PRPH2-Related Retinal Dystrophies: Mutational Spectrum in 103 Families from a Spanish Cohort
Abstract
1. Introduction
2. Results
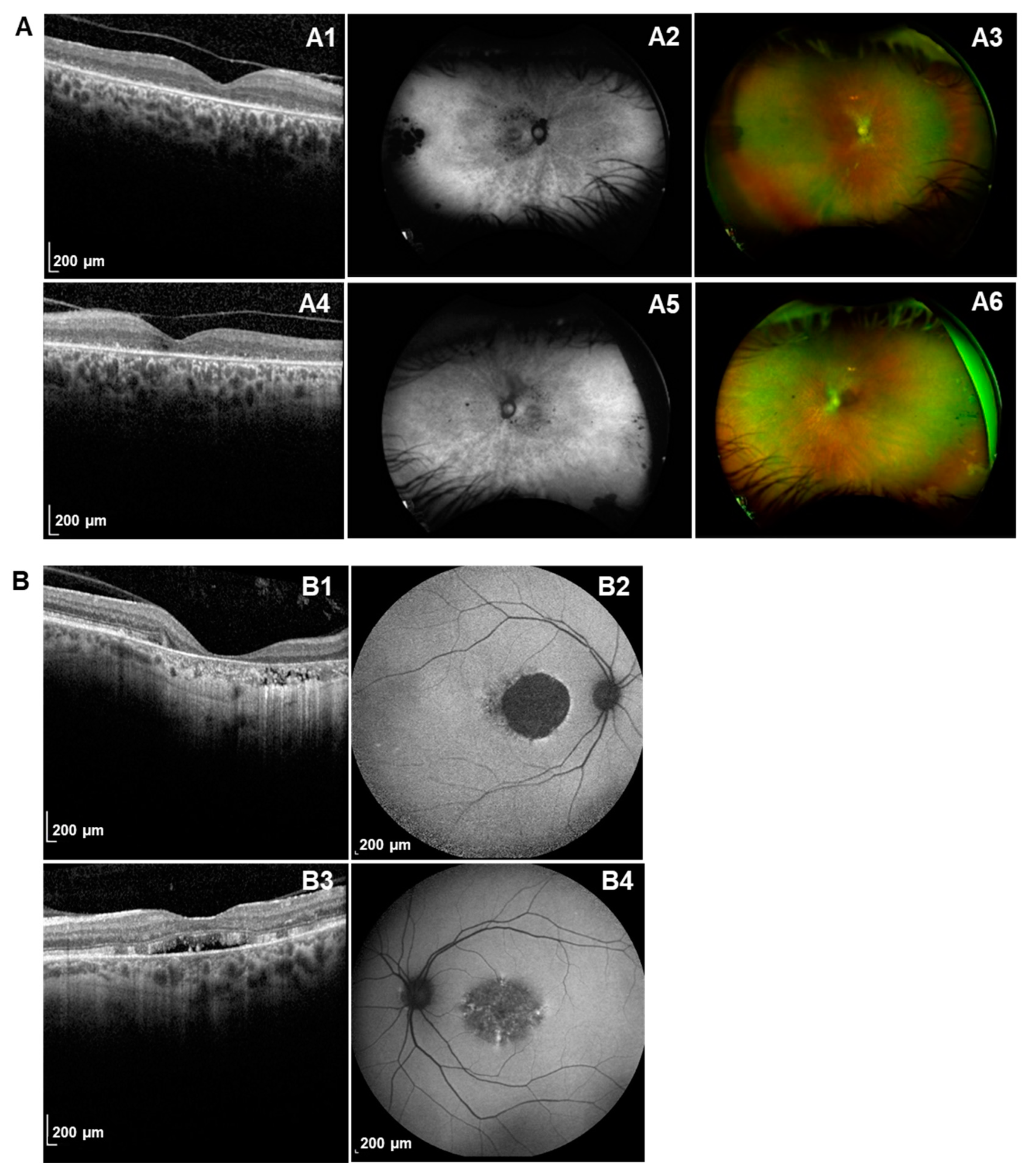
2.1. Ophthalmic Characteristics of PRPH2 Patients
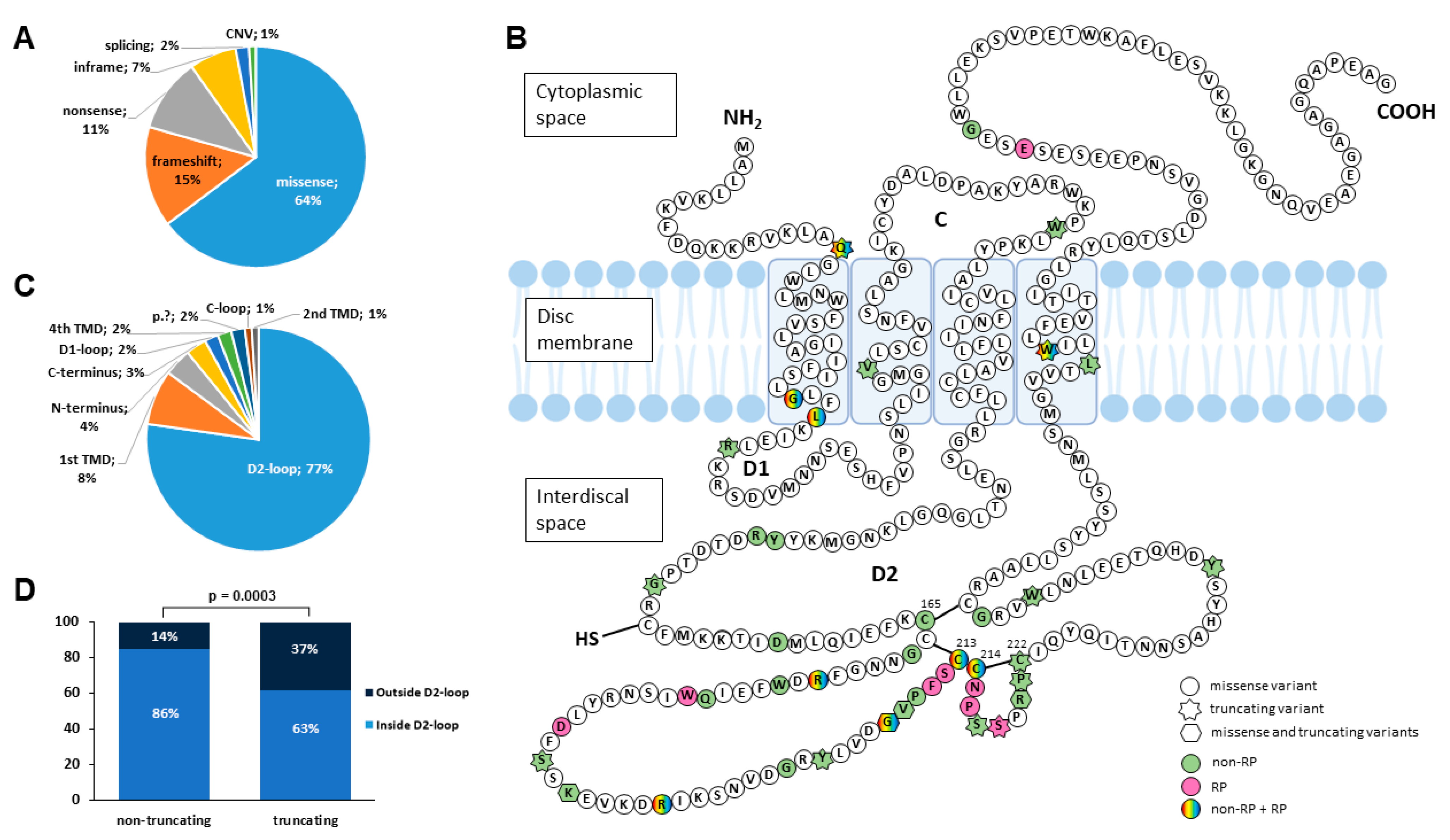
2.2. Mutational Spectrum of PRPH2 Variants in Our Spanish Cohort
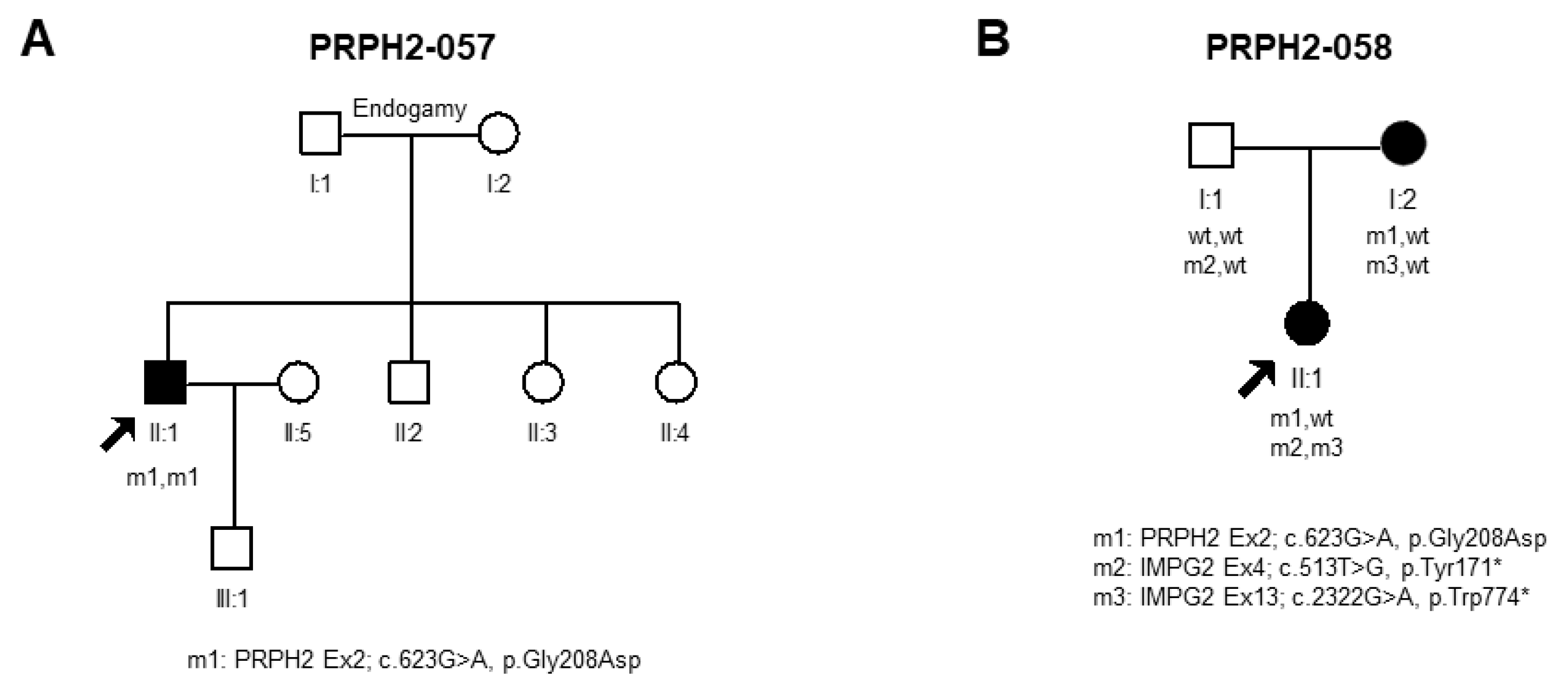
Novel Disease-Causing Variants and PRPH2-VUS Identified in Our Cohort
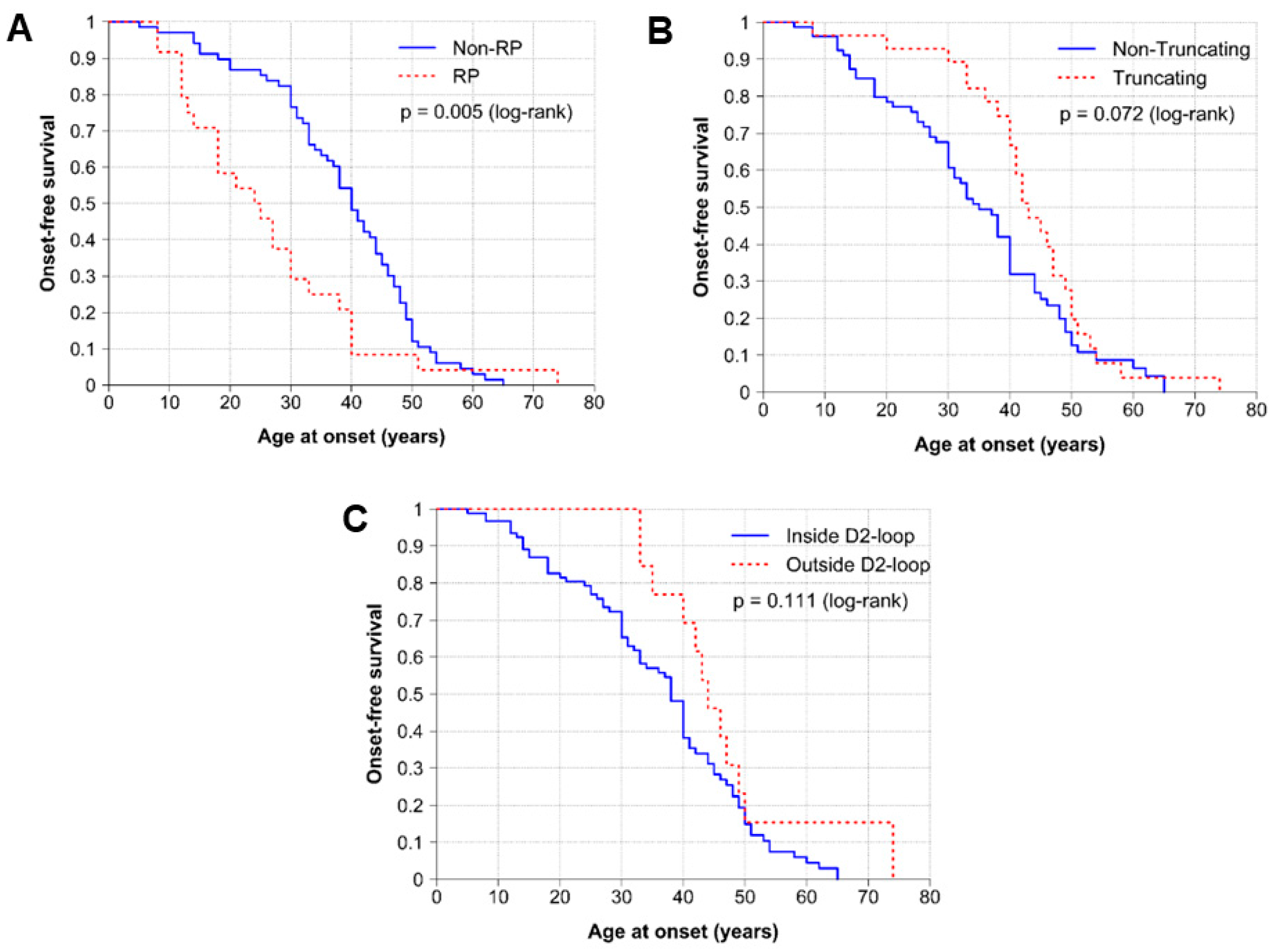
2.3. Genotype–Phenotype Correlations
3. Discussion
Study Limitations
4. Materials and Methods
4.1. Subjects
4.2. Clinical Classification
4.3. Molecular Diagnosis and Analysis of Variants
4.4. Haplotype Analysis
4.5. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wright, A.F.; Chakarova, C.F.; Abd El-Aziz, M.M.; Bhattacharya, S.S. Photoreceptor degeneration: Genetic and mechanistic dissection of a complex trait. Nat. Rev. Genet. 2010, 11, 273–284. [Google Scholar] [CrossRef]
- Vaidya, P. Retinitis Pigmentosa: Disease Encumbrance in the Eurozone. Int. J. Ophthalmol. Clin. Res. 2015, 2, 3. [Google Scholar]
- Hartong, D.T.; Berson, E.L.; Dryja, T.P. Retinitis pigmentosa. Lancet Lond. Engl. 2006, 368, 1795–1809. [Google Scholar] [CrossRef]
- Koyanagi, Y.; Akiyama, M.; Nishiguchi, K.M.; Momozawa, Y.; Kamatani, Y.; Takata, S.; Inai, C.; Iwasaki, Y.; Kumano, M.; Murakami, Y. Genetic characteristics of retinitis pigmentosa in 1204 Japanese patients. J. Med. Genet. 2019, 56, 662–670. [Google Scholar] [CrossRef]
- Goetz, K.E.; Reeves, M.J.; Gagadam, S.; Blain, D.; Bender, C.; Lwin, C.; Naik, A.; Tumminia, S.J.; Hufnagel, R.B. Genetic testing for inherited eye conditions in over 6000 individuals through the EYEGENE network. Am. J. Med. Genet. C Semin. Med. Genet. 2020, 184, 828–837. [Google Scholar] [CrossRef] [PubMed]
- Pontikos, N.; Arno, G.; Jurkute, N.; Schiff, E.; Ba-Abbad, R.; Malka, S.; Gimenez, A.; Georgiou, M.; Wright, G.; Armengol, M.; et al. Genetic Basis of Inherited Retinal Disease in a Molecularly Characterized Cohort of More Than 3000 Families from the United Kingdom. Ophthalmology 2020, 127, 1384–1394. [Google Scholar] [CrossRef] [PubMed]
- Perea-Romero, I.; Gordo, G.; Iancu, I.F.; Del Pozo-Valero, M.; Almoguera, B.; Blanco-Kelly, F.; Carreño, E.; Jimenez-Rolando, B.; Lopez-Rodriguez, R.; Lorda-Sanchez, I.; et al. Genetic landscape of 6089 inherited retinal dystrophies affected cases in Spain and their therapeutic and extended epidemiological implications. Sci. Rep. 2021, 11, 1526. [Google Scholar] [CrossRef] [PubMed]
- Connell, G.J.; Molday, R.S. Molecular cloning, primary structure, and orientation of the vertebrate photoreceptor cell protein peripherin in the rod outer segment disk membrane. Biochemistry 1990, 29, 4691–4698. [Google Scholar] [CrossRef] [PubMed]
- Sanyal, S.; Jansen, H.G. Absence of receptor outer segments in the retina of rds mutant mice. Neurosci. Lett. 1981, 21, 23–26. [Google Scholar] [CrossRef] [PubMed]
- Arikawa, K.; Molday, L.L.; Molday, R.S.; Williams, D.S. Localization of peripherin/rds in the disk membranes of cone and rod photoreceptors: Relationship to disk membrane morphogenesis and retinal degeneration. J. Cell Biol. 1992, 116, 659–667. [Google Scholar] [CrossRef]
- Cheng, T.; Peachey, N.S.; Li, S.; Goto, Y.; Cao, Y.; Naash, M.I. The effect of peripherin/rds haploinsufficiency on rod and cone photoreceptors. J. Neurosci. Off. J. Soc. Neurosci. 1997, 17, 8118–8128. [Google Scholar] [CrossRef]
- Rahman, N.; Georgiou, M.; Khan, K.N.; Michaelides, M. Macular dystrophies: Clinical and imaging features, molecular genetics and therapeutic options. Br. J. Ophthalmol. 2020, 104, 451–460. [Google Scholar] [CrossRef]
- Georgiou, M.; Ali, N.; Yang, E.; Grewal, P.S.; Rotsos, T.; Pontikos, N.; Robson, A.G.; Michaelides, M. Extending the phenotypic spectrum of PRPF8, PRPH2, RP1 and RPGR, and the genotypic spectrum of early-onset severe retinal dystrophy. Orphanet. J. Rare Dis. 2021, 16, 128. [Google Scholar] [CrossRef] [PubMed]
- Boon, C.J.F.; den Hollander, A.I.; Hoyng, C.B.; Cremers, F.P.M.; Klevering, B.J.; Keunen, J.E.E. The spectrum of retinal dystrophies caused by mutations in the peripherin/RDS gene. Prog. Retin. Eye Res. 2008, 27, 213–235. [Google Scholar] [CrossRef] [PubMed]
- Coco-Martin, R.M.; Sanchez-Tocino, H.T.; Desco, C.; Usategui-Martín, R.; Tellería, J.J. PRPH2-Related Retinal Diseases: Broadening the Clinical Spectrum and Describing a New Mutation. Genes 2020, 11, 773. [Google Scholar] [CrossRef] [PubMed]
- Peeters, M.H.C.A.; Khan, M.; Rooijakkers, A.A.M.B.; Mulders, T.; Haer-Wigman, L.; Boon, C.J.F.; Klaver, C.C.W.; van den Born, L.I.; Hoyng, C.B.; Cremers, F.P.M.; et al. PRPH2 mutation update: In silico assessment of 245 reported and 7 novel variants in patients with retinal disease. Hum. Mutat. 2021, 42, 1521–1547. [Google Scholar] [CrossRef] [PubMed]
- Martin-Merida, I.; Aguilera-Garcia, D.; Fernandez-San Jose, P.; Blanco-Kelly, F.; Zurita, O.; Almoguera, B.; Garcia-Sandoval, B.; Avila-Fernandez, A.; Arteche, A.; Minguez, P.; et al. Toward the Mutational Landscape of Autosomal Dominant Retinitis Pigmentosa: A Comprehensive Analysis of 258 Spanish Families. Investig. Opthalmol. Vis. Sci. 2018, 59, 2345. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-San Jose, P.; Corton, M.; Blanco-Kelly, F.; Avila-Fernandez, A.; Lopez-Martinez, M.A.; Sanchez-Navarro, I.; Sanchez-Alcudia, R.; Perez-Carro, R.; Zurita, O.; Sanchez-Bolivar, N.; et al. Targeted Next-Generation Sequencing Improves the Diagnosis of Autosomal Dominant Retinitis Pigmentosa in Spanish Patients. Investig. Opthalmol. Vis. Sci. 2015, 56, 2173. [Google Scholar] [CrossRef] [PubMed]
- Martin-Merida, I.; Avila-Fernandez, A.; Del Pozo-Valero, M.; Blanco-Kelly, F.; Zurita, O.; Perez-Carro, R.; Aguilera-Garcia, D.; Riveiro-Alvarez, R.; Arteche, A.; Trujillo-Tiebas, M.J.; et al. Genomic Landscape of Sporadic Retinitis Pigmentosa: Findings from 877 Spanish Cases. Ophthalmology 2019, 126, 1181–1188. [Google Scholar] [CrossRef] [PubMed]
- Del Pozo-Valero, M.; Riveiro-Alvarez, R.; Martin-Merida, I.; Blanco-Kelly, F.; Swafiri, S.; Lorda-Sanchez, I.; Trujillo-Tiebas, M.J.; Carreño, E.; Jimenez-Rolando, B.; Garcia-Sandoval, B.; et al. Impact of Next Generation Sequencing in Unraveling the Genetics of 1036 Spanish Families with Inherited Macular Dystrophies. Investig. Ophthalmol. Vis. Sci. 2022, 63, 11. [Google Scholar] [CrossRef]
- Bianco, L.; Arrigo, A.; Antropoli, A.; Saladino, A.; Spiga, I.; Patricelli, M.G.; Bandello, F.; Carrera, P.; Battaglia Parodi, M. PRPH2-Associated Retinopathy: Novel Variants and Genotype-Phenotype Correlations. Ophthalmol. Retina. 2023, 7, 450–461. [Google Scholar] [CrossRef]
- Meins, M.; Grüning, G.; Blankenagel, A.; Krastel, H.; Reck, B.; Fuchs, S.; Schwinger, E.; Gal, A. Heterozygous ‘null allele’ mutation in the human peripherin/RDS gene. Hum. Mol. Genet. 1993, 2, 2181–2182. [Google Scholar] [CrossRef]
- Manes, G.; Guillaumie, T.; Vos, W.L.; Devos, A.; Audo, I.; Zeitz, C.; Marquette, V.; Zanlonghi, X.; Defoort-Dhellemmes, S.; Puech, B.; et al. High prevalence of PRPH2 in autosomal dominant retinitis pigmentosa in France and characterization of biochemical and clinical features. Am. J. Ophthalmol. 2015, 159, 302–314. [Google Scholar] [CrossRef]
- Antonelli, G.; Parravano, M.; Barbano, L.; Costanzo, E.; Bertelli, M.; Medori, M.C.; Parisi, V.; Ziccardi, L. Multimodal Study of PRPH2 Gene-Related Retinal Phenotypes. Diagnostics 2022, 12, 1851. [Google Scholar] [CrossRef]
- Trujillo, M.J.; Martinez-Gimeno, M.; Giménez, A.; Lorda, I.; Bueno, J.; García-Sandoval, B.; Ramos, C.; Carballo, M.; Ayuso, C. Two novel mutations (Y141H; C214Y) and previously published mutation (R142W) in the RDS-peripherin gene in autosomal dominant macular dystrophies in Spanish families. Hum. Mutat. 2001, 17, 80. [Google Scholar] [CrossRef]
- Hoyng, C.B.; Heutink, P.; Testers, L.; Pinckers, A.; Deutman, A.F.; Oostra, B.A. Autosomal dominant central areolar choroidal dystrophy caused by a mutation in codon 142 in the peripherin/RDS gene. Am. J. Ophthalmol. 1996, 121, 623–629. [Google Scholar] [CrossRef]
- Trujillo, M.J.; Bueno, J.; Osorio, A.; Sanz, R.; Garcia-Sandoval, B.; Ramos, C.; Ayuso, C. Three novel RDS-peripherin mutations (689delT, 857del17, G208D) in Spanish families affected with autosomal dominant retinal degenerations. Human Mutat. 1998, 12, 70. [Google Scholar] [CrossRef]
- Jacobson, S.G.; Cideciyan, A.V.; Maguire, A.M.; Bennett, J.; Sheffield, V.C.; Stone, E.M. Preferential rod and cone photoreceptor abnormalities in heterozygotes with point mutations in the RDS gene. Exp. Eye Res. 1996, 63, 603–608. [Google Scholar] [CrossRef] [PubMed]
- Simonelli, F.; Testa, F.; Marini, V.; Interlandi, E.; Rossi, S.; Pognuz, D.R.; Virgili, G.; Garrè, C.; Bandello, F. Intrafamilial clinical heterogeneity associated with a novel mutation of the retinal degeneration slow/peripherin gene. Ophthalmic Res. 2007, 39, 255–259. [Google Scholar] [CrossRef] [PubMed]
- Testa, F.; Marini, V.; Rossi, S.; Interlandi, E.; Nesti, A.; Rinaldi, M.; Varano, M.; Garré, C.; Simonelli, F. A novel mutation in the RDS gene in an Italian family with pattern dystrophy. Br. J. Ophthalmol. 2005, 89, 1066–1068. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wells, J.; Wroblewski, J.; Keen, J.; Inglehearn, C.; Jubb, C.; Eckstein, A.; Jay, M.; Arden, G.; Bhattacharya, S.; Fitzke, F. Mutations in the human retinal degeneration slow (RDS) gene can cause either retinitis pigmentosa or macular dystrophy. Nat. Genet. 1993, 3, 213–218. [Google Scholar] [CrossRef]
- Sohocki, M.M.; Daiger, S.P.; Bowne, S.J.; Rodriquez, J.A.; Northrup, H.; Heckenlively, J.R.; Birch, D.G.; Mintz-Hittner, H.; Ruiz, R.S.; Lewis, R.A.; et al. Prevalence of mutations causing retinitis pigmentosa and other inherited retinopathies. Hum. Mut. 2001, 17, 42–51. [Google Scholar] [CrossRef]
- Kitiratschky, V.B.; Glöckner, C.J.; Kohl, S. Mutation screening of the GUCA1B gene in patients with autosomal dominant cone and cone rod dystrophy. Ophthalmic Genet. 2011, 32, 151–155. [Google Scholar] [CrossRef] [PubMed]
- Falsini, B.; Placidi, G.; De Siena, E.; Chiurazzi, P.; Minnella, A.M.; Savastano, M.C.; Ziccardi, L.; Parisi, V.; Iarossi, G.; Percio, M.; et al. Genetic characteristics of 234 Italian patients with macular and cone/cone-rod dystrophy. Sci. Rep. 2022, 12, 3774. [Google Scholar] [CrossRef] [PubMed]
- Weisschuh, N.; Obermaier, C.D.; Battke, F.; Bernd, A.; Kuehlewein, L.; Nasser, F.; Zobor, D.; Zrenner, E.; Weber, E.; Wissinger, B.; et al. Genetic architecture of inherited retinal degeneration in Germany: A large cohort study from a single diagnostic center over a 9-year period. Hum. Mut. 2020, 41, 1514–1527. [Google Scholar] [CrossRef]
- Alapati, A.; Goetz, K.; Suk, J.; Navani, M.; Al-Tarouti, A.; Jayasundera, T.; Tumminia, S.J.; Lee, P.; Ayyagari, R. Molecular diagnostic testing by eyeGENE: Analysis of patients with hereditary retinal dystrophy phenotypes involving central vision loss. Investig. Ophthalmol. Vis. Sci. 2014, 55, 5510–5521. [Google Scholar] [CrossRef] [PubMed]
- Yanagihashi, S.; Nakazawa, M.; Kurotaki, J.; Sato, M.; Miyagawa, Y.; Ohguro, H. Autosomal dominant central areolar choroidal dystrophy and a novel Arg195Leu mutation in the peripherin/RDS gene. Arch Ophthalmol. 2003, 121, 1458–1461. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Maggi, J.; Koller, S.; Bähr, L.; Feil, S.; Kivrak Pfiffner, F.; Hanson, J.V.M.; Maspoli, A.; Gerth-Kahlert, C.; Berger, W. Long-Range PCR-Based NGS Applications to Diagnose Mendelian Retinal Diseases. Int. J. Mol. Sci. 2021, 22, 1508. [Google Scholar] [CrossRef] [PubMed]
- Kohl, S.; Christ-Adler, M.; Apfelstedt-Sylla, E.; Kellner, U.; Eckstein, A.; Zrenner, E.; Wissinger, B. RDS/peripherin gene mutations are frequent causes of central retinal dystrophies. J. Med. Genet. 1997, 34, 620–626. [Google Scholar] [CrossRef]
- Coco, R.M.; Tellería, J.J.; Sanabria, M.R.; Rodríguez-Rúa, E.; García, M.T. PRPH2 (Peripherin/RDS) mutations associated with different macular dystrophies in a Spanish population: A new mutation. Eur. J. Ophthalmol. 2021, 20, 724–732. [Google Scholar] [CrossRef]
- Kemp, C.M.; Jacobson, S.G.; Cideciyan, A.V.; Kimura, A.E.; Sheffield, V.C.; Stone, E.M. RDS gene mutations causing retinitis pigmentosa or macular degeneration lead to the same abnormality in photoreceptor function. Invest Ophthalmol Vis Sci. 1994, 35, 3154–3162. [Google Scholar] [PubMed]
- Farrar, G.J.; Kenna, P.; Jordan, S.A.; Kumar-Singh, R.; Humphries, M.M.; Sharp, E.M.; Sheils, D.; Humphries, P. Autosomal dominant retinitis pigmentosa: A novel mutation at the peripherin/RDS locus in the original 6p-linked pedigree. Genomics 1993, 15, 466. [Google Scholar] [PubMed]
- Payne, A.M.; Downes, S.M.; Bessant, D.A.; Bird, A.C.; Bhattacharya, S.S. Founder effect, seen in the British population, of the 172 peripherin/RDS mutation-and further refinement of genetic positioning of the peripherin/RDS gene. Am. J. Hum. Genet. 1998, 62, 192–195. [Google Scholar] [CrossRef]
- Villaverde, C.; Trujillo-Tiebas, M.J.; Gallego-Merlo, J.; Vallespin, E.; Cantalapiedra, D.; Riveiro-Alvarez, R.; Carballo, M.; Ayuso, C. Novel human pathological mutations. Gene symbol: RDS. Disease: Macular dystrophy. Hum. Genet. 2007, 122, 555. [Google Scholar] [PubMed]
- Fishman, G.A.; Stone, E.; Gilbert, L.D.; Vandenburgh, K.; Sheffield, V.C.; Heckenlively, J.R. Clinical features of a previously undescribed codon 216 (proline to serine) mutation in the peripherin/retinal degeneration slow gene in autosomal dominant retinitis pigmentosa. Ophthalmology 1994, 101, 1409–1421. [Google Scholar] [CrossRef]
- Reeves, M.J.; Goetz, K.E.; Guan, B.; Ullah, E.; Blain, D.; Zein, W.M.; Tumminia, S.J.; Hufnagel, R.B. Genotype–phenotype associations in a large PRPH2-related retinopathy cohort. Hum. Mutat. 2020, 41, 1528–1539. [Google Scholar] [CrossRef]
- Boon, C.J.; van Schooneveld, M.J.; den Hollander, A.I.; van Lith-Verhoeven, J.J.; Zonneveld-Vrieling, M.N.; Theelen, T.; Cremers, F.P.; Hoyng, C.B.; Klevering, B.J. Mutations in the peripherin/RDS gene are an important cause of multifocal pattern dystrophy simulating STGD1/fundus flavimaculatus. Br. J. Ophthalmol. 2007, 91, 1504–1511. [Google Scholar] [CrossRef]
- Strom, S.P.; Gao, Y.Q.; Martinez, A.; Ortube, C.; Chen, Z.; Nelson, S.F.; Nusinowitz, S.; Farber, D.B.; Gorin, M.B. Molecular diagnosis of putative Stargardt Disease probands by exome sequencing. BMC Med. Genet. 2012, 13, 67. [Google Scholar] [CrossRef]
- Felbor, U.; Schilling, H.; Weber, B.H. Adult vitelliform macular dystrophy is frequently associated with mutations in the peripherin/RDS gene. Human Mut. 1997, 10, 301–309. [Google Scholar] [CrossRef]
- Diñeiro, M.; Capín, R.; Cifuentes, G.Á.; Fernández-Vega, B.; Villota, E.; Otero, A.; Santiago, A.; Pruneda, P.C.; Castillo, D.; Viejo-Díaz, M.; et al. Comprehensive genomic diagnosis of inherited retinal and optical nerve disorders reveals hidden syndromes and personalized therapeutic options. Acta Ophthalmol. 2020, 98, e1034–e1048. [Google Scholar] [CrossRef]
- Renner, A.B.; Fiebig, B.S.; Weber, B.H.; Wissinger, B.; Andreasson, S.; Gal, A.; Cropp, E.; Kohl, S.; Kellner, U. Phenotypic variability and long-term follow-up of patients with known and novel PRPH2/RDS gene mutations. Am. J. Ophthalmol. 2009, 147, 518–530.e1. [Google Scholar] [CrossRef]
- Soucy, M.; Kolesnikova, M.; Kim, A.H.; Tsang, S.H. Phenotypic variability in PRPH2 as demonstrated by a family with incomplete penetrance of autosomal dominant cone-rod dystrophy. Clin. Case Rep. 2023, 146, 267–272. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, J.; Jiang, Y.; Zhu, D.; Ouyang, J.; Yi, Z.; Li, S.; Jia, X.; Xiao, X.; Sun, W.; et al. New Insight into the Genotype-Phenotype Correlation of PRPH2-Related Diseases Based on a Large Chinese Cohort and Literature Review. Int. J. Mol. Sci. 2023, 24, 6728. [Google Scholar] [CrossRef]
- Gamundi, M.J.; Hernan, I.; Muntanyola, M.; Trujillo, M.J.; García-Sandoval, B.; Ayuso, C.; Baiget, M.; Carballo, M. High prevalence of mutations in peripherin/RDS in autosomal dominant macular dystrophies in a Spanish population. Mol. Vis. 2007, 13, 1031–1037. [Google Scholar] [PubMed]
- Birtel, J.; Eisenberger, T.; Gliem, M.; Müller, P.L.; Herrmann, P.; Betz, C.; Zahnleiter, D.; Neuhaus, C.; Lenzner, S.; Holz, F.G.; et al. Clinical and genetic characteristics of 251 consecutive patients with macular and cone/cone-rod dystrophy. Sci. Rep. 2018, 8, 4824. [Google Scholar] [CrossRef] [PubMed]
- Oishi, A.; Fujinami, K.; Mawatari, G.; Naoi, N.; Ikeda, Y.; Ueno, S.; Kuniyoshi, K.; Hayashi, T.; Kondo, H.; Mizota, A.; et al. Genetic and Phenotypic Landscape of PRPH2-Associated Retinal Dystrophy in Japan. Genes 2021, 12, 1817. [Google Scholar] [CrossRef] [PubMed]
- Michaelides, M.; Holder, G.E.; Bradshaw, K.; Hunt, D.M.; Moore, A.T. Cone-rod dystrophy, intrafamilial variability, and incomplete penetrance associated with the R172W mutation in the peripherin/RDS gene. Ophthalmology 2005, 112, 1592–1598. [Google Scholar] [CrossRef] [PubMed]
- Ekström, U.; Ponjavic, V.; Andréasson, S.; Ehinger, B.; Nilsson-Ehle, P.; Abrahamson, M. Detection of alterations in all three exons of the peripherin/RDS gene in Swedish patients with retinitis pigmentosa using an efficient DGGE system. Mol. Pathol. 1998, 51, 287–291. [Google Scholar] [CrossRef]
- Zulliger, R.; Conley, S.M.; Mwoyosvi, M.L.; Al-Ubaidi, M.R.; Naash, M.I. Oligomerization of Prph2 and Rom1 is essential for photoreceptor outer segment formation. Hum. Mol. Genet. 2018, 27, 3507–3518. [Google Scholar] [CrossRef] [PubMed]
- Conley, S.M.; Stuck, M.W.; Watson, J.N.; Zulliger, R.; Burnett, J.L.; Naash, M.I. Prph2 initiates outer segment morphogenesis but maturation requires Prph2/Rom1 oligomerization. Hum. Mol. Genet. 2019, 28, 459–475. [Google Scholar] [CrossRef]
- Goldberg, A.F.; Loewen, C.J.; Molday, R.S. Cysteine residues of photoreceptor peripherin/rds: Role in subunit assembly and autosomal dominant retinitis pigmentosa. Biochemistry 1998, 37, 680–685. [Google Scholar] [CrossRef]
- Ikelle, L.; Makia, M.; Lewis, T.; Crane, R.; Kakakhel, M.; Conley, S.M.; Birtley, J.R.; Arshavsky, V.Y.; Al-Ubaidi, M.R.; Naash, M.I. Comparative study of PRPH2 D2 loop mutants reveals divergent disease mechanism in rods and cones. Cell Mol. Life Sci. 2023, 80, 214. [Google Scholar] [CrossRef]
- Wang, X.; Wang, H.; Sun, V.; Tuan, H.F.; Keser, V.; Wang, K.; Ren, H.; Lopez, I.; Zaneveld, J.E.; Siddiqui, S.; et al. Comprehensive molecular diagnosis of 179 Leber congenital amaurosis and juvenile retinitis pigmentosa patients by targeted next generation sequencing. J. Med. Genet. 2013, 50, 674–688. [Google Scholar] [CrossRef] [PubMed]
- Bandah-Rozenfeld, D.; Collin, R.W.; Banin, E.; van den Born, L.I.; Coene, K.L.; Siemiatkowska, A.M.; Zelinger, L.; Khan, M.I.; Lefeber, D.J.; Erdinest, I.; et al. Mutations in IMPG2, encoding interphotoreceptor matrix proteoglycan 2, cause autosomal-recessive retinitis pigmentosa. Am. J. Hum. Genet. 2010, 87, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. Off. J. Am. Coll. Med. Genet. 2015, 17, 405–424. [Google Scholar] [CrossRef] [PubMed]
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.; Thormann, A.; Flicek, P.; Cunningham, F. The Ensembl Variant Effect Predictor. Genome Biol. 2016, 17, 122. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, C.H.M.; Pires, D.E.V.; Ascher, D.B. DynaMut2: Assessing changes in stability and flexibility upon single and multiple point missense mutations. Protein Sci. Publ. Protein Soc. 2021, 30, 60–69. [Google Scholar] [CrossRef]
- Rios, D.; McLaren, W.M.; Chen, Y.; Birney, E.; Stabenau, A.; Flicek, P.; Cunningham, F. A database and API for variation, dense genotyping and resequencing data. BMC Bioinform. 2010, 11, 238. [Google Scholar] [CrossRef]
| Patients’ Characteristics | Non-RP | RP | A | |
|---|---|---|---|---|
| MD | CD/CRD | |||
| TOTAL (%) | 103 (57) | 28 (15) | 35 (19) | 16 (9) |
| Male (no. total) | 43 | 14 | 11 | 6 |
| Female (no. total)c | 60 | 14 | 24 | 9 |
| AAO (no. patients with data/total) | 51/103 | 16/28 | 24/35 | - |
| Median | 40 | 41 | 24.5 | - |
| Range | 8–65 | 5–62 | 8–74 | - |
| VAL, no. patients with data | 41 | 13 | 16 | - |
| Median | 40 | 42 | 36.5 | - |
| Range, years | 8–65 | 5–62 | 12–74 | - |
| VFL, no. patients with data | 13 | 10 | 19 | - |
| Median | 45 | 46.5 | 35 | - |
| Range, years. | 25–60 | 5–65 | 12–74 | - |
| NB, no. patients with data | 16 | 11 | 19 | - |
| Median | 40.5 | 35 | 24 | - |
| Range, years. | 20–65 | 25–62 | 8–51 | - |
| Exon | Nucleotide Change | Protein Change | Protein Domain | ACMG Classification | Type of Variant | Allele Count (%) | Het. | Homo. | Double Diagnosis | References |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | c.52C>T | p.Gln18* | N-terminus | LP (PVS1, PM2) | nonsense | 4 (3.85) | 4 | 0 | 0 | Del Pozo-Valero, 2022 [20] |
| 1 | c.112G>A | p.Gly38Arg | 1st TMD | LP (PM2, PP1, PP2, PP3) | missense | 1 (0.96) | 1 | 0 | 0 | This study |
| 1 | c.113G>A | p.Gly38Glu | 1st TMD | LP (PM2, PP2, PP3) | missense | 2 (1.92) | 2 | 0 | 0 | This study |
| 1 | c.122T>C | p.Leu41Pro | 1st TMD | LP (PM2, PP3, PP2) | missense | 5 (4.81) | 5 | 0 | 0 | Peeters, 2021 [16]; Bianco, 2023 [21] |
| 1 | c.136C>T | p.Arg46* | D1-loop | P (PVS1, PM2, PP5) | nonsense | 2 (1.92) | 2 | 0 | 0 | Meins, 1993 [22] |
| 1 | c.205del | p.Val69Cysfs*30 | 2nd TMD | P (PVS1, PM2, PP5) | frameshift | 1 (0.96) | 1 | 0 | 0 | Manes, 2015 [23] |
| 1 | c.290G>A | p.Trp97* | C-loop | P (PVS1, PM2, PP5) | nonsense | 1 (0.96) | 1 | 0 | 0 | Antonelli, 2022 [24] |
| 1 | c.421T>C | p.Tyr141His | D2-loop | P (PM1, PM2, PM5, PP2, PP3, PP5) | missense | 1 (0.96) | 1 | 0 | 0 | Trujillo, 2001 [25] |
| 1 | c.424C>T | p.Arg142Trp | D2-loop | LP (PM1, PM2, PM5, PP2, PP5) | missense | 4 (3.85) | 4 | 0 | 0 | Hoyng, 1996 [26] |
| 1 | c.441del | p.Gly148Alafs*5 | D2-loop | P (PVS1, PM2, PP5) | frameshift | 1 (0.96) | 1 | 0 | 0 | Trujillo, 1998 [27] |
| 1 | c.469G>A | p.Asp157Asn | D2-loop | P (PM1, PM2, PM5, PP2, PP3, PP5) | missense | 1 (0.96) | 1 | 0 | 0 | Jacobson, 1996 [28] |
| 1 | c.493T>C | p.Cys165Arg | D2-loop | P (PM1, PM2, PM5, PP2, PP3, PP5) | missense | 1 (0.96) | 1 | 0 | 0 | Simonelli, 2007 [29] |
| 1 | c.499G>A | p.Gly167Ser | D2-loop | P (PM1, PM2, PM5, PP2, PP3, PP5) | missense | 7 (6.73) | 7 | 0 | 0 | Testa, 2005 [30] |
| 1 | c.514C>T | p.Arg172Trp | D2-loop | P (PM2, PM5, PP3, PP2, PP5) | missense | 2 (1.92) | 2 | 0 | 0 | Wells, 1993 [31] |
| 1 | c.515G>A | p.Arg172Gln | D2-loop | LP (PM1, PM2, PM5, PP2, PP5) | missense | 2 (1.92) | 2 | 0 | 0 | Wells, 1993 [31] |
| 1 | c.520T>A | p.Trp174Arg | D2-loop | P (PM1, PM2, PM5, PP2, PP3, PP5) | missense | 1 (0.96) | 1 | 0 | 0 | Peeters, 2021 [16] |
| 1 | c.533A>G | p.Gln178Arg | D2-loop | LP (PM1, PM2, PP2, PP3, PP5) | missense | 1 (0.96) | 1 | 0 | 0 | Sohocki, 2001 [32] |
| 1 | c.536G>T | p.Trp179Leu | D2-loop | P (PM1, PM2, PM5, PP2, PP3, PP5) | missense | 1 (0.96) | 1 | 0 | 0 | Fernandez-San Jose, 2015 [18] |
| 1 | c.556G>A | p.Asp186Asn | D2-loop | LP (PM1, PM2, PM5, PP2, PP5) | missense | 1 (0.96) | 1 | 0 | 0 | Kitiratschky, 2011 [33] |
| 1 | c.562del | p.Ser188Profs*68 | D2-loop | LP (PVS1, PM2) | frameshift | 4 (3.85) | 4 | 0 | 0 | This study |
| 1 | c.567dupC | p.Lys190Glnfs*28 | D2-loop | LP (PVS1, PM2) | frameshift | 1 (0.96) | 1 | 0 | 0 | This study |
| 1 | c.568A>G | p.Lys190Glu | D2-loop | LP (PM1, PM2, PP2) | missense | 1 (0.96) | 1 | 0 | 0 | Falsini, 2022 [34] |
| IVS1 | c.582-1G>A | p.? | - | P (PVS1, PM2, PP5) | splicing | 2 (1.92) | 2 | 0 | 0 | Fernandez-San Jose, 2015 [18] |
| 2 | c.(581+1_582-1)_(828+1_829-1)del | p.? | D2-loop to 4th TMD | LP (PVS1, PM2) | CNV | 1 (0.96) | 1 | 0 | 0 | Weisschuh, 2020 [35] |
| 2 | c.584G>A | p.Arg195Gln | D2-loop | P (PM1, PM2, PM5, PP2, PP3, PP5) | missense | 1 (0.96) | 1 | 0 | 0 | Alapati, 2014 [36] |
| 2 | c.584G>T | p.Arg195Leu | D2-loop | P (PM1, PM2, PM5, PP1, PP2, PP5) | missense | 4 (3.85) | 4 | 0 | 0 | Yanagihashi, 2003 [37] |
| 2 | c.605G>A | p.Gly202Glu | D2-loop | LP (PM1, PM2, PP2, PP5) | missense | 1 (0.96) | 1 | 0 | 0 | Maggi, 2021 [38] |
| 2 | c.609_625del | p.Tyr204Profs*8 | D2-loop | P (PVS1, PM2, PP5) | frameshift | 1 (0.96) | 1 | 0 | 0 | Trujillo, 1998 [27] |
| 2 | c.623del | p.Gly208Alafs*48 | D2-loop | LP (PVS1, PM2) | frameshift | 1 (0.96) | 1 | 0 | 0 | This study |
| 2 | c.623G>A | p.Gly208Asp | D2-loop | P (PM1, PM2, PM5, PP2, PP3, PP5, PS4) | missense | 7 (6.73) | 4 | 2 | 1 | Kohl, 1997 [39] |
| 2 | c.625G>A | p.Val209Ile | D2-loop | LP (PM1, PM2, PM5, PP2, PP5) | missense | 1 (0.96) | 1 | 0 | 0 | Coco, 2010 [40] |
| 2 | c.626del | p.Val209Alafs*47 | D2-loop | P (PVS1, PM2, PP5) | frameshift | 1 (0.96) | 1 | 0 | 0 | Martin-Merida, 2019 [19] |
| 2 | c.628C>T | p.Pro210Ser | D2-loop | P (PM1, PM2, PM5, PP2, PP3, PP5) | missense | 1 (0.96) | 1 | 0 | 0 | Kemp, 1994 [41] |
| 2 | c.631T>C | p.Phe211Leu | D2-loop | LP (PM1, PM2, PP2, PP3, PP5, PS1) | missense | 1 (0.96) | 1 | 0 | 0 | Manes, 2015 [23] |
| 2 | c.633_656del | p.Phe211_Pro219delinsLeu | D2-loop | LP (PM1, PM2, PM4) | inframe | 1 (0.96) | 1 | 0 | 0 | This study |
| 2 | c.634A>G | p.Ser212Gly | D2-loop | P (PM1, PM2, PM5, PP2, PP3, PP5, PS4) | missense | 4 (3.85) | 4 | 0 | 0 | Farrar, 1992 [42] |
| 2 | c.637T>C | p.Cys213Arg | D2-loop | P (PM1, PM2, PM5, PP2, PP3, PP5) | missense | 1 (0.96) | 1 | 0 | 0 | Payne, 1998 [43] |
| 2 | c.637T>G | p.Cys213Gly | D2-loop | LP (PM1, PM2, PM5, PP2, PP3) | missense | 1 (0.96) | 1 | 0 | 0 | This study |
| 2 | c.638G>T | p.Cys213Phe | D2-loop | P (PM1, PM2, PM5, PP2, PP3, PP5) | missense | 2 (1.92) | 2 | 0 | 0 | Villaverde, 2007 [44] |
| 2 | c.641G>A | p.Cys214Tyr | D2-loop | P (PM1, PM2, PM5, PP2, PP3, PP5) | missense | 2 (1.92) | 2 | 0 | 0 | Trujillo, 2001 [25] |
| 2 | c.643A>T | p.Asn215Tyr | D2-loop | P (PM1, PM2, PM5, PP2, PP3, PP5) | missense | 1 (0.96) | 1 | 0 | 0 | Martin-Merida, 2018 [17] |
| 2 | c.646C>T | p.Pro216Ser | D2-loop | P (PM1, PM2, PM5, PP2, PP3, PP5) | missense | 1 (0.96) | 1 | 0 | 0 | Fishman, 1994 [45] |
| 2 | c.649_650insTAGCTGCTGCAATCCTA | p.Ser217Ilefs*45 | D2-loop | LP (PVS1, PM2) | frameshift | 3 (2.91) | 3 | 0 | 0 | This study |
| 2 | c.653C>A | p.Ser218* | D2-loop | P (PVS1, PM2, PP5) | nonsense | 1 (0.96) | 1 | 0 | 0 | Reeves, 2020 [46] |
| 2 | c.658C>G | p.Arg220Gly | D2-loop | P (PM1, PM2, PM5, PP2, PP3, PP5) | missense | 1 (0.96) | 1 | 0 | 0 | ClinVar |
| 2 | c.658C>T | p.Arg220Trp | D2-loop | P (PM1, PM2, PM5, PP2, PP3, PP5) | missense | 3 (2.91) | 3 | 0 | 0 | Payne, 1998 [43] |
| 2 | c.658del | p.Arg220Glyfs36* | D2-loop | P (PVS1, PM2, PP5) | frameshift | 1 (0.96) | 1 | 0 | 0 | Boon, 2007 [47] |
| 2 | c.660_665del | p.Pro221_Cys222del | D2-loop | LP (PM1, PM2, PM4, PP5) | inframe | 6 (5.77) | 6 | 0 | 0 | Martin-Merida, 2019 [19] |
| 2 | c.708C>A | p.Tyr236* | D2-loop | LP (PVS1, PM2) | nonsense | 1 (0.96) | 1 | 0 | 0 | LOVD |
| 2 | c.708C>G | p.Tyr236* | D2-loop | P (PVS1, PM2, PP5) | nonsense | 1 (0.96) | 1 | 0 | 0 | Strom, 2012 [48] |
| 2 | c.734_737dupTGTG | p.Trp246Cysfs*56 | D2-loop | LP (PVS1, PM2) | frameshift | 1 (0.96) | 1 | 0 | 0 | Del Pozo-Valero, 2022 [20] |
| 2 | c.745G>C | p.Gly249Arg | D2-loop | LP (PM1, PM2, PM5, PP1, PP2, PP3) | missense | 1 (0.96) | 1 | 0 | 0 | This study |
| 2 | c.809_817delinsCCTTCGAGGTA | p.Leu270Profs*9 | 4th TMD | LP (PVS1, PM2) | frameshift | 1 (0.96) | 1 | 0 | 0 | This study |
| 2 | c.818G>A | p.Trp273* | 4th TMD | LP (PVS1, PM2, PP1) | nonsense | 1 (0.96) | 1 | 0 | 0 | This study |
| 3 | c.904G>A | p.Glu302Lys | C-terminus | VUS (PM2, PP2, BP4) | missense | 1 (0.96) | 1 | 0 | 0 | ClinVar |
| 3 | c.914G>A | p.Gly305Asp | C-terminus | LP (PM2, PP2, PP5) | missense | 2 (1.92) | 2 | 0 | 0 | Felbor, 1997 [49] |
| Variant | Chinese Cohort (Wang, 2023) [53] (n = 15) | Japanese Cohort (Oishi, 2021) [56] (n = 30) | USA Cohort (Reeves, 2020) [46] (n = 161) | Spanish Cohort (This Study) (n = 103) |
|---|---|---|---|---|
| c.122T>C (p.Leu41Pro) | Not reported | Not reported | Not reported | 4.8% |
| c.424C>T (p.Arg142Trp) | Not reported | 13.3% | 3.1% | 3.9% |
| c.499G>A (p.Gly167Ser) | Not reported | 6.7% | 0.6% | 6.7% |
| c.514C>T (p.Arg172Trp) | 6.7% | 13.3% | 5.6% | 1.9% |
| c.599T>A (p.Val200Glu) | Not reported | 10.0% | Not reported | Not found |
| c.623G>A (p.Gly208Asp) | Not reported | Not reported | 0.6% | 6.7% |
| c.660_665del (p.Pro221_Cys222del) | Not reported | Not reported | Not reported | 5.8% |
| c.828+3A>T (p.?) | Not reported | Not reported | 17.4% | Not found |
| c.914G>A (p.Gly305Alafs*19) | 33.3% | Not reported | Not reported | 1.9% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernández-Caballero, L.; Martín-Merida, I.; Blanco-Kelly, F.; Avila-Fernandez, A.; Carreño, E.; Fernandez-San Jose, P.; Irigoyen, C.; Jimenez-Rolando, B.; Lopez-Grondona, F.; Mahillo, I.; et al. PRPH2-Related Retinal Dystrophies: Mutational Spectrum in 103 Families from a Spanish Cohort. Int. J. Mol. Sci. 2024, 25, 2913. https://doi.org/10.3390/ijms25052913
Fernández-Caballero L, Martín-Merida I, Blanco-Kelly F, Avila-Fernandez A, Carreño E, Fernandez-San Jose P, Irigoyen C, Jimenez-Rolando B, Lopez-Grondona F, Mahillo I, et al. PRPH2-Related Retinal Dystrophies: Mutational Spectrum in 103 Families from a Spanish Cohort. International Journal of Molecular Sciences. 2024; 25(5):2913. https://doi.org/10.3390/ijms25052913
Chicago/Turabian StyleFernández-Caballero, Lidia, Inmaculada Martín-Merida, Fiona Blanco-Kelly, Almudena Avila-Fernandez, Ester Carreño, Patricia Fernandez-San Jose, Cristina Irigoyen, Belen Jimenez-Rolando, Fermina Lopez-Grondona, Ignacio Mahillo, and et al. 2024. "PRPH2-Related Retinal Dystrophies: Mutational Spectrum in 103 Families from a Spanish Cohort" International Journal of Molecular Sciences 25, no. 5: 2913. https://doi.org/10.3390/ijms25052913
APA StyleFernández-Caballero, L., Martín-Merida, I., Blanco-Kelly, F., Avila-Fernandez, A., Carreño, E., Fernandez-San Jose, P., Irigoyen, C., Jimenez-Rolando, B., Lopez-Grondona, F., Mahillo, I., Martin-Gutierrez, M. P., Minguez, P., Perea-Romero, I., Del Pozo-Valero, M., Riveiro-Alvarez, R., Rodilla, C., Rodriguez-Peña, L., Sánchez-Barbero, A. I., Swafiri, S. T., ... Ayuso, C. (2024). PRPH2-Related Retinal Dystrophies: Mutational Spectrum in 103 Families from a Spanish Cohort. International Journal of Molecular Sciences, 25(5), 2913. https://doi.org/10.3390/ijms25052913








