Biodegradation of Polystyrene by Galleria mellonella: Identification of Potential Enzymes Involved in the Degradative Pathway
Abstract
1. Introduction
2. Results
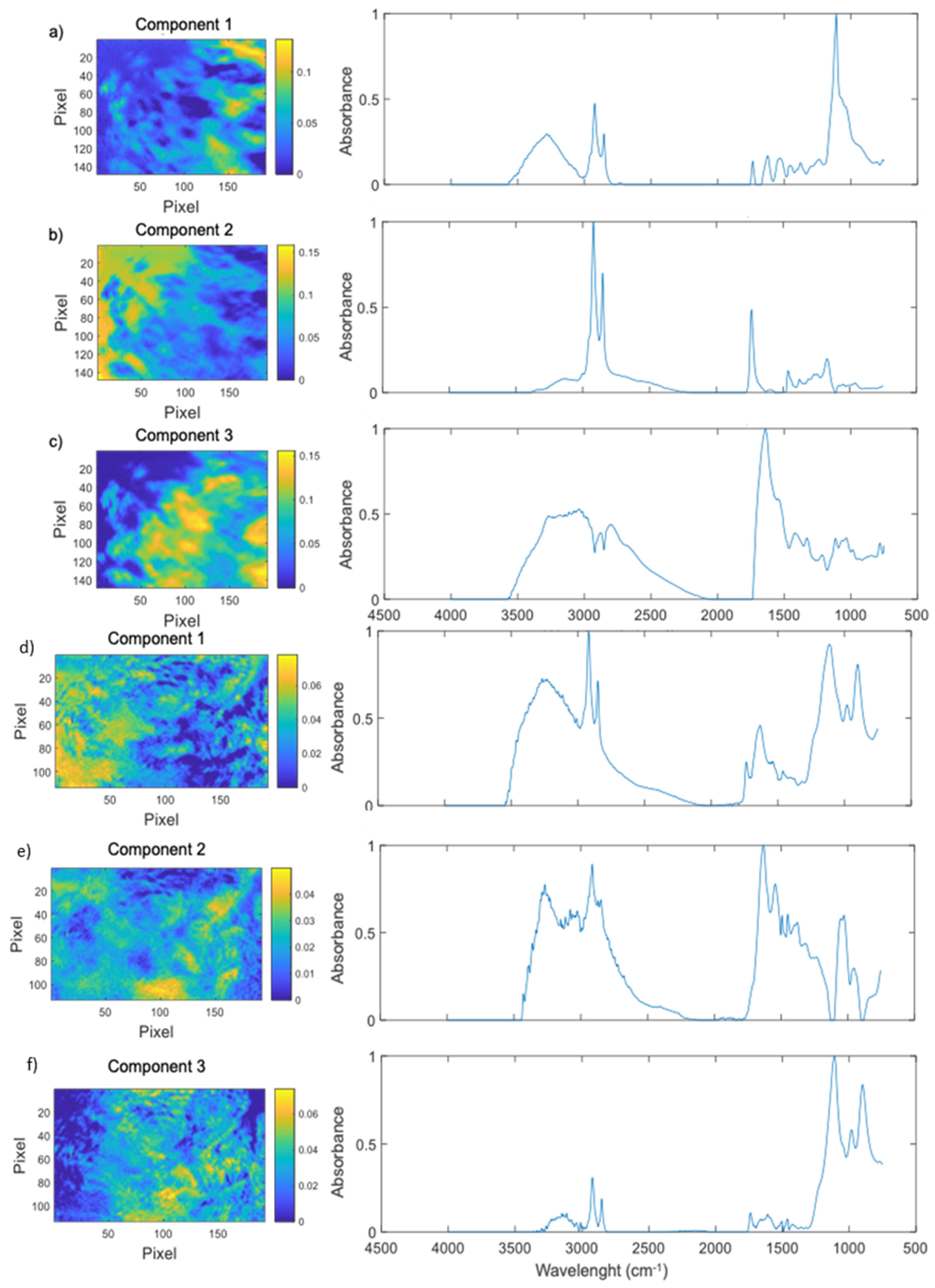
2.1. Degradation of Polystyrene by G. mellonella Larvae
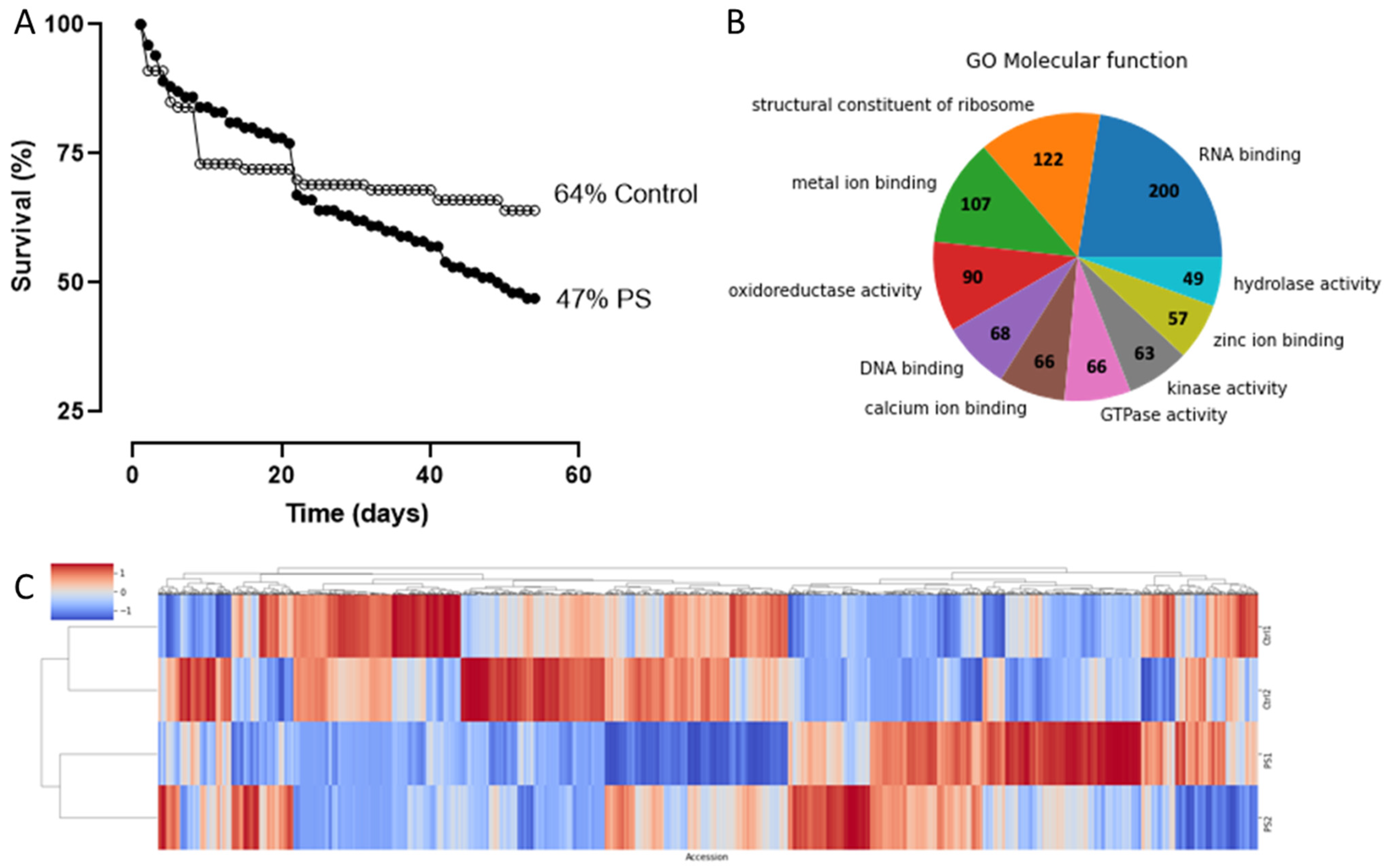
2.2. Effect of PS Feeding on Larvae Survival Rate
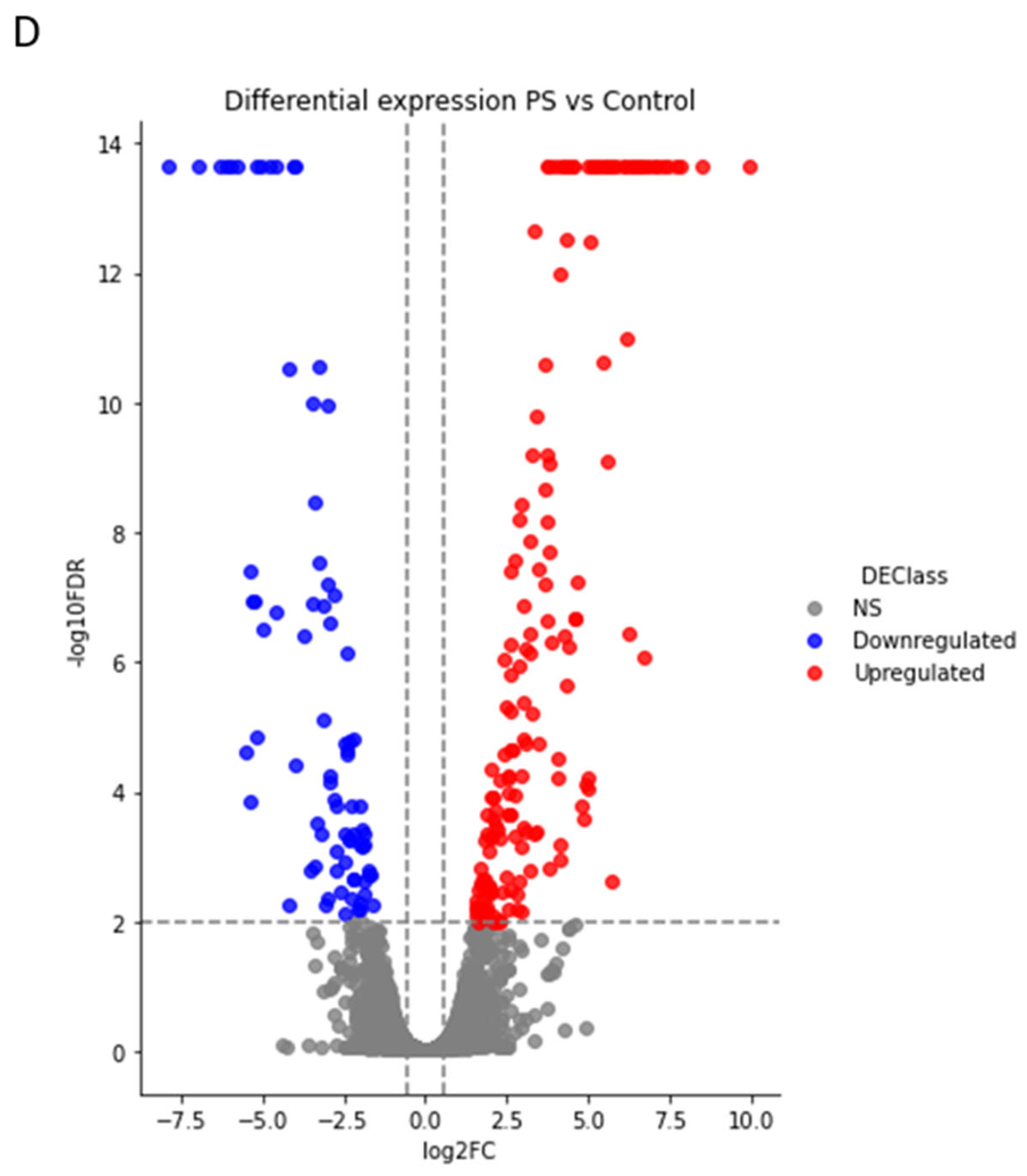
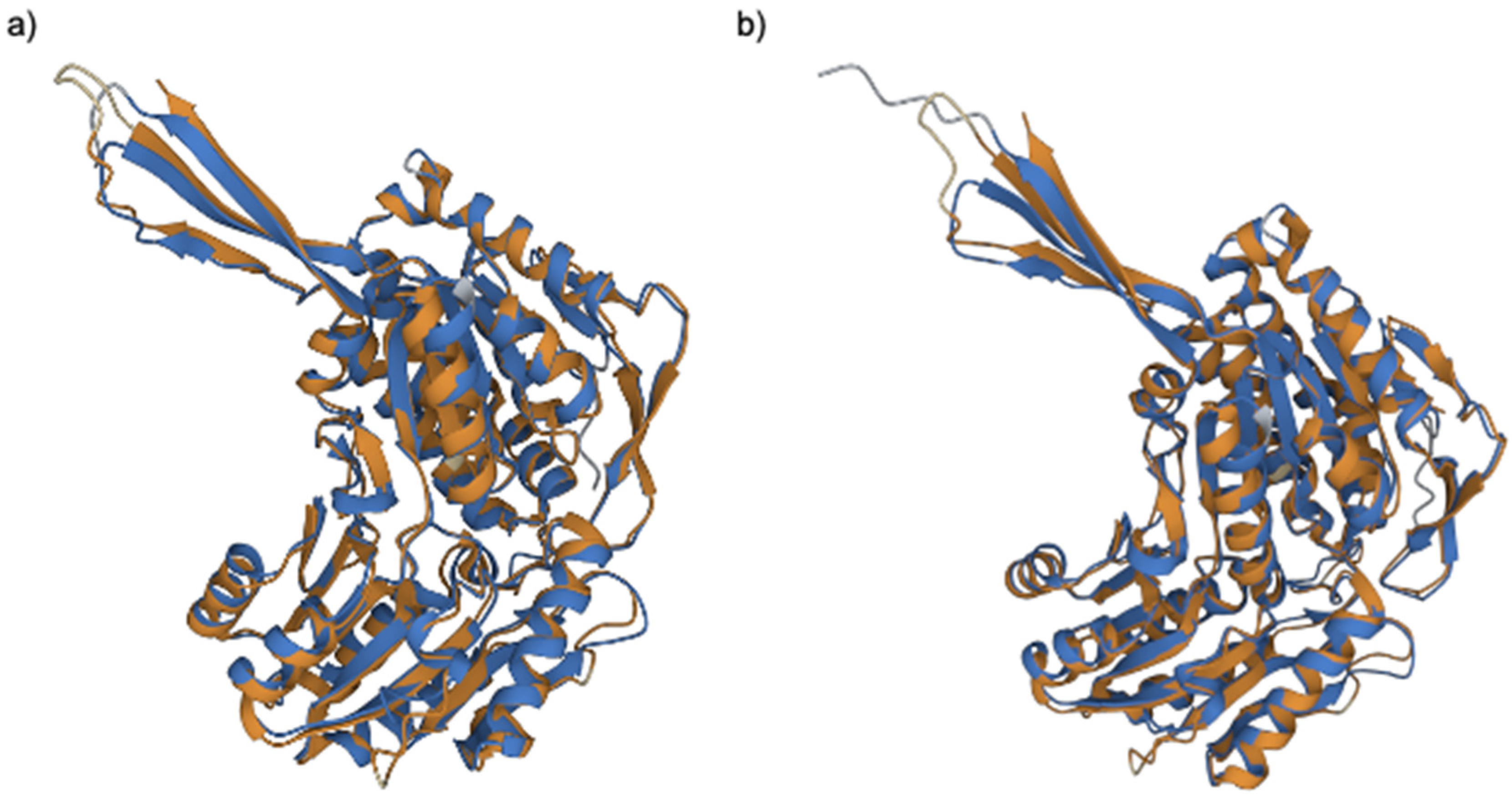
2.3. Comparative Proteomic Analysis after PS Degradation by G. mellonella
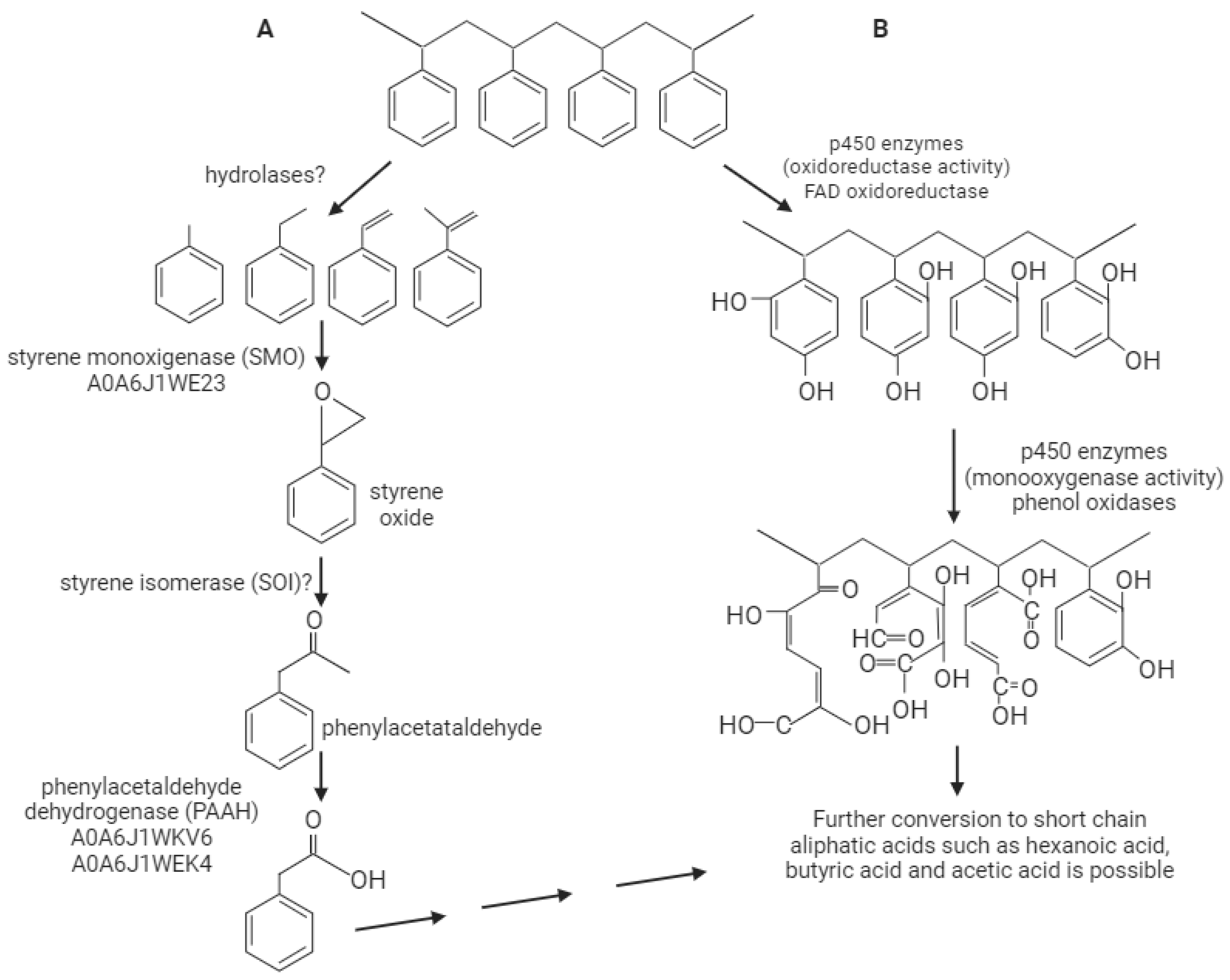
3. Discussion
4. Materials and Methods
4.1. Plastic Material
4.2. Polystyrene Degradation Assay
4.3. Larvae Feeding and Survival Assay
4.4. Fourier-Transform Infrared Microscopy (FTIR) and Chemometric Analysis
4.5. Larvae Dissection
4.6. Protein Extraction
4.7. LC-MS/MS Preparation
4.8. LC-MS/MS
4.9. Proteomic Data Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Danso, D.; Chow, J.; Streit, W.R. Plastics: Environmental and Biotechnological Perspectives on Microbial Degradation. Appl. Environ. Microbiol. 2019, 85, e01095-19. [Google Scholar] [CrossRef] [PubMed]
- Chamas, A.; Moon, H.; Zheng, J.; Qiu, Y.; Tabassum, T.; Jang, J.H.; Abu-Omar, M.; Scott, S.L.; Suh, S. Degradation Rates of Plastics in the Environment. ACS Sustain. Chem. Eng. 2020, 8, 3494–3511. [Google Scholar] [CrossRef]
- Jiang, S.; Su, T.; Zhao, J.; Wang, Z. Biodegradation of Polystyrene by Tenebrio molitor, Galleria mellonella, and Zophobas atratus Larvae and Comparison of Their Degradation Effects. Polymers 2021, 13, 3539. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Su, T.; Zhao, J.; Wang, Z. Isolation, Identification, and Characterization of Polystyrene-Degrading Bacteria From the Gut of Galleria mellonella (Lepidoptera: Pyralidae) Larvae. Front. Bioeng. Biotechnol. 2021, 9, 736062. [Google Scholar] [CrossRef] [PubMed]
- Bombelli, P.; Howe, C.J.; Bertocchini, F. Polyethylene bio-degradation by caterpillars of the wax moth Galleria mellonella. Curr. Biol. 2017, 27, R292–R293. [Google Scholar] [CrossRef] [PubMed]
- Lou, Y.; Ekaterina, P.; Yang, S.-S.; Lu, B.; Liu, B.; Ren, N.; Corvini, P.F.X.; Xing, D. Biodegradation of Polyethylene and Polystyrene by Greater Wax Moth Larvae (Galleria mellonella L.) and the Effect of Co-diet Supplementation on the Core Gut Microbiome. Environ. Sci. Technol. 2020, 54, 2821–2831. [Google Scholar] [CrossRef] [PubMed]
- Tsochatzis, E.D.; Berggreen, I.E.; Nørgaard, J.V.; Theodoridis, G.; Dalsgaard, T.K. Biodegradation of expanded polystyrene by mealworm larvae under different feeding strategies evaluated by metabolic profiling using GC-TOF-MS. Chemosphere 2021, 281, 130840. [Google Scholar] [CrossRef]
- Wang, S.; Shi, W.; Huang, Z.; Zhou, N.; Xie, Y.; Tang, Y.; Hu, F.; Liu, G.; Zheng, H. Complete digestion/biodegradation of polystyrene microplastics by greater wax moth (Galleria mellonella) larvae: Direct in vivo evidence, gut microbiota independence, and potential metabolic pathways. J. Hazard Mater. 2022, 423 Pt B, 127213. [Google Scholar] [CrossRef]
- Sanluis-Verdes, A.; Colomer-Vidal, P.; Rodríguez-Ventura, F.; Bello-Villarino, M.; Spinola-Amilibia, M.; Ruiz-López, E.; Illanes-Vicioso, R.; Castroviejo, P.; Cigliano, R.A.; Montoya, M.; et al. Wax worm saliva and the enzymes therein are the key to polyethylene degradation by Galleria mellonella. Nat. Commun. 2022, 13, 1–11. [Google Scholar] [CrossRef]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef]
- Ukaegbu, U.E.; Kantz, A.; Beaton, M.; Gassner, G.T.; Rosenzweig, A.C. Structure and ligand binding properties of the epoxidase component of styrene monooxygenase. Biochemistry 2010, 49, 1678–1688. [Google Scholar] [CrossRef] [PubMed]
- Crabo, A.G.; Singh, B.; Nguyen, T.; Emami, S.; Gassner, G.T.; Sazinsky, M.H. Structure and biochemistry of phenylacetaldehyde dehydrogenase from the Pseudomonas putida S12 styrene catabolic pathway. Arch. Biochem. Biophys. 2017, 616, 47–58. [Google Scholar] [CrossRef] [PubMed]
- Oelschlägel, M.; Zimmerling, J.; Tischler, D. A Review: The Styrene Metabolizing Cascade of Side-Chain Oxygenation as Biotechnological Basis to Gain Various Valuable Compounds. Front. Microbiol. 2018, 9, 490. [Google Scholar] [CrossRef] [PubMed]
- Cox, H.H.; Faber, B.W.; Van Heiningen, W.N.; Radhoe, H.; Doddema, H.J.; Harder, W. Styrene metabolism in Exophiala jeanselmei and involvement of a cytochrome P-450-dependent styrene monooxygenase. Appl. Environ. Microbiol. 1996, 62, 1471–1474. [Google Scholar] [CrossRef] [PubMed]
- Donoso, R.A.; Ruiz, D.; Gárate-Castro, C.; Villegas, P.; González-Pastor, J.E.; de Lorenzo, V.; González, B.; Pérez-Pantoja, D. Identification of a self-sufficient cytochrome P450 monooxygenase from Cupriavidus pinatubonensis JMP134 involved in 2-hydroxyphenylacetic acid catabolism, via homogentisate pathway. Microb. Biotechnol. 2021, 14, 1944–1960. [Google Scholar] [CrossRef] [PubMed]
- Mingot, J.M.; Peñalva, M.A.; Fernández-Cañón, J.M. Disruption of Aspergillus nidulans Gene Encoding a Novel Cytochrome P450 Monooxygenase Catalyzing Phenylacetate 2-Hydroxylation, Results in Penicillin Overproduction. J. Biol. Chem. 1999, 274, 14545–14550. [Google Scholar] [CrossRef]
- Calla, B.; Noble, K.; Johnson, R.M.; Walden, K.K.O.; Schuler, M.A.; Robertson, H.M.; Berenbaum, M.R. Cytochrome P450 diversification and hostplant utilization patterns in specialist and generalist moths: Birth, death and adaptation. Mol. Ecol. 2017, 26, 6021–6035. [Google Scholar] [CrossRef]
- Baggi, G.; Boga, M.M.; Catelani, D.; Galli, E.; Treccani, V. Styrene Catabolism by a Strain of Pseudomonas fluoresc. Syst. Appl. Microbiol. 1983, 4, 141–147. [Google Scholar] [CrossRef]
- Hou, L.; Majumder, E.L. Potential for and Distribution of Enzymatic Biodegradation of Polystyrene by Environmental Microorganisms. Materials 2021, 14, 503. [Google Scholar] [CrossRef]
- Jaumot, J.; Gargallo, R.; de Juan, A.; Tauler, R. A graphical user-friendly interface for MCR-ALS: A new tool for multivariate curve resolution in MATLAB. Chemom. Intell. Lab. Syst. 2005, 76, 101–110. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Sanhueza, S.; Vidal, M.A.; Hernandez, M.A.; Henriquez-Beltran, M.E.; Cabrera, C.; Quiroga, R.; Antilef, B.E.; Aguilar, K.P.; Castillo, D.A.; Llerena, F.J.; et al. Clinical and pulmonary function analysis in long-COVID revealed that long-term pulmonary dysfunction is associated with vascular inflammation pathways and metabolic syndrome. Front. Med. 2023, 10, 1271863. [Google Scholar] [CrossRef] [PubMed]
- Kong, H.G.; Kim, H.H.; Chung, J.H.; Jun, J.; Lee, S.; Kim, H.M.; Jeon, S.; Park, S.G.; Bhak, J.; Ryu, C.M. The Galleria mellonella Hologenome Supports Microbiota-Independent Metabolism of Long-Chain Hydrocarbon Beeswax. Cell Rep. 2019, 26, 2451–2464.e5. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Smits, A.H.; van Tilburg, G.B.A.; Ovaa, H.; Huber, W.; Vermeulen, M. Proteome-wide identification of ubiquitin interactions using UbIA-MS. Nat. Protoc. 2018, 13, 530–550. [Google Scholar] [CrossRef] [PubMed]
- Törönen, P.; Medlar, A.; Holm, L. PANNZER2: A rapid functional annotation web server. Nucleic Acids Res. 2018, 46, W84–W88. [Google Scholar] [CrossRef] [PubMed]
- Apweiler, R.; Bairoch, A.; Wu, C.H.; Barker, W.C.; Boeckmann, B.; Ferro, S.; Gasteiger, E.; Huang, H.; Lopez, R.; Magrane, M.; et al. UniProt: The Universal Protein knowledgebase. Nucleic Acids Res. 2004, 32 (Suppl. S1), D115–D119. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Venegas, S.; Alarcón, C.; Araya, J.; Gatica, M.; Morin, V.; Tarifeño-Saldivia, E.; Uribe, E. Biodegradation of Polystyrene by Galleria mellonella: Identification of Potential Enzymes Involved in the Degradative Pathway. Int. J. Mol. Sci. 2024, 25, 1576. https://doi.org/10.3390/ijms25031576
Venegas S, Alarcón C, Araya J, Gatica M, Morin V, Tarifeño-Saldivia E, Uribe E. Biodegradation of Polystyrene by Galleria mellonella: Identification of Potential Enzymes Involved in the Degradative Pathway. International Journal of Molecular Sciences. 2024; 25(3):1576. https://doi.org/10.3390/ijms25031576
Chicago/Turabian StyleVenegas, Sebastián, Carolina Alarcón, Juan Araya, Marcell Gatica, Violeta Morin, Estefanía Tarifeño-Saldivia, and Elena Uribe. 2024. "Biodegradation of Polystyrene by Galleria mellonella: Identification of Potential Enzymes Involved in the Degradative Pathway" International Journal of Molecular Sciences 25, no. 3: 1576. https://doi.org/10.3390/ijms25031576
APA StyleVenegas, S., Alarcón, C., Araya, J., Gatica, M., Morin, V., Tarifeño-Saldivia, E., & Uribe, E. (2024). Biodegradation of Polystyrene by Galleria mellonella: Identification of Potential Enzymes Involved in the Degradative Pathway. International Journal of Molecular Sciences, 25(3), 1576. https://doi.org/10.3390/ijms25031576




