Retinal Ciliopathies and Potential Gene Therapies: A Focus on Human iPSC-Derived Organoid Models
Abstract
1. Introduction
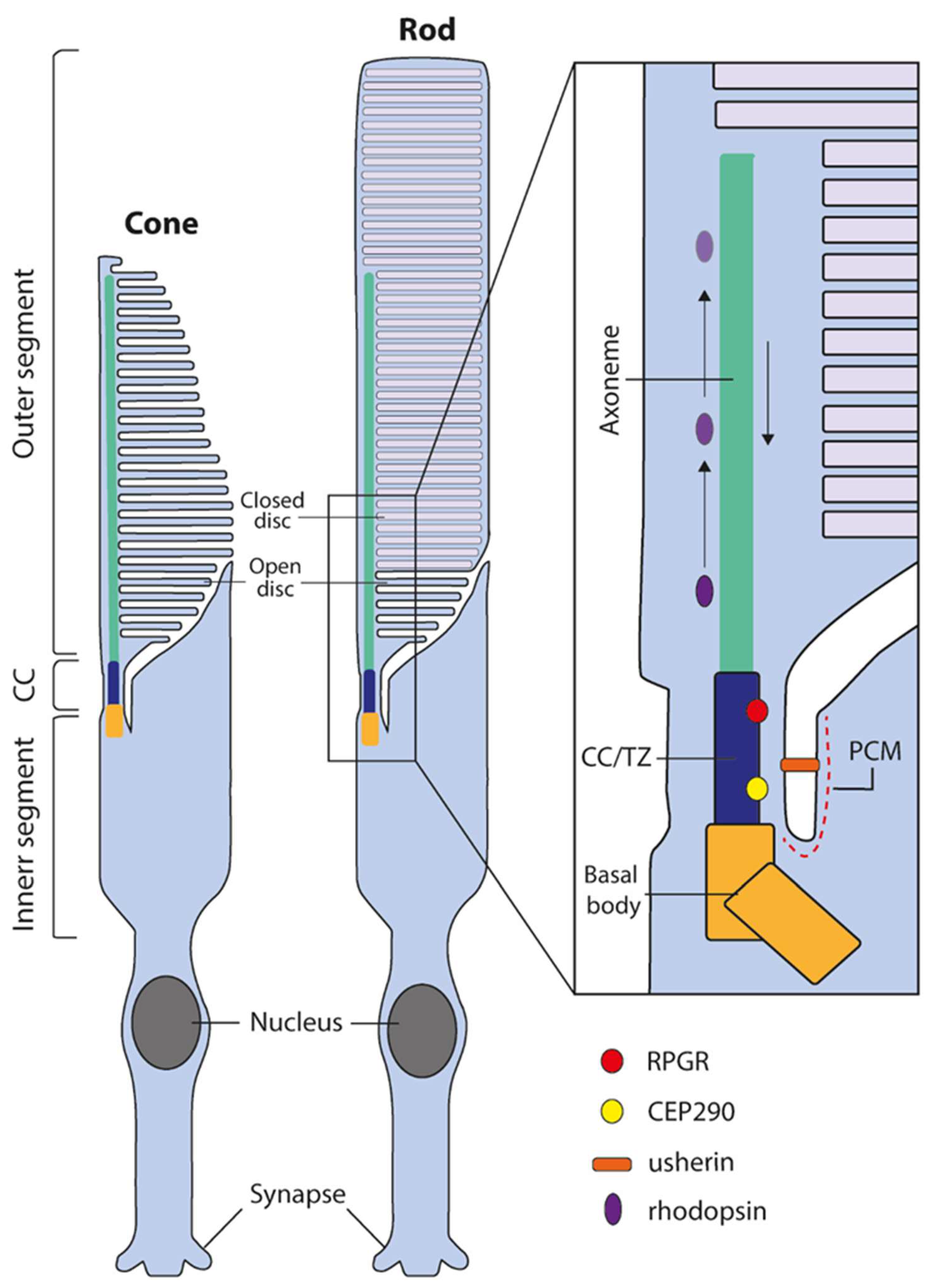
2. iPSC-Derived Retinal Organoids as Models for Human Retinal Ciliopathies
3. Retinal Organoid Ciliopathy Models
3.1. RPGR
3.2. CEP290
3.3. Usher Syndrome
3.3.1. MYO7A
3.3.2. USH2A
4. Retinal Gene Therapy
4.1. AAV-Mediated Gene Supplementation
4.2. Retinal Organoids as Platforms for Developing Virally Delivered Gene Therapies
4.3. Targeting of Gene Therapies to the Photoreceptor
4.4. AAV-Mediated Rescue of Cilial Genes in Retinal Organoid Models
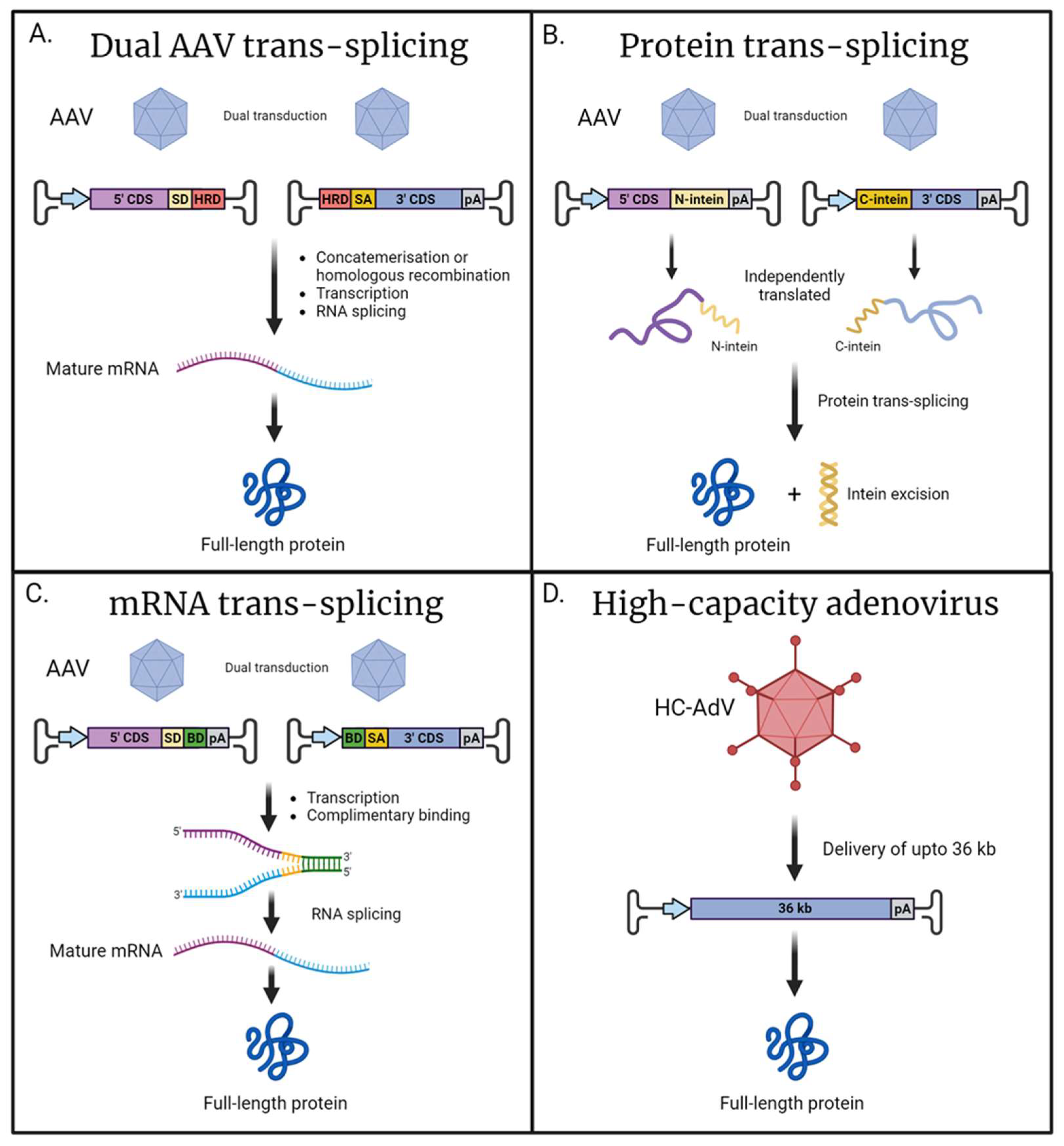
5. Alternative Approaches for the Delivery of Large Cilial Genes
6. Gene-Editing Approaches in the Retina
6.1. Prime Editing
6.2. Homology-Independent Targeted Integration
7. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wensel, T.G.; Potter, V.L.; Moye, A.; Zhang, Z.; Robichaux, M.A. Structure and dynamics of photoreceptor sensory cilia. Pflug. Arch. 2021, 473, 1517–1537. [Google Scholar] [CrossRef]
- Goldberg, A.F.; Moritz, O.L.; Williams, D.S. Molecular basis for photoreceptor outer segment architecture. Prog. Retin. Eye Res. 2016, 55, 52–81. [Google Scholar] [CrossRef]
- Bujakowska, K.M.; Liu, Q.; Pierce, E.A. Photoreceptor Cilia and Retinal Ciliopathies. Cold Spring Harb. Perspect. Biol. 2017, 9, a028274. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.Y.; Kelley, R.A.; Li, T.; Swaroop, A. Primary cilia biogenesis and associated retinal ciliopathies. Semin. Cell Dev. Biol. 2021, 110, 70–88. [Google Scholar] [CrossRef]
- Bachmann-Gagescu, R.; Neuhauss, S.C. The photoreceptor cilium and its diseases. Curr. Opin. Genet. Dev. 2019, 56, 22–33. [Google Scholar] [CrossRef]
- Adams, N.A.; Awadein, A.; Toma, H.S. The retinal ciliopathies. Ophthalmic Genet. 2007, 28, 113–125. [Google Scholar] [CrossRef] [PubMed]
- Waters, A.M.; Beales, P.L. Ciliopathies: An expanding disease spectrum. Pediatr. Nephrol. 2011, 26, 1039–1056. [Google Scholar] [CrossRef] [PubMed]
- Slijkerman, R.W.; Song, F.; Astuti, G.D.; Huynen, M.A.; van Wijk, E.; Stieger, K.; Collin, R.W. The pros and cons of vertebrate animal models for functional and therapeutic research on inherited retinal dystrophies. Prog. Retin. Eye Res. 2015, 48, 137–159. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.; Gutierrez, C.; Xue, T.; Hampton, C.; Vergara, M.N.; Cao, L.H.; Peters, A.; Park, T.S.; Zambidis, E.T.; Meyer, J.S.; et al. Generation of three-dimensional retinal tissue with functional photoreceptors from human iPSCs. Nat. Commun. 2014, 5, 4047. [Google Scholar] [CrossRef] [PubMed]
- Wahlin, K.J.; Maruotti, J.A.; Sripathi, S.R.; Ball, J.; Angueyra, J.M.; Kim, C.; Grebe, R.; Li, W.; Jones, B.W.; Zack, D.J. Photoreceptor Outer Segment-like Structures in Long-Term 3D Retinas from Human Pluripotent Stem Cells. Sci. Rep. 2017, 7, 766. [Google Scholar] [CrossRef] [PubMed]
- Dorgau, B.; Georgiou, M.; Chaudhary, A.; Moya-Molina, M.; Collin, J.; Queen, R.; Hilgen, G.; Davey, T.; Hewitt, P.; Schmitt, M.; et al. Human Retinal Organoids Provide a Suitable Tool for Toxicological Investigations: A Comprehensive Validation Using Drugs and Compounds Affecting the Retina. Stem Cells Transl. Med. 2022, 11, 159–177. [Google Scholar] [CrossRef]
- Kelley, R.A.; Wu, Z. Utilization of the retinal organoid model to evaluate the feasibility of genetic strategies to ameliorate retinal disease(s). Vision. Res. 2023, 210, 108269. [Google Scholar] [CrossRef]
- Eiraku, M.; Takata, N.; Ishibashi, H.; Kawada, M.; Sakakura, E.; Okuda, S.; Sekiguchi, K.; Adachi, T.; Sasai, Y. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature 2011, 472, 51–56. [Google Scholar] [CrossRef]
- Kim, S.; Lowe, A.; Dharmat, R.; Lee, S.; Owen, L.A.; Wang, J.; Shakoor, A.; Li, Y.; Morgan, D.J.; Hejazi, A.A.; et al. Generation, transcriptome profiling, and functional validation of cone-rich human retinal organoids. Proc. Natl. Acad. Sci. USA 2019, 116, 10824–10833. [Google Scholar] [CrossRef] [PubMed]
- Kaya, K.D.; Chen, H.Y.; Brooks, M.J.; Kelley, R.A.; Shimada, H.; Nagashima, K.; de Val, N.; Drinnan, C.T.; Gieser, L.; Kruczek, K.; et al. Transcriptome-based molecular staging of human stem cell-derived retinal organoids uncovers accelerated photoreceptor differentiation by 9-cis retinal. Mol. Vis. 2019, 25, 663–678. [Google Scholar]
- Kallman, A.; Capowski, E.E.; Wang, J.; Kaushik, A.M.; Jansen, A.D.; Edwards, K.L.; Chen, L.; Berlinicke, C.A.; Joseph Phillips, M.; Pierce, E.A.; et al. Investigating cone photoreceptor development using patient-derived NRL null retinal organoids. Commun. Biol. 2020, 3, 82. [Google Scholar] [CrossRef]
- Sridhar, A.; Hoshino, A.; Finkbeiner, C.R.; Chitsazan, A.; Dai, L.; Haugan, A.K.; Eschenbacher, K.M.; Jackson, D.L.; Trapnell, C.; Bermingham-McDonogh, O.; et al. Single-Cell Transcriptomic Comparison of Human Fetal Retina, hPSC-Derived Retinal Organoids, and Long-Term Retinal Cultures. Cell Rep. 2020, 30, 1644–1659. [Google Scholar] [CrossRef]
- Parfitt, D.A.; Lane, A.; Ramsden, C.M.; Carr, A.F.; Munro, P.M.; Jovanovic, K.; Schwarz, N.; Kanuga, N.; Muthiah, M.N.; Hull, S.; et al. Identification and Correction of Mechanisms Underlying Inherited Blindness in Human iPSC-Derived Optic Cups. Cell Stem Cell 2016, 18, 769–781. [Google Scholar] [CrossRef]
- Lowe, A.; Harris, R.; Bhansali, P.; Cvekl, A.; Liu, W. Intercellular Adhesion-Dependent Cell Survival and ROCK-Regulated Actomyosin-Driven Forces Mediate Self-Formation of a Retinal Organoid. Stem Cell Rep. 2016, 6, 743–756. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Cordero, A.; Kruczek, K.; Naeem, A.; Fernando, M.; Kloc, M.; Ribeiro, J.; Goh, D.; Duran, Y.; Blackford, S.J.I.; Abelleira-Hervas, L.; et al. Recapitulation of Human Retinal Development from Human Pluripotent Stem Cells Generates Transplantable Populations of Cone Photoreceptors. Stem Cell Rep. 2017, 9, 820–837. [Google Scholar] [CrossRef]
- Decembrini, S.; Hoehnel, S.; Brandenberg, N.; Arsenijevic, Y.; Lutolf, M.P. Hydrogel-based milliwell arrays for standardized and scalable retinal organoid cultures. Sci. Rep. 2020, 10, 10275. [Google Scholar] [CrossRef] [PubMed]
- West, E.L.; Majumder, P.; Naeem, A.; Fernando, M.; O’Hara-Wright, M.; Lanning, E.; Kloc, M.; Ribeiro, J.; Ovando-Roche, P.; Shum, I.O.; et al. Antioxidant and lipid supplementation improve the development of photoreceptor outer segments in pluripotent stem cell-derived retinal organoids. Stem Cell Rep. 2022, 17, 775–788. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.L.; Gao, M.L.; Lei, X.L.; Lv, J.N.; Zhao, H.; He, K.W.; Xia, X.X.; Li, L.Y.; Chen, Y.C.; Li, Y.P.; et al. Gene Correction Reverses Ciliopathy and Photoreceptor Loss in iPSC-Derived Retinal Organoids from Retinitis Pigmentosa Patients. Stem Cell Rep. 2018, 10, 2005. [Google Scholar] [CrossRef]
- Kruczek, K.; Qu, Z.; Welby, E.; Shimada, H.; Hiriyanna, S.; English, M.A.; Zein, W.M.; Brooks, B.P.; Swaroop, A. In vitro modeling and rescue of ciliopathy associated with IQCB1/NPHP5 mutations using patient-derived cells. Stem Cell Rep. 2022, 17, 2172–2186. [Google Scholar] [CrossRef] [PubMed]
- Chahine Karam, F.; Loi, T.H.; Ma, A.; Nash, B.M.; Grigg, J.R.; Parekh, D.; Riley, L.G.; Farnsworth, E.; Bennetts, B.; Gonzalez-Cordero, A.; et al. Human iPSC-Derived Retinal Organoids and Retinal Pigment Epithelium for Novel Intronic RPGR Variant Assessment for Therapy Suitability. J. Pers. Med. 2022, 12, 502. [Google Scholar] [CrossRef] [PubMed]
- Hallam, D.; Hilgen, G.; Dorgau, B.; Zhu, L.; Yu, M.; Bojic, S.; Hewitt, P.; Schmitt, M.; Uteng, M.; Kustermann, S.; et al. Human-Induced Pluripotent Stem Cells Generate Light Responsive Retinal Organoids with Variable and Nutrient-Dependent Efficiency. Stem Cells 2018, 36, 1535–1551. [Google Scholar] [CrossRef] [PubMed]
- Saha, A.; Capowski, E.; Fernandez Zepeda, M.A.; Nelson, E.C.; Gamm, D.M.; Sinha, R. Cone photoreceptors in human stem cell-derived retinal organoids demonstrate intrinsic light responses that mimic those of primate fovea. Cell Stem Cell 2022, 29, 460–471.e3. [Google Scholar] [CrossRef]
- Wagstaff, E.L.; Heredero Berzal, A.; Boon, C.J.F.; Quinn, P.M.J.; Ten Asbroek, A.; Bergen, A.A. The Role of Small Molecules and Their Effect on the Molecular Mechanisms of Early Retinal Organoid Development. Int. J. Mol. Sci. 2021, 22, 7081. [Google Scholar] [CrossRef]
- Walia, V.; Cuenca, A.; Vetter, M.; Insinna, C.; Perera, S.; Lu, Q.; Ritt, D.A.; Semler, E.; Specht, S.; Stauffer, J.; et al. Akt Regulates a Rab11-Effector Switch Required for Ciliogenesis. Dev. Cell 2019, 50, 229–246.e7. [Google Scholar] [CrossRef] [PubMed]
- Kevany, B.M.; Palczewski, K. Phagocytosis of retinal rod and cone photoreceptors. Physiology 2010, 25, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Achberger, K.; Probst, C.; Haderspeck, J.; Bolz, S.; Rogal, J.; Chuchuy, J.; Nikolova, M.; Cora, V.; Antkowiak, L.; Haq, W.; et al. Merging organoid and organ-on-a-chip technology to generate complex multi-layer tissue models in a human retina-on-a-chip platform. Elife 2019, 8, 46188. [Google Scholar] [CrossRef] [PubMed]
- Chichagova, V.; Hilgen, G.; Ghareeb, A.; Georgiou, M.; Carter, M.; Sernagor, E.; Lako, M.; Armstrong, L. Human iPSC differentiation to retinal organoids in response to IGF1 and BMP4 activation is line- and method-dependent. Stem Cells 2020, 38, 195–201. [Google Scholar] [CrossRef]
- Vaz, I.M.; Borgonovo, T.; Kasai-Brunswick, T.H.; Santos, D.S.D.; Mesquita, F.C.P.; Vasques, J.F.; Gubert, F.; Rebelatto, C.L.K.; Senegaglia, A.C.; Brofman, P.R.S. Chromosomal aberrations after induced pluripotent stem cells reprogramming. Genet. Mol. Biol. 2021, 44, e20200147. [Google Scholar] [CrossRef] [PubMed]
- Popp, B.; Krumbiegel, M.; Grosch, J.; Sommer, A.; Uebe, S.; Kohl, Z.; Plotz, S.; Farrell, M.; Trautmann, U.; Kraus, C.; et al. Need for high-resolution Genetic Analysis in iPSC: Results and Lessons from the ForIPS Consortium. Sci. Rep. 2018, 8, 17201. [Google Scholar] [CrossRef]
- Poetsch, M.S.; Strano, A.; Guan, K. Human Induced Pluripotent Stem Cells: From Cell Origin, Genomic Stability, and Epigenetic Memory to Translational Medicine. Stem Cells 2022, 40, 546–555. [Google Scholar] [CrossRef] [PubMed]
- Afanasyeva, T.A.V.; Corral-Serrano, J.C.; Garanto, A.; Roepman, R.; Cheetham, M.E.; Collin, R.W.J. A look into retinal organoids: Methods, analytical techniques, and applications. Cell. Mol. Life Sci. 2021, 78, 6505–6532. [Google Scholar] [CrossRef]
- Kruczek, K.; Swaroop, A. Pluripotent stem cell-derived retinal organoids for disease modeling and development of therapies. Stem Cells 2020, 38, 1206–1215. [Google Scholar] [CrossRef]
- Boon, N.; Lu, X.; Andriessen, C.A.; Moustakas, I.; Buck, T.M.; Freund, C.; Arendzen, C.H.; Bohringer, S.; Boon, C.J.F.; Mei, H.; et al. AAV-mediated gene augmentation therapy of CRB1 patient-derived retinal organoids restores the histological and transcriptional retinal phenotype. Stem Cell Rep. 2023, 18, 1388. [Google Scholar] [CrossRef]
- Boon, N.; Lu, X.; Andriessen, C.A.; Orlova, M.; Quinn, P.M.J.; Boon, C.J.F.; Wijnholds, J. Characterization and AAV-mediated CRB gene augmentation in human-derived CRB1(KO) and CRB1(KO)CRB2(+/−) retinal organoids. Mol. Ther. Methods Clin. Dev. 2023, 31, 101128. [Google Scholar] [CrossRef]
- Onyak, J.R.; Vergara, M.N.; Renna, J.M. Retinal organoid light responsivity: Current status and future opportunities. Transl. Res. 2022, 250, 98–111. [Google Scholar] [CrossRef] [PubMed]
- Quinn, P.M.; Buck, T.M.; Mulder, A.A.; Ohonin, C.; Alves, C.H.; Vos, R.M.; Bialecka, M.; van Herwaarden, T.; van Dijk, E.H.C.; Talib, M.; et al. Human iPSC-Derived Retinas Recapitulate the Fetal CRB1 CRB2 Complex Formation and Demonstrate that Photoreceptors and Muller Glia Are Targets of AAV5. Stem Cell Rep. 2019, 12, 906–919. [Google Scholar] [CrossRef]
- Gao, M.L.; Lei, X.L.; Han, F.; He, K.W.; Jin, S.Q.; Zhang, Y.Y.; Jin, Z.B. Patient-Specific Retinal Organoids Recapitulate Disease Features of Late-Onset Retinitis Pigmentosa. Front. Cell Dev. Biol. 2020, 8, 128. [Google Scholar] [CrossRef] [PubMed]
- Lane, A.; Jovanovic, K.; Shortall, C.; Ottaviani, D.; Panes, A.B.; Schwarz, N.; Guarascio, R.; Hayes, M.J.; Palfi, A.; Chadderton, N.; et al. Modeling and Rescue of RP2 Retinitis Pigmentosa Using iPSC-Derived Retinal Organoids. Stem Cell Rep. 2020, 15, 67–79. [Google Scholar] [CrossRef] [PubMed]
- Kruczek, K.; Qu, Z.; Gentry, J.; Fadl, B.R.; Gieser, L.; Hiriyanna, S.; Batz, Z.; Samant, M.; Samanta, A.; Chu, C.J.; et al. Gene Therapy of Dominant CRX-Leber Congenital Amaurosis using Patient Stem Cell-Derived Retinal Organoids. Stem Cell Rep. 2021, 16, 252–263. [Google Scholar] [CrossRef]
- Megaw, R.; Abu-Arafeh, H.; Jungnickel, M.; Mellough, C.; Gurniak, C.; Witke, W.; Zhang, W.; Khanna, H.; Mill, P.; Dhillon, B.; et al. Gelsolin dysfunction causes photoreceptor loss in induced pluripotent cell and animal retinitis pigmentosa models. Nat. Commun. 2017, 8, 271. [Google Scholar] [CrossRef]
- Buskin, A.; Zhu, L.; Chichagova, V.; Basu, B.; Mozaffari-Jovin, S.; Dolan, D.; Droop, A.; Collin, J.; Bronstein, R.; Mehrotra, S.; et al. Disrupted alternative splicing for genes implicated in splicing and ciliogenesis causes PRPF31 retinitis pigmentosa. Nat. Commun. 2018, 9, 4234. [Google Scholar] [CrossRef]
- Shimada, H.; Lu, Q.; Insinna-Kettenhofen, C.; Nagashima, K.; English, M.A.; Semler, E.M.; Mahgerefteh, J.; Cideciyan, A.V.; Li, T.; Brooks, B.P.; et al. In Vitro Modeling Using Ciliopathy-Patient-Derived Cells Reveals Distinct Cilia Dysfunctions Caused by CEP290 Mutations. Cell Rep. 2017, 20, 384–396. [Google Scholar] [CrossRef]
- Chen, H.Y.; Swaroop, M.; Papal, S.; Mondal, A.K.; Song, H.B.; Campello, L.; Tawa, G.J.; Regent, F.; Shimada, H.; Nagashima, K.; et al. Reserpine maintains photoreceptor survival in retinal ciliopathy by resolving proteostasis imbalance and ciliogenesis defects. Elife 2023, 12, e83205. [Google Scholar] [CrossRef] [PubMed]
- Dulla, K.; Aguila, M.; Lane, A.; Jovanovic, K.; Parfitt, D.A.; Schulkens, I.; Chan, H.L.; Schmidt, I.; Beumer, W.; Vorthoren, L.; et al. Splice-Modulating Oligonucleotide QR-110 Restores CEP290 mRNA and Function in Human c.2991+1655A>G LCA10 Models. Mol. Ther. Nucleic Acids 2018, 12, 730–740. [Google Scholar] [CrossRef] [PubMed]
- Corral-Serrano, J.C.; Sladen, P.E.; Ottaviani, D.; Rezek, O.F.; Athanasiou, D.; Jovanovic, K.; van der Spuy, J.; Mansfield, B.C.; Cheetham, M.E. Eupatilin Improves Cilia Defects in Human CEP290 Ciliopathy Models. Cells 2023, 12, 1575. [Google Scholar] [CrossRef] [PubMed]
- Afanasyeva, T.A.V.; Athanasiou, D.; Perdigao, P.R.L.; Whiting, K.R.; Duijkers, L.; Astuti, G.D.N.; Bennett, J.; Garanto, A.; van der Spuy, J.; Roepman, R.; et al. CRISPR-Cas9 correction of a nonsense mutation in LCA5 rescues lebercilin expression and localization in human retinal organoids. Mol. Ther. Methods Clin. Dev. 2023, 29, 522–531. [Google Scholar] [CrossRef]
- Guo, Y.; Wang, P.; Ma, J.H.; Cui, Z.; Yu, Q.; Liu, S.; Xue, Y.; Zhu, D.; Cao, J.; Li, Z.; et al. Modeling Retinitis Pigmentosa: Retinal Organoids Generated From the iPSCs of a Patient With the USH2A Mutation Show Early Developmental Abnormalities. Front. Cell Neurosci. 2019, 13, 361. [Google Scholar] [CrossRef]
- Su, T.; Liang, L.; Zhang, L.; Wang, J.; Chen, L.; Su, C.; Cao, J.; Yu, Q.; Deng, S.; Chan, H.F.; et al. Retinal organoids and microfluidic chip-based approaches to explore the retinitis pigmentosa with USH2A mutations. Front. Bioeng. Biotechnol. 2022, 10, 939774. [Google Scholar] [CrossRef]
- Sanjurjo-Soriano, C.; Jimenez-Medina, C.; Erkilic, N.; Cappellino, L.; Lefevre, A.; Nagel-Wolfrum, K.; Wolfrum, U.; Van Wijk, E.; Roux, A.F.; Meunier, I.; et al. USH2A variants causing retinitis pigmentosa or Usher syndrome provoke differential retinal phenotypes in disease-specific organoids. HGG Adv. 2023, 4, 100229. [Google Scholar] [CrossRef] [PubMed]
- Leong, Y.C.; Di Foggia, V.; Pramod, H.; Bitner-Glindzicz, M.; Patel, A.; Sowden, J.C. Molecular pathology of Usher 1B patient-derived retinal organoids at single cell resolution. Stem Cell Rep. 2022, 17, 2421–2437. [Google Scholar] [CrossRef] [PubMed]
- Megaw, R.D.; Soares, D.C.; Wright, A.F. RPGR: Its role in photoreceptor physiology, human disease, and future therapies. Exp. Eye Res. 2015, 138, 32–41. [Google Scholar] [CrossRef] [PubMed]
- Khanna, H.; Hurd, T.W.; Lillo, C.; Shu, X.; Parapuram, S.K.; He, S.; Akimoto, M.; Wright, A.F.; Margolis, B.; Williams, D.S.; et al. RPGR-ORF15, which is mutated in retinitis pigmentosa, associates with SMC1, SMC3, and microtubule transport proteins. J. Biol. Chem. 2005, 280, 33580–33587. [Google Scholar] [CrossRef] [PubMed]
- Murga-Zamalloa, C.A.; Atkins, S.J.; Peranen, J.; Swaroop, A.; Khanna, H. Interaction of retinitis pigmentosa GTPase regulator (RPGR) with RAB8A GTPase: Implications for cilia dysfunction and photoreceptor degeneration. Hum. Mol. Genet. 2010, 19, 3591–3598. [Google Scholar] [CrossRef] [PubMed]
- Gakovic, M.; Shu, X.; Kasioulis, I.; Carpanini, S.; Moraga, I.; Wright, A.F. The role of RPGR in cilia formation and actin stability. Hum. Mol. Genet. 2011, 20, 4840–4850. [Google Scholar] [CrossRef]
- Megaw, R.; Moye, A.; Zhang, X.; Newton, F.; McPhie, F.; Murphy, L.C.; McKie, L.; He, F.; Jungnickel, M.K.; von Kriegsheim, A.; et al. Ciliary tip actin dynamics regulate the cadence of photoreceptor disc formation. bioRxiv 2022. [CrossRef]
- Vervoort, R.; Lennon, A.; Bird, A.C.; Tulloch, B.; Axton, R.; Miano, M.G.; Meindl, A.; Meitinger, T.; Ciccodicola, A.; Wright, A.F. Mutational hot spot within a new RPGR exon in X-linked retinitis pigmentosa. Nat. Genet. 2000, 25, 462–466. [Google Scholar] [CrossRef] [PubMed]
- Pusch, C.M.; Broghammer, M.; Jurklies, B.; Besch, D.; Jacobi, F.K. Ten novel ORF15 mutations confirm mutational hot spot in the RPGR gene in European patients with X-linked retinitis pigmentosa. Hum. Mutat. 2002, 20, 405. [Google Scholar] [CrossRef]
- Breuer, D.K.; Yashar, B.M.; Filippova, E.; Hiriyanna, S.; Lyons, R.H.; Mears, A.J.; Asaye, B.; Acar, C.; Vervoort, R.; Wright, A.F.; et al. A comprehensive mutation analysis of RP2 and RPGR in a North American cohort of families with X-linked retinitis pigmentosa. Am. J. Hum. Genet. 2002, 70, 1545–1554. [Google Scholar] [CrossRef]
- Cehajic-Kapetanovic, J.; Martinez-Fernandez de la Camara, C.; Birtel, J.; Rehman, S.; McClements, M.E.; Charbel Issa, P.; Lotery, A.J.; MacLaren, R.E. Impaired glutamylation of RPGR(ORF15) underlies the cone-dominated phenotype associated with truncating distal ORF15 variants. Proc. Natl. Acad. Sci. USA 2022, 119, e2208707119. [Google Scholar] [CrossRef]
- Sun, X.; Park, J.H.; Gumerson, J.; Wu, Z.; Swaroop, A.; Qian, H.; Roll-Mecak, A.; Li, T. Loss of RPGR glutamylation underlies the pathogenic mechanism of retinal dystrophy caused by TTLL5 mutations. Proc. Natl. Acad. Sci. USA 2016, 113, E2925–E2934. [Google Scholar] [CrossRef]
- Thompson, D.A.; Khan, N.W.; Othman, M.I.; Chang, B.; Jia, L.; Grahek, G.; Wu, Z.; Hiriyanna, S.; Nellissery, J.; Li, T.; et al. Rd9 is a naturally occurring mouse model of a common form of retinitis pigmentosa caused by mutations in RPGR-ORF15. PLoS ONE 2012, 7, e35865. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Shahani, U.; Reilly, J.; Shu, X. Disease mechanisms and neuroprotection by tauroursodeoxycholic acid in Rpgr knockout mice. J. Cell Physiol. 2019, 234, 18801–18812. [Google Scholar] [CrossRef] [PubMed]
- Hong, D.H.; Pawlyk, B.S.; Shang, J.; Sandberg, M.A.; Berson, E.L.; Li, T. A retinitis pigmentosa GTPase regulator (RPGR)-deficient mouse model for X-linked retinitis pigmentosa (RP3). Proc. Natl. Acad. Sci. USA 2000, 97, 3649–3654. [Google Scholar] [CrossRef] [PubMed]
- Copeland, J. Actin-based regulation of ciliogenesis—The long and the short of it. Semin. Cell Dev. Biol. 2020, 102, 132–138. [Google Scholar] [CrossRef]
- Rao, K.N.; Anand, M.; Khanna, H. The carboxyl terminal mutational hotspot of the ciliary disease protein RPGRORF15 (retinitis pigmentosa GTPase regulator) is glutamylated in vivo. Biol. Open 2016, 5, 424–428. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Conlon, T.J.; Deng, W.T.; Coleman, K.E.; Zhu, P.; Plummer, C.; Mandapati, S.; Van Hoosear, M.; Green, K.B.; Sonnentag, P.; et al. Toxicology and Pharmacology of an AAV Vector Expressing Codon-Optimized RPGR in RPGR-Deficient Rd9 Mice. Hum. Gene Ther. Clin. Dev. 2018, 29, 188–197. [Google Scholar] [CrossRef] [PubMed]
- Andersen, J.S.; Wilkinson, C.J.; Mayor, T.; Mortensen, P.; Nigg, E.A.; Mann, M. Proteomic characterization of the human centrosome by protein correlation profiling. Nature 2003, 426, 570–574. [Google Scholar] [CrossRef] [PubMed]
- Barbelanne, M.; Hossain, D.; Chan, D.P.; Peranen, J.; Tsang, W.Y. Nephrocystin proteins NPHP5 and Cep290 regulate BBSome integrity, ciliary trafficking and cargo delivery. Hum. Mol. Genet. 2015, 24, 2185–2200. [Google Scholar] [CrossRef]
- Conkar, D.; Culfa, E.; Odabasi, E.; Rauniyar, N.; Yates, J.R., 3rd; Firat-Karalar, E.N. The centriolar satellite protein CCDC66 interacts with CEP290 and functions in cilium formation and trafficking. J. Cell Sci. 2017, 130, 1450–1462. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Pang, N.; Zhang, Y.; Chen, H.; Peng, Y.; Fu, J.; Wei, Q. CEP290 is essential for the initiation of ciliary transition zone assembly. PLoS Biol. 2020, 18, e3001034. [Google Scholar] [CrossRef] [PubMed]
- Feldhaus, B.; Weisschuh, N.; Nasser, F.; den Hollander, A.I.; Cremers, F.P.M.; Zrenner, E.; Kohl, S.; Zobor, D. CEP290 Mutation Spectrum and Delineation of the Associated Phenotype in a Large German Cohort: A Monocentric Study. Am. J. Ophthalmol. 2020, 211, 142–150. [Google Scholar] [CrossRef]
- Leroy, B.P.; Birch, D.G.; Duncan, J.L.; Lam, B.L.; Koenekoop, R.K.; Porto, F.B.O.; Russell, S.R.; Girach, A. Leber congenital amaurosis due to cep290 mutations—Severe vision impairment with a high unmet medical need: A Review. Retina 2021, 41, 898–907. [Google Scholar] [CrossRef]
- den Hollander, A.I.; Koenekoop, R.K.; Yzer, S.; Lopez, I.; Arends, M.L.; Voesenek, K.E.; Zonneveld, M.N.; Strom, T.M.; Meitinger, T.; Brunner, H.G.; et al. Mutations in the CEP290 (NPHP6) gene are a frequent cause of Leber congenital amaurosis. Am. J. Hum. Genet. 2006, 79, 556–561. [Google Scholar] [CrossRef]
- Drivas, T.G.; Wojno, A.P.; Tucker, B.A.; Stone, E.M.; Bennett, J. Basal exon skipping and genetic pleiotropy: A predictive model of disease pathogenesis. Sci. Transl. Med. 2015, 7, 291ra97. [Google Scholar] [CrossRef] [PubMed]
- Cideciyan, A.V.; Jacobson, S.G.; Ho, A.C.; Swider, M.; Sumaroka, A.; Roman, A.J.; Wu, V.; Russell, R.C.; Viarbitskaya, I.; Garafalo, A.V.; et al. Durable vision improvement after a single intravitreal treatment with antisense oligonucleotide in CEP290-LCA: Replication in two eyes. Am. J. Ophthalmol. Case Rep. 2023, 32, 101873. [Google Scholar] [CrossRef] [PubMed]
- Garanto, A.; Duijkers, L.; Collin, R.W. Species-dependent splice recognition of a cryptic exon resulting from a recurrent intronic CEP290 mutation that causes congenital blindness. Int. J. Mol. Sci. 2015, 16, 5285–5298. [Google Scholar] [CrossRef] [PubMed]
- Garanto, A.; van Beersum, S.E.; Peters, T.A.; Roepman, R.; Cremers, F.P.; Collin, R.W. Unexpected CEP290 mRNA splicing in a humanized knock-in mouse model for Leber congenital amaurosis. PLoS ONE 2013, 8, e79369. [Google Scholar] [CrossRef] [PubMed]
- Toms, M.; Pagarkar, W.; Moosajee, M. Usher syndrome: Clinical features, molecular genetics and advancing therapeutics. Ther. Adv. Ophthalmol. 2020, 12, 2515841420952194. [Google Scholar] [CrossRef]
- Nisenbaum, E.; Thielhelm, T.P.; Nourbakhsh, A.; Yan, D.; Blanton, S.H.; Shu, Y.; Koehler, K.R.; El-Amraoui, A.; Chen, Z.; Lam, B.L.; et al. Review of Genotype-Phenotype Correlations in Usher Syndrome. Ear Hear. 2022, 43, 1–8. [Google Scholar] [CrossRef]
- Fuster-Garcia, C.; Garcia-Bohorquez, B.; Rodriguez-Munoz, A.; Aller, E.; Jaijo, T.; Millan, J.M.; Garcia-Garcia, G. Usher Syndrome: Genetics of a Human Ciliopathy. Int. J. Mol. Sci. 2021, 22, 6723. [Google Scholar] [CrossRef] [PubMed]
- Castiglione, A.; Moller, C. Usher Syndrome. Audiol. Res. 2022, 12, 42–65. [Google Scholar] [CrossRef]
- Sahly, I.; Dufour, E.; Schietroma, C.; Michel, V.; Bahloul, A.; Perfettini, I.; Pepermans, E.; Estivalet, A.; Carette, D.; Aghaie, A.; et al. Localization of Usher 1 proteins to the photoreceptor calyceal processes, which are absent from mice. J. Cell Biol. 2012, 199, 381–399. [Google Scholar] [CrossRef]
- Sorusch, N.; Bauss, K.; Plutniok, J.; Samanta, A.; Knapp, B.; Nagel-Wolfrum, K.; Wolfrum, U. Characterization of the ternary Usher syndrome SANS/ush2a/whirlin protein complex. Hum. Mol. Genet. 2017, 26, 1157–1172. [Google Scholar] [CrossRef] [PubMed]
- Siemens, J.; Kazmierczak, P.; Reynolds, A.; Sticker, M.; Littlewood-Evans, A.; Muller, U. The Usher syndrome proteins cadherin 23 and harmonin form a complex by means of PDZ-domain interactions. Proc. Natl. Acad. Sci. USA 2002, 99, 14946–14951. [Google Scholar] [CrossRef] [PubMed]
- Schietroma, C.; Parain, K.; Estivalet, A.; Aghaie, A.; Boutet de Monvel, J.; Picaud, S.; Sahel, J.A.; Perron, M.; El-Amraoui, A.; Petit, C. Usher syndrome type 1-associated cadherins shape the photoreceptor outer segment. J. Cell Biol. 2017, 216, 1849–1864. [Google Scholar] [CrossRef]
- Gibson, F.; Walsh, J.; Mburu, P.; Varela, A.; Brown, K.A.; Antonio, M.; Beisel, K.W.; Steel, K.P.; Brown, S.D. A type VII myosin encoded by the mouse deafness gene shaker-1. Nature 1995, 374, 62–64. [Google Scholar] [CrossRef]
- El-Amraoui, A.; Petit, C. The retinal phenotype of Usher syndrome: Pathophysiological insights from animal models. Comptes Rendus Biol. 2014, 337, 167–177. [Google Scholar] [CrossRef]
- Jouret, G.; Poirsier, C.; Spodenkiewicz, M.; Jaquin, C.; Gouy, E.; Arndt, C.; Labrousse, M.; Gaillard, D.; Doco-Fenzy, M.; Lebre, A.S. Genetics of Usher Syndrome: New Insights From a Meta-analysis. Otol. Neurotol. 2019, 40, 121–129. [Google Scholar] [CrossRef]
- Heissler, S.M.; Manstein, D.J. Functional characterization of the human myosin-7a motor domain. Cell Mol. Life Sci. 2012, 69, 299–311. [Google Scholar] [CrossRef][Green Version]
- Delmaghani, S.; El-Amraoui, A. The genetic and phenotypic landscapes of Usher syndrome: From disease mechanisms to a new classification. Hum. Genet. 2022, 141, 709–735. [Google Scholar] [CrossRef] [PubMed]
- Finkbeiner, C.; Ortuno-Lizaran, I.; Sridhar, A.; Hooper, M.; Petter, S.; Reh, T.A. Single-cell ATAC-seq of fetal human retina and stem-cell-derived retinal organoids shows changing chromatin landscapes during cell fate acquisition. Cell Rep. 2022, 38, 110294. [Google Scholar] [CrossRef] [PubMed]
- Verbakel, S.K.; van Huet, R.A.C.; Boon, C.J.F.; den Hollander, A.I.; Collin, R.W.J.; Klaver, C.C.W.; Hoyng, C.B.; Roepman, R.; Klevering, B.J. Non-syndromic retinitis pigmentosa. Prog. Retin. Eye Res. 2018, 66, 157–186. [Google Scholar] [CrossRef] [PubMed]
- van Wijk, E.; Pennings, R.J.; te Brinke, H.; Claassen, A.; Yntema, H.G.; Hoefsloot, L.H.; Cremers, F.P.; Cremers, C.W.; Kremer, H. Identification of 51 novel exons of the Usher syndrome type 2A (USH2A) gene that encode multiple conserved functional domains and that are mutated in patients with Usher syndrome type II. Am. J. Hum. Genet. 2004, 74, 738–744. [Google Scholar] [CrossRef] [PubMed]
- Maerker, T.; van Wijk, E.; Overlack, N.; Kersten, F.F.; McGee, J.; Goldmann, T.; Sehn, E.; Roepman, R.; Walsh, E.J.; Kremer, H.; et al. A novel Usher protein network at the periciliary reloading point between molecular transport machineries in vertebrate photoreceptor cells. Hum. Mol. Genet. 2008, 17, 71–86. [Google Scholar] [CrossRef] [PubMed]
- Toualbi, L.; Toms, M.; Moosajee, M. USH2A-retinopathy: From genetics to therapeutics. Exp. Eye Res. 2020, 201, 108330. [Google Scholar] [CrossRef] [PubMed]
- Meunier, A.; Zanlonghi, X.; Roux, A.F.; Fils, J.F.; Caspers, L.; Migeotte, I.; Abramowicz, M.; Meunier, I. Natural history of Usher type 2 with the c.2299delG mutation of USH2A in a large cohort. Ophthalmic Genet 2022, 43, 470–475. [Google Scholar] [CrossRef]
- Liu, X.; Bulgakov, O.V.; Darrow, K.N.; Pawlyk, B.; Adamian, M.; Liberman, M.C.; Li, T. Usherin is required for maintenance of retinal photoreceptors and normal development of cochlear hair cells. Proc. Natl. Acad. Sci. USA 2007, 104, 4413–4418. [Google Scholar] [CrossRef]
- Dona, M.; Slijkerman, R.; Lerner, K.; Broekman, S.; Wegner, J.; Howat, T.; Peters, T.; Hetterschijt, L.; Boon, N.; de Vrieze, E.; et al. Usherin defects lead to early-onset retinal dysfunction in zebrafish. Exp. Eye Res. 2018, 173, 148–159. [Google Scholar] [CrossRef]
- Han, S.; Liu, X.; Xie, S.; Gao, M.; Liu, F.; Yu, S.; Sun, P.; Wang, C.; Archacki, S.; Lu, Z.; et al. Knockout of ush2a gene in zebrafish causes hearing impairment and late onset rod-cone dystrophy. Hum. Genet. 2018, 137, 779–794. [Google Scholar] [CrossRef]
- Monahan, P.E.; Samulski, R.J. Adeno-associated virus vectors for gene therapy: More pros than cons? Mol. Med. Today 2000, 6, 433–440. [Google Scholar] [CrossRef]
- Fuller-Carter, P.I.; Basiri, H.; Harvey, A.R.; Carvalho, L.S. Focused Update on AAV-Based Gene Therapy Clinical Trials for Inherited Retinal Degeneration. BioDrugs 2020, 34, 763–781. [Google Scholar] [CrossRef]
- Cheng, S.Y.; Punzo, C. Update on Viral Gene Therapy Clinical Trials for Retinal Diseases. Hum. Gene Ther. 2022, 33, 865–878. [Google Scholar] [CrossRef]
- Garita-Hernandez, M.; Routet, F.; Guibbal, L.; Khabou, H.; Toualbi, L.; Riancho, L.; Reichman, S.; Duebel, J.; Sahel, J.A.; Goureau, O.; et al. AAV-Mediated Gene Delivery to 3D Retinal Organoids Derived from Human Induced Pluripotent Stem Cells. Int. J. Mol. Sci. 2020, 21, 994. [Google Scholar] [CrossRef]
- Volkner, M.; Pavlou, M.; Buning, H.; Michalakis, S.; Karl, M.O. Optimized Adeno-Associated Virus Vectors for Efficient Transduction of Human Retinal Organoids. Hum. Gene Ther. 2021, 32, 694–706. [Google Scholar] [CrossRef]
- Gonzalez-Cordero, A.; Goh, D.; Kruczek, K.; Naeem, A.; Fernando, M.; Kleine Holthaus, S.M.; Takaaki, M.; Blackford, S.J.I.; Kloc, M.; Agundez, L.; et al. Assessment of AAV Vector Tropisms for Mouse and Human Pluripotent Stem Cell-Derived RPE and Photoreceptor Cells. Hum. Gene Ther. 2018, 29, 1124–1139. [Google Scholar] [CrossRef]
- McClements, M.E.; Steward, H.; Atkin, W.; Goode, E.A.; Gandara, C.; Chichagova, V.; MacLaren, R.E. Tropism of AAV Vectors in Photoreceptor-Like Cells of Human iPSC-Derived Retinal Organoids. Transl. Vis. Sci. Technol. 2022, 11, 3. [Google Scholar] [CrossRef]
- Keng, C.T.; Guo, K.; Liu, Y.C.; Shen, K.Y.; Lim, D.S.; Lovatt, M.; Ang, H.P.; Mehta, J.S.; Chew, W.L. Multiplex viral tropism assay in complex cell populations with single-cell resolution. Gene Ther. 2022, 29, 555–565. [Google Scholar] [CrossRef]
- Rodrigues, A.; Slembrouck-Brec, A.; Nanteau, C.; Terray, A.; Tymoshenko, Y.; Zagar, Y.; Reichman, S.; Xi, Z.; Sahel, J.A.; Fouquet, S.; et al. Modeling PRPF31 retinitis pigmentosa using retinal pigment epithelium and organoids combined with gene augmentation rescue. NPJ Regen. Med. 2022, 7, 39. [Google Scholar] [CrossRef] [PubMed]
- Schon, C.; Biel, M.; Michalakis, S. Retinal gene delivery by adeno-associated virus (AAV) vectors: Strategies and applications. Eur. J. Pharm. Biopharm. 2015, 95 Pt B, 343–352. [Google Scholar] [CrossRef]
- Vandenberghe, L.H.; Bell, P.; Maguire, A.M.; Cearley, C.N.; Xiao, R.; Calcedo, R.; Wang, L.; Castle, M.J.; Maguire, A.C.; Grant, R.; et al. Dosage thresholds for AAV2 and AAV8 photoreceptor gene therapy in monkey. Sci. Transl. Med. 2011, 3, 88ra54. [Google Scholar] [CrossRef] [PubMed]
- Manfredi, A.; Marrocco, E.; Puppo, A.; Cesi, G.; Sommella, A.; Della Corte, M.; Rossi, S.; Giunti, M.; Craft, C.M.; Bacci, M.L.; et al. Combined rod and cone transduction by adeno-associated virus 2/8. Hum. Gene Ther. 2013, 24, 982–992. [Google Scholar] [CrossRef]
- Boye, S.E.; Alexander, J.J.; Boye, S.L.; Witherspoon, C.D.; Sandefer, K.J.; Conlon, T.J.; Erger, K.; Sun, J.; Ryals, R.; Chiodo, V.A.; et al. The human rhodopsin kinase promoter in an AAV5 vector confers rod- and cone-specific expression in the primate retina. Hum. Gene Ther. 2012, 23, 1101–1115. [Google Scholar] [CrossRef] [PubMed]
- Wiley, L.A.; Burnight, E.R.; Kaalberg, E.E.; Jiao, C.; Riker, M.J.; Halder, J.A.; Luse, M.A.; Han, I.C.; Russell, S.R.; Sohn, E.H.; et al. Assessment of Adeno-Associated Virus Serotype Tropism in Human Retinal Explants. Hum. Gene Ther. 2018, 29, 424–436. [Google Scholar] [CrossRef]
- Xi, Z.; Öztürk, B.E.; Johnson, M.E.; Turunç, S.; Stauffer, W.R.; Byrne, L.C. Quantitative single-cell transcriptome-based ranking of engineered AAVs in human retinal explants. Mol. Ther. Methods Clin. Dev. 2022, 25, 476–489. [Google Scholar] [CrossRef]
- Dalkara, D.; Byrne, L.C.; Klimczak, R.R.; Visel, M.; Yin, L.; Merigan, W.H.; Flannery, J.G.; Schaffer, D.V. In vivo-directed evolution of a new adeno-associated virus for therapeutic outer retinal gene delivery from the vitreous. Sci. Transl. Med. 2013, 5, 189ra76. [Google Scholar] [CrossRef]
- Achberger, K.; Cipriano, M.; Düchs, M.J.; Schön, C.; Michelfelder, S.; Stierstorfer, B.; Lamla, T.; Kauschke, S.G.; Chuchuy, J.; Roosz, J.; et al. Human stem cell-based retina on chip as new translational model for validation of AAV retinal gene therapy vectors. Stem Cell Rep. 2021, 16, 2242–2256. [Google Scholar] [CrossRef]
- Ozturk, B.E.; Johnson, M.E.; Kleyman, M.; Turunc, S.; He, J.; Jabalameli, S.; Xi, Z.; Visel, M.; Dufour, V.L.; Iwabe, S.; et al. scAAVengr, a transcriptome-based pipeline for quantitative ranking of engineered AAVs with single-cell resolution. Elife 2021, 10, e64175. [Google Scholar] [CrossRef]
- Beltran, W.A.; Cideciyan, A.V.; Lewin, A.S.; Iwabe, S.; Khanna, H.; Sumaroka, A.; Chiodo, V.A.; Fajardo, D.S.; Roman, A.J.; Deng, W.T.; et al. Gene therapy rescues photoreceptor blindness in dogs and paves the way for treating human X-linked retinitis pigmentosa. Proc. Natl. Acad. Sci. USA 2012, 109, 2132–2137. [Google Scholar] [CrossRef]
- Young, J.E.; Vogt, T.; Gross, K.W.; Khani, S.C. A short, highly active photoreceptor-specific enhancer/promoter region upstream of the human rhodopsin kinase gene. Investig. Ophthalmol. Vis. Sci. 2003, 44, 4076–4085. [Google Scholar] [CrossRef] [PubMed]
- Khani, S.C.; Pawlyk, B.S.; Bulgakov, O.V.; Kasperek, E.; Young, J.E.; Adamian, M.; Sun, X.; Smith, A.J.; Ali, R.R.; Li, T. AAV-mediated expression targeting of rod and cone photoreceptors with a human rhodopsin kinase promoter. Investig. Ophthalmol. Vis. Sci. 2007, 48, 3954–3961. [Google Scholar] [CrossRef]
- Pawlyk, B.S.; Bulgakov, O.V.; Sun, X.; Adamian, M.; Shu, X.; Smith, A.J.; Berson, E.L.; Ali, R.R.; Khani, S.; Wright, A.F.; et al. Photoreceptor rescue by an abbreviated human RPGR gene in a murine model of X-linked retinitis pigmentosa. Gene Ther. 2016, 23, 196–204. [Google Scholar] [CrossRef] [PubMed]
- Cehajic-Kapetanovic, J.; Xue, K.; Martinez-Fernandez de la Camara, C.; Nanda, A.; Davies, A.; Wood, L.J.; Salvetti, A.P.; Fischer, M.D.; Aylward, J.W.; Barnard, A.R.; et al. Initial results from a first-in-human gene therapy trial on X-linked retinitis pigmentosa caused by mutations in RPGR. Nat. Med. 2020, 26, 354–359. [Google Scholar] [CrossRef]
- Trapani, I.; Tornabene, P.; Auricchio, A. Large gene delivery to the retina with AAV vectors: Are we there yet? Gene Ther. 2021, 28, 220–222. [Google Scholar] [CrossRef] [PubMed]
- Riedmayr, L.M.; Hinrichsmeyer, K.S.; Thalhammer, S.B.; Mittas, D.M.; Karguth, N.; Otify, D.Y.; Bohm, S.; Weber, V.J.; Bartoschek, M.D.; Splith, V.; et al. mRNA trans-splicing dual AAV vectors for (epi)genome editing and gene therapy. Nat. Commun. 2023, 14, 6578. [Google Scholar] [CrossRef]
- Yan, Z.; Zhang, Y.; Duan, D.; Engelhardt, J.F. Trans-splicing vectors expand the utility of adeno-associated virus for gene therapy. Proc. Natl. Acad. Sci. USA 2000, 97, 6716–6721. [Google Scholar] [CrossRef]
- Trapani, I. Adeno-Associated Viral Vectors as a Tool for Large Gene Delivery to the Retina. Genes 2019, 10, 287. [Google Scholar] [CrossRef]
- Trapani, I.; Colella, P.; Sommella, A.; Iodice, C.; Cesi, G.; de Simone, S.; Marrocco, E.; Rossi, S.; Giunti, M.; Palfi, A.; et al. Effective delivery of large genes to the retina by dual AAV vectors. EMBO Mol. Med. 2014, 6, 194–211. [Google Scholar] [CrossRef]
- Maddalena, A.; Tornabene, P.; Tiberi, P.; Minopoli, R.; Manfredi, A.; Mutarelli, M.; Rossi, S.; Simonelli, F.; Naggert, J.K.; Cacchiarelli, D.; et al. Triple Vectors Expand AAV Transfer Capacity in the Retina. Mol. Ther. 2018, 26, 524–541. [Google Scholar] [CrossRef] [PubMed]
- Volkmann, G.; Iwai, H. Protein trans-splicing and its use in structural biology: Opportunities and limitations. Mol. Biosyst. 2010, 6, 2110–2121. [Google Scholar] [CrossRef]
- Mills, K.V.; Johnson, M.A.; Perler, F.B. Protein splicing: How inteins escape from precursor proteins. J. Biol. Chem. 2014, 289, 14498–14505. [Google Scholar] [CrossRef]
- Tornabene, P.; Trapani, I.; Centrulo, M.; Marrocco, E.; Minopoli, R.; Lupo, M.; Iodice, C.; Gesualdo, C.; Simonelli, F.; Surace, E.M.; et al. Inclusion of a degron reduces levelsof undesired inteins after AAV-mediated proteintrans-splicing in the retina. Mol. Ther. Methods Clin. Dev. 2021, 23, 448–459. [Google Scholar] [CrossRef] [PubMed]
- Tornabene, P.; Trapani, I.; Minopoli, R.; Centrulo, M.; Lupo, M.; de Simone, S.; Tiberi, P.; Dell’Aquila, F.; Marrocco, E.; Iodice, C.; et al. Intein-mediated protein trans-splicing expands adeno-associated virus transfer capacity in the retina. Sci. Transl. Med. 2019, 11, aav4523. [Google Scholar] [CrossRef]
- Cashman, S.M.; McCullough, L.; Kumar-Singh, R. Improved retinal transduction in vivo and photoreceptor-specific transgene expression using adenovirus vectors with modified penton base. Mol. Ther. 2007, 15, 1640–1646. [Google Scholar] [CrossRef]
- Mallam, J.N.; Hurwitz, M.Y.; Mahoney, T.; Chevez-Barrios, P.; Hurwitz, R.L. Efficient gene transfer into retinal cells using adenoviral vectors: Dependence on receptor expression. Investig. Ophthalmol. Vis. Sci. 2004, 45, 1680–1687. [Google Scholar] [CrossRef]
- Sweigard, J.H.; Cashman, S.M.; Kumar-Singh, R. Adenovirus vectors targeting distinct cell types in the retina. Investig. Ophthalmol. Vis. Sci. 2010, 51, 2219–2228. [Google Scholar] [CrossRef]
- Zhang, S.H.; Wu, J.H.; Wu, X.B.; Dong, X.Y.; Liu, X.J.; Li, C.Y.; Qian, H. Distinctive gene transduction efficiencies of commonly used viral vectors in the retina. Curr. Eye Res. 2008, 33, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Han, I.C.; Burnight, E.R.; Kaalberg, E.E.; Boyce, T.M.; Stone, E.M.; Fingert, J.H.; Mullins, R.F.; Tucker, B.A.; Wiley, L.A. Chimeric Helper-Dependent Adenoviruses Transduce Retinal Ganglion Cells and Muller Cells in Human Retinal Explants. J. Ocul. Pharmacol. Ther. 2021, 37, 575–579. [Google Scholar] [CrossRef]
- Puppo, A.; Cesi, G.; Marrocco, E.; Piccolo, P.; Jacca, S.; Shayakhmetov, D.M.; Parks, R.J.; Davidson, B.L.; Colloca, S.; Brunetti-Pierri, N.; et al. Retinal transduction profiles by high-capacity viral vectors. Gene Ther. 2014, 21, 855–865. [Google Scholar] [CrossRef] [PubMed]
- Goncalves, M.A.; de Vries, A.A. Adenovirus: From foe to friend. Rev. Med. Virol. 2006, 16, 167–186. [Google Scholar] [CrossRef] [PubMed]
- Han, I.C.; Burnight, E.R.; Ulferts, M.J.; Worthington, K.S.; Russell, S.R.; Sohn, E.H.; Mullins, R.F.; Stone, E.M.; Tucker, B.A.; Wiley, L.A. Helper-Dependent Adenovirus Transduces the Human and Rat Retina but Elicits an Inflammatory Reaction When Delivered Subretinally in Rats. Hum. Gene Ther. 2019, 30, 1371–1384. [Google Scholar] [CrossRef] [PubMed]
- Ricobaraza, A.; Gonzalez-Aparicio, M.; Mora-Jimenez, L.; Lumbreras, S.; Hernandez-Alcoceba, R. High-Capacity Adenoviral Vectors: Expanding the Scope of Gene Therapy. Int. J. Mol. Sci. 2020, 21, 3643. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; East, A.; Cheng, A.; Lin, S.; Ma, E.; Doudna, J. RNA-programmed genome editing in human cells. Elife 2013, 2, e00471. [Google Scholar] [CrossRef]
- Doudna, J.A.; Charpentier, E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef] [PubMed]
- Sander, J.D.; Joung, J.K. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 2014, 32, 347–355. [Google Scholar] [CrossRef]
- Anzalone, A.V.; Randolph, P.B.; Davis, J.R.; Sousa, A.A.; Koblan, L.W.; Levy, J.M.; Chen, P.J.; Wilson, C.; Newby, G.A.; Raguram, A.; et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 2019, 576, 149–157. [Google Scholar] [CrossRef]
- Yee, T.; Wert, K.J. Base and Prime Editing in the Retina-From Preclinical Research toward Human Clinical Trials. Int. J. Mol. Sci. 2022, 23, 12375. [Google Scholar] [CrossRef]
- Maeder, M.L.; Mepani, R.; Gloskowski, S.; Skor, M.; Collins, M.; Shen, S.; Gotta, G.; Marco, E.; Barrera, L.; Jayaram, H.; et al. 124. Therapeutic Correction of an LCA-Causing Splice Defect in the CEP290 Gene by CRISPR/Cas-Mediated Gene Editing. Mol. Ther. 2016, 24, S51–S52. [Google Scholar] [CrossRef]
- Newby, G.A.; Liu, D.R. In vivo somatic cell base editing and prime editing. Mol. Ther. 2021, 29, 3107–3124. [Google Scholar] [CrossRef]
- Jang, H.; Jo, D.H.; Cho, C.S.; Shin, J.H.; Seo, J.H.; Yu, G.; Gopalappa, R.; Kim, D.; Cho, S.R.; Kim, J.H.; et al. Application of prime editing to the correction of mutations and phenotypes in adult mice with liver and eye diseases. Nat. Biomed. Eng. 2022, 6, 181–194. [Google Scholar] [CrossRef] [PubMed]
- She, K.; Liu, Y.; Zhao, Q.; Jin, X.; Yang, Y.; Su, J.; Li, R.; Song, L.; Xiao, J.; Yao, S.; et al. Dual-AAV split prime editor corrects the mutation and phenotype in mice with inherited retinal degeneration. Signal Transduct. Target. Ther. 2023, 8, 57. [Google Scholar] [CrossRef]
- Zhi, S.; Chen, Y.; Wu, G.; Wen, J.; Wu, J.; Liu, Q.; Li, Y.; Kang, R.; Hu, S.; Wang, J.; et al. Dual-AAV delivering split prime editor system for in vivo genome editing. Mol. Ther. 2022, 30, 283–294. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.S.; Li, Q.C.; Yin, C.Q.; Xue, W.; Song, C.Q. Advances in CRISPR/Cas-based Gene Therapy in Human Genetic Diseases. Theranostics 2020, 10, 4374–4382. [Google Scholar] [CrossRef] [PubMed]
- Gaj, T.; Gersbach, C.A.; Barbas, C.F., 3rd. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013, 31, 397–405. [Google Scholar] [CrossRef] [PubMed]
- Orthwein, A.; Noordermeer, S.M.; Wilson, M.D.; Landry, S.; Enchev, R.I.; Sherker, A.; Munro, M.; Pinder, J.; Salsman, J.; Dellaire, G.; et al. A mechanism for the suppression of homologous recombination in G1 cells. Nature 2015, 528, 422–426. [Google Scholar] [CrossRef] [PubMed]
- Lieber, M.R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 2010, 79, 181–211. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Tsunekawa, Y.; Hernandez-Benitez, R.; Wu, J.; Zhu, J.; Kim, E.J.; Hatanaka, F.; Yamamoto, M.; Araoka, T.; Li, Z.; et al. In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature 2016, 540, 144–149. [Google Scholar] [CrossRef] [PubMed]
- Maddalena, A.; Kleinlogel, S. CRISPR-mediated optogene expression from a cell-specific endogenous promoter in retinal ON-bipolar cells to restore vision. Front. Drug Deliv. 2023, 3, 934394. [Google Scholar] [CrossRef]
- Tornabene, P.; Ferla, R.; Llado-Santaeularia, M.; Centrulo, M.; Dell’Anno, M.; Esposito, F.; Marrocco, E.; Pone, E.; Minopoli, R.; Iodice, C.; et al. Therapeutic homology-independent targeted integration in retina and liver. Nat. Commun. 2022, 13, 1963. [Google Scholar] [CrossRef] [PubMed]
| Associated Disorder | Gene | Mutation | Reference |
|---|---|---|---|
| X-linked retinitis pigmentosa | RPGR | c.ORF15+689−692del4 | [45] |
| X-linked retinitis pigmentosa | RPGR | c.1685_1686delAT c.2234_2235delGA c.2403_2404delAG | [23] |
| X-linked retinitis pigmentosa | RPGR | c.ORF15+652_653delAG c.ORF15+594_595delGA | [22] |
| X-linked retinitis pigmentosa | RPGR | c.1415 − 9A>G | [25] |
| X-linked retinitis pigmentosa | RP2 | c.358C>T c.371_378delAAGCTGGA | [43] |
| Retinitis pigmentosa | PRPF31 | c.1115_1125del | [46] |
| Leber congenital amaurosis | CEP290 | c.2991+1665A>G + c.2991+1665A>G | [18] |
| Leber congenital amaurosis | CEP290 | c.2991+1665A>G + c.2991+1665A>G c.2991+1655A>G + c.5668G>T | [47,48] |
| Leber congenital amaurosis | CEP290 | c.2991+1665A>G + c.2991+1665A>G | [49] |
| Leber congenital amaurosis | CEP290 | c.2991+1665A>G + c.2991+1665A>G c.315del + c.316del | [50] |
| Leber congenital amaurosis | LCA5 | c.835C>T + c.835C>T | [51] |
| Leber congenital amaurosis | NPHP5 | c.421_422delTT + c.1036G>T | [24] |
| Non-syndromic retinitis pigmentosa | USH2A | c.8559-2A>G + c.9127_9129del | [52,53] |
| Non-syndromic retinitis pigmentosa/Usher syndrome (Type 2) | USH2A | c.2299delG + c.2276G>T c.2299delG + c.2299delG | [54] |
| Usher syndrome (Type 1) | USH1B | c.6070C>T + c.223G>C c.1996C>T + c.133-2A>G | [55] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
McDonald, A.; Wijnholds, J. Retinal Ciliopathies and Potential Gene Therapies: A Focus on Human iPSC-Derived Organoid Models. Int. J. Mol. Sci. 2024, 25, 2887. https://doi.org/10.3390/ijms25052887
McDonald A, Wijnholds J. Retinal Ciliopathies and Potential Gene Therapies: A Focus on Human iPSC-Derived Organoid Models. International Journal of Molecular Sciences. 2024; 25(5):2887. https://doi.org/10.3390/ijms25052887
Chicago/Turabian StyleMcDonald, Andrew, and Jan Wijnholds. 2024. "Retinal Ciliopathies and Potential Gene Therapies: A Focus on Human iPSC-Derived Organoid Models" International Journal of Molecular Sciences 25, no. 5: 2887. https://doi.org/10.3390/ijms25052887
APA StyleMcDonald, A., & Wijnholds, J. (2024). Retinal Ciliopathies and Potential Gene Therapies: A Focus on Human iPSC-Derived Organoid Models. International Journal of Molecular Sciences, 25(5), 2887. https://doi.org/10.3390/ijms25052887







