendy, G.N.; Sugimoto, T. Involvement of the osteoinductive factors, Tmem119 and BMP-2, and the ER stress response PERK-eIF2alpha-ATF4 pathway in the commitment of myoblastic into osteoblastic cells. Calcif. Tissue Int. 2014, 94, 454–464. [Google Scholar] [CrossRef]
- Hisa, I.; Inoue, Y.; Hendy, G.N.; Canaff, L.; Kitazawa, R.; Kitazawa, S.; Komori, T.; Sugimoto, T.; Seino, S.; Kaji, H. Parathyroid hormone-responsive Smad3-related factor, Tmem119, promotes osteoblast differentiation and interacts with the bone morphogenetic protein-Runx2 pathway. J. Biol. Chem. 2011, 286, 9787–9796. [Google Scholar] [CrossRef]
- Davidson, M.; Mayer, M.; Habib, A.; Rashidi, N.; Filippone, R.T.; Fraser, S.; Prakash, M.D.; Sinnayah, P.; Tangalakis, K.; Mathai, M.L.; et al. Methamphetamine Induces Systemic Inflammation and Anxiety: The Role of the Gut-Immune-Brain Axis. Int. J. Mol. Sci. 2022, 23, 11224. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Bi, F.; Liu, Z.; Yang, Q. TMEM119 facilitates ovarian cancer cell proliferation, invasion, and migration via the PDGFRB/PI3K/AKT signaling pathway. J. Transl. Med. 2021, 19, 111. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.H.; Peng, J.; Yang, H.L.; Fu, X.-L.; Wang, J.-Z.; Liu, L.; Jiang, J.-N.; Tan, Y.-F.; Ge, Z.-J. Upregulation and biological function of transmembrane protein 119 in osteosarcoma. Exp. Mol. Med. 2017, 49, e329. [Google Scholar] [CrossRef] [PubMed]
- Zheng, P.; Wang, W.; Ji, M.; Zhu, Q.; Feng, Y.; Zhou, F.; He, Q. TMEM119 promotes gastric cancer cell migration and invasion through STAT3 signaling pathway. Oncotargets Ther. 2018, 11, 5835–5844. [Google Scholar] [CrossRef] [PubMed]
- Zheng, P.; Wang, W.; Ji, M.; Zhu, Q.; Feng, Y.; Zhou, F.; He, Q. TMEM119 silencing inhibits cell viability and causes the apoptosis of gastric cancer SGC-7901 cells. Oncol. Lett. 2018, 15, 8281–8286. [Google Scholar] [CrossRef] [PubMed]
- Mizuhashi, K.; Chaya, T.; Kanamoto, T.; Omori, Y.; Furukawa, T. Obif, a Transmembrane Protein, Is Required for Bone Mineralization and Spermatogenesis in Mice. PLoS ONE 2015, 10, e133704. [Google Scholar] [CrossRef] [PubMed]
- Mizuhashi, K.; Kanamoto, T.; Ito, M.; Moriishi, T.; Muranishi, Y.; Omori, Y.; Terada, K.; Komori, T.; Furukawa, T. OBIF, an osteoblast induction factor, plays an essential role in bone formation in association with osteoblastogenesis. Dev. Growth Differ. 2012, 54, 474–480. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.C.; Guntur, A.R.; Long, F.; Rosen, C.J. Energy Metabolism of the Osteoblast: Implications for Osteoporosis. Endocr. Rev. 2017, 38, 255–266. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.C.; Ji, X.; Nissim, I.; Long, F. Malic Enzyme Couples Mitochondria with Aerobic Glycolysis in Osteoblasts. Cell Rep. 2020, 32, 108108. [Google Scholar] [CrossRef]
- Esen, E.; Long, F. Aerobic glycolysis in osteoblasts. Curr. Osteoporos. Rep. 2014, 12, 433–438. [Google Scholar] [CrossRef]
- Gao, Z.; Dlamini, M.B.; Ge, H.; Jiang, L.; Geng, C.; Li, Q.; Shi, X.; Liu, Y.; Cao, J. ATF4-mediated autophagy-dependent glycolysis plays an important role in attenuating apoptosis induced by Cr (VI) in A549 cells. Toxicol. Lett. 2020, 331, 178–187. [Google Scholar] [CrossRef]
- Yang, X.; Matsuda, K.; Bialek, P.; Jacquot, S.; Masuoka, H.C.; Schinke, T.; Li, L.; Brancorsini, S.; Sassone-Corsi, P.; Townes, T.M.; et al. ATF4 is a substrate of RSK2 and an essential regulator of osteoblast biology; implication for Coffin-Lowry Syndrome. Cell 2004, 117, 387–398. [Google Scholar] [CrossRef] [PubMed]
- Xiao, G.; Jiang, D.; Ge, C.; Zhao, Z.; Lai, Y.; Boules, H.; Phimphilai, M.; Yang, X.; Karsenty, G.; Franceschi, R.T. Cooperative interactions between activating transcription factor 4 and Runx2/Cbfa1 stimulate osteoblast-specific osteocalcin gene expression. J. Biol. Chem. 2005, 280, 30689–30696. [Google Scholar] [CrossRef] [PubMed]

| Patient | Sex (F/M) | Clinical Feature of PFE | Mutation | |||
|---|---|---|---|---|---|---|
| Nucleotide | Codon | Type | Location | |||
| I-2,II-2 | F | Doubtful | Unknown | Unknown | Unknown | Unknown |
| II-3 | M | Doubtful | Unknown | Unknown | Unknown | Unknown |
| III-1 | F | Yes | 143G > A | S48L | Missense | Extra-membrane |
| IV-1 | F | Yes | 143G > A | S48L | Missense | Extra-membrane |
| Control 1 | F | No | Wild-type | Normal | Normal | Extra-membrane |
| Control 2 | M | No | Wild-type | Normal | Normal | Extra-membrane |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, M.; Wang, D.; Li, K.; Ma, T.; Wang, Y.; Xia, B. TMEM119 (c.G143A, p.S48L) Mutation Is Involved in Primary Failure of Eruption by Attenuating Glycolysis-Mediated Osteogenesis. Int. J. Mol. Sci. 2024, 25, 2821. https://doi.org/10.3390/ijms25052821
Xu M, Wang D, Li K, Ma T, Wang Y, Xia B. TMEM119 (c.G143A, p.S48L) Mutation Is Involved in Primary Failure of Eruption by Attenuating Glycolysis-Mediated Osteogenesis. International Journal of Molecular Sciences. 2024; 25(5):2821. https://doi.org/10.3390/ijms25052821
Chicago/Turabian StyleXu, Mindi, Dandan Wang, Kefan Li, Tianyu Ma, Yixiang Wang, and Bin Xia. 2024. "TMEM119 (c.G143A, p.S48L) Mutation Is Involved in Primary Failure of Eruption by Attenuating Glycolysis-Mediated Osteogenesis" International Journal of Molecular Sciences 25, no. 5: 2821. https://doi.org/10.3390/ijms25052821





