Abstract
Gastric cancer (GC) remains a significant contributor to cancer-related mortality. Novel high-throughput techniques have enlightened the epigenetic mechanisms governing gene-expression regulation. Epigenetic characteristics contribute to molecular taxonomy and give rise to cancer-specific epigenetic patterns. Helicobacter pylori (Hp) infection has an impact on aberrant DNA methylation either through its pathogenic CagA protein or by inducing chronic inflammation. The hypomethylation of specific repetitive elements generates an epigenetic field effect early in tumorigenesis. Epstein–Barr virus (EBV) infection triggers DNA methylation by dysregulating DNA methyltransferases (DNMT) enzyme activity, while persistent Hp-EBV co-infection leads to aggressive tumor behavior. Distinct histone modifications are also responsible for oncogene upregulation and tumor-suppressor gene silencing in gastric carcinomas. While histone methylation and acetylation processes have been extensively studied, other less prevalent alterations contribute to the development and migration of gastric cancer via a complex network of interactions. Enzymes, such as Nicotinamide N-methyltransferase (NNMT), which is involved in tumor’s metabolic reprogramming, interact with methyltransferases and modify gene expression. Non-coding RNA molecules, including long non-coding RNAs, circular RNAs, and miRNAs serve as epigenetic regulators contributing to GC development, metastasis, poor outcomes and therapy resistance. Serum RNA molecules hold the potential to serve as non-invasive biomarkers for diagnostic, prognostic or therapeutic applications. Gastric fluids represent a valuable source to identify potential biomarkers with diagnostic use in terms of liquid biopsy. Ongoing clinical trials are currently evaluating the efficacy of next-generation epigenetic drugs, displaying promising outcomes. Various approaches including multiple miRNA inhibitors or targeted nanoparticles carrying epigenetic drugs are being designed to enhance existing treatment efficacy and overcome treatment resistance.
1. Introduction
Gastric cancer (GC) ranks fourth in terms of cancer-related mortality and stands as the fifth most prevalent cancer worldwide. There is a pronounced gender disparity, with males experiencing a two-fold higher incidence rate compared to females [1]. While the incidence and mortality rates tend to decline in developed nations, there is evidence of rising incidence rates among younger individuals (under the age of 50) [2]. Given the intricate nature of the GC pathophysiology, individuals with elevated GC risks should be given extra care while monitoring. Age, gender, and ethnicity comprise pivotal risk factors for GC development augmented by factors such as obesity, lifestyle, gastrointestinal microbiota, as well as Helicobacter pylori (Hp), and Epstein–Barr virus (EBV) infections [3,4].
Gastric cancer is characterized by intestinal and diffuse-type adenocarcinomas. Lately, prompt technological advancements have allowed for an understanding of the molecular mechanisms that underlie the main histological subtypes. Genome-wide association studies and transcriptomic analyses build a combined histological–molecular approach leading to molecular taxonomy [5].
The Cancer Genome Atlas (TCGA) research network team, based on sequencing data, classified GC into four molecular subtypes: chromosomal instability (CIN), Epstein–Barr virus (EBV), microsatellite instability (MSI) and genomically stable (GS) [6]. Similarly, the Asian Cancer Research Group (ACRG) Network team implemented a transcriptome classifier to identify molecular subtypes including microsatellite instability (MSI), microsatellite stability/Epithelial-to-Mesenchymal Transition (MSS/EMT), MSS/TP53 active, and MSS/TP53 inactive [7]. So far, several studies have described molecular subtyping methodologies for GC, employing high-throughput profiling and multi-omics platforms that encompass genomic, proteomic, and epigenetic features [8,9]. Li et al. (2023), using multi-omics data and integrated optimum algorithms, proposed an advanced molecular classification system identifying three GC subtypes based on mRNA, microRNA, and DNA methylation data. Subtype 1 correlates with favorable outcomes and a high mutation rate of ARID1A and PIK3CA mutations, whereas subtypes 2 and 3 are linked with adverse prognostic outcomes exhibiting TP53, APOA1, and CDH1 mutations, respectively [10].
Beyond TCGA and ACRG classifications, several other classifications have been proposed. Recently, Weng et al. (2023) suggested an epigenetic-based classification. According to their proposal, the analysis of 1521 GC cases from GEO and TCGA databases in combination with miRNA-expression and DNA-methylation profiles led to four molecular GC clusters. The C1 cluster is related to the cell cycle, DNA replication and MSI and is accompanied by a better prognosis. In contrast, the C4 cluster involves immune-related processes, microsatellite stability, and TP53 mutations and relates to high-grade carcinomas and poor prognosis. Clusters 2 and 3 seem to correlate to a moderate prognosis, and advanced stages and are related to histone and DNA alterations, signaling pathways and immune invasion [11].
Epigenetic alterations exert regulatory control over gene expression without altering DNA sequence. Numerous epigenetic mechanisms, encompassing DNA methylation, histone modifications, chromatin regeneration, and miRNA interference, allow transcriptional control through regulatory proteins. Gene expression, DNA repair, and cell growth are influenced by epigenetic modifications. Possible malfunction of these mechanisms can result in the initiation of carcinogenesis. Hp infection is prevalent in over 80% of GC cases, with the resultant inflammatory milieu generated affecting both epigenetics and signaling pathways [12].
In this narrative review, we aim to present the currently available literature and enlighten the main epigenetic mechanisms that interplay in gastric cancer development, including potential biomarkers for diagnosis, prognosis, and treatment in clinical settings.
2. Methods
PubMed, Cochrane Library, Embase, and Google Scholar were initially searched to retrieve studies reporting data on epigenetics from 2001 to the present day. The following Medical Subject Heading [MeSH] terms were used alone or matched by the logical operators “OR” or “AND” in all possible combinations to obtain the maximal number of articles: “Gastric Cancer”, “Epigenetics”, “Stomach adenocarcinomas”, “Histone modifications”, “DNA methylation”, “miRNAs”, “Long non-coding RNAs”, “circular RNAs” “Epigenetic changes”. We excluded repetitive as well as non-English studies and we focused mainly on studies conducted after 2018. After an initial title and abstract screening, a full-text copy of each article was retrieved. Each relevant article was subsequently reviewed, and 197 representative scientific papers were finally selected.
3. Epigenetics Background (DNA Methylation, Histone Modifications, and Non-Coding RNAs)
Epigenetic alterations are evident in both early and advanced stages of gastric carcinomas. The rising interest in these epigenetic modifications aims to shed light on the underlying physiology of gastric cancer and unveil novel potential targets for precision medicine. Afterwards, we will highlight the main epigenetic mechanisms and their interplay during GC tumorigenesis (Figure 1).
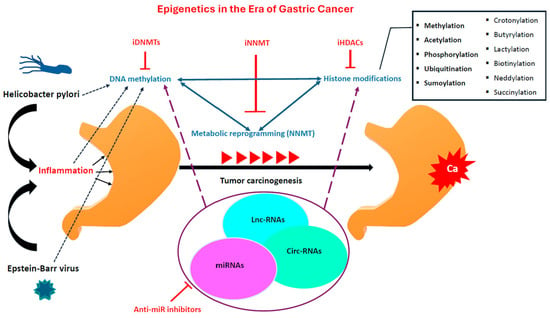
Figure 1.
The main epigenetic mechanisms and environmental factors that mediate during gastric carcinogenesis; anti-miR inhibitors: anti-microRNA inhibitors; circ-RNAs: circular RNAs; iDNMTs: DNA Methyltransferase inhibitors; iHDACs: Histone deacetylase inhibitors; iNNMT: Nicotinamide N-methyltransferase inhibitors; lnc-RNAS: long non-coding RNAs; miRNAs: micro-RNAs; NNMT: Nicotinamide N-methyltransferase.
3.1. DNA Methylation
Cytosine–Guanine-sequence rich (CpG) islands located within the promoter gene region play a crucial role in gene-expression regulation. DNA methyltransferases (DNMTs) catalyze the formation of 5-methylcytosine (5mC) within CpG dinucleotides. It is well established that the methylation of CpG islands within secondary promoter domains can be accelerated by factors such as aging, viral infections, and chronic inflammation. As previously reported, the analysis of the de novo methylation in gastric cell lines, exhibiting varying degrees of CpG-methylated islands, revealed elevated de novo methylation rates in cell lines containing numerous CpG methylated sites. This observation reinforces the concept that random methylation stimulates extensive CpG methylation, thus promoting gene silencing [13,14].
Previous studies have also documented the detection of aberrant methylation in DAPK, E-cadherin, GSTP1, p15, and p16 genes in paired-tissue and serum samples of GC patients. This underlines the potential utility of analyzing methylation status for assessing and monitoring gastric cancer [15,16].
The TCGA group, conducting a molecular analysis of gastric carcinomas, has identified two subgroups characterized by elevated methylation levels. These subgroups, referred to as gastric CpG-island methylator phenotypes (CIMP) and EBV-positive tumors, including the MSI subtype, exhibit unique methylation patterns [6].
Genetic and epigenetic alterations are the key elements of CDH1-suppressor gene depletion, evident in both intestinal and diffuse gastric carcinomas. CDH1 hypermethylation arises early during GC development and is detectable in nearly 50% of hereditary, diffuse gastric carcinomas (Table 1) [17,18].

Table 1.
Common epigenetically altered genes through DNA methylation in gastric carcinomas.
DNA-mismatch-repair (MMR) pathway genes play a crucial role in preserving genomic stability across various cancers, including sporadic GCs. As previously demonstrated, the methylation of both MLH1 and MLH2 promoters correlates to the onset and progression of GC. In patients with surgically resectable gastric carcinomas, MLH1 promoter methylation is associated with a favorable prognosis, whereas its absence highly correlates with tumors displaying MSI [6,19]. However, in early-stage gastric carcinomas exhibiting a papillary phenotype, microsatellite instability has been described, driven by MLH1-promoter hypermethylation [34].
The CDKN2A gene, which mediates in cell cycle arrest, is frequently methylated in GC and other gastrointestinal malignancies. Its presence in precancerous gastric lesions, associated with Hp and EBV infections, suggests its potential involvement in GC development [20,21]. DNMTs, the enzymes responsible for directing methylation processes, are notably upregulated in gastric carcinomas. As previously documented, the DNMT1 enzyme’s expression is linked to GC risk and unfavorable prognosis, particularly in stage-III and -IV GC patients [35]. The expression of the DNMT enzyme can also be influenced by the release of oncogenic proteins and inflammatory responses mediated by tumor-associated macrophages (TAMs) [36]. The APC gene also regulates the expression of the DNMT1 enzyme via the APC/β-catenin/TCF pathway. This hypothesis is also strengthened by the frequently observed hypermethylation of the APC promoter in GC patients, suggesting a possible mechanism for its inactivation [37].
3.2. Histone Modifications
Histones are responsible for nucleosome formation, the primary structural unit of chromatin. Histone modifications encompass a range of post-translational effects on synthesized proteins. Modifications that occur on specific amino acids (lysine, arginine, serine, and threonine), including acetylation, methylation, phosphorylation, and ubiquitination, have been described. Recently, processes like sumoylation, butyrylation, lactylation, succinylation, and crotonylation have also been added to the already-known histone modifications [38,39,40,41]. The majority of these modifications are evident in gastrointestinal tumors, potentially offering new insights into the detection and management of gastric carcinomas.
Histone methylation typically targets lysine or arginine residues in H3 and H4 proteins. The methylation of different amino acid residues is achieved through the interaction of Histone methyltransferases (HMTs) and Histone demethylases (HDMs), resulting in either gene activation or silencing [42]. The prognostic significance of EZH2 and H3k27me3 protein overexpression in gastric cancer has been elucidated, demonstrating a negative correlation between H3K27me3 levels and overall survival in GC patients [43]. Cytotoxin-associated gene A (CagA), a virulent component of Hp, promotes the expression of Myc, DNMT3B, and EZH2, inducing both H3K27me3 and DNA methylation on the let-7 promoter [44,45]. Elevated levels of H3K9me3 have been linked to tumor stage, the recurrence of gastric cancer (GC), and a worse prognosis in a cohort of 261 GC patients [46].
Recently, Reyes et al. (2021), conducted a meta-analysis study describing 10 HMTs (PRDM14, PRDM9, SUV39H2, NSD2, SMYD5, SETDB1, PRDM12, SUV39H1, NSD3, and EHMT2) harboring various alterations in gastric adenocarcinomas, with an impact on specific residues of histone proteins. Computational analysis revealed reduced HMT SUV39H2 expression in patients with GC progression, suggesting that HMTs could serve as potential biomarkers for treatment approaches [47]. In another study, HMT DOT1L was found to be involved in the methylation of H3K79 in patients with familial GC, suggesting a potential role in gastric carcinogenesis [48].
Histone demethylases (HDMs) exhibit the potential for both up and downregulation. Previous studies have shown that the upregulation of LSD1 suppresses p21 transcription by constraining its H3K4 promoter methylation, thereby resulting in GC progression. On the other hand, the downregulation of HDMs DPY300 and KDM5A was detected to enhance H3K4 methylation, consequently inhibiting GC’s growth [49,50]. Nishikawaji et al. (2016) demonstrated that GC cell growth and invasion undergo a marked reduction, via the knockdown of HDM SETDB2, which stimulates tumor-suppressor CADM1 and WWOX-expression levels [51].
Histone acetylation is intricately linked to DNA repair, chromatin remodeling, and gene-expression regulatory mechanisms. Histone acetylases (HAT) and deacetylases (HAD) are the primary enzymes involved in the regulation of gene transcription, by altering chromatin structure and controlling the binding of specific transcription factors. Several studies have established associations between acetylation/deacetylation processes, tumor TNM staging, GC development, and poor prognosis [52,53]. Elevated HDAC2-expression levels correlate with unfavorable outcomes in gastric cancer [54]. Similarly, another study presents a significant correlation between elevated HDACs 1-3-expression levels coexisting with lymph-node invasion and poor OS in GC patients [55].
Helicobacter pylori induces p21WAP1/CIP1 expression via the acetylation of H4 in the p21 promoter. Hp also promotes gastric carcinogenesis through the upregulation of JMJD2B expression, which, in turn, enhances COX-2 expression via NF-κΒ and correlates with the tumor stage, GC cell expansion, and recurrence [56,57,58]. Previous studies have suggested a strong correlation between H3K9 acetylation and GC poor prognosis, whereas reduced acetylation levels on H3K9 and H4K16 histones have been associated with poorly differentiated GC tumors [53,58].
The KAT2B and EP300 genes, which encode HATs, frequently undergo mutations or silencing in GC cases. Hp infection hinders HAT-p300 binding to the p27 promoter, leading to H4 hypoacetylation and ultimately to GC development [59].
Histone phosphorylation controls various cellular processes, such as cell signaling, apoptosis, chromosomal folding, compression, segregation, and DNA damage repair via kinases and phosphatases interaction [60]. Histones H3 and H4 are predominantly phosphorylated. In gastric carcinomas, H3S10 is less expressed in surgical resection margins, suggesting a significant prognostic role of phosphorylation in defining negative resection margins [61]. The Ras ERK1/2 signaling pathway also contributes to GC progression by inhibiting the phosphorylation of histone 1.4 at Serine 27 residue through Aurora B. Takahashi et al., in their cohort study involving 122 GC patients, demonstrated that the overexpression of phosphorylated histone H3 is indicative of poor prognosis [62]. H3S10 phosphorylation is catalyzed by Aurora kinase A (AURKA) [63].
AURKA is overexpressed in premalignant lesions such as gastric inflammation and intestinal metaplasia and is also negatively correlated with survival in gastric-cancer patients, suggesting a potential prognostic role [64,65,66].
Previous studies report that H3S10 phosphorylation is also induced by Hp infection and has been implicated in promoting gastric carcinogenesis [67]. Conversely, Hp infection in gastric epithelial cell lines diminishes H3S10 and H3T3 phosphorylation via a type-IV secretion system-dependent approach [68].
Histone ubiquitination in gastric carcinomas typically occurs subsequently to histone acetylation and/or methylation, or as a result of alterations in enzymes’ stability and activity, thereby exerting a synergistic effect on the cell cycle, DNA damage, and apoptosis [69]. The ubiquitination of mainly H2A and H2B histones impacts chromosome structure and can lead to degradation through the proteasome pathway [70]. H2B ubiquitination levels vary between differentiated carcinomas and the significantly lower levels detected in malignant tissues, strengthening the potential therapeutic role of the ubiquitination process in gastric cancer [71]. The ubiquitination of H2AK119 is consistently accompanied by H3K27me3 via the polycomb repressive complex 2 (PRC2) [72].
The Cullin4B-RING E3 ligase complex (CRL4B) is also capable of catalyzing H2AK119 ubiquitination and in cooperation with the PRC2 complex induces tumorigenesis [73]. The methylation of H3K4 and H3K79 histones via COMPASS and DOT1L enzymes, respectively, presupposes prior H2B ubiquitination [74]. The interplay between distinct histone modifications or the crosstalk between enzymes that modify histones requires further elucidation [75]. The observation that the expression of ubiquitinated H2B is notably reduced in gastric cancer cases suggests a potential therapeutic role for the histone-ubiquitination process [71].
Recently, a spectrum of novel histone modifications has been identified, like crotonylation, sumoylation, butyrylation, lactylation, biotinylation, neddylation, and succinylation [76,77]. In 2021, Fang et al. unveiled that during human embryonic stem cells’ meso-/endodermal differentiation, histone-crotonylation levels surged, resulting in a meso-/endoderm commitment. This finding suggests a connection between histone crotonylation and development processes [78]. Histone butyrylation and crotonylation seem to affect cancer proliferation and metastasis. Recently, the identification and analysis of the histone isobutyrylation process revealed the involvement of HAT1 and p300 [79,80,81].
SUMO family proteins catalyze the sumoylation of histone H4 and mediate transcriptional repression. H4K12 sumoylation dampens transcriptional activity by suppressing acetylation and methylation processes [82]. Contemporary investigations support that H4K12 sumoylation mediates to histone acetylation and methylation processes via p-300 and SET1/COMPASS suppression, respectively [82]. While the precise role of histone lactylation in cancer remains unknown, it is acknowledged to affect gene expression and metabolic regulation [80,81]. Less frequently observed histone modifications have been reported to interfere with the methylation or acetylation of histone proteins. Yet, the impact of these rare histone modifications in cancer biology remains unclear; although, evidence suggests their involvement in gene regulation, DNA damage repair, and metabolic regulation [76].
Metabolism reprogramming stands as a fundamental hallmark of cancer and intricately interacts with post-translational modifications and epigenetic processes via the crosstalk of signaling pathways [83,84,85]. Ulanovskaya et al. (2013), revealed that Nicotinamide N-methyltransferase (NNMT), a metabolic enzyme overexpressed in numerous human cancers, influences the methylation landscape of cancer cells [86].
The NNMT enzyme functions as a methyltransferase, catalyzing the conversion of nicotinamide to 1-methyl nicotinamide, utilizing the cofactor S-adenosyl-l-methionine (SAM) as the methyl-group donor. The removal of a methyl group converts SAM to S-adenosyl-L-homocysteine (SAH), known to inhibit methyltransferases. The NNMT enzyme appears to have the capacity to control the methylation status of cancer cells. Elevated NNMT levels have been described to correlate with the induction of EMT. They seem to alter gene expression not only via histone-dependent mechanisms but also through interactions with other proteins. Liang et al. (2017) conducted in vitro experiments on GC cell lines, demonstrating that heightened NNMT-expression levels activate TGF-β1/Smad signaling, thereby promoting the occurrence of EMT. Additionally, Wang et al. (2022) illustrated that NNMT activity mediates various intracellular processes by regulating the equilibrium of NAD+/NADH [87,88].
In gastric adenocarcinomas, NNMT is overexpressed, hinting at a potential role in GC tumorigenesis. In GC tumor cells, the elevated enzyme levels correlate positively with the TNM stage, tumor size, lymph-node infiltration, and distant metastasis, whereas reduced levels are linked to enhanced survival rates [89]. In GC cases exhibiting high expression levels of the enzyme, variations in the immune-cell composition within the tumor microenvironment suggest that NNMT could promote immune infiltration, potentially serving as a prognostic biomarker [90,91,92]. Furthermore, Wang et al. (2022), performing single-cell RNA sequencing analysis on over 95,000 cells originating from early gastric cardia adenocarcinoma cases, revealed that NNMT expression levels rose progressively throughout the malignant progression correlating with a worsened outcome [93].
3.3. Non-Coding RNAs
microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs) play pivotal roles in various processes implicated in gastric carcinogenesis (cell cycle, apoptosis, proliferation, migration, and invasion), and correlate with chemo- or radiosensitivity [94]. miRNAs function by binding to the 3′ (UTR) of the mRNA target, resulting in gene silencing [95]. Yu et al., conducting miRNA microarray experiments on an early gastric-cancer mouse model, unveiled the significant involvement of the miR200 family in gastric carcinogenesis initiation. These miRNA molecules prove to be efficient predictors of overall survival in early gastric carcinomas (Table 2) [94,95].

Table 2.
miRNAs as candidate biomarkers in gastric cancer.
lncRNAs mediate the regulation of chromatin remodeling, transcription, and post-transcriptional gene-expression processes [96]. HOTAIR, a frequently overexpressed lncRNA, may contribute to the dissemination of gastric-cancer cells. Previous studies suggest its involvement in specific signaling pathways, including Wnt/β-catenin and PI3K/Akt pathways, as well as its role in either silencing or upregulating HOXD and miR34a or miR-330 and miR-331-3p, respectively (Table 3) [105,106,107]. lncRNAs like H19, MNX1-AS1, MALAT1, HULC, and UCA1, have been characterized as playing an oncogenic role in gastric carcinomas [108], whereas others like CRNDE appear to inhibit GC development [109].

Table 3.
Lnc-RNAs as candidate biomarkers in gastric cancer.
Another interesting lncRNA is the X-inactive specific transcript (XIST) that has been described to be overexpressed in various cancers, including gastric cancer. As previously described, lncRNA XIST acts via miR-101 and regulates EZH2, supporting the lncRNA-miRNA crosstalk that has been already mentioned in various cancers. In gastric cancer, XIST lncRNA-expression levels were found to be overexpressed in both GC patients and cell lines, and its upregulation was linked to tumor size, positive lymph nodes, and the TNM stage. It was also proposed that lncRNA XIST targets oncogene MACC1, which mediates the HGF/c-Met pathway, via its interaction with miR-497 and promotes gastric tumor growth and invasion [114]. Recent studies on the role of lncRNA XIST in gastric carcinogenesis have proposed an interplay between XIST lncRNA and Janus kinase 2 (JAK2) transcription factor that leads to GC cell migration [115]. Furthermore, another research study demonstrated that XIST lncRNA alters miR-132 and paxillin (PXN) expression levels. The last one is an adhesion molecule that has been linked to GC invasiveness. miR-132 targets the PXN gene, thus the interplay between XIST lncRNA and PXN leads to the regulation of the latter. Given the information provided, one might propose that XIST lncRNA could serve as a potential therapeutic target in gastric carcinomas [116].
circRNAs, as previously described, contribute to tumor development, metastasis, recurrence, and therapeutic resistance [117]. Sequencing data analysis of GC cases revealed numerous circRNA molecules with pro- or antitumor activity. Elevated levels of ciRS-7 are related to GC progression [118]. Acting as an oncogene, ciRS-7 counteracts the mirR-7-mediated inhibition of the PTEN/PI3K/AKT pathway in GC (Table 4). Recently, Lin et al. presented the correlation between circRIMS and gastric-cancer metastasis. In this study, circRIMS expression levels were significantly elevated in T3N3M0 and T3N1M0 gastric cancer cases, suggesting a potential diagnostic and therapeutic role of circRIMS in gastric carcinomas [119].

Table 4.
circ-RNAs as candidate biomarkers in gastric cancer.
Overexpressed hsa_circ_0005092 and hsa_circ_0002647 molecules are positively correlated with recurrence-free survival (RFS) and OS in patients with gastric adenocarcinomas [120]. Given their high stability in body fluid, circular RNAs could serve as effective biomarkers for cancer diagnosis, prognosis, and monitoring responses to treatment [138]. Based on research conducted by Han et al., a notable decrease in circRNAs hsa_circ_0021087 and hsa_circ_0005051 was observed in tissue and liquid biopsies from GC patients, indicating their potential as a non-invasive diagnostic biomarker [121]. A recent meta-analysis in gastric adenocarcinomas revealed that hsa_circ_0002019 and hsa_circ_0074736 molecules could regulate post-transcriptionally the expression levels of several genes (ERBB4, PRTG, SLITRK2, and GUCY1A2) linked to gastric-cancer survival. Therefore, circRNAs could serve as prognostic biomarkers [122].
4. Gastric-Cancer Clinical Management
Gastric carcinomas exhibit notable inter- and intratumoral heterogeneity, which may partially be responsible for the unfavorable outcome of this condition. Understanding tumor biology is expected to enhance treatment efficacy and accelerate the discovery of novel prognostic and predictive biomarkers [139]. Radical surgery remains the main treatment approach for localized gastric cancer. Various therapeutic strategies, including perioperative chemotherapy and adjuvant chemo- or adjuvant chemoradiotherapy, have been proposed to decrease the recurrence risk and prolong lifespan [140,141,142,143]. Targeted therapy with antihuman epidermal receptor 2 (anti-HER2) and anti-vascular endothelial growth factor (anti-VEGF) perioperatively remains under investigation.
Prior studies have shown that perioperative chemotherapy, comparative to surgery alone, can improve patients’ prognosis, establishing it as a widely adopted practice in various countries. Numerous clinical trials have shown improved survival rates in stage II/III GC patients undergoing primary surgery, whilst the data about adjuvant radiotherapy remains controversial [140]. The precise role of targeted therapies (anti-HER2, anti-VEGF), along with immunotherapy using Immune Checkpoint Inhibitors (ICIs), remains unclear, with several phase-II/III clinical trials seeking clarity.
Molecular biomarkers such as HER2, MSI, and programmed cell death ligand 1 (PD-L1) could distinguish patients with metastatic disease who may benefit from targeted therapy or immunotherapy. Nonetheless, early detection methods and preventing-recurrence measures remain an urgent need. In the context of liquid biopsy, circulating molecules have the potential to predict recurrence [141,142]. Resistance to ICI treatment is another issue that remains to be addressed. A comprehensive investigation into epigenetics, metabolism, microbiomes, and the immune system is necessary to elucidate the underlying mechanisms of immune modulation and resistance [143,144,145].
5. Discussion
Gastric cancer is commonly diagnosed worldwide and exerts a significant impact on healthcare. The absence of specific clinical symptoms often leads to the diagnosis of the disease at late stages. Inter- and intratumoral heterogeneity contributes to poor prognosis. Given the unclear effectiveness of targeted therapies and immunotherapy, it is essential to establish reliable biomarkers for prognostic and diagnostic purposes. Epigenetics emerges as a promising field for acquiring comprehensive insights into the molecular processes underlying gastric cancer, as well as for developing potential prognostic and predictive biomarkers.
Promoter hyper- or hypomethylation is a common mechanism of tumor-suppressor gene silencing or oncogene activation, respectively. Tumor-suppressor genes, silenced by improper DNA methylation, serve as drivers in tumorigenesis. Widespread methylation in cancer cells also leads to non-core region methylation in surrounding tissues [14]. Previous studies have proposed an in vivo model for aberrant DNA methylation. According to this model, several inflammatory mediators, such as IL-1, IL-6, and TNF, activate DNMT1 and EZH2, causing dense methylation in promoter CpG regions. This process predisposes cancer development as it generates an “epigenetic field” effect even in noncancerous or precancerous cases [146,147]. However, in gastric carcinomas, hypomethylation, focused on the decreased methylation of repetitive DNA sequences (ALU, LINE1), as we progress from chronic to cancerous lesions, has also been described. This fact establishes hypomethylation as an early event in GC carcinogenesis and suggests that LINE1 hypomethylation could serve as a marker for “epigenetic field cancerization” [148].
Hp and EBV infections exert an impact on aberrant DNA methylation. As previously mentioned, EBV infection, although a rare event, triggers DNA methylation through the direct dysregulation of DNMT enzymes. On the other hand, Hp induces DNA methylation in two ways: directly, through the effect of its pathogenic product CagA protein, and indirectly, via chronic inflammation and the CpG-island methylation of tumor-suppressor genes such as CDH1, p16, and IL1β [147]. Additionally, previous studies have suggested that EBV could also trigger Hp infection, resulting in increased bacterial pathogenic CagA-protein activity [146]. Furthermore, as previously pointed out, the synergistic effects of Hp and EBV coinfection via oncoprotein gankyrin overexpression dysregulate migration, apoptotic, and DNA damage-repair pathways, boosting GC aggressiveness [149].
We are aware that CDKN2A and MLH1 silencing are common epigenetic events in gastric carcinomas and are associated with cell-cycle regulation and DNA repair mechanisms (see Table 1). The methylation-induced silencing of p16 has been reported in 25–42% of gastric carcinomas, thus potentially serving as a biomarker for primary cancer diagnosis by predicting the malignancy of dysplastic lesions [150,151]. Approximately 30% of gastric carcinomas exhibit CDKN2A methylation, potentially contributing to the malignant transformation of gastric lesions [152,153]. The methylation of the MLH1 promoter has been observed in 31–67% of gastric carcinomas and has been identified as an early event in GC cases, primarily in the papillary subtype [34]. Furthermore, in half of GC cases, promoter hypermethylation, as well as hemizygous deletion, lead to RUNX3 tumor-suppressor gene silencing [154].
EBV infection-mediated methylation may occur in CpG islands as well as in low-CpG regions [155]. TCGA analysis has shown that EBV-positive tumors present a distinct CpG-island methylator phenotype (CIMP), harboring CDKN2A (p16) gene-promoter methylation but not MLH1 methylation [6]. The CIMP-high phenotype is related to a more favorable prognosis, diffuse histological subtype, and earlier disease stages than a CIMP-negative phenotype [156]. EBV-positive gastric adenocarcinomas exhibit high immunogenicity and therefore could be utilized for additional studies on immunotherapy, leading to new potential targets for personalized treatment [157]. In EBV-positive GC cases, almost 270 genes are methylated, including CXXC4, TIMP2, and PLXND1, while COL9A2, EYA1, and ZNF365 are highly methylated in both EBV-positive and EBV-negative/MSI-high subtypes. The MLH1 gene promoter is frequently methylated (46%) in EBV-negative/MSI-high subtypes but not in EBV-positive gastric carcinomas. Moreover, DNA hypermethylation in the MSI subgroup strengthens the idea that specific “epimutations” serve as a compass for subsequent alterations [154].
In patients with primary gastric carcinomas, Laminin a4 subunit (LAMA4) expression is associated with high-grade tumors and predicts poor OS. LAMA4 gene expression is upregulated in gastric-cancer cells through the binding of the ZEB1 (Zinc finger E-box-binding homeobox 1) transcription factor to the LAMA4 promoter. It has been reported that LAMA4 upregulation correlates with invasion and metastasis [22]. Additional molecules acting as transcription factors like caudal-type homeobox 1/2(CDX1/2), and Kruppel-like factor 5 (KLF5) are involved in the Sonic Hedgehog pathway (SHH). CDX1/CDX2 genes seem to have a key role in intestinal dysplasia reprogramming. Previous studies have mentioned a remarkably high CDX2 expression but decreased CDX1/KLF5 expression in gastric tissue samples mainly due to methylation. Small molecules show antiproliferative activity in colon cancer through KLF5-expression inhibition, thus SHH implication in gastric carcinogenesis could be further evaluated for targeted therapy [158].
Kim et al. (2018) demonstrated that the hypermethylation of Insulin-like growth factor-binding protein 7 (IGFBP7) exon 1 induces gene downregulation, thereby supporting tumor-suppressor activities in gastric cancer. IGFBP7 protein-expression levels correlate with poor clinical outcomes, and therefore could be a potential therapeutic target, particularly in patients with poor prognosis [26].
FBXO32 is a tumor-suppressor gene that encodes an F-box protein, constituting one of the four subunits of the ubiquitin protein ligase complex. The hypermethylation of FBXO32 leads to the downregulation or loss of function. The reactivation of the FBXO32 gene is suggested to possess prognostic significance and may offer therapeutic advantages [23]. The CDH11 tumor-suppressor gene is also silenced through promoter hypermethylation in gastric adenocarcinomas and is considered a potential prognostic biomarker associated with malignant behavior [24,25].
The prognosis of advanced gastric adenocarcinomas can be predicted by evaluating the Claudin-3-promoter methylation status, which results in decreased protein levels. Conversely, claudin-3-promoter hypomethylation appears to contribute to the genesis of intestinal-type GC cases. Zhang et al. (2018) demonstrated that promoter hypermethylation and low claudin-3-expression levels indicate a poorly differentiated phenotype and a higher metastatic status of gastric carcinoma with lymph-node spread. The methylation profile of claudin-3 could serve as a prognostic/predictive biomarker as well as a promising therapeutic target for advanced GC cases [27].
Recently, Heo et al. (2023) showed that in both early- and advanced-stage primary gastric carcinomas, the Tensin 4 gene (TNS4) was overexpressed. Based on TCGA molecular subtypes, TNS4-expression levels were higher in the EBV-positive subgroup. The TNS4 gene encodes a protein located in focal adhesion sites, facilitating interaction between the cytoskeletal network and the extracellular matrix. Elevated TNS4 expression has been shown to promote the EMT process through AKT/GSK-3β signaling and is proposed to correlate with the development and progression of gastric adenomas, a process purportedly triggered by Hp infection [159].
In gastric carcinomas, the elevated methylation of the hTERT promoter has also been observed. To identify patients at risk of gastric-carcinoma development, hTERT hypermethylation is crucial. Gastric-cancer proliferation as well as distant and lymphatic metastasis are correlated to hTERT-promoter methylation, thus the latter could be used as a diagnostic marker of gastric carcinomas and classify patients who are at high risk of GC development [28,29].
The SRBC gene (serum-deprivation response factor-related gene product that binds to the c-kinase) encodes a protein that is downregulated in various cancer cell lines, suggesting a possible tumor-suppressor role. Normally, the protein regulates the traffic and/or budding of caveolae and plays a role in caveolae formation in a tissue-specific manner. The inactivation of SRBC is implicated in tumor resistance against chemotherapeutic agents such as oxaliplatin, as previously reported [160]. Research studies have reported that inactivation is achieved mainly through CpG-island DNA hypermethylation, while gene somatic mutations or biallelic deletion seem to be less frequent events. However, the DNA methylation of the SRBC gene promoter is not the only inactivation mechanism. Histone modifications were also found to play a role in gene inactivation. EZH2 trimethylates histone H3 at lysine 27, leading to gene-expression control and contributing to SRBC gene downregulation in gastric-cancer tissues [161].
Furthermore, the CD274 and PDCD1LG2 genes encode the immunosuppressive molecules PD-L1 and PD-L2. According to the TCGA group classification, only the EBV (+) GC molecular subtype exhibits elevated PD-L1- and/or PD-L2-expression levels. Recently, Zhu et al. (2020) have shown that the PD-L1 promoter was mostly hypermethylated in gastric carcinoma patients who have already received anti-PD-1 therapy. PD-L1 promoter methylation was elevated after therapy with pembrolizumab. However, surgery alone after recurrence did not have an impact on the methylation status of PD-L1. PD-L1 seems to exert an oncogenic activity and promotes cancer development and infiltration through RAS/MAP and AKT signaling pathways. Resistance to anti-PD-1 immunotherapy might develop because of PD-L1 promoter methylation [30].
PD-L2 is a second ligand of PD-1 and can be expressed via tumor cells. In gastric carcinomas, EBV status, CIMP phenotype, as well as MLH1 and CDKN2A methylation status, were strongly associated with methylated PD-L2. The TCGA group found that PD-L2 methylation is associated with EBV infection, CD8+ T cell infiltration, microsatellite instability, and high tumor mutational load. PD-L2 hypermethylation appears to be a promising biomarker for the prediction of responses in GC patients after anti-PD-1 immunotherapy [31].
DNMTs are pivotal contributors to the process of DNA methylation. Previous studies have shown elevated levels of DNMT-1- and DNMTs-3A/3B-protein expression in both HP-induced gastritis and gastric carcinomas [162,163]. Hedayati et al. (2022), presented a correlation between Hp pathogenicity and increased DNMT1-expression levels [164], whereas Song et al. (2020) identified a novel epigenetic signature in gastrointestinal adenocarcinomas. They scrutinized their findings along with TCGA datasets for DNA methylation and RNA sequencing and identified a total of five methylation-driven genes, capable of predicting overall survival (OS) in GC patients. Three of those, HENMT1, GRIN2A, and STC2, seem to relate to processes such as hypoxia response and hormone-metabolism regulation, thereby contributing to the development and progression of several carcinomas, including gastric adenocarcinomas [165].
Purkait et al. (2020), have documented the interplay between DNA methylation and histone modifications in the pathogenesis of gastric carcinomas. DNMT-1/3A/3B and EZH2 expression were significantly upregulated in Hp-associated gastritis and carcinomas. EZH2-expression levels were notably higher in cases of metaplasia and exhibited a positive correlation with DNMT-expression levels. Epigenetic modifications can be reversed, thus targeting DNMTs and EZH2 could offer a promising therapeutic approach for gastric carcinoma [163]. Numerous inhibitors targeting DNMTs and EZH2 are examined to assess their potential efficacy in treating various malignancies [166,167].
Takeshima et al. revealed that chromatin remodelers like SMARCA1 present a distinct methylation status in GC patients, compared to noncancerous tissues. That claim it itself supports that chromatin remodeler disorders occur early during tumorigenesis and contribute to the production of an epigenetic field effect [168].
Numerous in vitro and in vivo studies have suggested that Hp infection causes DNA double-strand breaks (DSBs) and activates the ATM (Ataxia-Telangiectasia-Mutated serine/threonine kinase) response. ATM is involved in the DSB-repair pathway and phosphorylates other DSB-implicated proteins. Santos et al. (2018) demonstrated that ATM gene transcription is epigenetically regulated via promoter hypomethylation as well as the hyperacetylation of H3/H4 histones. Chromatin immunoprecipitation-quantitative PCR experiments unveiled that H3 and H4 histones were highly acetylated four hours after Hp infection [169].
In certain cancer types, the chromatin landscape of various genes and their interactions with polycomb repressive complex 2 (PRC2) and acetyltransferase complexes can significantly influence the transcriptional status of the entire genome. It is well recognized that histone modifications including H3/H4 acetylations and H3 trimethylation are linked to gene upregulation, while gene downregulation is mainly achieved through H3 lysine 9 di-/trimethylation as well as H3 lysine 27 trimethylation [146]. Members of the MLLs family serve as tumor-suppressor genes and mediate in gene activation through histone H3 methylation at lysine 4 amino acid (H3K4). Lysine-specific demethylase 6A (KDM6A) also regulates gene expression through H3K27 demethylation. Previous studies in GC cases have highlighted the association between PCAF (P300/CBP-associated factor) loss and poor outcomes. Brasacchio et al. (2018), have demonstrated that PCAF loss has an impact on gastric adenocarcinoma initiation, thus PCAF could serve as a candidate acetylation factor [32,33].
Hp and EBV infections alter histone modifications as previously described. EBV infection converts H3K9me3+ heterochromatin into a state characterized by H3K4me1+/H3K27ac+ bivalency. This fact leads to the activation of latent enhancers that stimulate the expression of genes implicated in gastric carcinogenesis like TGFBR2 and MZT1 [170]. Hp upregulates the expression of both the p21 WAP/CIP1 tumor-suppressor gene as well as the JMJD2B gene, which promotes GC carcinogenesis through histone acetylation.
Non-coding RNAs, miRNAs, and circRNAs have been extensively studied as one of the three pillars of epigenetics. Fan et al. (2020) elucidated elevated expression levels of the CCDC144NL-AS1 gene, which are related to unfavorable prognosis, invasion, migration, and apoptosis inhibition in GC cells [110]. CCDC144NL-AS1 likely acts as a competing endogenous RNA (ceRNA), occupying common miRNA binding regions. Therefore, it could serve as a promising diagnostic and therapeutic target [171]. HOTAIR has been described as being overexpressed in gastric cancer, playing a pivotal role in metastasis and survival. The specific lncRNA molecule engages in the interplay with miRNAs and acts as a sponge for miR331-3p. Moreover, it can trigger the HER2 target’s expression levels [172].
Liu et al. (2021), analyzed LINC01232 in gastric-cancer cell lines and the TCGA dataset. Their findings unveiled that KLF2 expression is inhibited through H3K27me3 histone methylation. The knockdown of LINC01232 is shown to suppress the proliferation of gastric-cancer cells both in vivo and in vitro. This finding suggests LINC01232’s potential as an effective therapeutic target for gastric cancer (Table 2) [111].
Recently, Peng et al. (2022), performing RNA-Seq data analysis in stomach adenocarcinomas, demonstrated that the lncRNA TM4SF1-AS1 inhibits the immune killing capacity mediated by T cells and serves as a prognostic marker for assessing the immune response to anti-PD1 therapy [112].
The zinc finger antisense 1 (ZFAS1) is another lncRNA demonstrating oncogenic activity in gastric cancer. RNA sequencing analysis has revealed that upregulated ZFAS1 expression correlates with diminished levels of hypoxia-inducible factors 1 and 2 (HIF1, HIF2) suggesting a potential involvement of ZFAS1 in HIF1A epigenetic silencing. ZFAS1 upregulates the HIF2 protein levels under both hypoxia and normoxia conditions. The knockdown of ZFAS1 via siRNA in gastric-cancer cell lines results in decreased cell migration, invasion, and proliferation, providing evidence that ZFAS1 may serve as an independent prognostic biomarker as well as a therapeutic target in gastric cancer [113].
Lately, a comprehensive analysis of immune-related long non-coding RNAs (Jin et al., 2023) identified nine significant lncRNA molecules. Gastric cancer was stratified into five subtypes, with cluster C3 being the most versus cluster C5 being the least immunogenic. This study documented that the specific lncRNA subtypes possess the ability to identify patients who may derive benefit from receiving chemotherapy and immunotherapy [173].
Regarding miRNAs in gastric-cancer development, their role has been extensively examined. Data analysis conducted by Kipkeeva et al. (2020), revealed that miRNAs associated with the spread of cancer cells are commonly engaged with the Wnt/-catenin pathway or influence genes that trigger the EMT process through interaction with other pathways. miRNAs implicated in chemotherapy resistance mostly target apoptotic regulators and are linked to the PI3K/AKT/mTOR pathway. Their potential as biomarkers holds considerable promise for detecting and monitoring gastric cancer [174].
As previously described, the direct targeting of the nuclear protein Forkhead box M1 (FoxM1) triggers pathways such as MEK/ERK, NF-κB, and PI3K/AKT, via mir-320d in GC cases, suggesting its tumor-suppressive properties and positioning mir-320d as a potential biomarker for cancer prognosis and treatment (Table 3) [97]. Additionally, miR-942 is implicated in invasion and migration processes. miR 942 3p functions as an oncogene, while miR 942 5p acts as a tumor-suppressor gene. Moreover, serum miR-942 expression levels are linked to disease progression and low survival rates, suggesting its utility as a prognostic biomarker [98].
miR-141 and miR-1269b are also implicated in gastric carcinomas. The first one affects MEK/ERK- and MAPK-signaling pathways and has been demonstrated to inhibit proliferation and trigger apoptosis in gastric-adenocarcinoma cell lines. miR-1269b, through its regulatory control over methyltransferase-like 3 (METTL3), exerts inhibitory effects on gastric-cancer development. The overexpression of miR-1269b suppresses the migration and invasion of GC cells, whereas its inhibition leads to the opposite effects [99,175]. Reduced levels of miR-203a/b, accompanied by the methylation of their proximal promoters, indicate their role as tumor-suppressive microRNAs in GC cases. Furthermore, miR-3196, miR-1244, miR-135b-5p, and miR-628-3p are associated with GC differentiation, while miR-196a-5p appears to correlate with the age of GC onset.
In another study, the performance of a miRNA microarray analysis on 353 Japanese patients with primary gastric tumors identified a GC miRNA signature consisting of 22 up- and 13 downregulated miRNAs. In this study, a “histotype” miRNA panel was developed, capable of distinguishing diffuse and intestinal-type gastric tumors. Moreover, miR-let7g emerged as an independent predictive biomarker for disease-free survival (DFS) [176]. The let-7 family negatively regulates HMGA2, whose elevated expression levels are associated with tumor invasiveness and unfavorable outcomes, suggesting a potential prognostic role in gastric cancer [104].
Qu et al. (2020), utilizing miRNA-expression data from 386 GC cases reported from the TCGA group, demonstrated that miR-30a-3p and miR-105-5p have the potential to serve as biomarkers for MSI-H gastric adenocarcinoma, likely due to their ability to modify the expression of DNA damage-repair genes [102]. The GEO database analysis of miRNA- and mRNA-expression data from GC cases identified hsa-miR-196b-3p and four important nodal genes (CALML4, SMAD6, PITX2, and TGFB2) as prognostic GC biomarkers in Hp-positive cases. It was suggested that hsa-miR-196b-3p may serve as a reliable biomarker for predicting GC prognosis [100].
Gilani et al. (2022) analyzed the GSE106817 dataset with 2.566 miRNAs, utilizing the Boruta machine learning variable-selection approach, and discovered a total of 30 miRNAs capable of serving as biomarkers in diagnosing GC. hsa-miR-1343-3p presented the highest ranking among them. Utilizing artificial intelligence technology, they identified hsa-miR-1343-3 as a strong candidate for biomarker analysis in GC diagnosis, as well as hsa-miR-8073 and hsa-miR-1228-5p, which exhibit significant contributions to GC prediction [101].
Additionally, miR-1228 via the downregulation of the macrophage migration inhibitory factor (MIF) serves as a negative regulator of gastric-cancer growth and angiogenesis, supporting its use as a potential therapeutic target for anti-angiogenic therapy against gastric cancer [103].
Concerning the role of circular RNAs, numerous studies have been conducted. Currently, traditional tumor markers like CEA and CA19-9 have limited clinical utility due to their decreased sensitivity and specificity. Research studies have established the presence of circRNAs not only within tissues but also in human serum, plasma, and other body fluids [177].
Previous studies have highlighted elevated levels of CircAKT3 and circLMO7 in gastric-cancer cells, suggesting potential implications in GC development [123]. The elevated expression levels of serum circSHKBP1 (hsa_circ_0000936) are associated with diminished survival rates and advanced TNM stages, as reported by Xie et al. (2020) (Table 4). Furthermore, the plasma levels of hsa_circ_0000745 correlate with TNM staging. The study conducted by Reisdas-Mercês et al. (2022) demonstrated that hsa_circ_0000211, hsa_circ_0000284, and hsa_circ_0004771 maintain consistent expression profiles across various techniques (RNA-Seq and RTqPCR) and distinct sample types (tissue and blood) [124,178].
Several circRNAs exhibit potential for early gastric-cancer screening in combination with other tumor markers, though their sensitivity remains limited when used alone. For instance, the co-detection of hsa_circ_0001017 and hsa_circ_0061276 in both gastric-cancer tissues and patient plasma exhibits a significant diagnostic potential, with a sensitivity and specificity of 95.5% and 95.7%, respectively [179].
Zheng et al. (2022) observed a significant reduction in the expression levels of exosomal hsa_circ_0015286 in GC patients shortly after the surgical treatment. This noteworthy finding indicates that exosomal hsa_circ_0015286 has the potential to serve as a non-invasive biomarker for the diagnosis and prognostic evaluation of GC [125].
Zhang et al. (2017) demonstrated that the decreased expression of circLARP4 in GC tissues serves as an independent prognostic indicator for the overall survival of GC patients [126].
The differential expression of serum hsa_circ_0007507 among post-operative GC, gastritis, intestinal metaplasia, and relapsed patients indicates its potential utility as a novel diagnostic and dynamic-monitoring biomarker for GC [127]. Significantly distinct levels of hsa_-circ_002059 were also observed in plasma samples collected post-operatively compared to those collected pre-operatively. The lower expression levels exhibited a significant association with distant metastasis, the TNM stage, and patient age [128]. Prior studies have demonstrated a close relation between hsa_circ_0000467-expression levels and TNM staging, while the downregulation of hsa_circ_KIAA1244 in GC patients suggests a possible role as a diagnostic marker for GC, considering its correlation to TNM stage, metastatic potential, and shorter survival rates [129].
Zhang et al. identified several circRNAs (circDLST, circCACTIN, and circNRIP1) pivotal in tumor growth, migration, and invasion [130,131,132]. Wang et al. (2019), reported elevated levels of CircLMTK2 in GC tissues, with its presence correlating with unfavorable prognosis and advanced TNM stages. circOSBPL10 (hsa_circ_0008549) exhibits a significant upregulation in GC tissues and promotes gastric-cancer cell growth. Therefore, circOSBPL10 (hsa_circ_0008549) was proposed as a novel prognostic biomarker in gastric carcinomas [133,134].
CircRNAs can also impact drug resistance. Shi and Wang (2022) demonstrated that Circ_AKT3 knockdown results in increased cisplatin sensitivity in cisplatin-resistant GC cells via the miR-206/PTPN14 axis. Furthermore, Fan et al. (2022) found that the METTL14-mediated m6A alteration of circORC5 inhibits gastric-cancer progression. Fang et al. (2022) discovered that circCPM plays a crucial role in regulating GC autophagy and 5-Fluoro- Uracil resistance, while Chen et al. (2021) revealed that CircDLG1 was highly increased in distant metastatic lesions and anti-PD-1-resistant gastric-cancer tissues. CircDLG1 was also linked with an aggressive tumor phenotype and poor prognosis in GC patients treated with anti-PD-1 drugs [135,136,137,180].
The linkage between DNA methylation and miRNAs has been characterized in an expanded mass of the literature as an additional tier in the control of gene expression. Several studies have mentioned the crosstalk between DNA methylation and miRNAs. This interplay happens via the methylation process of miRNA promoters leading to the control of their expression, or through the inhibition of enzymes (Dicer, Drosha) that participate in the miRNA process. On the other hand, miRNA molecules alter gene methylation profiles by modifying enzymes or co-factors that take part in the DNA methylation process [181]. A study conducted in GC patients (2016) reported that miR-106a promoter hypomethylation resulted in its overexpression, and plasma miR-106a expression levels were decreased after gastrectomy, illustrating a potential diagnostic role for miR-106a [182].
Another interesting interaction is the one between miRNAs and histone-modifying enzymes. Previous studies report direct and indirect interactions with enzymes or mechanisms mediating chromatin remodeling. The major components of the miRNA-chromatin-remodeling network include signaling pathways, transcription factors, DNA methylation, long non-coding RNAs, and various chromatin-remodeling complexes. These elements are crucial for the regulation of processes such as apoptosis, cell proliferation, and differentiation [183].
Epigenetic drugs are compounds that target and repair various post-translational modifications of enzymes implicated in histone remodeling and DNA methylation processes [184]. They are classified into five groups: DNA methyltransferase inhibitors (iDNMTs), histone methyltransferase inhibitors (iHMTs), histone demethylase inhibitors (iHDMs), histone acetyltransferase/de-acetyltransferase inhibitors, (iHATs/iHDACs), and miRNA inhibitors (anti-miRs). However, only few iDNMTs and iHDACs have been approved by the FDA for use in several malignancies including cancer. Both iHMTs and iHDMs as well as anti-miRs are under investigation and numerous clinical studies are still ongoing [185].
Epigenetics and epigenetic treatment options represent rapidly advancing fields. Novel pharmaceutical compounds that address pharmacokinetic and instability problems in epigenetic treatment hold promise for surpassing previous generations of DNMT and HDAC inhibitors [186]. In addition, recent research has assessed the synergistic effects of iDNMTs and iHDACs, which provide further support for the encouraging findings. However, the utilization of these compounds in clinical practice is diminishing due to their lack of specificity and potential to induce mutations that contribute to additional carcinogenesis [185]. Therefore, the pharmaceutical research focuses not only on the development of next-generation iDNMTs and iHDACs, but also in other compounds such as NNMT inhibitors and anti-miRs.
Lately, Gao et al. (2021) investigated NNMT inhibitors and discovered the GYZ-319 compound as a potent iNNMT [187]. However, when it was used against several cancer cell lines, it showed poor cell permeability. The lack of cellular activity was rendered to the presence of two highly polar functional groups, which are found in all effective bisubstrate NNMT inhibitors: the carboxylic acid and amine groups of the amino acid motif. Therefore, van Haren et al., from the previous study group, conducted further research and utilizing the RaPID mRNA display methodology identified a group of macrocyclic peptides with NNMT-affinity properties [188]. Van Haren et al. (2021) conducted a study where they chemically modified the iNNMT GYZ-319 molecule to enhance its cellular activity. The process included converting carboxylic acid into several esters, followed by their assessment. Their experiments resulted in the conclusion that the isopropyl ester 12e and the isopropyl ester/TML dual prodrug 14e were the most promising molecules according to stability and cellular activity evaluation [189].
In recent years, several studies have focused on the development of epigenome-targeted therapies. Moro et al. (2020) examined epigenetic priming in multiple gastric cell lines, evaluating the efficacy of SN38, CDDP, PTX, and 5-FU cytotoxic drugs for gastric cancer. Their findings demonstrated that epigenetic priming is effective in cell lines resistant to SN38 and CDDP, as well as that genes involved in apoptosis and cell death were activated [190].
Furthermore, another study conducted by Hu et al. (2022) revealed that nanoparticles could be produced in a manner that could be able to deliver epigenetic drugs, suggesting a possible therapeutic tool in epigenome therapy [191].
Additional studies confirm that epigenetic agents, in conjunction with specific compounds, may boost antitumor immunity. Ongoing clinical studies investigate the combined use of epigenetic drugs and immune checkpoint inhibitors to evaluate their therapeutic benefits and potential adverse effects [192]
Finally, understanding the role of miRNAs in gastric-cancer epigenetics has spurred the development of anticancer miRNA-replacement therapy. miRNAs exhibiting oncogenic activity are considered therapeutic targets. Anti-miR oligonucleotides could be used to bind the guide strand of miRNAs. Thus, a multiple-miRNA-inhibition therapy in combination with other chemical compounds could achieve a synergistic antitumor effect [193].
6. Conclusions
Given the intricate nature of the mechanisms underlying the onset and progression of gastric carcinomas, identifying pivotal contributors to cancer pathogenesis holds immense significance. It is imperative to comprehend the precise mechanisms that underlie significant changes in epigenetic drivers. The identification and application of non-invasive epigenetic biomarkers will ensure the early detection and monitoring of both early- and advanced-stage gastric-cancer patients, facilitating tailored treatment strategies based on their epigenome profile. However, so far, we are lacking a specific and validated epigenetic signature. Therefore, numerous clinical trials involving the use of various epigenetic-modulating agents alone or in a multi-therapy approach with encouraging outcomes are ongoing. Deciphering the epigenetic network provides tremendous potential for precision medicine and will allow the implementation of revolutionary epigenetic approaches (anti-miR therapy and nanoparticles delivering epigenetic drugs) in routine clinical practice.
Author Contributions
Conceptualization, M.S., E.T. and G.C.; methodology, M.S. and G.C.; validation, M.S., E.T. and G.C.; investigation, M.S., K.-E.K. and M.-N.K.; writing—original draft preparation, M.S., E.T., G.C., K.-E.K. and M.-N.K.; writing—review and editing, M.S., G.C., K.-E.K. and M.-N.K.; supervision, M.S.; project administration, M.S., E.T. and G.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Arnold, M.; Park, J.Y.; Camargo, M.C.; Lunet, N.; Forman, D.; Soerjomataram, I. Is Gastric Cancer Becoming a Rare Disease? A Global Assessment of Predicted Incidence Trends to 2035. Gut 2020, 69, 823–829. [Google Scholar] [CrossRef]
- Rawla, P.; Barsouk, A. Epidemiology of Gastric Cancer: Global Trends, Risk Factors and Prevention. Prz. Gastroenterol. 2019, 14, 26–38. [Google Scholar] [CrossRef]
- Toh, J.W.T.; Wilson, R.B. Pathways of Gastric Carcinogenesis, Helicobacter Pylori Virulence and Interactions with Antioxidant Systems, Vitamin C and Phytochemicals. Int. J. Mol. Sci. 2020, 21, 6451. [Google Scholar] [CrossRef]
- Puneet; Kazmi, H.R.; Kumari, S.; Tiwari, S.; Khanna, A.; Narayan, G. Epigenetic Mechanisms and Events in Gastric Cancer-Emerging Novel Biomarkers. Pathol. Oncol. Res. 2018, 24, 757–770. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network. Comprehensive Molecular Characterization of Gastric Adenocarcinoma. Nature 2014, 513, 202–209. [Google Scholar] [CrossRef]
- Cristescu, R.; Lee, J.; Nebozhyn, M.; Kim, K.-M.; Ting, J.C.; Wong, S.S.; Liu, J.; Yue, Y.G.; Wang, J.; Yu, K.; et al. Molecular Analysis of Gastric Cancer Identifies Subtypes Associated with Distinct Clinical Outcomes. Nat. Med. 2015, 21, 449–456. [Google Scholar] [CrossRef]
- Hu, X.; Wang, Z.; Wang, Q.; Chen, K.; Han, Q.; Bai, S.; Du, J.; Chen, W. Molecular Classification Reveals the Diverse Genetic and Prognostic Features of Gastric Cancer: A Multi-Omics Consensus Ensemble Clustering. Biomed. Pharmacother. 2021, 144, 112222. [Google Scholar] [CrossRef]
- Wang, H.; Ding, Y.; Chen, Y.; Jiang, J.; Chen, Y.; Lu, J.; Kong, M.; Mo, F.; Huang, Y.; Zhao, W.; et al. A Novel Genomic Classification System of Gastric Cancer via Integrating Multidimensional Genomic Characteristics. Gastric. Cancer 2021, 24, 1227–1241. [Google Scholar] [CrossRef]
- Li, B.; Zhang, F.; Niu, Q.; Liu, J.; Yu, Y.; Wang, P.; Zhang, S.; Zhang, H.; Wang, Z. A Molecular Classification of Gastric Cancer Associated with Distinct Clinical Outcomes and Validated by an XGBoost-Based Prediction Model. Mol. Ther. Nucleic Acids 2023, 31, 224–240. [Google Scholar] [CrossRef]
- Weng, S.; Li, M.; Deng, J.; Xu, H.; Ren, Y.; Zhou, Z.; Wang, L.; Zhang, Y.; Xing, Z.; Li, L.; et al. Epigenetically regulated gene expression profiles decipher four molecular subtypes with prognostic and therapeutic implications in gastric cancer. Clin. Epigenetics 2023, 15, 64. [Google Scholar] [CrossRef]
- Yousefi, B.; Mohammadlou, M.; Abdollahi, M.; Salek Farrokhi, A.; Karbalaei, M.; Keikha, M.; Kokhaei, P.; Valizadeh, S.; Rezaiemanesh, A.; Arabkari, V.; et al. Epigenetic Changes in Gastric Cancer Induction by Helicobacter pylori. J. Cell. Physiol. 2019, 234, 21770–21784. [Google Scholar] [CrossRef]
- Ushijima, T.; Watanabe, N.; Shimizu, K.; Miyamoto, K.; Sugimura, T.; Kaneda, A. Decreased fidelity in replicating CpG methylation patterns in cancer cells. Cancer Res. 2005, 65, 11–17. [Google Scholar] [CrossRef]
- Ushijima, T.; Okochi-Takada, E. Aberrant methylations in cancer cells: Where do they come from? Cancer Sci. 2005, 96, 206–211. [Google Scholar] [CrossRef]
- Lee, T.L.; Leung, W.K.; Chan, M.W.; Ng, E.K.; Tong, J.H.; Lo, K.W.; Chung, S.C.; Sung, J.J.; To, K.F. Detection of gene promoter hypermethylation in the tumor and serum of patients with gastric carcinoma. Clin. Cancer Res. 2002, 8, 1761–1766. [Google Scholar]
- Miyamoto, K.; Ushijima, T. Diagnostic and therapeutic applications of epigenetics. Jpn. J. Clin. Oncol. 2005, 35, 293–301. [Google Scholar] [CrossRef]
- Yamada, H.; Shinmura, K.; Goto, M.; Iwaizumi, M.; Konno, H.; Kataoka, H.; Yamada, M.; Ozawa, T.; Tsuneyoshi, T.; Tanioka, F.; et al. Absence of germline mono-allelic promoter hypermethylation of the CDH1 gene in gastric cancer patients. Mol. Cancer 2009, 8, 63. [Google Scholar] [CrossRef]
- Zeng, W.; Zhu, J.; Shan, L.; Han, Z.; Aerxiding, P.; Quhai, A.; Zeng, F.; Wang, Z.; Li, H. The Clinicopathological Significance of CDH1 in Gastric Cancer: A Meta-Analysis and Systematic Review. Drug Des. Devel. Ther. 2015, 9, 2149–2157. [Google Scholar] [CrossRef]
- Li, Y.; Yang, Y.; Lu, Y.; Herman, J.G.; Brock, M.V.; Zhao, P.; Guo, M. Predictive Value of CHFR and MLH1 Methylation in Human Gastric Cancer. Gastric. Cancer 2015, 18, 280–287. [Google Scholar] [CrossRef]
- Dong, C.-X.; Deng, D.-J.; Pan, K.-F.; Zhang, L.; Zhang, Y.; Zhou, J.; You, W.-C. Promoter Methylation of P16 Associated with Helicobacter pylori Infection in Precancerous Gastric Lesions: A Population-Based Study. Int. J. Cancer 2009, 124, 434–439. [Google Scholar] [CrossRef]
- He, D.; Zhang, Y.; Zhang, N.; Zhou, L.; Chen, J.; Jiang, Y.; Shao, C. Aberrant Gene Promoter Methylation of P16, FHIT, CRBP1, WWOX, and DLC-1 in Epstein-Barr Virus-Associated Gastric Carcinomas. Med. Oncol. 2015, 32, 92. [Google Scholar] [CrossRef]
- Wang, X.; Hou, Q.; Zhou, X. LAMA4 Expression Is Activated by Zinc Finger E-box-binding Homeobox 1 and Independently Predicts Poor Overall Survival in Gastric Cancer. Oncol. Rep. 2018, 40, 1725–1733. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Zhang, M.; Guo, Y.; Shen, S.; Guo, X.; Dong, Z. FBXO32, a New TGF-β/Smad Signaling Pathway Target Gene, Is Epigenetically Inactivated in Gastric Cardia Adenocarcinoma. Neoplasma 2015, 62, 646–657. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yuan, S.; Li, L.; Xiang, S.; Jia, H.; Luo, T. Cadherin-11 Is Inactivated Due to Promoter Methylation and Functions in Colorectal Cancer as a Tumour Suppressor. Cancer Manag. Res. 2019, 11, 2517–2529. [Google Scholar] [CrossRef]
- Lin, Y.-L.; Gui, S.-L.; Ma, J.-G. Aberrant Methylation of CDH11 Predicts a Poor Outcome for Patients with Bladder Cancer. Oncol. Lett. 2015, 10, 647–652. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kim, J.; Kim, W.H.; Byeon, S.-J.; Lee, B.L.; Kim, M.A. Epigenetic Downregulation and Growth Inhibition of IGFBP7 in Gastric Cancer. Asian Pac. J. Cancer Prev. 2018, 19, 667–675. [Google Scholar] [CrossRef]
- Zhang, Z.; Yu, W.; Chen, S.; Chen, Y.; Chen, L.; Zhang, S. Methylation of the Claudin-3 Promoter Predicts the Prognosis of Advanced Gastric Adenocarcinoma. Oncol. Rep. 2018, 40, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Li, G.; He, D.; Yang, F.; He, G.; He, L.; Zhang, H.; Deng, Y.; Fan, M.; Shen, L.; et al. Telomerase Reverse Transcriptase Methylation Predicts Lymph Node Metastasis and Prognosis in Patients with Gastric Cancer. Onco Targets Ther. 2016, 9, 279–286. [Google Scholar] [CrossRef][Green Version]
- Vahidi, S.; Norollahi, S.E.; Agah, S.; Samadani, A.A. DNA Methylation Profiling of hTERT Gene Alongside with the Telomere Performance in Gastric Adenocarcinoma. J. Gastrointest. Cancer 2020, 51, 788–799. [Google Scholar] [CrossRef]
- Zhu, T.; Hu, Z.; Wang, Z.; Ding, H.; Li, R.; Sun, J.; Wang, G. Epigenetically Silenced PD-L1 Confers Drug Resistance to Anti-PD1 Therapy in Gastric Cardia Adenocarcinoma. Int. Immunopharmacol. 2020, 82, 106245. [Google Scholar] [CrossRef]
- Lingohr, P.; Dohmen, J.; Semaan, A.; Branchi, V.; Dietrich, J.; Bootz, F.; Kalff, J.C.; Matthaei, H.; Dietrich, D. Clinicopathological, Immune and Molecular Correlates of PD-L2 Methylation in Gastric Adenocarcinomas. Epigenomics 2019, 11, 639–653. [Google Scholar] [CrossRef] [PubMed]
- Brasacchio, D.; Busuttil, R.A.; Noori, T.; Johnstone, R.W.; Boussioutas, A.; Trapani, J.A. Down-Regulation of a pro-Apoptotic Pathway Regulated by PCAF/ADA3 in Early Stage Gastric Cancer. Cell Death Dis. 2018, 9, 442. [Google Scholar] [CrossRef] [PubMed]
- Ramezankhani, R.; Solhi, R.; Es, H.A.; Vosough, M.; Hassan, M. Novel Molecular Targets in Gastric Adenocarcinoma. Pharmacol. Ther. 2021, 220, 107714. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.J.; Arai, H.; Kitayama, Y.; Igarashi, H.; Hemmi, H.; Arai, T.; Hanai, H.; Sugimura, H. Microsatellite instability of papillary subtype of human gastric adenocarcinoma and hMLH1 promoter hypermethylation in the surrounding mucosa. Pathol. Int. 2001, 51, 240–247. [Google Scholar] [CrossRef] [PubMed]
- Ma, T.; Li, H.; Sun, M.; Yuan, Y.; Sun, L.-P. DNMT1 Overexpression Predicting Gastric Carcinogenesis, Subsequent Progression and Prognosis: A Meta and Bioinformatic Analysis. Oncotarget 2017, 8, 96396–96408. [Google Scholar] [CrossRef] [PubMed]
- Fattahi, S.; Kosari-Monfared, M.; Ghadami, E.; Golpour, M.; Khodadadi, P.; Ghasemiyan, M.; Akhavan-Niaki, H. Infection-Associated Epigenetic Alterations in Gastric Cancer: New Insight in Cancer Therapy. J. Cell. Physiol. 2018, 233, 9261–9270. [Google Scholar] [CrossRef]
- Zhou, X.; Jiao, D.; Dou, M.; Zhang, W.; Hua, H.; Chen, J.; Li, Z.; Li, L.; Han, X. Association of APC Gene Promoter Methylation and the Risk of Gastric Cancer: A Meta-Analysis and Bioinformatics Study. Medicine 2020, 99, e19828. [Google Scholar] [CrossRef]
- Du, L.; Fakih, M.G.; Rosen, S.T.; Chen, Y. SUMOylation of E2F1 Regulates Expression of EZH2. Cancer Res. 2020, 80, 4212–4223. [Google Scholar] [CrossRef]
- Kishore, C. Epigenetic Regulation and Promising Therapies in Colorectal Cancer. Curr. Mol. Pharmacol. 2021, 14, 838–852. [Google Scholar] [CrossRef]
- Wan, J.; Liu, H.; Ming, L. Lysine Crotonylation Is Involved in Hepatocellular Carcinoma Progression. Biomed. Pharmacother. 2019, 111, 976–982. [Google Scholar] [CrossRef]
- Yang, G.; Yuan, Y.; Yuan, H.; Wang, J.; Yun, H.; Geng, Y.; Zhao, M.; Li, L.; Weng, Y.; Liu, Z.; et al. Histone Acetyltransferase 1 Is a Succinyltransferase for Histones and Non-Histones and Promotes Tumorigenesis. EMBO Rep. 2021, 22, e50967. [Google Scholar] [CrossRef]
- Jarrold, J.; Davies, C.C. PRMTs and Arginine Methylation: Cancer’s Best-Kept Secret? Trends Mol. Med. 2019, 25, 993–1009. [Google Scholar] [CrossRef]
- He, L.-J.; Cai, M.-Y.; Xu, G.-L.; Li, J.-J.; Weng, Z.-J.; Xu, D.-Z.; Luo, G.-Y.; Zhu, S.-L.; Xie, D. Prognostic Significance of Overexpression of EZH2 and H3k27me3 Proteins in Gastric Cancer. Asian Pac. J. Cancer Prev. 2012, 13, 3173–3178. [Google Scholar] [CrossRef]
- Hatakeyama, M. Oncogenic mechanisms of the Helicobacter pylori CagA protein. Nat. Rev. Cancer 2004, 4, 688–694. [Google Scholar] [CrossRef]
- Hayashi, Y.; Tsujii, M.; Wang, J.; Kondo, J.; Akasaka, T.; Jin, Y.; Li, W.; Nakamura, T.; Nishida, T.; Lijima, H.; et al. CagA mediates epigenetic regulation to attenuate let-7 expression in Helicobacter pylori-related carcinogenesis. Gut 2013, 62, 1536–1546. [Google Scholar] [CrossRef]
- Herz, H.M.; Garruss, A.; Shilatifard, A. SET for life: Biochemical activities and biological functions of SET domain-containing proteins. Trends Biochem. Sci. 2013, 38, 621–639. [Google Scholar] [CrossRef]
- Reyes, D.A.; Sarría, V.M.S.; Salazar-Viedma, M.; D’Afonseca, V. Histone Methyltransferases Useful in Gastric Cancer Research. Cancer Inform. 2021, 20, 11769351211039862. [Google Scholar] [CrossRef]
- Donner, I.; Kiviluoto, T.; Ristimäki, A.; Aaltonen, L.A.; Vahteristo, P. Exome sequencing reveals three novel candidate predisposition genes for diffuse gastric cancer. Fam. Cancer 2015, 14, 241–246. [Google Scholar] [CrossRef] [PubMed]
- Torres, I.O.; Kuchenbecker, K.M.; Nnadi, C.I.; Fletterick, R.J.; Kelly, M.J.; Fujimori, D.G. Histone demethylase KDM5A is regulated by its reader domain through a positive-feedback mechanism. Nat. Commun. 2015, 6, 6204. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.W.; Xia, R.; Lu, K.; Xie, M.; Yang, F.; Sun, M.; De, W.; Wang, C.; Ji, G. LincRNAFEZF1-AS1 represses p21 expression to promote gastric cancer proliferation through LSD1-Mediated H3K4me2 demethylation. Mol. Cancer 2017, 16, 39. [Google Scholar] [CrossRef] [PubMed]
- Nishikawaji, T.; Akiyama, Y.; Shimada, S.; Kojima, K.; Kawano, T.; Eishi, Y.; Yuasa, Y.; Tanaka, S. Oncogenic roles of the SETDB2 histone methyltransferase in gastric cancer. Oncotarget 2016, 7, 67251–67265. [Google Scholar] [CrossRef]
- Wisnieski, F.; Calcagno, D.Q.; Leal, M.F.; Santos, L.C.; Gigek, C.O.; Chen, E.S.; Demachki, S.; Artigiani, R.; Assumpção, P.P.; Lourenço, L.G.; et al. CDKN1A Histone Acetylation and Gene Expression Relationship in Gastric Adenocarcinomas. Clin. Exp. Med. 2017, 17, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Wisnieski, F.; Leal, M.F.; Calcagno, D.Q.; Santos, L.C.; Gigek, C.O.; Chen, E.S.; Artigiani, R.; Demachki, S.; Assumpção, P.P.; Lourenço, L.G.; et al. BMP8B Is a Tumor Suppressor Gene Regulated by Histone Acetylation in Gastric Cancer. J. Cell. Biochem. 2017, 118, 869–877. [Google Scholar] [CrossRef]
- Song, J.; Noh, J.H.; Lee, J.H.; Eun, J.W.; Ahn, Y.M.; Kim, S.Y.; Lee, S.H.; Park, W.S.; Yoo, N.J.; Lee, J.Y.; et al. Increased expression of histone deacetylase 2 is found in human gastric cancer. APMIS 2005, 113, 264–268. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Noh, J.H.; Lee, J.H.; Eun, J.W.; Ahn, Y.M.; Kim, S.Y.; Lee, S.H.; Park, W.S.; Yoo, N.J.; Lee, J.Y.; et al. Association of patterns of class I histone deacetylase expression with patient prognosis in gastric cancer: A retrospective analysis. Lancet Oncol. 2008, 9, 139–148. [Google Scholar]
- Xia, G.; Schneider-Stock, R.; Diestel, A.; Habold, C.; Krueger, S.; Roessner, A.; Naumann, M.; Lendeckel, U. Helicobacter pylori regulates p21(WAF1) by histone H4 acetylation. Biochem. Biophys. Res. Commun. 2008, 369, 526–531. [Google Scholar] [CrossRef] [PubMed]
- Han, F.; Ren, J.; Zhang, J.; Sun, Y.; Ma, F.; Liu, Z.; Yu, H.; Jia, J.; Li, W. JMJD2B is required for Helicobacter pylori-induced gas-tric carcinogenesis via regulating COX-2 expression. Oncotarget 2016, 7, 38626–38637. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.S.; Jin, M.Y.; Kim, Y.J.; Yook, J.H.; Kim, B.S.; Jang, S.J. The global histone modification pattern correlates with cancer recurrence and overall survival in gastric adenocarcinoma. Ann. Surg. Oncol. 2008, 15, 1968–1976. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.Y.; Zhou, P.J.; Meng, Y.; Zeng, F.R.; Deng, G.T. Gastric cancer: An epigenetic view. World J. Gastrointest. Oncol. 2022, 14, 90–109. [Google Scholar] [CrossRef]
- Murakami, Y. Phosphorylation of Repressive Histone Code Readers by Casein Kinase 2 Plays Diverse Roles in Heterochromatin Regulation. J. Biochem. 2019, 166, 3–6. [Google Scholar] [CrossRef]
- Khan, S.A.; Amnekar, R.; Khade, B.; Barreto, S.G.; Ramadwar, M.; Shrikhande, S.V.; Gupta, S. P38-MAPK/MSK1-Mediated Overexpression of Histone H3 Serine 10 Phosphorylation Defines Distance-Dependent Prognostic Value of Negative Resection Margin in Gastric Cancer. Clin. Epigenetics 2016, 8, 88. [Google Scholar] [CrossRef]
- Takahashi, H.; Murai, Y.; Tsuneyama, K.; Nomoto, K.; Okada, E.; Fujita, H.; Takano, Y. Overexpression of Phosphorylated Histone H3 Is an Indicator of Poor Prognosis in Gastric Adenocarcinoma Patients. Appl. Immunohistochem. Mol. Morphol. 2006, 14, 296–302. [Google Scholar] [CrossRef]
- Kim, S.R.; Kim, K.B.; Chae, Y.C.; Park, J.W.; Seo, S.B. H3S10 phosphorylation-mediated transcriptional regulation by Aurora kinase, A. Biochem. Biophys. Res. Commun. 2016, 469, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Dar, A.A.; Zaika, A.; Piazuelo, M.B.; Correa, P.; Koyama, T.; Belkhiri, A.; Washington, K.; Castells, A.; Pera, M.; El-Rifai, W. Frequent overexpression of Aurora Kinase A in upper gastrointestinal adenocarcinomas correlates with potent antiapoptotic functions. Cancer 2008, 112, 1688–1698. [Google Scholar] [CrossRef] [PubMed]
- Kamran, M.; Long, Z.J.; Xu, D.; Lv, S.S.; Liu, B.; Wang, C.L.; Xu, J.; Lam, E.W.-F.; Liu, Q. Aurora kinase A regulates survivin stability through targeting FBXL7 in gastric cancer drug resistance and prognosis. Oncogenesis 2017, 6, e298. [Google Scholar] [CrossRef]
- Xu, X.; Xiao, J.; Wei, S.; Xie, X.; Huang, Y.; Tian, X.; Wang, C. Relationship between expression of Aurka and clinicopathological characteristics in gastric cancer patients. Sci. Res. 2014, 6, 243–249. [Google Scholar] [CrossRef]
- Yang, T.; Cao, N.; Zhang, H.; Wei, J.; Song, X.; Yi, D.; Chao, S.; Zhang, L.; Kong, L.; Han, S.; et al. Helicobacter pylori infection-induced H3Ser10 phosphorylation in stepwise gastric carcinogenesis and its clinical implications. Helicobacter 2018, 23, e12486. [Google Scholar] [CrossRef]
- Fehri, L.F.; Rechner, C.; Janßen, S.; Mak, T.N.; Holland, C.; Bartfeld, S.; Brüggemann, H.; Meyer, T.F. Helicobacter pylori-induced modification of the histone H3 phosphorylation status in gastric epithelial cells reflects its impact on cell cycle regulation. Epigenetics 2009, 4, 577–586. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Qiu, Z.; Wu, Y. Ubiquitin Regulation: The Histone Modifying Enzyme’s Story. Cells 2018, 7, 118. [Google Scholar] [CrossRef]
- Swatek, K.N.; Komander, D. Ubiquitin Modifications. Cell Res. 2016, 26, 399–422. [Google Scholar] [CrossRef]
- Wang, Z.-J.; Yang, J.-L.; Wang, Y.-P.; Lou, J.-Y.; Chen, J.; Liu, C.; Guo, L.-D. Decreased Histone H2B Monoubiquitination in Malignant Gastric Carcinoma. World J. Gastroenterol. 2013, 19, 8099–8107. [Google Scholar] [CrossRef]
- van Mierlo, G.; Veenstra, G.J.C.; Vermeulen, M.; Marks, H. The complexity of PRC2 subcomplexes. Trends Cell Biol. 2019, 29, 660–671. [Google Scholar] [CrossRef]
- Hu, H.; Yang, Y.; Ji, Q.; Zhao, W.; Jiang, B.; Liu, R.; Yuan, J.; Liu, Q.; Li, X.; Zou, Y.; et al. CRL4B catalyzes H2AK119 monoubiquitination and coordinates with PRC2 to promote tumorigenesis. Cancer Cell 2012, 22, 781–795. [Google Scholar] [CrossRef] [PubMed]
- Serrano-Quilez, J.; Roig-Soucase, S.; Rodriguez-Navarro, S. Sharing marks: H3K4 methylation and H2B ubiquitination as features of meiotic recombination and transcription. Int. J. Mol. Sci. 2020, 21, 4510. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, M.; Wang, Y. The roles of histone modifications in tumorigenesis and associated inhibitors in cancer therapy. J. Natl. Cancer Cent. 2022, 2, 277–290. [Google Scholar] [CrossRef]
- Zhao, Z.; Shilatifard, A. Epigenetic modifications of histones in cancer. Genome Biol. 2019, 20, 1–16. [Google Scholar] [CrossRef]
- Chan, J.C.; Maze, I. Nothing is yet set in (hi)stone: Novel post-translational modifications regulating chromatin function. Trends Biochem. Sci. 2020, 45, 829–844. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Xu, X.; Ding, J.; Yang, L.; Doan, M.T.; Karmaus, P.W.F.; Snyder, N.W.; Zhao, Y.; Li, J.-L.; Li, X. Histone Crotonylation Promotes Mesoendodermal Commitment of Human Embryonic Stem Cells. Cell Stem Cell 2021, 28, 748–763.e7. [Google Scholar] [CrossRef]
- Zhu, Z.; Han, Z.; Halabelian, L.; Yang, X.; Ding, J.; Zhang, N.; Ngo, L.; Song, J.; Zeng, H.; He, M.; et al. Identification of lysine isobutyrylation as a new histone modification mark. Nucleic Acids Res. 2021, 49, 177–189. [Google Scholar] [CrossRef]
- Liberti, M.V.; Locasale, J.W. Histone Lactylation: A New Role for Glucose Metabolism. Trends Biochem. Sci. 2020, 45, 179–182. [Google Scholar] [CrossRef]
- Zhang, D.; Tang, Z.; Huang, H.; Zhou, G.; Cui, C.; Weng, Y.; Liu, W.; Kim, S.; Lee, S.; Perez-Neut, M.; et al. Metabolic Regulation of Gene Expression by Histone Lactylation. Nature 2019, 574, 575–580. [Google Scholar] [CrossRef]
- Leonen, C.J.A.; Shimada, M.; Weller, C.E.; Nakadai, T.; Hsu, P.L.; Tyson, E.L.; Mishra, A.; Shelton, P.M.; Sadilek, M.; Hawkins, R.D.; et al. Sumoylation of the Human Histone H4 Tail Inhibits P300-Mediated Transcription by RNA Polymerase II in Cellular Extracts. Elife 2021, 10, e67952. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg Robert, A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Wellen, K.E.; Thompson, C.B. A two-way street: Reciprocal regulation of metabolism and signaling. Nat. Rev. Mol. Cell Biol. 2012, 13, 270–276. [Google Scholar] [CrossRef]
- DeBerardinis, R.J.; Sayed, N.; Ditsworth, D.; Thompson, C.B. Brick by brick: Metabolism and tumor cell growth. Curr. Opin. Genet. Dev. 2008, 18, 54–61. [Google Scholar] [CrossRef]
- Ulanovskaya, O.A.; Zuhl, A.M.; Cravatt, B.F. NNMT promotes epigenetic remodeling in cancer by creating a metabolic methylation sink. Nat. Chem. Biol. 2013, 9, 300–306. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Zeng, M.; Pan, H.; Liu, H.; He, Y. Nicotinamide N-methyltransferase promotes epithelial-mesenchymal transition in gastric cancer cells by activating transforming growth factor-β1 expression. Oncol. Lett. 2018, 15, 4592–4598. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Yang, C.; Wang, T.; Deng, H. Complex roles of nicotinamide N-methyltransferase in cancer progression. Cell Death Dis. 2022, 13, 267. [Google Scholar] [CrossRef]
- Chen, C.; Wang, X.; Huang, X.; Yong, H.; Shen, J.; Tang, Q.; Zhu, J.; Ni, J.; Feng, Z. Nicotinamide N-methyltransferase: A potential biomarker for worse prognosis in gastric carcinoma. Am. J. Cancer Res. 2016, 6, 649–663. [Google Scholar] [PubMed]
- Wu, M.; Hu, W.; Wang, G.; Yao, Y.; Yu, X.F. Nicotinamide N-Methyltransferase Is a Prognostic Biomarker and Correlated with Immune Infiltrates in Gastric Cancer. Front. Genet. 2020, 11, 580299. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Song, M.; Zhang, F.; Yuan, H.; Chang, W.; Yu, G.; Niu, Y. Accumulation of Nicotinamide N-Methyltransferase (NNMT) in Cancer-associated Fibroblasts: A Potential Prognostic and Predictive Biomarker for Gastric Carcinoma. J. Histochem. Cytochem. 2021, 69, 165–176. [Google Scholar] [CrossRef]
- Pozzi, V.; Campagna, R.; Sartini, D.; Emanuelli, M. Nicotinamide N-Methyltransferase as Promising Tool for Management of Gastrointestinal Neoplasms. Biomolecules 2022, 12, 1173. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, Q.; Chen, C.; Zhao, X.; Wang, H.; Xu, L.; Fu, Y.; Huang, G.; Li, M.; Xu, J.; et al. NNMT enriches for AQP5+ cancer stem cells to drive malignant progression in early gastric cardia adenocarcinoma. Gut 2024, 73, 63–77. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Sun, J.; Wang, J.; Song, Y.; Gao, P.; Shi, J.; Chen, P.; Wang, Z. Long Noncoding RNAs in Gastric Cancer: Functions and Clinical Applications. Onco Targets Ther. 2016, 9, 681–697. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Wu, D.; Gao, H.; Balic, J.J.; Tsykin, A.; Han, T.-S.; Liu, Y.D.; Kennedy, C.L.; Li, J.K.; Mao, J.Q.; et al. Clinical Utility of a STAT3-Regulated miRNA-200 Family Signature with Prognostic Potential in Early Gastric Cancer. Clin. Cancer Res. 2018, 24, 1459–1472. [Google Scholar] [CrossRef] [PubMed]
- Marchese, F.P.; Huarte, M. Long Non-Coding RNAs and Chromatin Modifiers: Their Place in the Epigenetic Code. Epigenetics 2014, 9, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Gao, S.; Zhao, Z.; Liang, G.; Kong, J.; Feng, X. MicroRNA-320d Regulates Tumor Growth and Invasion by Promoting FoxM1 and Predicts Poor Outcome in Gastric Cardiac Adenocarcinoma. Cell Biosci. 2020, 10, 80. [Google Scholar] [CrossRef] [PubMed]
- Yadegar, N.; Dadashi, Z.; Shams, K.; Mohammadi, M.; Abyar, M.; Rafat, M. The Prominent Role of miR-942 in Carcinogenesis of Tumors. Adv. Biomed. Res. 2022, 11, 63. [Google Scholar] [CrossRef]
- Abdi, E.; Latifi-Navid, S.; Zahri, S.; Yazdanbod, A.; Pourfarzi, F. Risk Factors Predisposing to Cardia Gastric Adenocarcinoma: Insights and New Perspectives. Cancer Med. 2019, 8, 6114–6126. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; Liu, J.; Yang, T.; Zhang, L.; Gong, P.; Li, B.; Zhou, X. Construction and Investigation of MicroRNA-mRNA Regulatory Network of Gastric Cancer with Helicobacter Pylori Infection. Biochem. Res. Int. 2020, 2020, 6285987. [Google Scholar] [CrossRef]
- Gilani, N.; Arabi Belaghi, R.; Aftabi, Y.; Faramarzi, E.; Edgünlü, T.; Somi, M.H. Identifying Potential miRNA Biomarkers for Gastric Cancer Diagnosis Using Machine Learning Variable Selection Approach. Front. Genet. 2021, 12, 779455. [Google Scholar] [CrossRef]
- Qu, X.; Zhao, L.; Zhang, R.; Wei, Q.; Wang, M. Differential microRNA Expression Profiles Associated with Microsatellite Status Reveal Possible Epigenetic Regulation of Microsatellite Instability in Gastric Adenocarcinoma. Ann. Transl. Med. 2020, 8, 484. [Google Scholar] [CrossRef]
- Jia, L.; Chen, J.; Xie, C.; Shao, L.; Xu, Z.; Zhang, L. microRNA-1228⁎ impairs the pro-angiogenic activity of gastric cancer cells by targeting macrophage migration inhibitory factor. Life Sci. 2017, 180, 9–16. [Google Scholar] [CrossRef]
- Motoyama, K.; Inoue, H.; Nakamura, Y.; Uetake, H.; Sugihara, K.; Mori, M. Clinical significance of high mobility group A2 in human gastric cancer and its relationship to let-7 microRNA family. Clin. Cancer Res. 2008, 14, 2334–2340. [Google Scholar] [CrossRef]
- Bie, L.; Luo, S.; Li, D.; Wei, Y.; Mu, Y.; Chen, X.; Wang, S.; Guo, P.; Lu, X. HOTAIR Competitively Binds MiRNA330 as a Molecular Sponge to Increase the Resistance of Gastric Cancer to Trastuzumab. Curr. Cancer Drug Targets 2020, 20, 700–709. [Google Scholar] [CrossRef] [PubMed]
- Endo, H.; Shiroki, T.; Nakagawa, T.; Yokoyama, M.; Tamai, K.; Yamanami, H.; Fujiya, T.; Sato, I.; Yamaguchi, K.; Tanaka, N.; et al. Enhanced Expression of Long Non-Coding RNA HOTAIR Is Associated with the Development of Gastric Cancer. PLoS ONE 2013, 8, e77070. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Qin, Y.; Zhi, Q.; Wang, J.; Qin, C. Knockdown of Long Non-Coding RNA HOTAIR Inhibits Cisplatin Resistance of Gastric Cancer Cells through Inhibiting the PI3K/Akt and Wnt/β-Catenin Signaling Pathways by up-Regulating miR-34a. Int. J. Biol. Macromol. 2018, 107, 2620–2629. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Li, J.; Yan, W.; Yao, Z.; Wang, R.; Zhou, X.; Wu, H.; Zhang, G.; Shi, T.; Chen, W. H19 Promotes Aerobic Glycolysis, Proliferation, and Immune Escape of Gastric Cancer Cells through the microRNA-519d-3p/Lactate Dehydrogenase A Axis. Cancer Sci. 2021, 112, 2245–2259. [Google Scholar] [CrossRef] [PubMed]
- Xin, L.; Zhou, L.-Q.; Liu, C.; Zeng, F.; Yuan, Y.-W.; Zhou, Q.; Li, S.-H.; Wu, Y.; Wang, J.-L.; Wu, D.-Z.; et al. Transfer of LncRNA CRNDE in TAM-Derived Exosomes Is Linked with Cisplatin Resistance in Gastric Cancer. EMBO Rep. 2021, 22, e52124. [Google Scholar] [CrossRef]
- Fan, H.; Ge, Y.; Ma, X.; Li, Z.; Shi, L.; Lin, L.; Xiao, J.; Chen, W.; Ni, P.; Yang, L.; et al. Long Non-Coding RNA CCDC144NL-AS1 Sponges miR-143-3p and Regulates MAP3K7 by Acting as a Competing Endogenous RNA in Gastric Cancer. Cell Death Dis. 2020, 11, 521. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, Z.; Yu, G.; Wang, T.; Qu, G.; Wang, Y. LINC01232 Promotes Gastric Cancer Proliferation through Interacting with EZH2 to Inhibit the Transcription of KLF2. J. Microbiol. Biotechnol. 2021, 31, 1358–1365. [Google Scholar] [CrossRef]
- Peng, L.; Peng, J.-Y.; Cai, D.-K.; Qiu, Y.-T.; Lan, Q.-S.; Luo, J.; Yang, B.; Xie, H.-T.; Du, Z.-P.; Yuan, X.-Q.; et al. Immune Infiltration and Clinical Outcome of Super-Enhancer-Associated lncRNAs in Stomach Adenocarcinoma. Front. Oncol. 2022, 12, 780493. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.; Wang, Z.; Wang, G.; Hu, Z.; Ding, H.; Li, R.; Sun, J. Long Non-Coding RNA ZFAS1 Promotes the Expression of EPAS1 in Gastric Cardia Adenocarcinoma. J. Adv. Res. 2021, 28, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Zhou, Y.; Luo, X.; Gao, H.; Deng, X.; Jiang, Y. Long non-coding RNA XIST promotes cell growth and invasion through regulating miR-497/MACC1 axis in gastric cancer. Oncotarget 2017, 8, 4125–4135. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Li, J.; Zhou, X.; Cui, L.; Wang, Y. The lncRNA XIST promotes proliferation, migration and invasion of gastric cancer cells by targeting miR-337. Arab. J. Gastroenterol. 2020, 21, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Wang, L.; Li, P.; Hu, F.; Cao, Y.; Tang, D.; Ye, G.; Li, H.; Wang, D. Silencing lncRNA XIST exhibits antiproliferative and proapoptotic effects on gastric cancer cells by up-regulating microRNA-132 and down-regulating PXN. Aging 2020, 13, 14469–14481. [Google Scholar] [CrossRef]
- Guarnerio, J.; Bezzi, M.; Jeong, J.C.; Paffenholz, S.V.; Berry, K.; Naldini, M.M.; Lo-Coco, F.; Tay, Y.; Beck, A.H.; Pandolfi, P.P. Oncogenic Role of Fusion-circRNAs Derived from Cancer-Associated Chromosomal Translocations. Cell 2016, 166, 1055–1056. [Google Scholar] [CrossRef] [PubMed]
- Pan, H.; Li, T.; Jiang, Y.; Pan, C.; Ding, Y.; Huang, Z.; Yu, H.; Kong, D. Overexpression of Circular RNA ciRS-7 Abrogates the Tumor Suppressive Effect of miR-7 on Gastric Cancer via PTEN/PI3K/AKT Signaling Pathway. J. Cell. Biochem. 2018, 119, 440–446. [Google Scholar] [CrossRef]
- Lin, J.; Zhang, Y.; Zeng, X.; Xue, C.; Lin, X. CircRNA CircRIMS Acts as a MicroRNA Sponge to Promote Gastric Cancer Metastasis. ACS Omega 2020, 5, 23237–23246. [Google Scholar] [CrossRef]
- Liu, J.; Zhu, X.; Yan, M.; Li, H. Development of a Two-Circular RNA Panel as Potential Prognostic Biomarker for Gastric Cancer. J. Transl. Med. 2021, 19, 412. [Google Scholar] [CrossRef]
- Han, L.; Zhang, X.; Wang, A.; Ji, Y.; Cao, X.; Qin, Q.; Yu, T.; Huang, H.; Yin, L. A Dual-Circular RNA Signature as a Non-Invasive Diagnostic Biomarker for Gastric Cancer. Front. Oncol. 2020, 10, 184. [Google Scholar] [CrossRef] [PubMed]
- Yesharim, L.; Talebi, S.; Mojbafan, M.; Alemrajabi, M.; Teimourian, S. An Evaluation of Gastric Adenocarcinoma-Associated CircRNAs Based on Microarray Meta-Analysis and ceRNA Networks. Transl. Oncol. 2023, 28, 101611. [Google Scholar] [CrossRef]
- Cao, J.; Zhang, X.; Xu, P.; Wang, H.; Wang, S.; Zhang, L.; Li, Z.; Xie, L.; Sun, G.; Xia, Y.; et al. Circular RNA circLMO7 Acts as a microRNA-30a-3p Sponge to Promote Gastric Cancer Progression via the WNT2/β-Catenin Pathway. J. Exp. Clin. Cancer Res. 2021, 40, 6. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Yu, T.; Jing, X.; Ma, L.; Fan, Y.; Yang, F.; Ma, P.; Jiang, H.; Wu, X.; Shu, Y.; et al. Exosomal circSHKBP1 Promotes Gastric Cancer Progression via Regulating the miR-582-3p/HUR/VEGF Axis and Suppressing HSP90 Degradation. Mol. Cancer 2020, 19, 112. [Google Scholar] [CrossRef] [PubMed]
- Zheng, P.; Gao, H.; Xie, X.; Lu, P. Plasma Exosomal Hsa_circ_0015286 as a Potential Diagnostic and Prognostic Biomarker for Gastric Cancer. Pathol. Oncol. Res. 2022, 28, 1610446. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, H.; Hou, L.; Wang, G.; Zhang, R.; Huang, Y.; Chen, X.; Zhu, J. Circular RNA_LARP4 Inhibits Cell Proliferation and Invasion of Gastric Cancer by Sponging miR-424-5p and Regulating LATS1 Expression. Mol. Cancer 2017, 16, 151. [Google Scholar] [CrossRef]
- Zhang, W.; Zheng, M.; Kong, S.; Li, X.; Meng, S.; Wang, X.; Wang, F.; Tang, C.; Ju, S. Circular RNA Hsa_circ_0007507 May Serve as a Biomarker for the Diagnosis and Prognosis of Gastric Cancer. Front. Oncol. 2021, 11, 699625. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Chen, S.; Chen, H.; Mo, X.; Li, T.; Shao, Y.; Xiao, B.; Guo, J. Using Circular RNA as a Novel Type of Biomarker in the Screening of Gastric Cancer. Clin. Chim. Acta 2015, 444, 132–136. [Google Scholar] [CrossRef]
- PubMed. CircRNA Microarray Profiling Identifies a Novel Circulating Biomarker for Detection of Gastric Cancer. Available online: https://pubmed.ncbi.nlm.nih.gov/30236115/ (accessed on 28 November 2023).
- Zhang, J.; Hou, L.; Liang, R.; Chen, X.; Zhang, R.; Chen, W.; Zhu, J. CircDLST Promotes the Tumorigenesis and Metastasis of Gastric Cancer by Sponging miR-502-5p and Activating the NRAS/MEK1/ERK1/2 Signaling. Mol. Cancer 2019, 18, 80. [Google Scholar] [CrossRef]
- Zhang, L.; Song, X.; Chen, X.; Wang, Q.; Zheng, X.; Wu, C.; Jiang, J. Circular RNA CircCACTIN Promotes Gastric Cancer Progression by Sponging MiR-331-3p and Regulating TGFBR1 Expression. Int. J. Biol. Sci. 2019, 15, 1091–1103. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, S.; Wang, H.; Cao, J.; Huang, X.; Chen, Z.; Xu, P.; Sun, G.; Xu, J.; Lv, J.; et al. Circular RNA circNRIP1 Acts as a microRNA-149-5p Sponge to Promote Gastric Cancer Progression via the AKT1/mTOR Pathway. Mol. Cancer 2019, 18, 20. [Google Scholar] [CrossRef]
- Wang, S.; Tang, D.; Wang, W.; Yang, Y.; Wu, X.; Wang, L.; Wang, D. circLMTK2 Acts as a Sponge of miR-150-5p and Promotes Proliferation and Metastasis in Gastric Cancer. Mol. Cancer 2019, 18, 162. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhang, X.; Li, Z.; Wang, W.; Li, B.; Huang, X.; Sun, G.; Xu, J.; Li, Q.; Xu, Z.; et al. Circular RNA Profile Identifies circOSBPL10 as an Oncogenic Factor and Prognostic Marker in Gastric Cancer. Oncogene 2019, 38, 6985–7001. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Wang, F. circ_AKT3 Knockdown Suppresses Cisplatin Resistance in Gastric Cancer. Open Med. 2022, 17, 280–291. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.-N.; Chen, Z.-Y.; Chen, X.-Y.; Chen, M.; Yi, Y.-C.; Zhu, J.-S.; Zhang, J. METTL14-Mediated m6A Modification of circORC5 Suppresses Gastric Cancer Progression by Regulating miR-30c-2-3p/AKT1S1 Axis. Mol. Cancer 2022, 21, 51. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.-L.; Sheng, H.; Zhang, D.-S.; Jin, Y.; Zhao, B.-T.; Chen, N.; Song, K.; Xu, R.-H. The Circular RNA circDLG1 Promotes Gastric Cancer Progression and Anti-PD-1 Resistance through the Regulation of CXCL12 by Sponging miR-141-3p. Mol. Cancer 2021, 20, 166. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, Y.; Su, X.; Wang, P.; Lin, W. The Value of Circulating Circular RNA in Cancer Diagnosis, Monitoring, Prognosis, and Guiding Treatment. Front. Oncol. 2021, 11, 736546. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Xiu, J.; Baca, Y.; Battaglin, F.; Arai, H.; Kawanishi, N.; Soni, S.; Zhang, W.; Millstein, J.; Salhia, B.; et al. Large-Scale Analysis of KMT2 Mutations Defines a Distinctive Molecular Subset with Treatment Implication in Gastric Cancer. Oncogene 2021, 40, 4894–4905. [Google Scholar] [CrossRef]
- Lordick, F.; Carneiro, F.; Cascinu, S.; Fleitas, T.; Haustermans, K.; Piessen, G.; Vogel, A.; Smyth, E.C.; on behalf of the ESMO Guidelines Committee. Gastric Cancer: ESMO Clinical Practice Guideline for Diagnosis, Treatment and Follow-Up. Ann. Oncol. 2022, 33, 1005–1020. [Google Scholar] [CrossRef]
- Wang, F.-H.; Zhang, X.-T.; Li, Y.-F.; Tang, L.; Qu, X.-J.; Ying, J.-E.; Zhang, J.; Sun, L.-Y.; Lin, R.-B.; Qiu, H.; et al. The Chinese Society of Clinical Oncology (CSCO): Clinical Guidelines for the Diagnosis and Treatment of Gastric Cancer, 2021. Cancer Commun. 2021, 41, 747–795. [Google Scholar] [CrossRef]
- Ajani, J.A.; D’Amico, T.A.; Bentrem, D.J.; Chao, J.; Cooke, D.; Corvera, C.; Das, P.; Enzinger, P.C.; Enzler, T.; Fanta, P.; et al. Gastric Cancer, Version 2.2022, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Canc. Netw. 2022, 20, 167–192. [Google Scholar] [CrossRef]
- Guan, W.-L.; He, Y.; Xu, R.-H. Gastric Cancer Treatment: Recent Progress and Future Perspectives. J. Hematol. Oncol. 2023, 16, 57. [Google Scholar] [CrossRef]
- Leal, A.; van Grieken, N.C.T.; Palsgrove, D.N.; Phallen, J.; Medina, J.E.; Hruban, C.; Broeckaert, M.A.M.; Anagnostou, V.; Adleff, V.; Bruhm, D.C.; et al. White Blood Cell and Cell-Free DNA Analyses for Detection of Residual Disease in Gastric Cancer. Nat. Commun. 2020, 11, 525. [Google Scholar] [CrossRef]
- Maron, S.B.; Chase, L.M.; Lomnicki, S.; Kochanny, S.; Moore, K.L.; Joshi, S.S.; Landron, S.; Johnson, J.; Kiedrowski, L.A.; Nagy, R.J.; et al. Circulating Tumor DNA Sequencing Analysis of Gastroesophageal Adenocarcinoma. Clin. Cancer Res. 2019, 25, 7098–7112. [Google Scholar] [CrossRef]
- Chiba, T.; Marusawa, H.; Ushijima, T. Inflammation-associated cancer development in digestive organs: Mechanisms and roles for genetic and epigenetic modulation. Gastroenterology 2012, 143, 550–563. [Google Scholar] [CrossRef]
- Hattori, N.; Ushijima, T. Epigenetic impact of infection on carcinogenesis: Mechanisms and applications. Genome Med. 2016, 8, 10. [Google Scholar] [CrossRef]
- Baba, Y.; Ishimoto, T.; Kurashige, J.; Iwatsuki, M.; Sakamoto, Y.; Yoshida, N.; Watanabe, M.; Baba, H. Epigenetic field cancerization in gastrointestinal cancers. Cancer Lett. 2016, 375, 360–366. [Google Scholar] [CrossRef]
- Kashyap, D.; Baral, B.; Jakhmola, S.; Singh, A.K.; Jha, H.C. Helicobacter pylori and Epstein-Barr virus coinfection stimulates aggressiveness in gastric cancer through the regulation of gankyrin. mSphere 2021, 6, e00751-21. [Google Scholar] [CrossRef]
- Jiang, C.; Pugh, B.F. Nucleosome Positioning and Gene Regulation: Advances through Genomics. Nat. Rev. Genet. 2009, 10, 161–172. [Google Scholar] [CrossRef]
- Matsusaka, K.; Funata, S.; Fukayama, M.; Kaneda, A. DNA methylation in gastric cancer, related to Helicobacter pylori and Epstein-Barr virus. World J. Gastroenterol. 2014, 20, 3916–3926. [Google Scholar] [CrossRef]
- Tan, P.; Yeoh, K.-G. Genetics and Molecular Pathogenesis of Gastric Adenocarcinoma. Gastroenterology 2015, 149, 1153–1162.e3. [Google Scholar] [CrossRef]
- Saletore, Y.; Meyer, K.; Korlach, J.; Vilfan, I.D.; Jaffrey, S.; Mason, C.E. The Birth of the Epitranscriptome: Deciphering the Function of RNA Modifications. Genome Biol. 2012, 13, 175. [Google Scholar] [CrossRef]
- Padmanabhan, N.; Ushijima, T.; Tan, P. How to stomach an epigenetic insult: The gastric cancer epigenome. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 467–478. [Google Scholar] [CrossRef]
- Kaneda, A.; Matsusaka, K.; Aburatani, H.; Fukayama, M. Epstein-Barr virus infection as an epigenetic driver of tumorigenesis. Cancer Res. 2012, 72, 3445–3450. [Google Scholar] [CrossRef]
- Jácome, A.A.D.A.; Lima, E.M.; Kazzi, A.I.; Chaves, G.F.; de Mendonça, D.C.; Maciel, M.M.; Santos, J.S.D. Epstein-Barr Virus-Positive Gastric Cancer: A Distinct Molecular Subtype of the Disease? Rev. Soc. Bras. Med. Trop. 2016, 49, 150–157. [Google Scholar] [CrossRef]
- Ignatova, E.; Seriak, D.; Fedyanin, M.; Tryakin, A.; Pokataev, I.; Menshikova, S.; Vakhabova, Y.; Smirnova, K.; Tjulandin, S.; Ajani, J.A. Epstein-Barr Virus-Associated Gastric Cancer: Disease That Requires Special Approach. Gastric. Cancer 2020, 23, 951–960. [Google Scholar] [CrossRef]
- Samadani, A.A.; Nikbakhsh, N.; Taheri, H.; Shafaee, S.; Fattahi, S.; Pilehchian Langroudi, M.; Hajian, K.; Akhavan-Niaki, H. CDX1/2 and KLF5 Expression and Epigenetic Modulation of Sonic Hedgehog Signaling in Gastric Adenocarcinoma. Pathol. Oncol. Res. 2019, 25, 1215–1222. [Google Scholar] [CrossRef]
- Heo, H.; Kim, H.-J.; Haam, K.; Sohn, H.A.; Shin, Y.-J.; Go, H.; Jung, H.-J.; Kim, J.-H.; Lee, S.-I.; Song, K.-S.; et al. Epigenetic Activation of Tensin 4 Promotes Gastric Cancer Progression. Mol. Cells 2023, 46, 298–308. [Google Scholar] [CrossRef]
- Moutinho, C.; Martinez-Cardús, A.; Santos, C.; Navarro-Pérez, V.; Martínez-Balibrea, E.; Musulen, E.; Carmona, F.J.; Sartore-Bianchi, A.; Cassingena, A.; Siena, S.; et al. Epigenetic Inactivation of the BRCA1 Interactor SRBC and Resistance to Oxaliplatin in Colorectal Cancer. J. Natl. Cancer Inst. 2014, 106, djt322. [Google Scholar] [CrossRef]
- Rezaei, S.; Hosseinpourfeizi, M.A.; Moaddab, Y.; Safaralizadeh, R. Contribution of DNA Methylation and EZH2 in SRBC Down-Regulation in Gastric Cancer. Mol. Biol. Rep. 2020, 47, 5721–5727. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.-Y.; Ma, H.-X.; Shang, Y.-H.; Jin, M.-S.; Kong, F.; Jia, Z.-F.; Cao, D.-H.; Wang, Y.-P.; Suo, J.; Jiang, J. DNA Methyltransferase3a Expression Is an Independent Poor Prognostic Indicator in Gastric Cancer. World J. Gastroenterol. 2014, 20, 8201–8208. [Google Scholar] [CrossRef]
- Purkait, S.; Patra, S.; Mitra, S.; Behera, M.M.; Panigrahi, M.K.; Kumar, P.; Kar, M.; Hallur, V.; Chandra Samal, S. Elevated Expression of DNA Methyltransferases and Enhancer of Zeste Homolog 2 in Helicobacter Pylori—Gastritis and Gastric Carcinoma. Dig. Dis. 2022, 40, 156–167. [Google Scholar] [CrossRef] [PubMed]
- Ahmadi Hedayati, M.; Ahmadi, A.; Khatooni, Z. DNMT1 Gene Expression in Patients with Helicobacter Pylori Infection. Sci. World J. 2022, 2022, 2386891. [Google Scholar] [CrossRef]
- Song, W.; Ren, J.; Wang, W.-J.; Wang, C.-T.; Fu, T. Genome-Wide Methylation and Expression Profiling Identify a Novel Epigenetic Signature in Gastrointestinal Pan-Adenocarcinomas. Epigenomics 2020, 12, 907–920. [Google Scholar] [CrossRef]
- Han, L.C.; Chen, Y. Targeting EZH2 for Cancer Therapy: Progress and Perspective. Curr. Protein Pept. Sci. 2015, 16, 559–570. [Google Scholar] [CrossRef]
- Fattahi, S.; Golpour, M.; Amjadi-Moheb, F.; Sharifi-Pasandi, M.; Khodadadi, P.; Pilehchian-Langroudi, M.; Ashrafi, G.H.; Akhavan-Niaki, H. DNA Methyltransferases and Gastric Cancer: Insight into Targeted Therapy. Epigenomics 2018, 10, 1477–1497. [Google Scholar] [CrossRef]
- Takeshima, H.; Niwa, T.; Takahashi, T.; Wakabayashi, M.; Yamashita, S.; Ando, T.; Inagawa, Y.; Taniguchi, H.; Katai, H.; Sugiyama, T.; et al. Frequent involvement of chromatin remodeler alterations in gastric field cancerization. Cancer Lett. 2015, 357, 328–338. [Google Scholar] [CrossRef]
- Santos, J.C.; Gambeloni, R.Z.; Roque, A.T.; Oeck, S.; Ribeiro, M.L. Epigenetic Mechanisms of ATM Activation after Helicobacter Pylori Infection. Am. J. Pathol. 2018, 188, 329–335. [Google Scholar] [CrossRef]
- Okabe, A.; Huang, K.K.; Matsusaka, K.; Fukuyo, M.; Xing, M.; Ong, X.; Hoshii, T.; Usui, G.; Seki, M.; Mano, Y.; et al. Cross-Species Chromatin Interactions Drive Transcriptional Rewiring in Epstein-Barr Virus-Positive Gastric Adenocarcinoma. Nat. Genet. 2020, 52, 919–930. [Google Scholar] [CrossRef]
- Abd El Fattah, Y.K.; Abulsoud, A.I.; AbdelHamid, S.G.; Hamdy, N.M. Interactome Battling of lncRNA CCDC144NL-AS1: Its Role in the Emergence and Ferocity of Cancer and Beyond. Int. J. Biol. Macromol. 2022, 222, 1676–1687. [Google Scholar] [CrossRef]
- Liu, X.-H.; Sun, M.; Nie, F.-Q.; Ge, Y.-B.; Zhang, E.-B.; Yin, D.-D.; Kong, R.; Xia, R.; Lu, K.-H.; Li, J.-H.; et al. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol. Cancer 2014, 13, 92. [Google Scholar] [CrossRef]
- Jin, Y.; Ma, J.; Wang, L. Comprehensive Analysis of Immune-Related lncRNAs and Their Clinical Relevance in Gastric Adenocarcinoma. Funct. Integr. Genom. 2023, 23, 28. [Google Scholar] [CrossRef]
- Kipkeeva, F.; Muzaffarova, T.; Korotaeva, A.; Nikulin, M.; Grishina, K.; Mansorunov, D.; Apanovich, P.; Karpukhin, A. MicroRNA in Gastric Cancer Development: Mechanisms and Biomarkers. Diagnostics 2020, 10, 891. [Google Scholar] [CrossRef]
- Kang, J.; Huang, X.; Dong, W.; Zhu, X.; Li, M.; Cui, N. MicroRNA-1269b Inhibits Gastric Cancer Development through Regulating Methyltransferase-like 3 (METTL3). Bioengineered 2021, 12, 1150–1160. [Google Scholar] [CrossRef]
- Ueda, T.; Volinia, S.; Okumura, H.; Shimizu, M.; Taccioli, C.; Rossi, S.; Alder, H.; Liu, C.G.; Oue, N.; Yasui, W.; et al. Relation between microRNA expression and progression and prognosis of gastric cancer: A microRNA expression analysis. Lancet Oncol. 2010, 11, 136–146. [Google Scholar] [CrossRef]
- Li, Y.; Zheng, Q.; Bao, C.; Li, S.; Guo, W.; Zhao, J.; Chen, D.; Gu, J.; He, X.; Huang, S. Circular RNA Is Enriched and Stable in Exosomes: A Promising Biomarker for Cancer Diagnosis. Cell Res. 2015, 25, 981–984. [Google Scholar] [CrossRef]
- Reis-das-Mercês, L.; Vinasco-Sandoval, T.; Pompeu, R.; Ramos, A.C.; Anaissi, A.K.M.; Demachki, S.; de Assumpção, P.P.; Vidal, A.F.; Ribeiro-Dos-Santos, Â.; Magalhães, L. CircRNAs as Potential Blood Biomarkers and Key Elements in Regulatory Networks in Gastric Cancer. Int. J. Mol. Sci. 2022, 23, 650. [Google Scholar] [CrossRef]
- Li, T.; Shao, Y.; Fu, L.; Xie, Y.; Zhu, L.; Sun, W.; Yu, R.; Xiao, B.; Guo, J. Plasma Circular RNA Profiling of Patients with Gastric Cancer and Their Droplet Digital RT-PCR Detection. J. Mol. Med. 2018, 96, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Fang, L.; Lv, J.; Xuan, Z.; Li, B.; Li, Z.; He, Z.; Li, F.; Xu, J.; Wang, S.; Xia, Y.; et al. Circular CPM Promotes Chemoresistance of Gastric Cancer via Activating PRKAA2-Mediated Autophagy. Clin. Transl. Med. 2022, 12, e708. [Google Scholar] [CrossRef] [PubMed]
- Saviana, M.; Le, P.; Micalo, L.; Del Valle-Morales, D.; Romano, G.; Acunzo, M.; Li, H.; Nana-Sinkam, P. Crosstalk between miRNAs and DNA Methylation in Cancer. Genes 2023, 14, 1075. [Google Scholar] [CrossRef]
- Yuan, R.; Wang, G.; Xu, Z.; Zhao, H.; Chen, H.; Han, Y.; Wang, B.; Zhou, J.; Hu, H.; Guo, Z.; et al. Up-regulated Circulating miR-106a by DNA Methylation Promised a Potential Diagnostic and Prognostic Marker for Gastric Cancer. Anticancer. Agents Med. Chem. 2016, 16, 1093–1100. [Google Scholar] [CrossRef]
- Szczepanek, J.; Tretyn, A. MicroRNA-Mediated Regulation of Histone-Modifying Enzymes in Cancer: Mechanisms and Therapeutic Implications. Biomolecules 2023, 13, 1590. [Google Scholar] [CrossRef]
- Qi, Y.; Wang, D.; Wang, D.; Jin, T.; Yang, L.; Wu, H.; Li, Y.; Zhao, J.; Du, F.; Song, M.; et al. HEDD: The human epigenetic drug database. Database 2016, 2016, baw159. [Google Scholar] [CrossRef]
- Sahafnejad, Z.; Ramazi, S.; Allahverdi, A. An Update of Epigenetic Drugs for the Treatment of Cancers and Brain Diseases: A Comprehensive Review. Genes 2023, 14, 873. [Google Scholar] [CrossRef]
- Abdelfatah, E.; Kerner, Z.; Nanda, N.; Ahuja, N. Epigenetic therapy in gastrointestinal cancer: The right combination. Ther. Adv. Gastroenterol. 2016, 9, 560–579. [Google Scholar] [CrossRef]
- Gao, Y.; van Haren, M.J.; Buijs, N.; Innocenti, P.; Zhang, Y.; Sartini, D.; Campagna, R.; Emanuelli, M.; Parsons, R.B.; Jespers, W.; et al. Potent Inhibition of Nicotinamide N-Methyltransferase by Alkene-Linked Bisubstrate Mimics Bearing Electron Deficient Aromatics. J. Med. Chem. 2021, 64, 12938–12963. [Google Scholar] [CrossRef]
- van Haren, M.J.; Zhang, Y.; Thijssen, V.; Buijs, N.; Gao, Y.; Mateuszuk, L.; Fedak, F.A.; Kij, A.; Campagna, R.; Sartini, D.; et al. Macrocyclic peptides as allosteric inhibitors of nicotinamide N-methyltransferase (NNMT). RSC Chem. Biol. 2021, 2, 1546–1555. [Google Scholar] [CrossRef] [PubMed]
- van Haren, M.J.; Gao, Y.; Buijs, N.; Campagna, R.; Sartini, D.; Emanuelli, M.; Mateuszuk, L.; Kij, A.; Chlopicki, S.; Escudé Martinez de Castilla, P.; et al. Esterase-Sensitive Prodrugs of a Potent Bisubstrate Inhibitor of Nicotinamide N-Methyltransferase (NNMT) Display Cellular Activity. Biomolecules 2021, 11, 1357. [Google Scholar] [CrossRef] [PubMed]
- Moro, H.; Hattori, N.; Nakamura, Y.; Kimura, K.; Imai, T.; Maeda, M.; Yashiro, M.; Ushijima, T. Epigenetic priming sensitizes gastric cancer cells to irinotecan and cisplatin by restoring multiple pathways. Gastric. Cancer 2020, 23, 105–115. [Google Scholar] [CrossRef] [PubMed]
- Hu, N.; Li, W.; Hong, Y.; Zeng, Z.; Zhang, J.; Wu, X.; Zhou, K.; Wu, F. A PD1 targeted nano-delivery system based on epigenetic alterations of T cell responses in the treatment of gastric cancer. Mol. Ther.-Oncolytics 2022, 24, 148–159. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Turcan, S. Epigenetic Drugs and Their Immune Modulating Potential in Cancers. Biomedicines 2022, 10, 211. [Google Scholar] [CrossRef]
- Miranda Furtado, C.L.; Dos Santos Luciano, M.C.; da Silva Santos, R.; Furtado, G.P.; Moraes, M.O.; Pessoa, C. Epidrugs: Targeting epigenetic marks in cancer treatment. Epigenetics 2019, 14, 1164–1176. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).