Functional Characterization of Endo- and Exo-Hydrolase Genes in Arabinan Degradation Gene Cluster of Bifidobacterium longum subsp. suis
Abstract
1. Introduction
2. Results
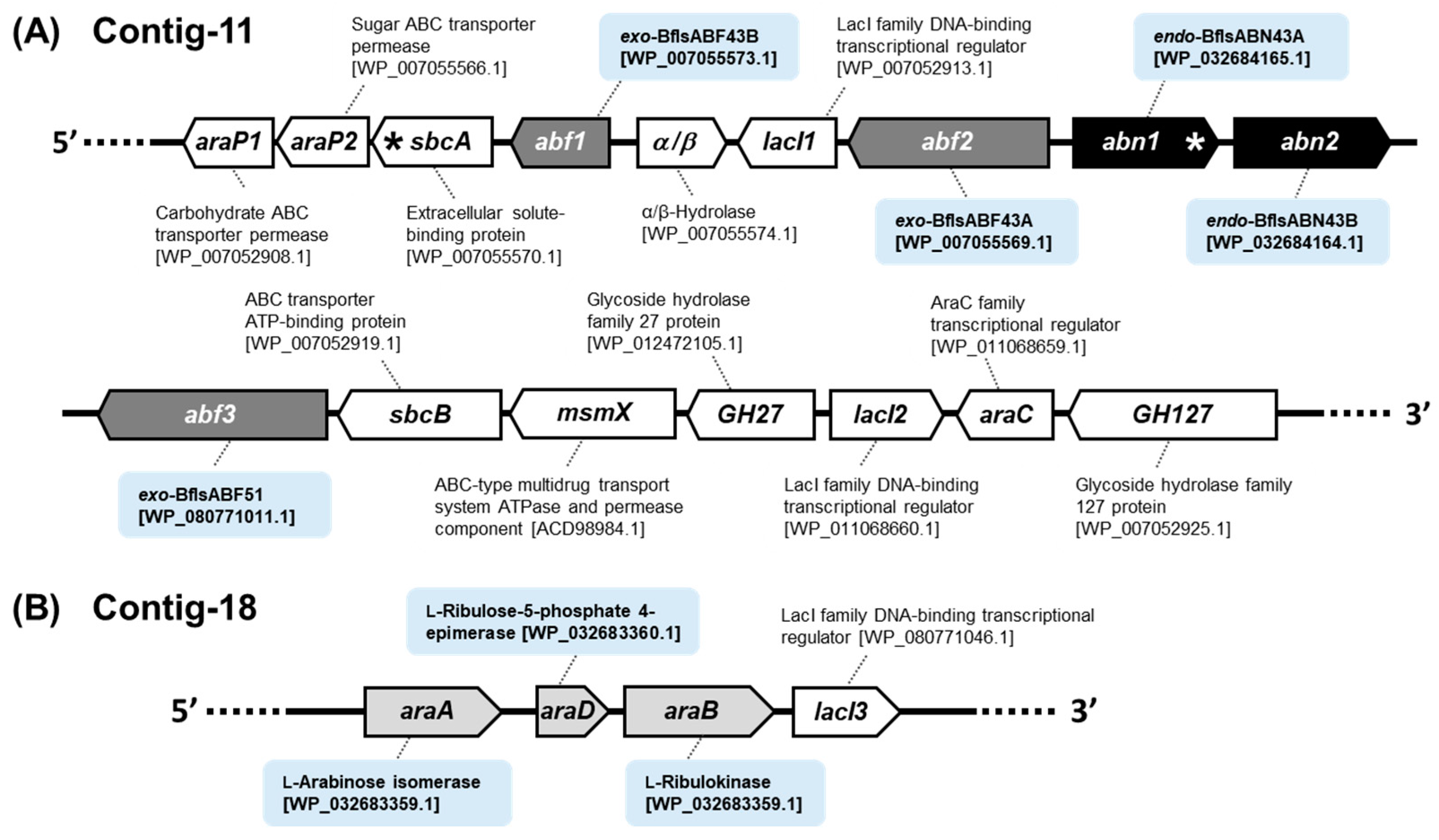
2.1. Gene Cluster Analysis for Arabinan Utilization in Bf. longum subsp. suis
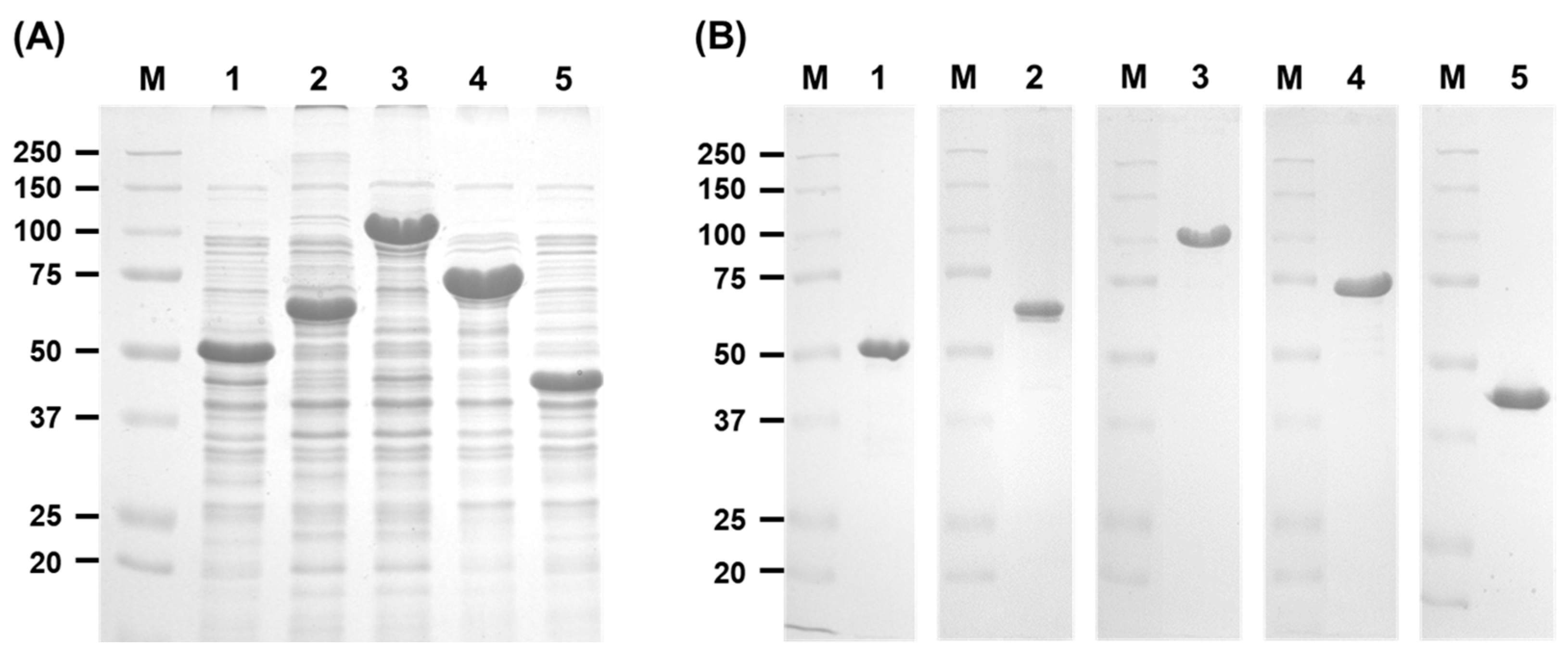
2.2. Gene Expression and Enzyme Purification of Arabinan Hydrolases
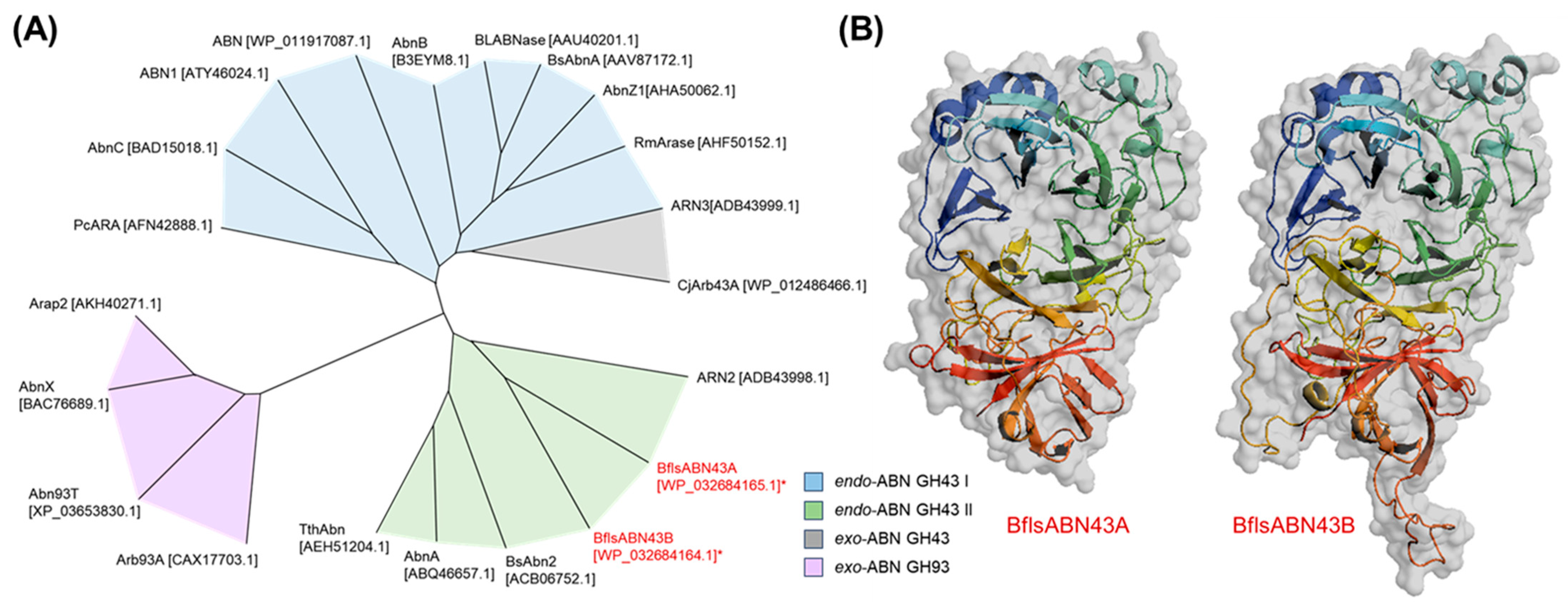
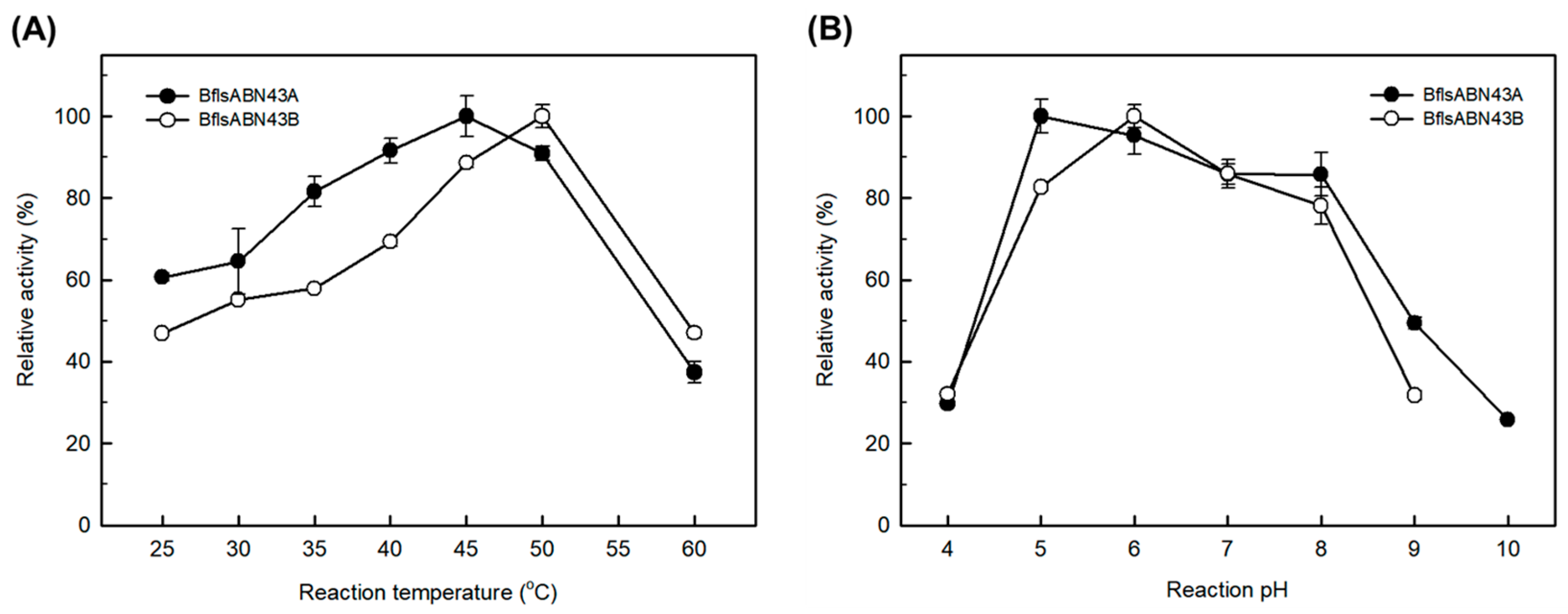
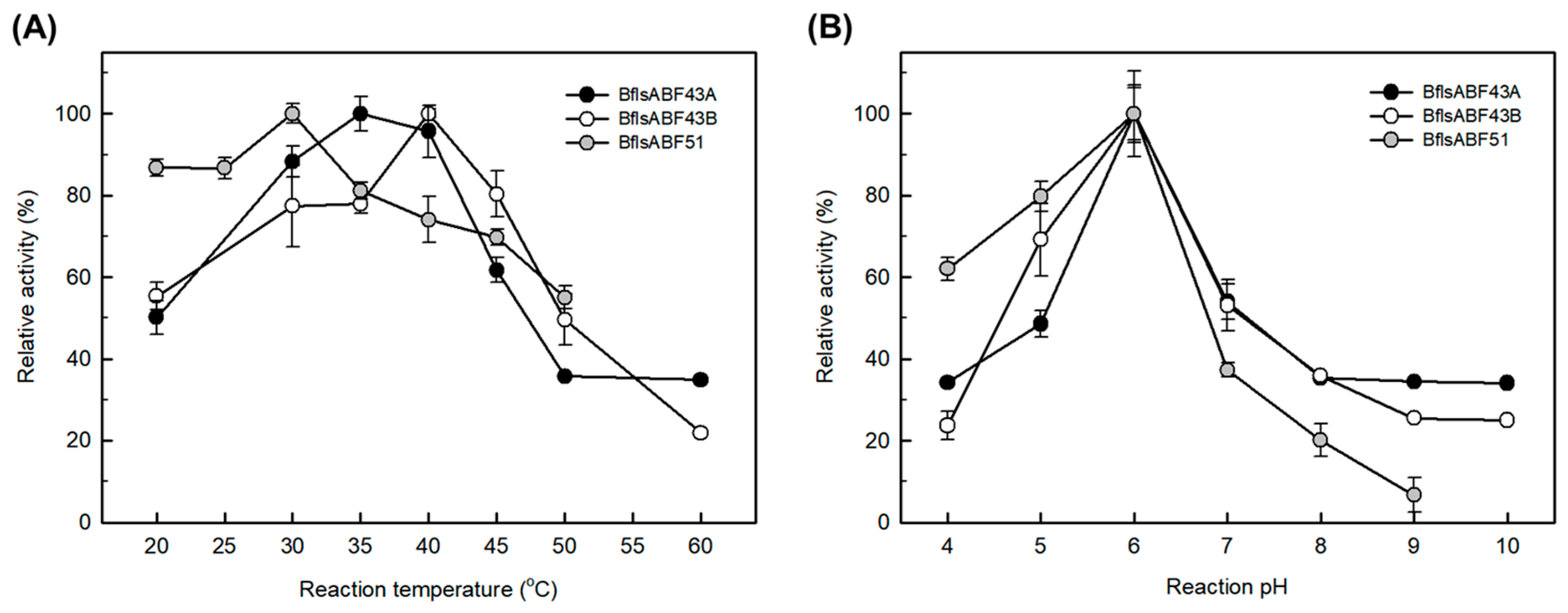
2.3. Enzymatic Characterization of Arabinan Hydrolases
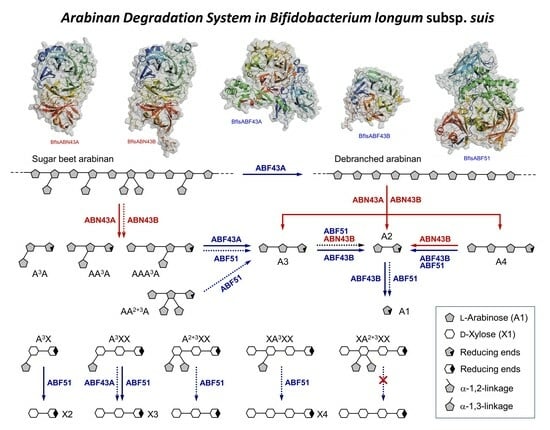
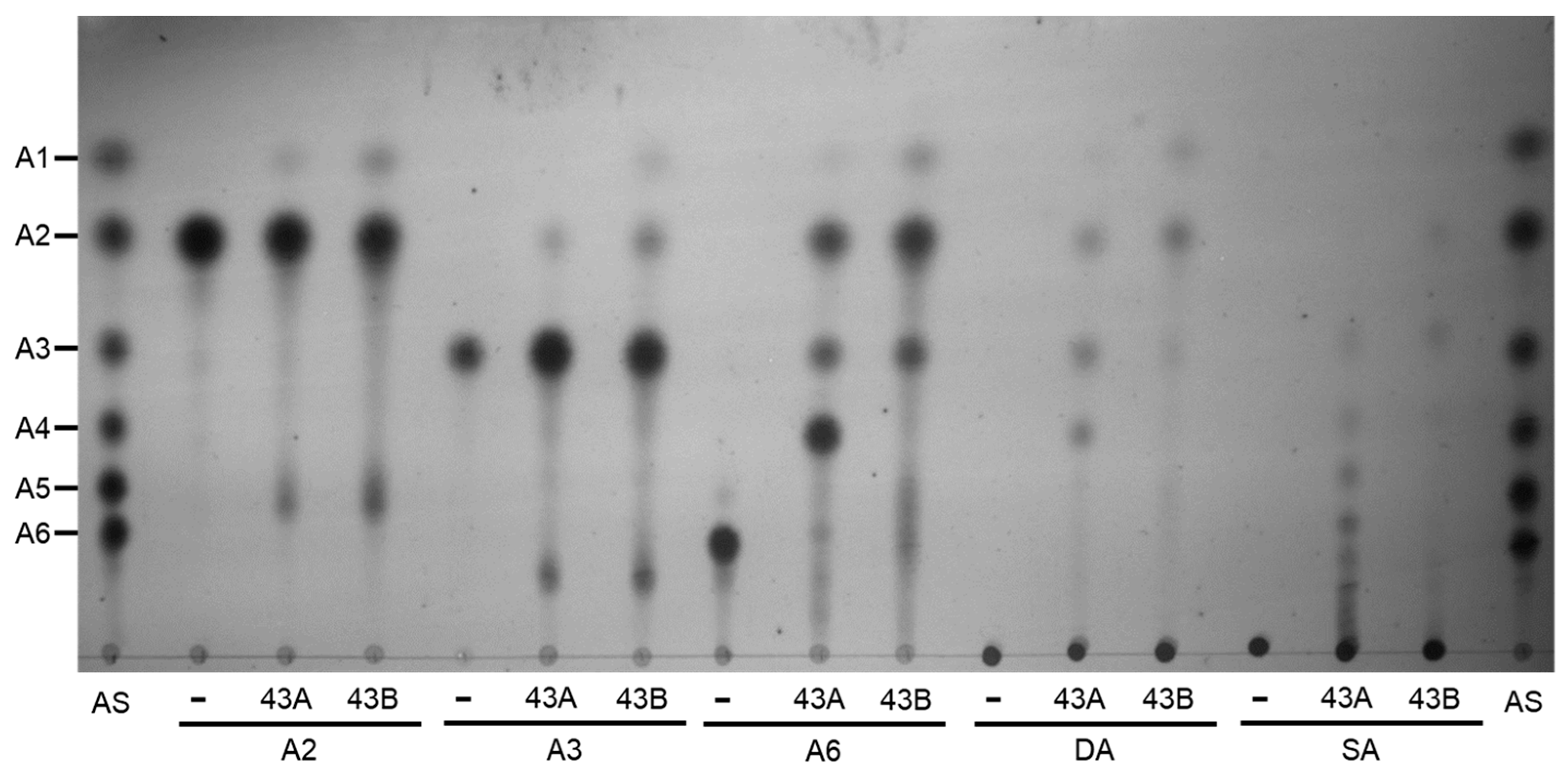
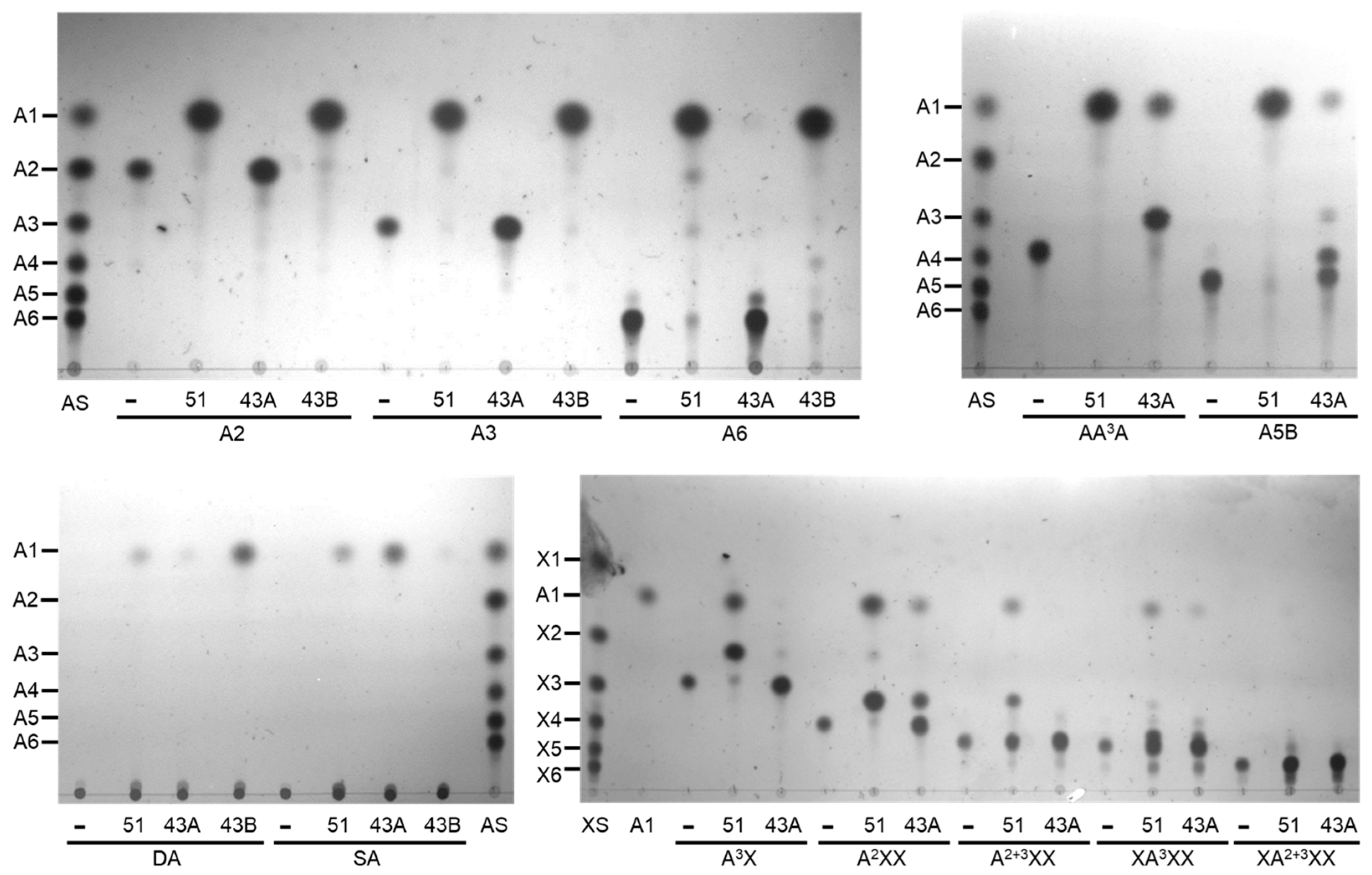
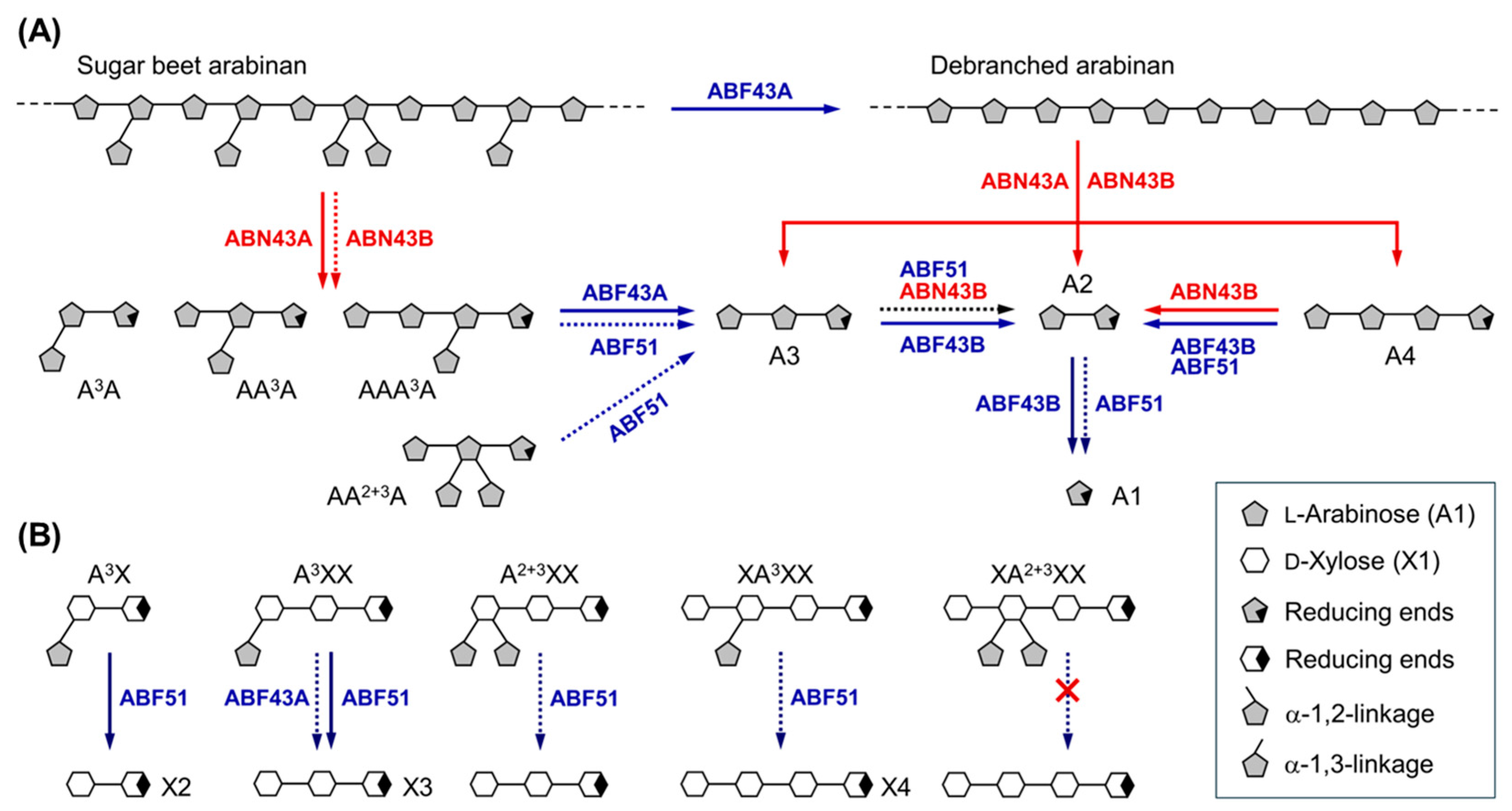
2.4. Hydrolytic Modes of Action of Arabinan Hydrolases
2.4.1. endo-α-1,5-l-Arabinanases (BflsABNs)
2.4.2. α-l-Arabinofuranosidases (BflsABFs)
3. Discussion
4. Materials and Methods
4.1. Microorganisms and Plasmids
4.2. Enzymes and Chemical Reagents
4.3. Gene Cloning of Arabinan Hydrolases
4.4. Gene Expression and Enzyme Purification
4.5. Enzyme Activity Assays
4.5.1. DNS (3,5-Dinitrosalicylic Acid) Reducing Sugar Assay
4.5.2. l-Arabinose Assay
4.6. Thin-Layer Chromatography (TLC) Analysis
4.7. Three-Dimensional Structure Modeling
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hidalgo-Cantabrana, C.; Delgado, S.; Ruiz, L.; Ruas-Madiedo, P.; Sanchez, B.; Margolles, A. Bifidobacteria and their health-promoting effects. Microbiol. Spectr. 2017, 5. [Google Scholar] [CrossRef]
- Gibson, G.R.; Hutkins, R.; Sanders, M.E.; Prescott, S.L.; Reimer, R.A.; Salminen, S.J.; Scott, K.; Stanton, C.; Swanson, K.S.; Cani, P.D.; et al. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 491–502. [Google Scholar] [CrossRef] [PubMed]
- Markowiak, P.; Slizewska, K. Effects of probiotics, prebiotics, and synbiotics on human health. Nutrients 2017, 9, 1021. [Google Scholar] [CrossRef] [PubMed]
- Al-Tamimi, M.A.; Palframan, R.J.; Cooper, J.M.; Gibson, G.R.; Rastall, R.A. In vitro fermentation of sugar beet arabinan and arabino-oligosaccharides by the human gut microflora. J. Appl. Microbiol. 2006, 100, 407–414. [Google Scholar] [CrossRef] [PubMed]
- Jana, U.K.; Kango, N.; Pletschke, B. Hemicellulose-derived oligosaccharides: Emerging prebiotics in disease alleviation. Front. Nutr. 2021, 8, 670817. [Google Scholar] [CrossRef] [PubMed]
- Moon, J.S.; Shin, S.Y.; Choi, H.S.; Joo, W.; Cho, S.K.; Li, L.; Kang, J.H.; Kim, T.J.; Han, N.S. In vitro digestion and fermentation properties of linear sugar-beet arabinan and its oligosaccharides. Carbohydr. Polym. 2015, 131, 50–56. [Google Scholar] [CrossRef] [PubMed]
- Sulek, K.; Vigsnaes, L.K.; Schmidt, L.R.; Holck, J.; Frandsen, H.L.; Smedsgaard, J.; Skov, T.H.; Meyer, A.S.; Licht, T.R. A combined metabolomic and phylogenetic study reveals putatively prebiotic effects of high molecular weight arabino-oligosaccharides when assessed by in vitro fermentation in bacterial communities derived from humans. Anaerobe 2014, 28, 68–77. [Google Scholar] [CrossRef] [PubMed]
- Vigsnaes, L.K.; Holck, J.; Meyer, A.S.; Licht, T.R. In vitro fermentation of sugar beet arabino-oligosaccharides by fecal microbiota obtained from patients with ulcerative colitis to selectively stimulate the growth of Bifidobacterium spp. and Lactobacillus spp. Appl. Environ. Microbiol. 2011, 77, 8336–8344. [Google Scholar] [CrossRef] [PubMed]
- Komeno, M.; Yoshihara, Y.; Kawasaki, J.; Nabeshima, W.; Maeda, K.; Sasaki, Y.; Fujita, K.; Ashida, H. Two α-L-arabinofuranosidases from Bifidobacterium longum subsp. longum are involved in arabinoxylan utilization. Appl. Microbiol. Biotechnol. 2022, 106, 1957–1965. [Google Scholar] [CrossRef]
- Saito, Y.; Shigehisa, A.; Watanabe, Y.; Tsukuda, N.; Moriyama-Ohara, K.; Hara, T.; Matsumoto, S.; Tsuji, H.; Matsuki, T. Multiple transporters and glycoside hydrolases are involved in arabinoxylan-derived oligosaccharide utilization in Bifidobacterium pseudocatenulatum. Appl. Environ. Microbiol. 2020, 86, e01782-20. [Google Scholar] [CrossRef]
- Tingirikari, J.M.R. In-vitro prebiotic analysis of microbiota accessible pectic polysaccharides. Curr. Microbiol. 2019, 76, 1452–1460. [Google Scholar] [CrossRef]
- Van Laere, K.M.; Hartemink, R.; Bosveld, M.; Schols, H.A.; Voragen, A.G. Fermentation of plant cell wall derived polysaccharides and their corresponding oligosaccharides by intestinal bacteria. J. Agric. Food Chem. 2000, 48, 1644–1652. [Google Scholar] [CrossRef] [PubMed]
- Park, J.M.; Jang, M.U.; Oh, G.W.; Lee, E.H.; Kang, J.H.; Song, Y.B.; Han, N.S.; Kim, T.J. Synergistic action modes of arabinan degradation by exo- and endo-arabinosyl hydrolases. J. Microbiol. Biotechnol. 2015, 25, 227–233. [Google Scholar] [CrossRef] [PubMed]
- Seri, K.; Sanai, K.; Matsuo, N.; Kawakubo, K.; Xue, C.; Inoue, S. L-Arabinose selectively inhibits intestinal sucrase in an uncompetitive manner and suppresses glycemic response after sucrose ingestion in animals. Metabolism 1996, 45, 1368–1374. [Google Scholar] [CrossRef]
- Komeno, M.; Hayamizu, H.; Fujita, K.; Ashida, H. Two novel α-L-arabinofuranosidases from Bifidobacterium longum subsp. longum belonging to glycoside hydrolase family 43 cooperatively degrade arabinan. Appl. Environ. Microbiol. 2019, 85, e02582-18. [Google Scholar] [CrossRef]
- Poria, V.; Saini, J.K.; Singh, S.; Nain, L.; Kuhad, R.C. Arabinofuranosidases: Characteristics, microbial production, and potential in waste valorization and industrial applications. Bioresour. Technol. 2020, 304, 123019. [Google Scholar] [CrossRef] [PubMed]
- Beylot, M.H.; McKie, V.A.; Voragen, A.G.; Doeswijk-Voragen, C.H.; Gilbert, H.J. The Pseudomonas cellulosa glycoside hydrolase family 51 arabinofuranosidase exhibits wide substrate specificity. Biochem. J. 2001, 358, 607–614. [Google Scholar] [CrossRef]
- Ichinose, H.; Yoshida, M.; Fujimoto, Z.; Kaneko, S. Characterization of a modular enzyme of exo-1,5-α-L-arabinofuranosidase and arabinan binding module from Streptomyces avermitilis NBRC14893. Appl. Microbiol. Biotechnol. 2008, 80, 399–408. [Google Scholar] [CrossRef]
- Dumbrepatil, A.; Park, J.M.; Jung, T.Y.; Song, H.N.; Jang, M.U.; Han, N.S.; Kim, T.J.; Woo, E.J. Structural analysis of α-L-arabinofuranosidase from Thermotoga maritima reveals characteristics for thermostability and substrate specificity. J. Microbiol. Biotechnol. 2012, 22, 1724–1730. [Google Scholar] [CrossRef]
- Michlmayr, H.; Hell, J.; Lorenz, C.; Bohmdorfer, S.; Rosenau, T.; Kneifel, W. Arabinoxylan oligosaccharide hydrolysis by family 43 and 51 glycosidases from Lactobacillus brevis DSM 20054. Appl. Environ. Microbiol. 2013, 79, 6747–6754. [Google Scholar] [CrossRef]
- Inacio, J.M.; Correia, I.L.; de Sa-Nogueira, I. Two distinct arabinofuranosidases contribute to arabino-oligosaccharide degradation in Bacillus subtilis. Microbiology 2008, 154, 2719–2729. [Google Scholar] [CrossRef]
- Oh, G.W.; Kang, Y.; Choi, C.Y.; Kang, S.Y.; Kang, J.H.; Lee, M.L.; Han, N.S.; Kim, T.J. Detailed mode of action of arabinan-debranching α-L-arabinofuranosidase GH51 from Bacillus velezensis. J. Microbiol. Biotechnol. 2019, 29, 37–43. [Google Scholar] [CrossRef]
- Lim, Y.R.; Yoon, R.Y.; Seo, E.S.; Kim, Y.S.; Park, C.S.; Oh, D.K. Hydrolytic properties of a thermostable α-L-arabinofuranosidase from Caldicellulosiruptor saccharolyticus. J. Appl. Microbiol. 2010, 109, 1188–1197. [Google Scholar] [CrossRef]
- Hovel, K.; Shallom, D.; Niefind, K.; Belakhov, V.; Shoham, G.; Baasov, T.; Shoham, Y.; Schomburg, D. Crystal structure and snapshots along the reaction pathway of a family 51 α-L-arabinofuranosidase. EMBO J. 2003, 22, 4922–4932. [Google Scholar] [CrossRef]
- Ahmed, S.; Luis, A.S.; Bras, J.L.; Ghosh, A.; Gautam, S.; Gupta, M.N.; Fontes, C.M.; Goyal, A. A novel α-L-arabinofuranosidase of family 43 glycoside hydrolase (Ct43Araf) from Clostridium thermocellum. PLoS ONE 2013, 8, e73575. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.J.; Kang, Y.; Son, B.S.; Kim, M.J.; Park, T.H.; Park, D.; Kim, T.J. Hydrolysis of Arabinoxylo-oligosaccharides by α-L-arabinofuranosidases and β-D-xylosidase from Bifidobacterium dentium. J. Microbiol. Biotechnol. 2022, 32, 187–194. [Google Scholar] [CrossRef] [PubMed]
- Lagaert, S.; Pollet, A.; Delcour, J.A.; Lavigne, R.; Courtin, C.M.; Volckaert, G. Substrate specificity of three recombinant α-L-arabinofuranosidases from Bifidobacterium adolescentis and their divergent action on arabinoxylan and arabinoxylan oligosaccharides. Biochem. Biophys. Res. Commun. 2010, 402, 644–650. [Google Scholar] [CrossRef] [PubMed]
- Linares-Pasten, J.A.; Falck, P.; Albasri, K.; Kjellstrom, S.; Adlercreutz, P.; Logan, D.T.; Karlsson, E.N. Three-dimensional structures and functional studies of two GH43 arabinofuranosidases from Weissella sp. strain 142 and Lactobacillus brevis. FEBS J. 2017, 284, 2019–2036. [Google Scholar] [CrossRef] [PubMed]
- Koseki, T.; Okuda, M.; Sudoh, S.; Kizaki, Y.; Iwano, K.; Aramaki, I.; Matsuzawa, H. Role of two α-L-arabinofuranosidases in arabinoxylan degradation and characteristics of the encoding genes from shochu koji molds, Aspergillus kawachii and Aspergillus awamori. J. Biosci. Bioeng. 2003, 96, 232–241. [Google Scholar] [CrossRef] [PubMed]
- Park, T.H.; Choi, C.Y.; Kim, H.J.; Song, J.R.; Park, D.; Kang, H.A.; Kim, T.J. Arabinoxylo- and arabino-oligosaccharides-specific α-L-arabinofuranosidase GH51 isozymes from the amylolytic yeast Saccharomycopsis fibuligera. J. Microbiol. Biotechnol. 2021, 31, 272–279. [Google Scholar] [CrossRef]
- Pitson, S.M.; Voragen, A.G.; Vincken, J.P.; Beldman, G. Action patterns and mapping of the substrate-binding regions of endo-(1→5)-α-l-arabinanases from Aspergillus niger and Aspergillus aculeatus. Carbohydr. Res. 1997, 303, 207–218. [Google Scholar] [CrossRef]
- Nurizzo, D.; Turkenburg, J.P.; Charnock, S.J.; Roberts, S.M.; Dodson, E.J.; McKie, V.A.; Taylor, E.J.; Gilbert, H.J.; Davies, G.J. Cellvibrio japonicus α-L-arabinanase 43A has a novel five-blade β-propeller fold. Nat. Struct. Biol. 2002, 9, 665–668. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, T.; Ihara, H.; Kozaki, S.; Kawasaki, H. A cold-adapted endo-arabinanase from Penicillium chrysogenum. Biochim. Biophys. Acta 2003, 1624, 70–75. [Google Scholar] [CrossRef] [PubMed]
- Park, J.M.; Jang, M.U.; Kang, J.H.; Kim, M.J.; Lee, S.W.; Song, Y.B.; Shin, C.S.; Han, N.S.; Kim, T.J. Detailed modes of action and biochemical characterization of endo-arabinanase from Bacillus licheniformis DSM13. J. Microbiol. 2012, 50, 1041–1046. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, A.; Tada, T.; Wada, K.; Nakaniwa, T.; Kitatani, T.; Sogabe, Y.; Takao, M.; Sakai, T.; Nishimura, K. Structural basis for thermostability of endo-1,5-α-L-arabinanase from Bacillus thermodenitrificans TS-3. J. Biochem. 2005, 137, 587–592. [Google Scholar] [CrossRef] [PubMed]
- Hong, M.R.; Park, C.S.; Oh, D.K. Characterization of a thermostable endo-1,5-α-L-arabinanase from Caldicellulorsiruptor saccharolyticus. Biotechnol. Lett. 2009, 31, 1439–1443. [Google Scholar] [CrossRef] [PubMed]
- Alhassid, A.; Ben-David, A.; Tabachnikov, O.; Libster, D.; Naveh, E.; Zolotnitsky, G.; Shoham, Y.; Shoham, G. Crystal structure of an inverting GH 43 1,5-α-L-arabinanase from Geobacillus stearothermophilus complexed with its substrate. Biochem. J. 2009, 422, 73–82. [Google Scholar] [CrossRef]
- Huy, N.D.; Thiyagarajan, S.; Choi, Y.E.; Kim, D.H.; Park, S.M. Cloning and characterization of a thermostable endo-arabinanase from Phanerochaete chrysosporium and its synergistic action with endo-xylanase. Bioprocess Biosyst. Eng. 2013, 36, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Squina, F.M.; Santos, C.R.; Ribeiro, D.A.; Cota, J.; de Oliveira, R.R.; Ruller, R.; Mort, A.; Murakami, M.T.; Prade, R.A. Substrate cleavage pattern, biophysical characterization and low-resolution structure of a novel hyperthermostable arabinanase from Thermotoga petrophila. Biochem. Biophys. Res. Commun. 2010, 399, 505–511. [Google Scholar] [CrossRef]
- Chen, Z.; Liu, Y.; Yan, Q.; Yang, S.; Jiang, Z. Biochemical characterization of a novel endo-1,5-α-L-arabinanase from Rhizomucor miehei. J. Agric. Food Chem. 2015, 63, 1226–1233. [Google Scholar] [CrossRef]
- Wefers, D.; Dong, J.; Abdel-Hamid, A.M.; Paul, H.M.; Pereira, G.V.; Han, Y.; Dodd, D.; Baskaran, R.; Mayer, B.; Mackie, R.I.; et al. Enzymatic mechanism for arabinan degradation and transport in the thermophilic bacterium Caldanaerobius polysaccharolyticus. Appl. Environ. Microbiol. 2017, 83, e00794-17. [Google Scholar] [CrossRef]
- Li, Q.; Ganzle, M.G. Characterization of two extracellular arabinanases in Lactobacillus crispatus. Appl. Microbiol. Biotechnol. 2020, 104, 10091–10103. [Google Scholar] [CrossRef]
- Leal, T.F.; de Sa-Nogueira, I. Purification, characterization and functional analysis of an endo-arabinanase (AbnA) from Bacillus subtilis. FEMS Microbiol. Lett. 2004, 241, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Inacio, J.M.; de Sa-Nogueira, I. Characterization of abn2 (yxiA), encoding a Bacillus subtilis GH43 arabinanase, Abn2, and its role in arabino-polysaccharide degradation. J. Bacteriol. 2008, 190, 4272–4280. [Google Scholar] [CrossRef]
- Akel, E.; Metz, B.; Seiboth, B.; Kubicek, C.P. Molecular regulation of arabinan and L-arabinose metabolism in Hypocrea jecorina (Trichoderma reesei). Eukaryot. Cell 2009, 8, 1837–1844. [Google Scholar] [CrossRef]
- Kawaguchi, H.; Sasaki, M.; Vertes, A.A.; Inui, M.; Yukawa, H. Identification and functional analysis of the gene cluster for L-arabinose utilization in Corynebacterium glutamicum. Appl. Environ. Microbiol. 2009, 75, 3419–3429. [Google Scholar] [CrossRef]
- Shulami, S.; Raz-Pasteur, A.; Tabachnikov, O.; Gilead-Gropper, S.; Shner, I.; Shoham, Y. The L-Arabinan utilization system of Geobacillus stearothermophilus. J. Bacteriol. 2011, 193, 2838–2850. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.Y.; Park, S.Y.; Sung, J.H.; Kim, D.H. Purification and characterization of α-L-arabinopyranosidase and α-L-arabinofuranosidase from Bifidobacterium breve K-110, a human intestinal anaerobic bacterium metabolizing ginsenoside Rb2 and Rc. Appl. Environ. Microbiol. 2003, 69, 7116–7123. [Google Scholar] [CrossRef] [PubMed]
- Fujita, K.; Takashi, Y.; Obuchi, E.; Kitahara, K.; Suganuma, T. Characterization of a novel β-L-arabinofuranosidase in Bifidobacterium longum: Functional elucidation of a DUF1680 protein family member. J. Biol. Chem. 2014, 289, 5240–5249. [Google Scholar] [CrossRef]
- Zhao, H.; Lu, L.; Xiao, M.; Wang, Q.; Lu, Y.; Liu, C.; Wang, P.; Kumagai, H.; Yamamoto, K. Cloning and characterization of a novel α-galactosidase from Bifidobacterium breve 203 capable of synthesizing Gal-α-1,4 linkage. FEMS Microbiol. Lett. 2008, 285, 278–283. [Google Scholar] [CrossRef]
- Goulas, T.K.; Goulas, A.K.; Tzortzis, G.; Gibson, G.R. Molecular cloning and comparative analysis of four β-galactosidase genes from Bifidobacterium bifidum NCIMB41171. Appl. Microbiol. Biotechnol. 2007, 76, 1365–1372. [Google Scholar] [CrossRef]
- Sakata, S.; Kitahara, M.; Sakamoto, M.; Hayashi, H.; Fukuyama, M.; Benno, Y. Unification of Bifidobacterium infantis and Bifidobacterium suis as Bifidobacterium longum. Int. J. Syst. Evol. Microbiol. 2002, 52, 1945–1951. [Google Scholar] [CrossRef]
- de Sanctis, D.; Inacio, J.M.; Lindley, P.F.; de Sa-Nogueira, I.; Bento, I. New evidence for the role of calcium in the glycosidase reaction of GH43 arabinanases. FEBS J. 2010, 277, 4562–4574. [Google Scholar] [CrossRef]
- Fujimoto, Z.; Ichinose, H.; Maehara, T.; Honda, M.; Kitaoka, M.; Kaneko, S. Crystal structure of an exo-1,5-α-L-arabinofuranosidase from Streptomyces avermitilis provides insights into the mechanism of substrate discrimination between exo- and endo-type enzymes in glycoside hydrolase family 43. J. Biol. Chem. 2010, 285, 34134–34143. [Google Scholar] [CrossRef]
- Hovel, K.; Shallom, D.; Niefind, K.; Baasov, T.; Shoham, G.; Shoham, Y.; Schomburg, D. Crystallization and preliminary X-ray analysis of a family 51 glycoside hydrolase, the α-L-arabinofuranosidase from Geobacillus stearothermophilus T-6. Acta Crystallogr. D Biol. Crystallogr. 2003, 59, 913–915. [Google Scholar] [CrossRef]
- Paes, G.; Skov, L.K.; O’Donohue, M.J.; Remond, C.; Kastrup, J.S.; Gajhede, M.; Mirza, O. The structure of the complex between a branched pentasaccharide and Thermobacillus xylanilyticus GH-51 arabinofuranosidase reveals xylan-binding determinants and induced fit. Biochemistry 2008, 47, 7441–7451. [Google Scholar] [CrossRef]
- Miyanaga, A.; Koseki, T.; Matsuzawa, H.; Wakagi, T.; Shoun, H.; Fushinobu, S. Expression, purification, crystallization and preliminary X-ray analysis of α-L-arabinofuranosidase B from Aspergillus kawachii. Acta Crystallogr. D Biol. Crystallogr. 2004, 60, 1286–1288. [Google Scholar] [CrossRef] [PubMed]
- Arzamasov, A.A.; van Sinderen, D.; Rodionov, D.A. Comparative genomics reveals the regulatory complexity of bifidobacterial arabinose and arabino-oligosaccharide utilization. Front. Microbiol. 2018, 9, 776. [Google Scholar] [CrossRef] [PubMed]
- Park, J.M.; Han, N.S.; Kim, T.J. Rapid detection and isolation of known and putative α-L-arabinofuranosidase genes using degenerate PCR primers. J. Microbiol. Biotechnol. 2007, 17, 481–489. [Google Scholar] [PubMed]

| Substrate 1 | Specific Activity (U/mg) 2 | ||||
|---|---|---|---|---|---|
| BflsABN43A | BflsABN43B | BflsABF43A | BflsABF43B | BflsABF51 | |
| Arabinobiose (A2) | NA | NA | NA | 439.65 ± 16.09 | 1.21 ± 0.08 |
| Arabinotriose (A3) | NA | 9.38 ± 1.08 | NA | 444.97 ± 21.46 | 0.47 ± 0.03 |
| BAOS (AA3A) | NA | NA | 46.00 ± 1.29 | NA | 2.16 ± 0.18 |
| Debranched arabinan (DA) | 51.19 ± 0.97 | 63.27 ± 1.86 | NA | 61.34 ± 0.63 | NA |
| Sugar beet arabinan (SA) | 9.85 ± 0.32 | 2.77 ± 0.06 | 20.87 ± 1.08 | NA | NA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kang, Y.; Choi, C.-Y.; Kang, J.; Ju, Y.-R.; Kim, H.B.; Han, N.S.; Kim, T.-J. Functional Characterization of Endo- and Exo-Hydrolase Genes in Arabinan Degradation Gene Cluster of Bifidobacterium longum subsp. suis. Int. J. Mol. Sci. 2024, 25, 3175. https://doi.org/10.3390/ijms25063175
Kang Y, Choi C-Y, Kang J, Ju Y-R, Kim HB, Han NS, Kim T-J. Functional Characterization of Endo- and Exo-Hydrolase Genes in Arabinan Degradation Gene Cluster of Bifidobacterium longum subsp. suis. International Journal of Molecular Sciences. 2024; 25(6):3175. https://doi.org/10.3390/ijms25063175
Chicago/Turabian StyleKang, Yewon, Chang-Yun Choi, Jihun Kang, Ye-Rin Ju, Hye Bin Kim, Nam Soo Han, and Tae-Jip Kim. 2024. "Functional Characterization of Endo- and Exo-Hydrolase Genes in Arabinan Degradation Gene Cluster of Bifidobacterium longum subsp. suis" International Journal of Molecular Sciences 25, no. 6: 3175. https://doi.org/10.3390/ijms25063175
APA StyleKang, Y., Choi, C.-Y., Kang, J., Ju, Y.-R., Kim, H. B., Han, N. S., & Kim, T.-J. (2024). Functional Characterization of Endo- and Exo-Hydrolase Genes in Arabinan Degradation Gene Cluster of Bifidobacterium longum subsp. suis. International Journal of Molecular Sciences, 25(6), 3175. https://doi.org/10.3390/ijms25063175