Binding Pattern and Structural Interactome of the Anticancer Drug 5-Fluorouracil: A Critical Review
Abstract
1. Introduction
2. Crystal Structures of 5-FU Complexes in the Protein Data Bank (PDB)
3. The Binding Mode of 5-FU
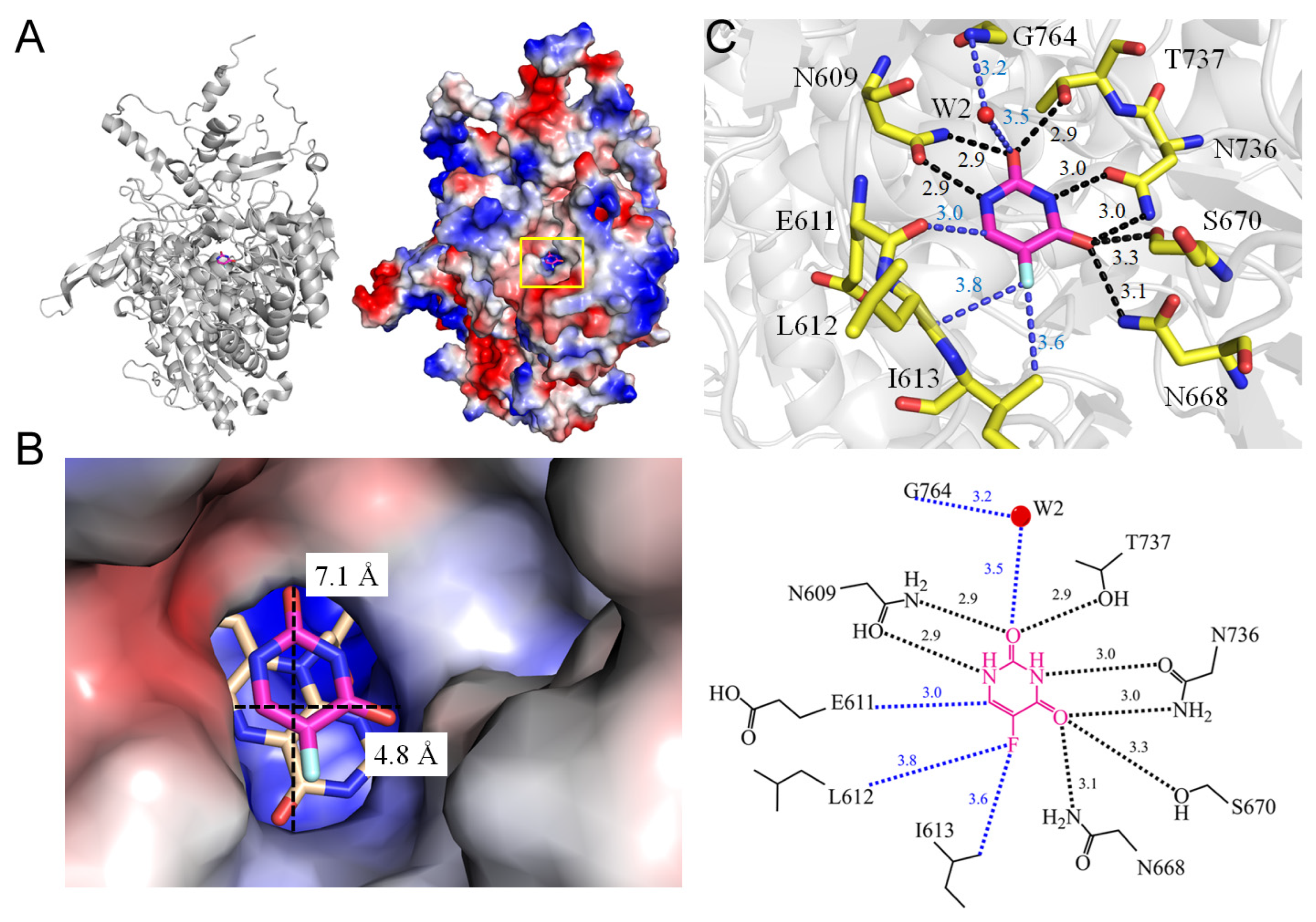
3.1. Dihydropyrimidine Dehydrogenase
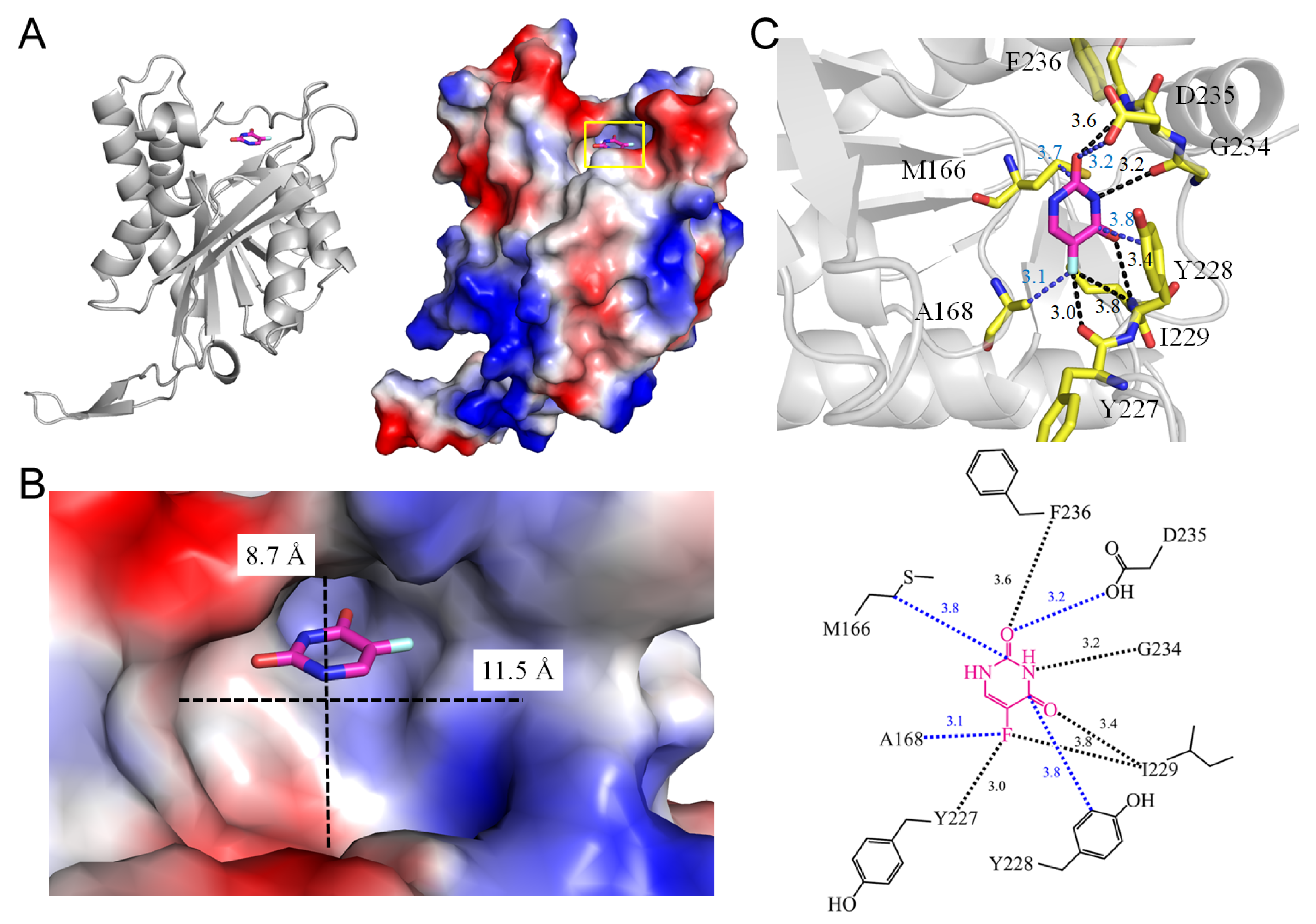
3.2. Uracil Phosphoribosyltransferase
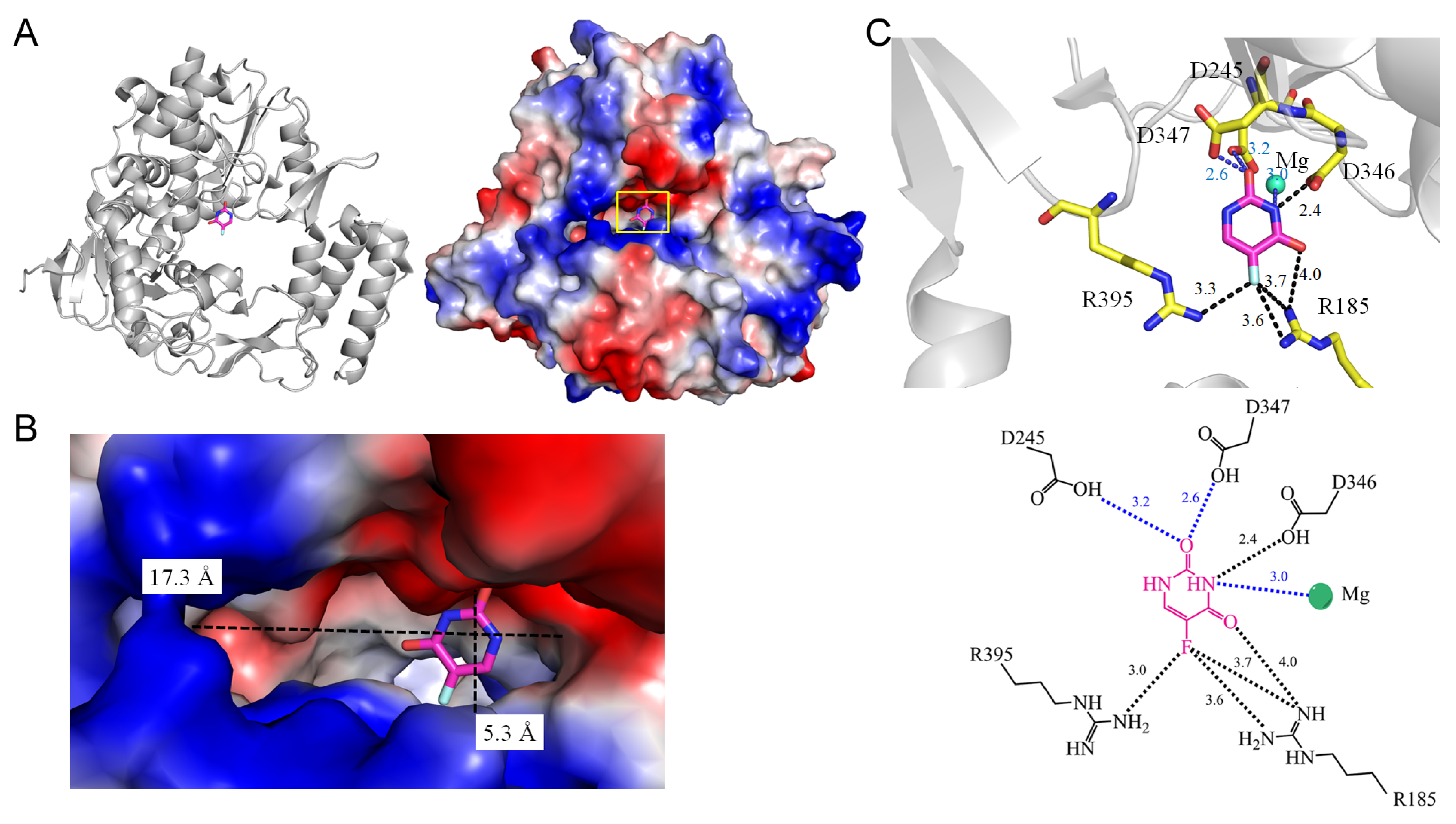
3.3. RNA-Dependent RNA Polymerase
3.4. Uridine Phosphorylase
3.5. rRNA N-Glycosidase
3.6. Uracil-DNA Glycosylase
3.7. Pyrimidine Operon Regulatory Protein PyrR
3.8. PI3Kα
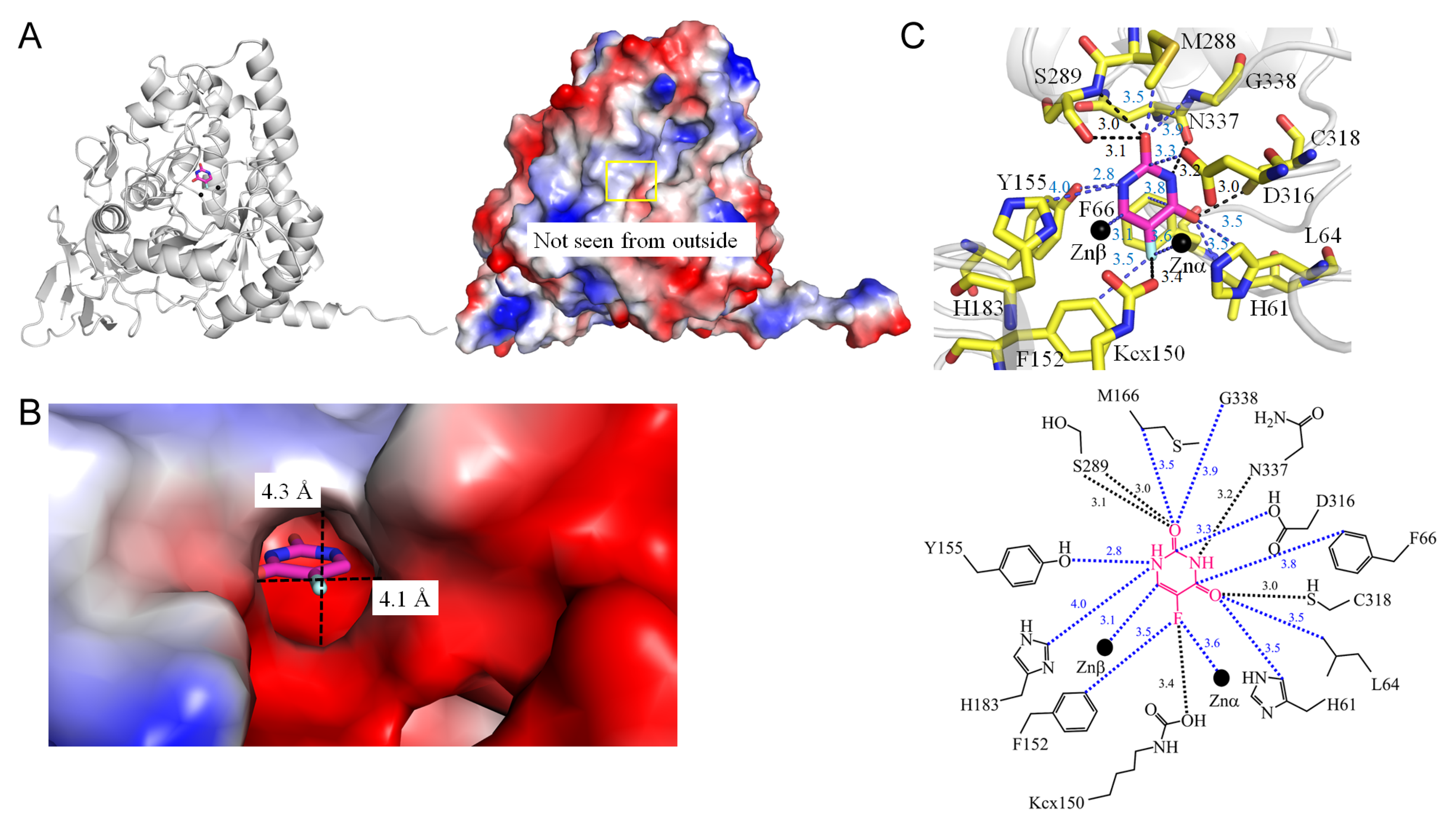
3.9. Dihydropyrimidinase
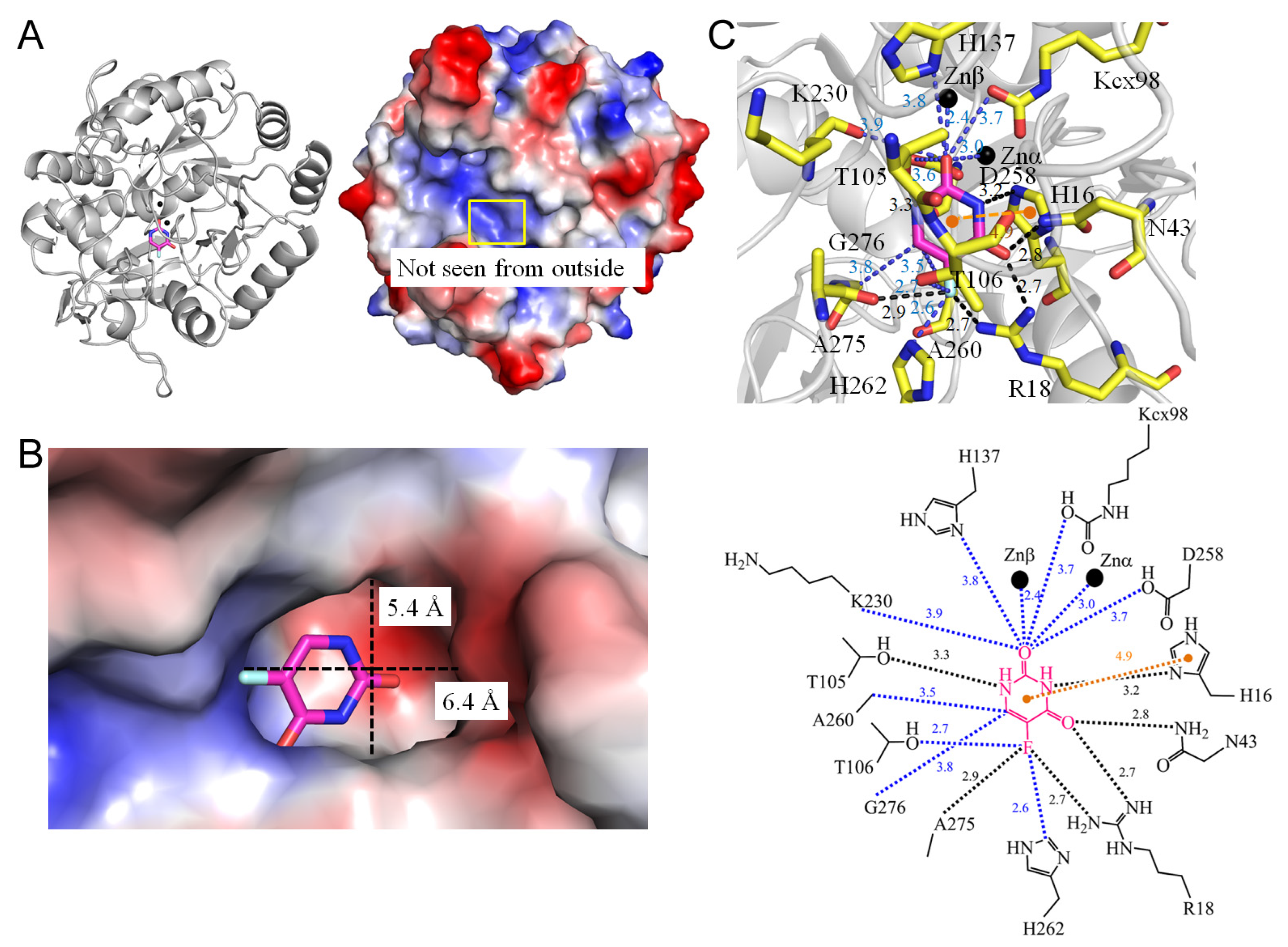
3.10. Dihydroorotases
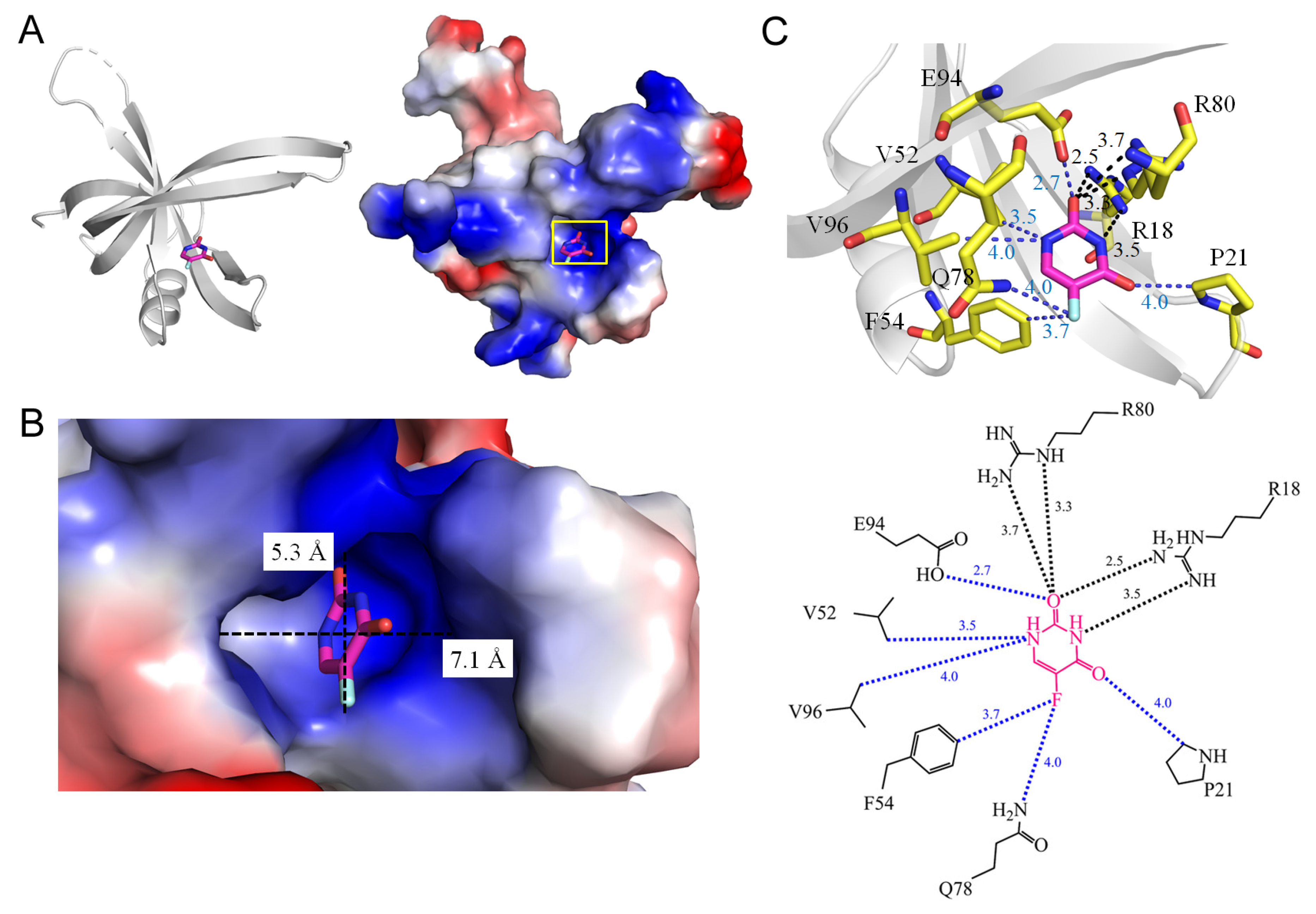
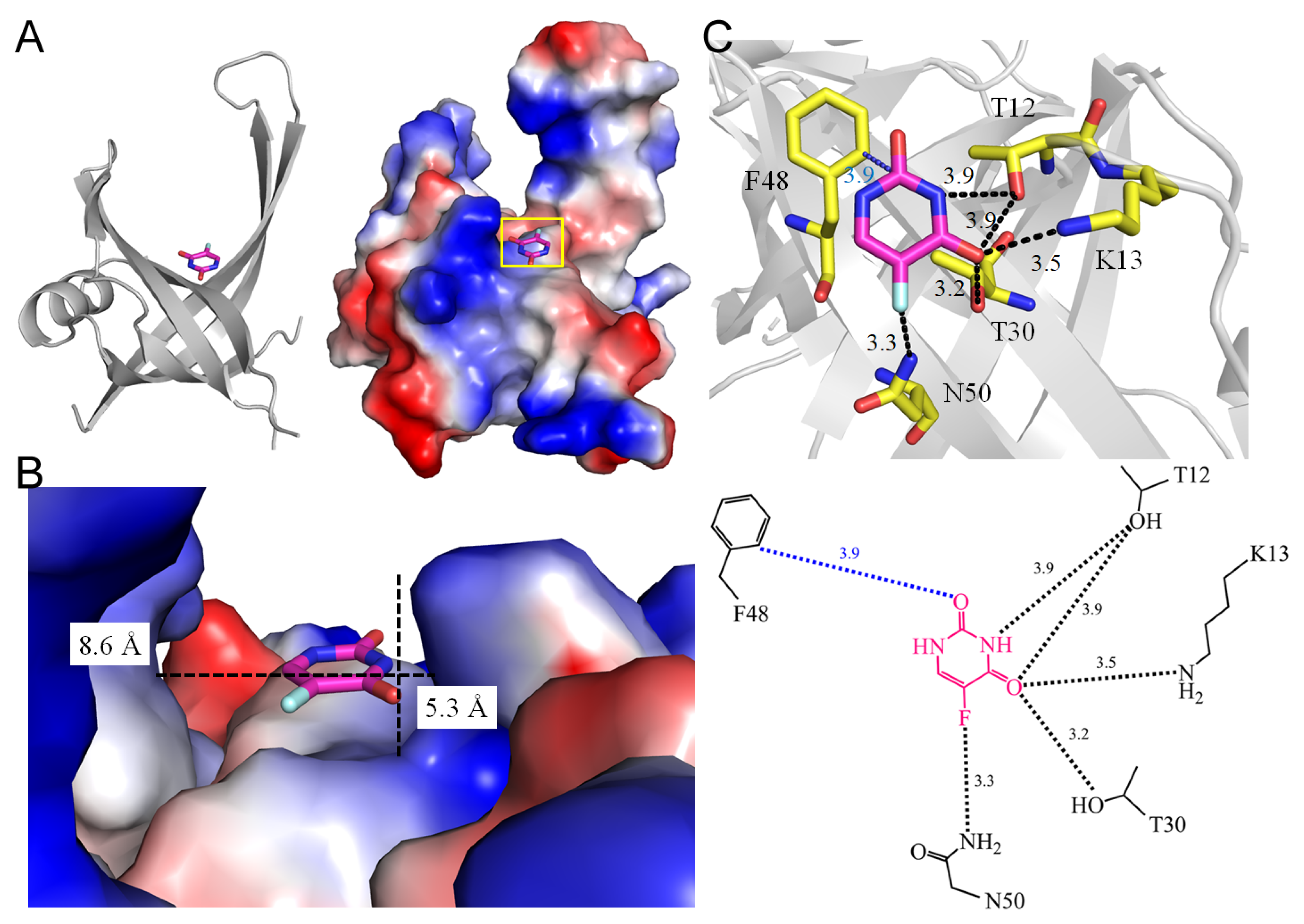
3.11. Single-Stranded DNA-Binding Proteins SsbA and SsbB
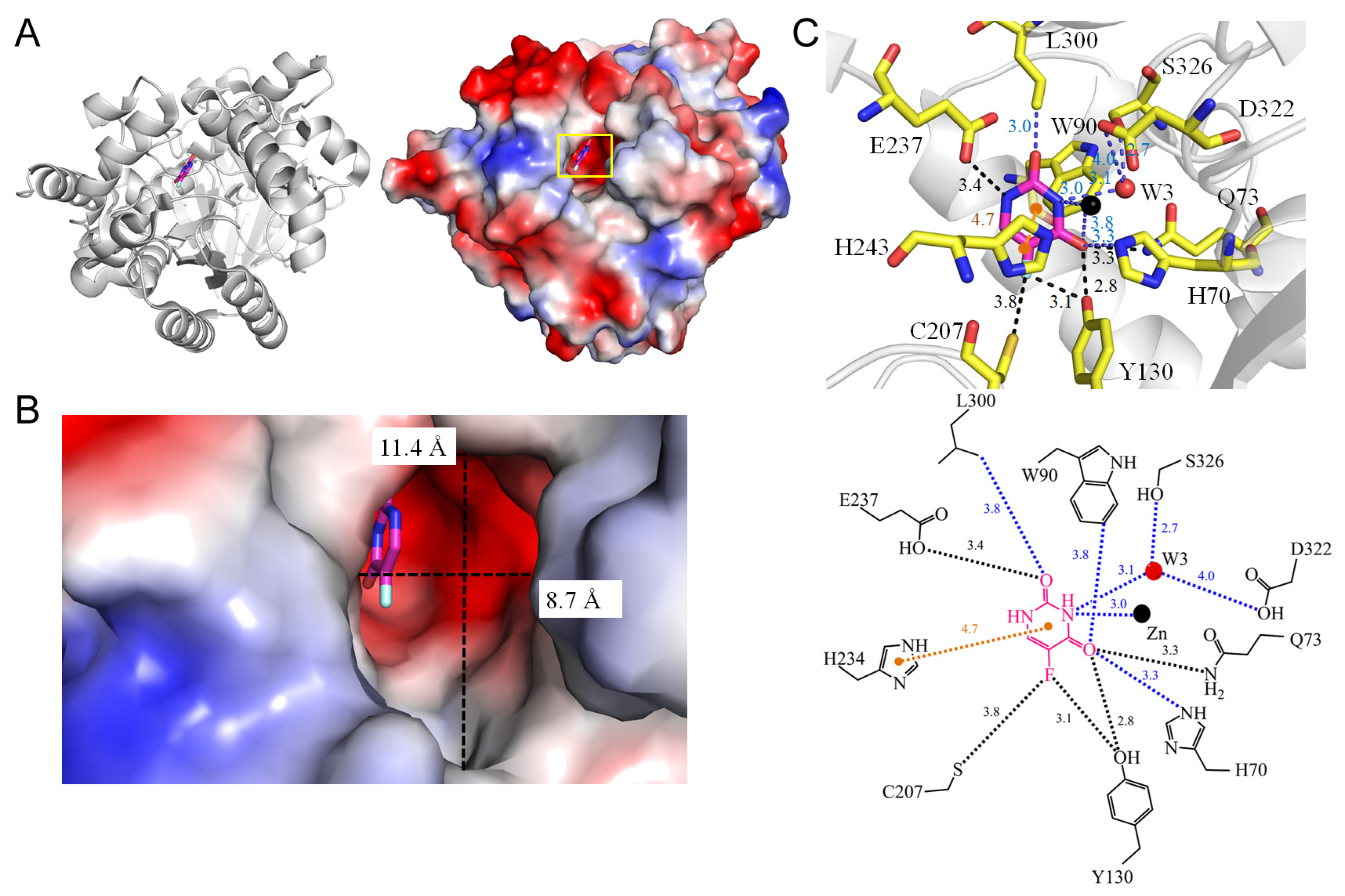
3.12. Hydroxydechloroatrazine Ethylaminohydrolase VCZ
4. Interaction Patterns
- Type 1 (P-R type): The contact distance involves residues from both P (Arg and Lys) and R (Phe, Tyr, Trp, and His) types.
- Type 2 (P type): The contact distance involves more than two P-type residues, Arg and/or Lys.
- Type 3 (R type): The contact distance involves R-type residues, Phe, Tyr, Trp, and/or His.
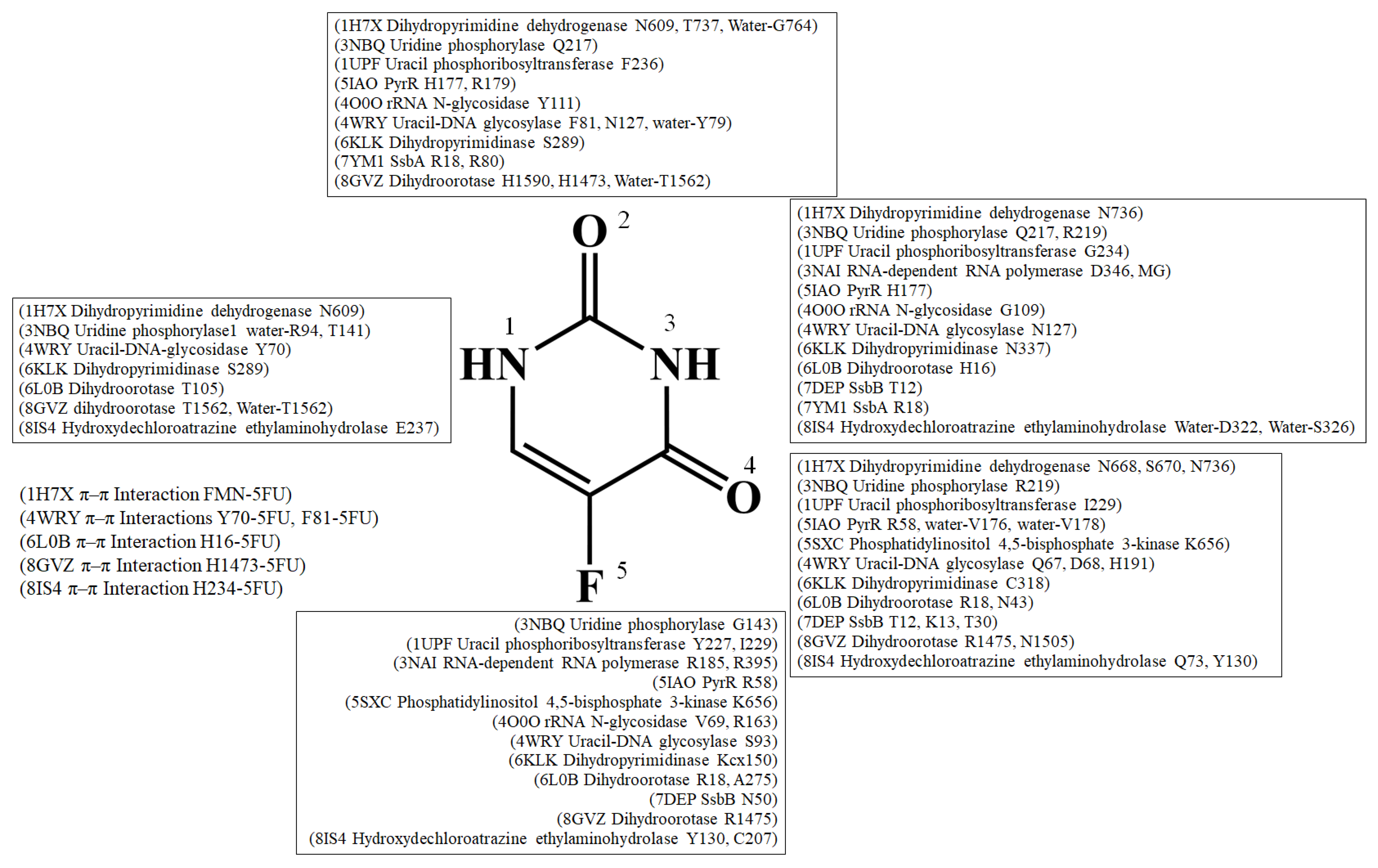
| Type | PDB | Interactive Residues | HB | WB | π-π Int. | F5 (HB) | F5 (CD) | N1 or N3 (HB) | N1 or N3 (CD) | N3 (HB) | N3 (CD) | O4 (HB) | O4 (CD) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
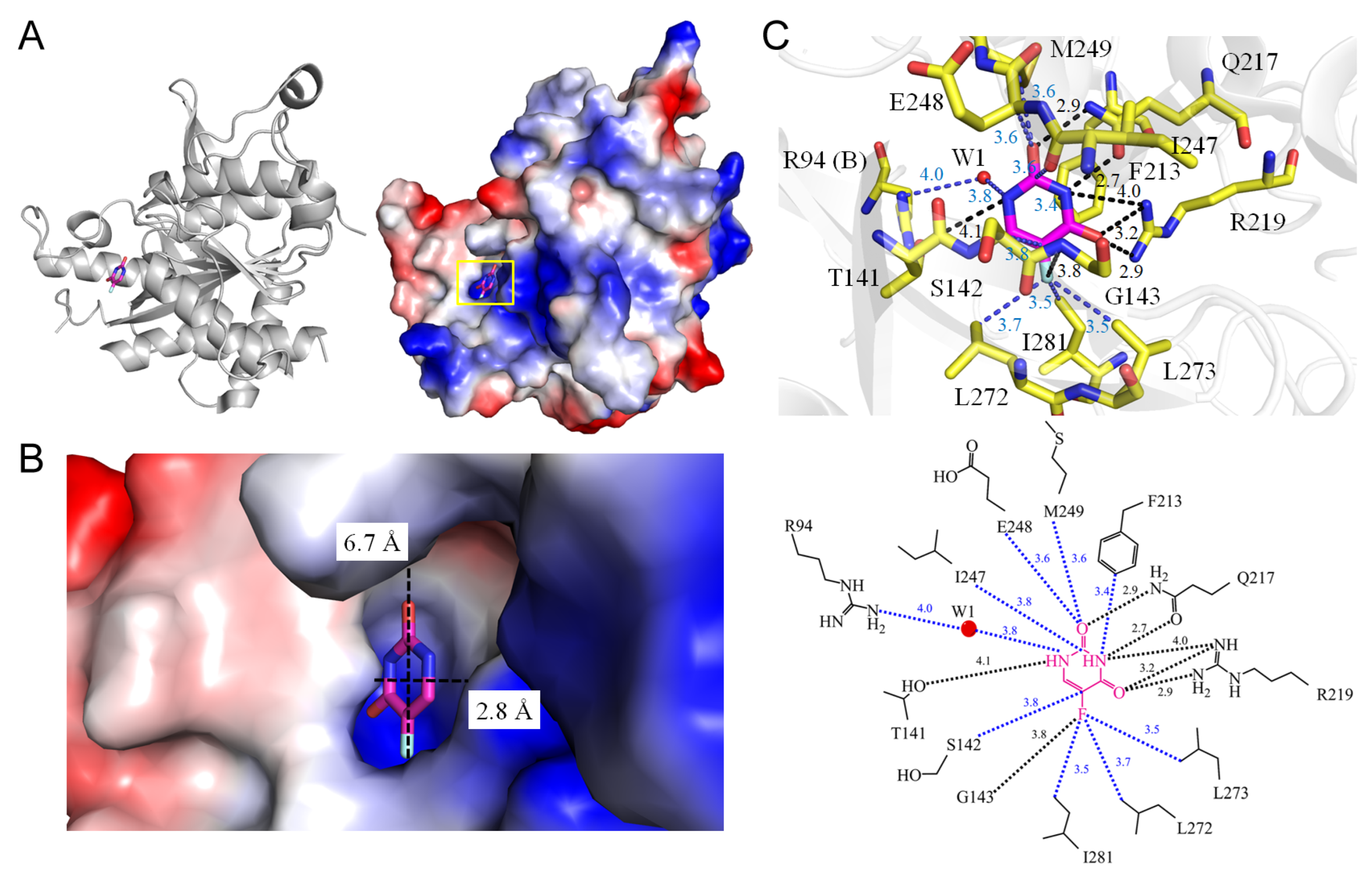
| 1 | 3NBQ | R94, T141, S142, G143, F213, Q217, R219, I247, E248, M249, L272, L273, I281, W1-R94(B) | T141, G143, Q217, R219 | W1-R94(B) | - | G143 [N] | I281, L272, L273 | T141 [OG1], Q217 [OE1], R219 [NH1] | F213 | Q217 [OE1], R219 [NH1] | F213 | R219 [NH1] [NH2] | - |
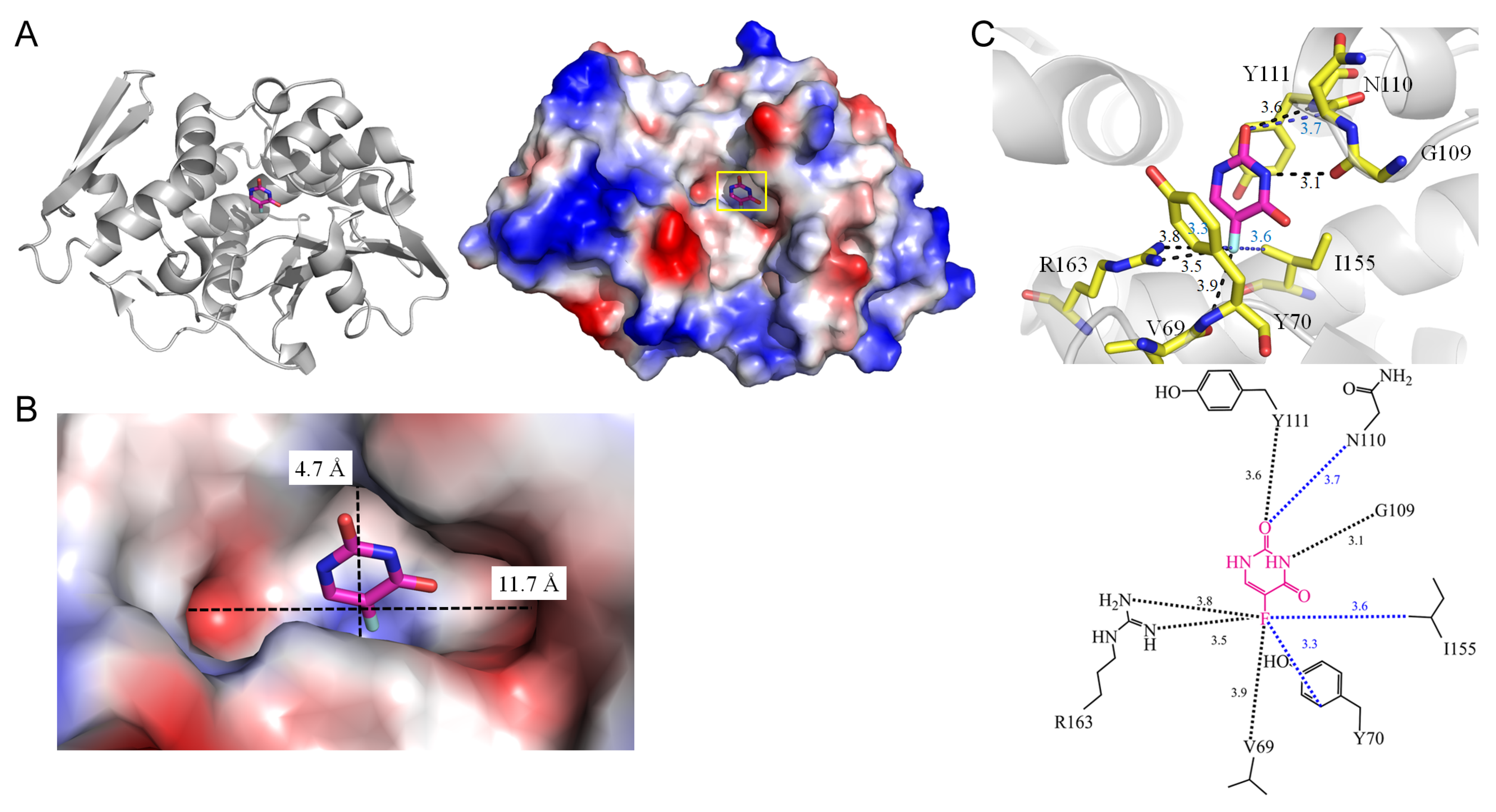
| 1 | 4O0O | V69, Y70, G109, N110, Y111, I155, R163 | V69, G109, Y111, R163 | - | - | V69 [O], R163 [NH1], R163 [NH2] | I155,Y70 | G109 [O] | - | G109 [O] | - | - | - |
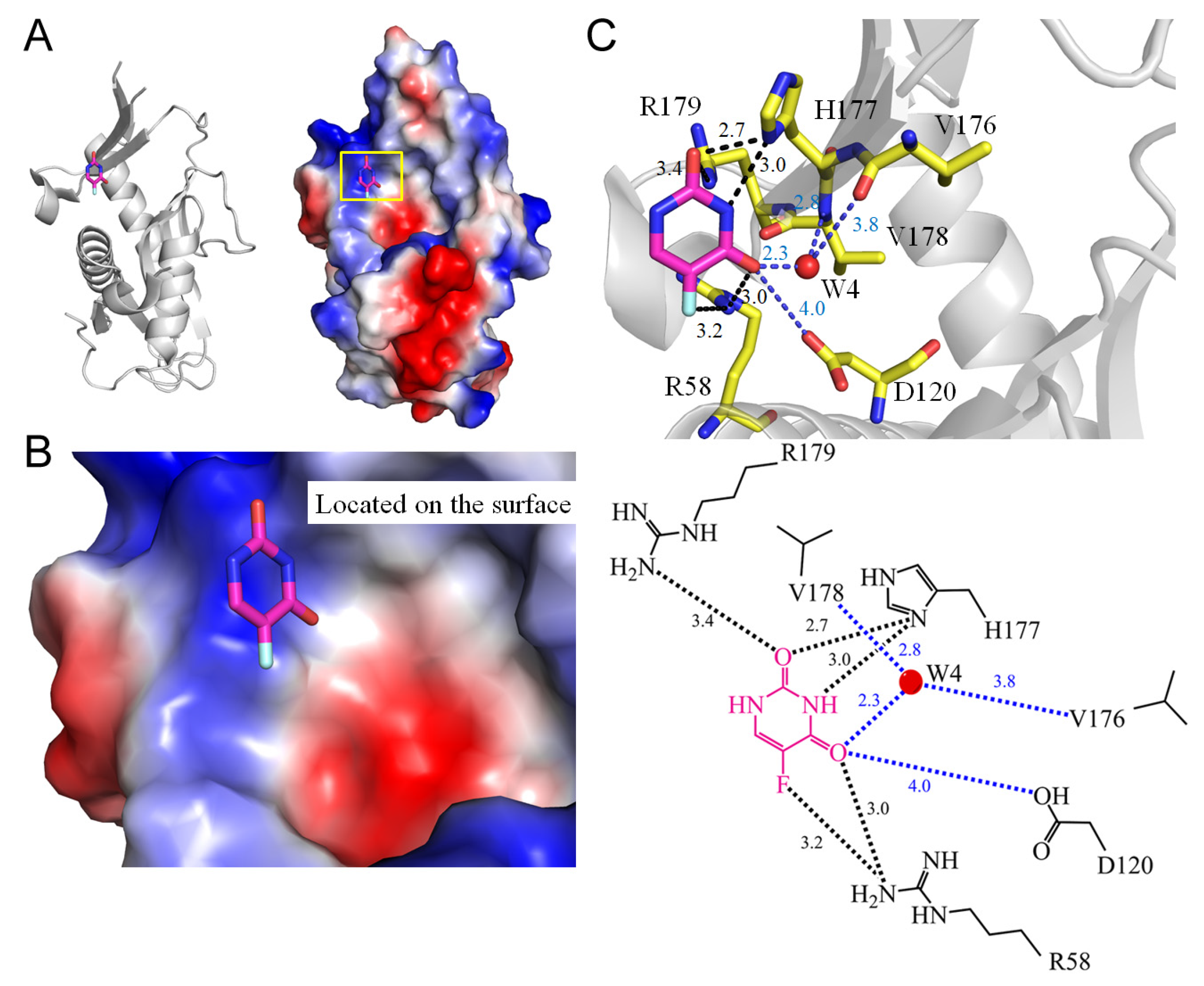
| 1 | 5IAO | R58, D120, V176, H177, R179, W4-V178 | R58, H177, R179, | W4-V178 | - | R58 [NH1], R58 [NE2] | D120 | H177 [ND1] | - | H177 [ND1] | - | R58 [NH1] | D120 |
| 1 | 6KLK | H61, L64, F66, Kcx150, F152, Y155, H183, M288, S289, D316, C318, N337, G338, | Kcx150, S289, D316, C318, N337, G338, | - | - | Kcx150 [OQ2] | F152 | S289 [O], N337 [O] | Y155, H183 | N337 [O] | C318 [SG] | H61, L64 | |
| 1 | 6L0B | H16, R18, N43, Kcx98, T105, T106, H137, K230, D258, A260, H262, G276, A275 | H16, R18, N43, T105 | - | H16 (4.9Å) | A275 [O] | H262 | T105 [OG1], H16 [ND1] | - | H16 [ND1] | - | R18 [NH1], N43 [ND2] | - |
| 1 | 7DEP | T12, K13, T30, F48, N50 | T12, K13, T30, N50 | - | - | N50 [ND2] | - | T12 [OG1], | - | T12 [OG1], | - | T12 [OG1], K13 [NZ], T30 [OG1] | - |
| 1 | 7YM1 | R18, P21, F54, V52, Q78, R80, E94, V96 | R18, R80 | - | - | Q78, F54 | R18 [NH2] | V52, V96 | R18 [NH2] | - | - | P21 | |
| 1 | 8GVZ | R1475, H1473, N1505, Kcx1556, T1562, F1563, H1590, R1661, D1686, A1688, H1690, P1702, | R1475, H1473, N1505, T1562, H1590 | W1- T1562 | H1473 (4.5 Å) | R1475 [NH2] | F1360, H1690 | T1562 [OG1], H1473 [ND1] | - | H1473 [ND1] | - | R1475 [NH1] [NH2], N1505 [ND2] | - |
| 2 | 3NAI | R185, D245, R395, D346, D347 | R185, D245, R395 | - | - | R185 [NH1], R185 [NH2], R395 [NH2] | - | D346 [OD2] | - | D346 [OD2] | - | - | - |
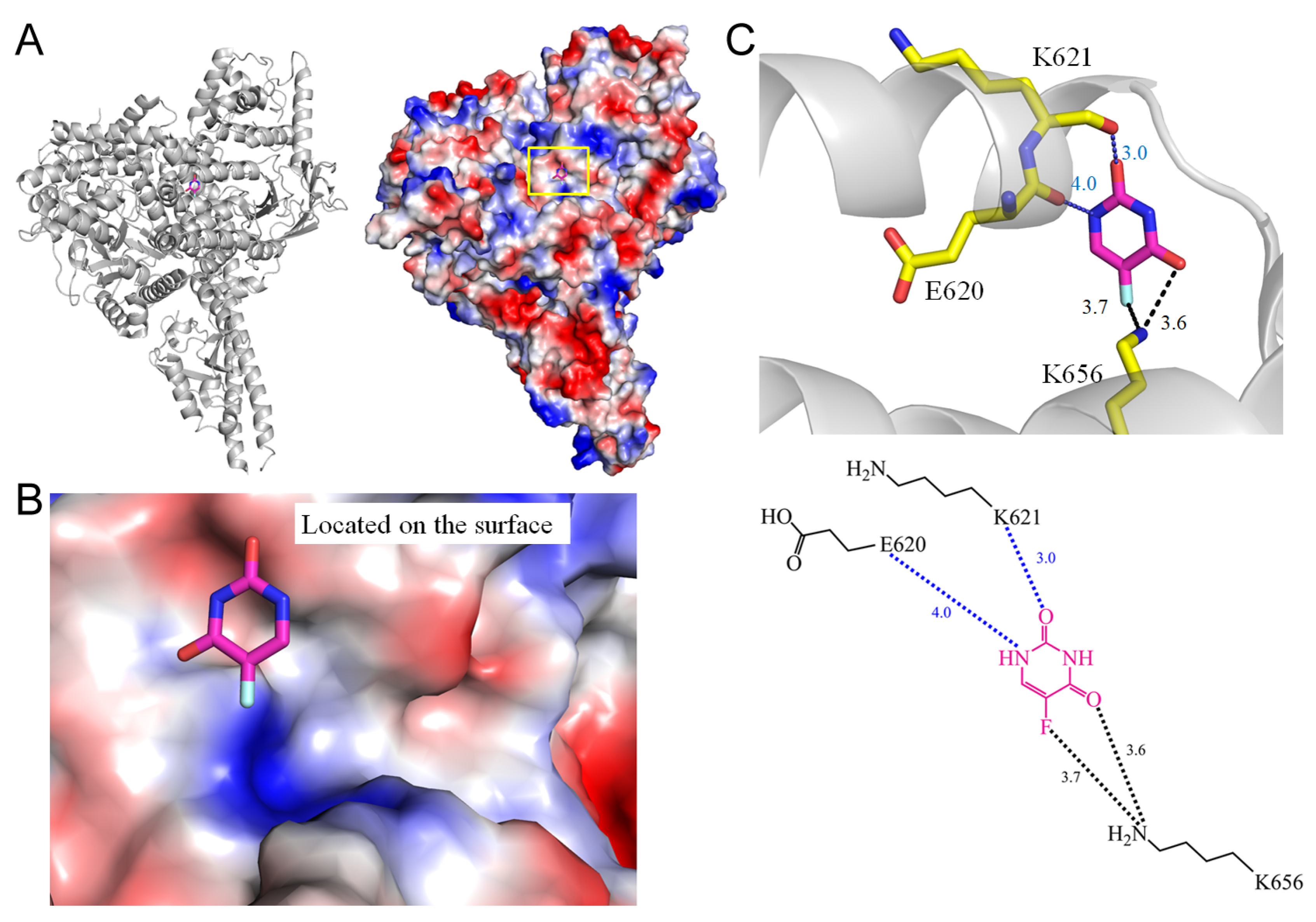
| 2 | 5SXC | E620, K621, K656 | K656 | - | - | K656 [NZ] | - | - | E620 | - | - | K656 [NZ] | - |
| 3 | 1H7X | N609, E611, L612, I613, N668, S670, N736, T737, W2-G764 | N609, N668, S670, N736, T737 | W2-G764 | - | - | L612, I613, FMN | N609 [OD1], N736 [OD1] | - | N609 [OD1], N736 [OD1] | - | N668 [ND2], S670 [OG], N736 [ND2] | - |
| 3 | 1UPF | M166, A168, Y227, Y228, I229, G234, F236, D316 | Y227, I229, G234, F236 | - | - | Y227 [O], I229 [N] | A168 | G234 [O] | - | G234 [O] | - | I229 [N] | - |
| 3 | 4WRY | G66, Q67, D68, Y70, S80, F81, S93, N127, H191, W2-L79 | G66, Q67, D68, Y70, S93, N127, H191 | W2-L79 | Y70 (4.7 Å) F81 (3.9 Å) | S93 [OG] | - | Y70 [N], N127 [OD1] | - | N127 [OD1] | - | Q67 [N], D68 [N], H191 [NE2] | - |
| 3 | 8IS4 | H70, Q73, W90, Y130, C207, E237, L300, D322, W3-S326 | Q73, Y130, C207, E237 | W3-S326 | H234 (4.7 Å) | C207 [SG], Y130 [OH] | - | - | - | - | - | Q73 [NE2], Y130 [OH] | W90 [CH2] H70 [NE2] |
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Sessler, J.L.; Lawrence, C.M.; Jayawickramarajah, J. Molecular recognition via base-pairing. Chem. Soc. Rev. 2007, 36, 314–325. [Google Scholar] [CrossRef]
- Zhu, J.; Thompson, C.B. Metabolic regulation of cell growth and proliferation. Nat. Rev. Mol. Cell Biol. 2019, 20, 436–450. [Google Scholar] [CrossRef] [PubMed]
- Fung, S.K.; Lok, A.S. Drug insight: Nucleoside and nucleotide analog inhibitors for hepatitis B. Nat. Clin. Pract. Gastroenterol. Hepatol. 2004, 1, 90–97. [Google Scholar] [CrossRef] [PubMed]
- Peters, H.L.; Ku, T.C.; Seley-Radtke, K.L. Flexibility as a Strategy in Nucleoside Antiviral Drug Design. Curr. Med. Chem. 2015, 22, 3910–3921. [Google Scholar] [CrossRef]
- Yssel, A.E.J.; Vanderleyden, J.; Steenackers, H.P. Repurposing of nucleoside- and nucleobase-derivative drugs as antibiotics and biofilm inhibitors. J. Antimicrob. Chemother. 2017, 72, 2156–2170. [Google Scholar] [CrossRef]
- Ku, S.K.; Baek, M.C.; Bae, J.S. Anti-inflammatory effects of methylthiouracil in vitro and in vivo. Toxicol. Appl. Pharmacol. 2015, 288, 374–386. [Google Scholar] [CrossRef]
- Gmeiner, W.H. Fluoropyrimidine Modulation of the Anti-Tumor Immune Response-Prospects for Improved Colorectal Cancer Treatment. Cancers 2020, 12, 1641. [Google Scholar] [CrossRef]
- Alvarez, P.; Marchal, J.A.; Boulaiz, H.; Carrillo, E.; Velez, C.; Rodriguez-Serrano, F.; Melguizo, C.; Prados, J.; Madeddu, R.; Aranega, A. 5-Fluorouracil derivatives: A patent review. Expert. Opin. Ther. Pat. 2012, 22, 107–123. [Google Scholar] [CrossRef]
- Longley, D.B.; Harkin, D.P.; Johnston, P.G. 5-Fluorouracil: Mechanisms of action and clinical strategies. Nat. Rev. Cancer 2003, 3, 330–338. [Google Scholar] [CrossRef]
- Wilson, P.M.; Danenberg, P.V.; Johnston, P.G.; Lenz, H.J.; Ladner, R.D. Standing the test of time: Targeting thymidylate biosyn- thesis in cancer therapy. Nat. Rev. Clin. Oncol. 2014, 11, 282–298. [Google Scholar] [CrossRef] [PubMed]
- Hertz, D.L.; Smith, D.M.; Scott, S.A.; Patel, J.N.; Hicks, J.K. Response to the FDA Decision Regarding DPYD Testing Prior to Fluoropyrimidine Chemotherapy. Clin. Pharmacol. Ther. 2023, 114, 768–779. [Google Scholar] [CrossRef]
- Vodenkova, S.; Buchler, T.; Cervena, K.; Veskrnova, V.; Vodicka, P.; Vymetalkova, V. 5-fluorouracil and other fluoropyrimidines in colorectal cancer: Past, present and future. Pharmacol. Ther. 2020, 206, 107447. [Google Scholar] [CrossRef]
- van Kuilenburg, A.B.; Meinsma, R.; Zonnenberg, B.A.; Zoetekouw, L.; Baas, F.; Matsuda, K.; Tamaki, N.; van Gennip, A.H. Dihy- dropyrimidinase deficiency and severe 5-fluorouracil toxicity. Clin. Cancer Res. 2003, 9, 4363–4367. [Google Scholar] [PubMed]
- Alexander, J.L.; Wilson, I.D.; Teare, J.; Marchesi, J.R.; Nicholson, J.K.; Kinross, J.M. Gut microbiota modulation of chemotherapy efficacy and toxicity. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 356–365. [Google Scholar] [CrossRef] [PubMed]
- Ghode, P.; Jobichen, C.; Ramachandran, S.; Bifani, P.; Sivaraman, J. Structural basis of mapping the spontaneous mutations with 5-flurouracil in uracil phosphoribosyltransferase from Mycobacterium tuberculosis. Biochem. Biophys. Res. Commun. 2015, 467, 577–582. [Google Scholar] [CrossRef] [PubMed]
- Shakya, N.; Srivastav, N.C.; Desroches, N.; Agrawal, B.; Kunimoto, D.Y.; Kumar, R. 3’-bromo analogues of pyrimidine nucleosides as a new class of potent inhibitors of Mycobacterium tuberculosis. J. Med. Chem. 2010, 53, 4130–4140. [Google Scholar] [CrossRef] [PubMed]
- Ezzeldin, H.; Diasio, R. Dihydropyrimidine dehydrogenase deficiency, a pharmacogenetic syndrome associated with potentially life-threatening toxicity following 5-fluorouracil administration. Clin. Colorectal. Cancer 2004, 4, 181–189. [Google Scholar] [CrossRef]
- Krissinel, E.; Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007, 372, 774–797. [Google Scholar] [CrossRef] [PubMed]
- Adasme, M.F.; Linnemann, K.L.; Bolz, S.N.; Kaiser, F.; Salentin, S.; Haupt, V.J.; Schroeder, M. PLIP 2021: Expanding the scope of the protein-ligand interaction profiler to DNA and RNA. Nucleic Acids Res. 2021, 49, W530–W534. [Google Scholar] [CrossRef] [PubMed]
- Tian, W.; Chen, C.; Lei, X.; Zhao, J.; Liang, J. CASTp 3.0: Computed atlas of surface topography of proteins. Nucleic Acids Res. 2018, 46, W363–W367. [Google Scholar] [CrossRef]
- Schnackerz, K.D.; Dobritzsch, D. Amidohydrolases of the reductive pyrimidine catabolic pathway purification, characterization, structure, reaction mechanisms and enzyme deficiency. Biochim. Biophys. Acta 2008, 1784, 431–444. [Google Scholar] [CrossRef] [PubMed]
- Schnackerz, K.D.; Dobritzsch, D.; Lindqvist, Y.; Cook, P.F. Dihydropyrimidine dehydrogenase: A flavoprotein with four iron-sulfur clusters. Biochim. Biophys. Acta 2004, 1701, 61–74. [Google Scholar] [CrossRef] [PubMed]
- Etienne-Grimaldi, M.C.; Pallet, N.; Boige, V.; Ciccolini, J.; Chouchana, L.; Barin-Le Guellec, C.; Zaanan, A.; Narjoz, C.; Taieb, J.; Thomas, F.; et al. Current diagnostic and clinical issues of screening for dihydropyrimidine dehydrogenase deficiency. Eur. J. Cancer 2023, 181, 3–17. [Google Scholar] [CrossRef] [PubMed]
- Dobritzsch, D.; Schneider, G.; Schnackerz, K.D.; Lindqvist, Y. Crystal structure of dihydropyrimidine dehydrogenase, a major determinant of the pharmacokinetics of the anti-cancer drug 5-fluorouracil. EMBO J. 2001, 20, 650–660. [Google Scholar] [CrossRef] [PubMed]
- Iltzsch, M.H.; Tankersley, K.O. Structure-activity relationship of ligands of uracil phosphoribosyltransferase from Toxoplasma gondii. Biochem. Pharmacol. 1994, 48, 781–792. [Google Scholar] [CrossRef]
- Carter, D.; Donald, R.G.; Roos, D.; Ullman, B. Expression, purification, and characterization of uracil phosphoribosyltransferase from Toxoplasma gondii. Mol. Biochem. Parasitol. 1997, 87, 137–144. [Google Scholar] [CrossRef]
- Schumacher, M.A.; Carter, D.; Scott, D.M.; Roos, D.S.; Ullman, B.; Brennan, R.G. Crystal structures of Toxoplasma gondii uracil phosphoribosyltransferase reveal the atomic basis of pyrimidine discrimination and prodrug binding. EMBO J. 1998, 17, 3219–3232. [Google Scholar] [CrossRef]
- Ferrer-Orta, C.; Arias, A.; Pérez-Luque, R.; Escarmís, C.; Domingo, E.; Verdaguer, N. Sequential structures provide insights into the fidelity of RNA replication. Proc. Natl. Acad. Sci. USA 2007, 104, 9463–9468. [Google Scholar] [CrossRef]
- Ng, K.K.; Arnold, J.J.; Cameron, C.E. Structure-function relationships among RNA-dependent RNA polymerases. Curr. Top. Microbiol. Immunol. 2008, 320, 137–156. [Google Scholar]
- Ferrer-Orta, C.; Arias, A.; Escarmís, C.; Verdaguer, N. A comparison of viral RNA-dependent RNA polymerases. Curr. Opin. Struct. Biol. 2006, 16, 27–34. [Google Scholar] [CrossRef]
- Lee, J.H.; Alam, I.; Han, K.R.; Cho, S.; Shin, S.; Kang, S.; Yang, J.M.; Kim, K.H. Crystal structures of murine norovirus-1 RNA-dependent RNA polymerase. J. Gen. Virol. 2011, 92, 1607–1616. [Google Scholar] [CrossRef]
- Deval, J.; Jin, Z.; Chuang, Y.C.; Kao, C.C. Structure(s), function(s), and inhibition of the RNA-dependent RNA polymerase of noroviruses. Virus Res. 2017, 234, 21–33. [Google Scholar] [CrossRef]
- Pugmire, M.J.; Ealick, S.E. Structural analyses reveal two distinct families of nucleoside phosphorylases. Biochem. J. 2002, 361, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Cao, D.; Pizzorno, G. Uridine phosophorylase: An important enzyme in pyrimidine metabolism and fluoropyrimidine activation. Drugs Today 2004, 40, 431–443. [Google Scholar] [CrossRef]
- Paul, D.; O’Leary, S.E.; Rajashankar, K.; Bu, W.; Toms, A.; Settembre, E.C.; Sanders, J.M.; Begley, T.P.; Ealick, S.E. Glycal formation in crystals of uridine phosphorylase. Biochemistry 2010, 49, 3499–3509. [Google Scholar] [CrossRef][Green Version]
- Caradoc-Davies, T.T.; Cutfield, S.M.; Lamont, I.L.; Cutfield, J.F. Crystal structures of Escherichia coli uridine phosphorylase in two native and three complexed forms reveal basis of substrate specificity, induced conformational changes and influence of potassium. J. Mol. Biol. 2004, 337, 337–354. [Google Scholar] [CrossRef]
- Roosild, T.P.; Castronovo, S. Active site conformational dynamics in human uridine phosphorylase 1. PLoS ONE 2010, 5, e12741. [Google Scholar] [CrossRef] [PubMed]
- Lashkov, A.A.; Sotnichenko, S.E.; Prokofiev, I.I.; Gabdulkhakov, A.G.; Agapov, I.I.; Shtil, A.A.; Betzel, C.; Mironov, A.S.; Mikhailov, A.M. X-ray structure of Salmonella typhimurium uridine phosphorylase complexed with 5-fluorouracil and molecular modelling of the complex of 5-fluorouracil with uridine phosphorylase from Vibrio cholerae. Acta Crystallogr. D Biol. Crystallogr. 2012, 68, 968–974. [Google Scholar] [CrossRef] [PubMed]
- da Silva Neto, A.M.; Torini de Souza, J.R.; Romanello, L.; Cassago, A.; Serrão, V.H.; DeMarco, R.; Brandão-Neto, J.; Garratt, R.C.; Pereira, H.D. Analysis of two Schistosoma mansoni uridine phosphorylases isoforms suggests the emergence of a protein with a non-canonical function. Biochimie 2016, 125, 12–22. [Google Scholar] [CrossRef]
- Larson, E.T.; Mudeppa, D.G.; Gillespie, J.R.; Mueller, N.; Napuli, A.J.; Arif, J.A.; Ross, J.; Arakaki, T.L.; Lauricella, A.; Detitta, G.; et al. The crystal structure and activity of a putative trypanosomal nucleoside phosphorylase reveal it to be a homodimeric uridine phosphorylase. J. Mol. Biol. 2010, 396, 1244–1259. [Google Scholar] [CrossRef]
- Puri, M.; Kaur, I.; Perugini, M.A.; Gupta, R.C. Ribosome-inactivating proteins: Current status and biomedical applications. Drug Discov. Today 2012, 17, 774–783. [Google Scholar] [CrossRef]
- Endo, Y.; Mitsui, K.; Motizuki, M.; Tsurugi, K. The mechanism of action of ricin and related toxic lectins on eukaryotic ribosomes. The site and the characteristics of the modification in 28 S ribosomal RNA caused by the toxins. J. Biol. Chem. 1987, 262, 5908–5912. [Google Scholar] [CrossRef]
- Endo, Y.; Tsurugi, K. RNA N-glycosidase activity of ricin A-chain. Mechanism of action of the toxic lectin ricin on eukaryotic ribosomes. J. Biol. Chem. 1987, 262, 8128–8130. [Google Scholar] [CrossRef] [PubMed]
- Yamini, S.; Pandey, S.N.; Kaur, P.; Sharma, S.; Singh, T.P. Binding and structural studies of the complexes of type 1 ribosome inactivating protein from Momordica balsamina with cytosine, cytidine, and cytidine diphosphate. Biochem. Biophys. Rep. 2015, 4, 134–140. [Google Scholar] [CrossRef][Green Version]
- Schormann, N.; Ricciardi, R.; Chattopadhyay, D. Uracil-DNA glycosylases-structural and functional perspectives on an essential family of DNA repair enzymes. Protein Sci. 2014, 23, 1667–1685. [Google Scholar] [CrossRef] [PubMed]
- Krokan, H.E.; Standal, R.; Slupphaug, G. DNA glycosylases in the base excision repair of DNA. Biochem. J. 1997, 325, 1–16. [Google Scholar] [CrossRef]
- Wyatt, M.D.; Wilson, D.M., 3rd. Participation of DNA repair in the response to 5-fluorouracil. Cell. Mol. Life Sci. 2009, 66, 788–799. [Google Scholar] [CrossRef]
- Weeks, L.D.; Zentner, G.E.; Scacheri, P.C.; Gerson, S.L. Uracil DNA glycosylase (UNG) loss enhances DNA double strand break formation in human cancer cells exposed to pemetrexed. Cell Death Dis. 2014, 5, e1045. [Google Scholar] [CrossRef]
- Savva, R. Targeting uracil-DNA glycosylases for therapeutic outcomes using insights from virus evolution. Future Med. Chem. 2019, 11, 1323–1344. [Google Scholar] [CrossRef] [PubMed]
- Raj, P.; Selvam, K.; Roy, K.; Mani Tripathi, S.; Kesharwani, S.; Gopal, B.; Varshney, U.; Sundriyal, S. Identification of a new and diverse set of Mycobacterium tuberculosis uracil-DNA glycosylase (MtUng) inhibitors using structure-based virtual screening: Experimental validation and molecular dynamics studies. Bioorg. Med. Chem. Lett. 2022, 76, 129008. [Google Scholar] [CrossRef]
- Arif, S.M.; Geethanandan, K.; Mishra, P.; Surolia, A.; Varshney, U.; Vijayan, M. Structural plasticity in Mycobacterium tuberculosis uracil-DNA glycosylase (MtUng) and its functional implications. Acta Crystallogr. D Biol. Crystallogr. 2015, 71, 1514–1527. [Google Scholar] [CrossRef]
- Hove-Jensen, B.; Andersen, K.R.; Kilstrup, M.; Martinussen, J.; Switzer, R.L.; Willemoës, M. Phosphoribosyl Diphosphate (PRPP): Biosynthesis, Enzymology, Utilization, and Metabolic Significance. Microbiol. Mol. Biol. Rev. 2017, 81, e00040-16. [Google Scholar] [CrossRef]
- Cole, S.T.; Brosch, R.; Parkhill, J.; Garnier, T.; Churcher, C.; Harris, D.; Gordon, S.V.; Eiglmeier, K.; Gas, S.; Barry, C.E., 3rd; et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 1998, 393, 537–544. [Google Scholar] [CrossRef]
- Houman, F.; Diaz-Torres, M.R.; Wright, A. Transcriptional antitermination in the bgl operon of E. coli is modulated by a specific RNA binding protein. Cell 1990, 62, 1153–1163. [Google Scholar] [CrossRef]
- Ghode, P.; Ramachandran, S.; Bifani, P.; Sivaraman, J. Structure and mapping of spontaneous mutational sites of PyrR from Mycobacterium tuberculosis. Biochem. Biophys. Res. Commun. 2016, 471, 409–415. [Google Scholar] [CrossRef]
- Vasan, N.; Cantley, L.C. At a crossroads: How to translate the roles of PI3K in oncogenic and metabolic signalling into improve- ments in cancer therapy. Nat. Rev. Clin. Oncol. 2022, 19, 471–485. [Google Scholar] [CrossRef]
- Vanhaesebroeck, B.; Perry, M.W.D.; Brown, J.R.; André, F.; Okkenhaug, K. PI3K inhibitors are finally coming of age. Nat. Rev. Drug Discov. 2021, 20, 741–769. [Google Scholar] [CrossRef]
- Knight, Z.A.; Gonzalez, B.; Feldman, M.E.; Zunder, E.R.; Goldenberg, D.D.; Williams, O.; Loewith, R.; Stokoe, D.; Balla, A.; Toth, B.; et al. A pharmacological map of the PI3-K family defines a role for p110alpha in insulin signaling. Cell 2006, 125, 733–747. [Google Scholar] [CrossRef]
- Foukas, L.C.; Claret, M.; Pearce, W.; Okkenhaug, K.; Meek, S.; Peskett, E.; Sancho, S.; Smith, A.J.; Withers, D.J.; Vanhaesebroeck, B. Critical role for the p110alpha phosphoinositide-3-OH kinase in growth and metabolic regulation. Nature 2006, 441, 366–370. [Google Scholar] [CrossRef]
- Miller, M.S.; Maheshwari, S.; McRobb, F.M.; Kinzler, K.W.; Amzel, L.M.; Vogelstein, B.; Gabelli, S.B. Identification of allosteric binding sites for PI3Kα oncogenic mutant specific inhibitor design. Bioorg. Med. Chem. 2017, 25, 1481–1486. [Google Scholar] [CrossRef]
- Huang, C.Y. Structure, catalytic mechanism, posttranslational lysine carbamylation, and inhibition of dihydropyrimidinases. Adv. Protein Chem. Struct. Biol. 2020, 122, 63–96. [Google Scholar] [PubMed]
- Cheng, J.H.; Huang, C.C.; Huang, Y.H.; Huang, C.Y. Structural Basis for pH-Dependent Oligomerization of Dihydropyrimidinase from Pseudomonas aeruginosa PAO1. Bioinorg. Chem. Appl. 2018, 2018, 9564391. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.Y. Inhibition of a putative dihydropyrimidinase from Pseudomonas aeruginosa PAO1 by flavonoids and substrates of cyclic amidohydrolases. PLoS ONE 2015, 10, e0127634. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cheng, J.H.; Huang, Y.H.; Lin, J.J.; Huang, C.Y. Crystal structures of monometallic dihydropyrimidinase and the human dihy- droorotase domain K1556A mutant reveal no lysine carbamylation within the active site. Biochem. Biophys. Res. Commun. 2018, 505, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Ho, Y.Y.; Huang, Y.H.; Huang, C.Y. Chemical rescue of the post-translationally carboxylated lysine mutant of allantoinase and dihydroorotase by metal ions and short-chain carboxylic acids. Amino Acids 2013, 44, 1181–1191. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.H.; Yang, P.C.; Lin, E.S.; Ho, Y.Y.; Peng, W.F.; Lu, H.P.; Huang, C.C.; Huang, C.Y. Crystal Structure of Allantoinase from Escherichia coli BL21: A Molecular Insight into a Role of the Active Site Loops in Catalysis. Molecules 2023, 28, 827. [Google Scholar] [CrossRef] [PubMed]
- Lin, E.S.; Huang, C.Y. Cytotoxic Activities and the Allantoinase Inhibitory Effect of the Leaf Extract of the Carnivorous Pitcher Plant Nepenthes miranda. Plants 2022, 11, 2265. [Google Scholar] [CrossRef]
- Peng, W.F.; Huang, C.Y. Allantoinase and dihydroorotase binding and inhibition by flavonols and the substrates of cyclic ami- dohydrolases. Biochimie 2014, 101, 113–122. [Google Scholar] [CrossRef]
- Huang, Y.H.; Ning, Z.J.; Huang, C.Y. Crystal structure of dihydropyrimidinase in complex with anticancer drug 5-fluorouracil. Biochem. Biophys. Res. Commun. 2019, 519, 160–165. [Google Scholar] [CrossRef]
- Lin, E.S.; Luo, R.H.; Yang, Y.C.; Huang, C.Y. Molecular Insights into How the Dimetal Center in Dihydropyrimidinase Can Bind the Thymine Antagonist 5-Aminouracil: A Different Binding Mode from the Anticancer Drug 5-Fluorouracil. Bioinorg. Chem. Appl. 2022, 2022, 1817745. [Google Scholar] [CrossRef] [PubMed]
- Sumi, S.; Imaeda, M.; Kidouchi, K.; Ohba, S.; Hamajima, N.; Kodama, K.; Togari, H.; Wada, Y. Population and family studies of dihydropyrimidinuria: Prevalence, inheritance mode, and risk of fluorouracil toxicity. Am. J. Med. Genet. 1998, 78, 336–340. [Google Scholar] [CrossRef]
- Huang, Y.H.; Lien, Y.; Chen, J.H.; Lin, E.S.; Huang, C.Y. Identification and characterization of dihydropyrimidinase inhibited by plumbagin isolated from Nepenthes miranda extract. Biochimie 2020, 171–172, 124–135. [Google Scholar] [CrossRef] [PubMed]
- Evans, D.R.; Guy, H.I. Mammalian pyrimidine biosynthesis: Fresh insights into an ancient pathway. J. Biol. Chem. 2004, 279, 33035–33038. [Google Scholar] [CrossRef] [PubMed]
- Grande-Garcia, A.; Lallous, N.; Diaz-Tejada, C.; Ramon-Maiques, S. Structure, functional characterization, and evolution of the dihydroorotase domain of human CAD. Structure 2014, 22, 185–198. [Google Scholar] [CrossRef] [PubMed]
- Del Cano-Ochoa, F.; Moreno-Morcillo, M.; Ramon-Maiques, S. CAD, A Multienzymatic Protein at the Head of de Novo Pyrimi- dine Biosynthesis. Subcell. Biochem. 2019, 93, 505–538. [Google Scholar] [PubMed]
- Guan, H.H.; Huang, Y.H.; Lin, E.S.; Chen, C.J.; Huang, C.Y. Structural basis for the interaction modes of dihydroorotase with the anticancer drugs 5-fluorouracil and 5-aminouracil. Biochem. Biophys. Res. Commun. 2021, 551, 33–37. [Google Scholar] [CrossRef] [PubMed]
- Lin, E.S.; Huang, Y.H.; Yang, P.C.; Peng, W.F.; Huang, C.Y. Complexed Crystal Structure of the Dihydroorotase Domain of Human CAD Protein with the Anticancer Drug 5-Fluorouracil. Biomolecules 2023, 13, 149. [Google Scholar] [CrossRef]
- Antony, E.; Lohman, T.M. Dynamics of E. coli single stranded DNA binding (SSB) protein-DNA complexes. Semin. Cell Dev. Biol. 2019, 86, 102–111. [Google Scholar] [CrossRef]
- Croft, L.V.; Bolderson, E.; Adams, M.N.; El-Kamand, S.; Kariawasam, R.; Cubeddu, L.; Gamsjaeger, R.; Richard, D.J. Human single-stranded DNA binding protein 1 (hSSB1, OBFC2B), a critical component of the DNA damage response. Semin. Cell Dev. Biol. 2019, 86, 121–128. [Google Scholar] [CrossRef]
- Byrne, B.M.; Oakley, G.G. Replication protein A, the laxative that keeps DNA regular: The importance of RPA phosphorylation in maintaining genome stability. Semin. Cell Dev. Biol. 2019, 86, 112–120. [Google Scholar] [CrossRef]
- Shereda, R.D.; Kozlov, A.G.; Lohman, T.M.; Cox, M.M.; Keck, J.L. SSB as an organizer/mobilizer of genome maintenance com- plexes. Crit. Rev. Biochem. Mol. Biol. 2008, 43, 289–318. [Google Scholar] [CrossRef]
- Dickey, T.H.; Altschuler, S.E.; Wuttke, D.S. Single-stranded DNA-binding proteins: Multiple domains for multiple functions. Structure 2013, 21, 1074–1084. [Google Scholar] [CrossRef]
- Huang, Y.H.; Huang, C.Y. The glycine-rich flexible region in SSB is crucial for PriA stimulation. RSC Adv. 2018, 8, 35280–35288. [Google Scholar] [CrossRef]
- Huang, Y.H.; Huang, C.Y. Characterization of a single-stranded DNA-binding protein from Klebsiella pneumoniae: Mutation at either Arg73 or Ser76 causes a less cooperative complex on DNA. Genes Cells 2012, 17, 146–157. [Google Scholar] [CrossRef] [PubMed]
- Jan, H.C.; Lee, Y.L.; Huang, C.Y. Characterization of a single-stranded DNA-binding protein from Pseudomonas aeruginosa PAO1. Protein J. 2011, 30, 20–26. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.H.; Lee, Y.L.; Huang, C.Y. Characterization of a single-stranded DNA binding protein from Salmonella enterica serovar Typhimurium LT2. Protein J. 2011, 30, 102–108. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.H.; Lin, E.S.; Huang, C.Y. Complexed crystal structure of SSB reveals a novel single-stranded DNA binding mode (SSB)3:1: Phe60 is not crucial for defining binding paths. Biochem. Biophys. Res. Commun. 2019, 520, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.H.; Chen, I.C.; Huang, C.Y. Characterization of an SSB-dT25 complex: Structural insights into the S-shaped ssDNA binding conformation. RSC Adv. 2019, 9, 40388–40396. [Google Scholar] [CrossRef] [PubMed]
- Dubiel, K.; Myers, A.R.; Kozlov, A.G.; Yang, O.; Zhang, J.; Ha, T.; Lohman, T.M.; Keck, J.L. Structural Mechanisms of Cooperative DNA Binding by Bacterial Single-Stranded DNA-Binding Proteins. J. Mol. Biol. 2019, 431, 178–195. [Google Scholar] [CrossRef] [PubMed]
- Raghunathan, S.; Kozlov, A.G.; Lohman, T.M.; Waksman, G. Structure of the DNA binding domain of E. coli SSB bound to ssDNA. Nat. Struct. Biol. 2000, 7, 648–652. [Google Scholar] [CrossRef] [PubMed]
- Lin, E.S.; Huang, Y.H.; Huang, C.Y. Characterization of the Chimeric PriB-SSBc Protein. Int. J. Mol. Sci. 2021, 22, 10854. [Google Scholar] [CrossRef]
- Huang, Y.H.; Lo, Y.H.; Huang, W.; Huang, C.Y. Crystal structure and DNA-binding mode of Klebsiella pneumoniae primosomal PriB protein. Genes Cells 2012, 17, 837–849. [Google Scholar] [CrossRef]
- Huang, C.Y.; Hsu, C.H.; Sun, Y.J.; Wu, H.N.; Hsiao, C.D. Complexed crystal structure of replication restart primosome protein PriB reveals a novel single-stranded DNA-binding mode. Nucleic Acids Res. 2006, 34, 3878–3886. [Google Scholar] [CrossRef]
- Lin, E.S.; Huang, C.Y. The inhibition activities of the fruit extract of Plinia cauliflora against melanoma cells and the single-stranded DNA-binding protein (SSB) from Klebsiella pneumoniae. Appl. Sci. 2023, 13, 11061. [Google Scholar] [CrossRef]
- Lin, E.S.; Huang, Y.H.; Chung, J.C.; Su, H.H.; Huang, C.Y. The Inhibitory Effects and Cytotoxic Activities of the Stem Extract of Nepenthes miranda against Single-Stranded DNA-Binding Protein and Oral Carcinoma Cells. Plants 2023, 12, 2188. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.W.; Chiang, W.Y.; Huang, Y.H.; Huang, C.Y. The Inhibitory Effects and Cytotoxic Activities of the Stem Extract of Sarra- cenia purpurea against Melanoma Cells and the SsbA Protein. Plants 2022, 11, 3164. [Google Scholar] [CrossRef] [PubMed]
- Lin, E.S.; Luo, R.H.; Huang, C.Y. A Complexed Crystal Structure of a Single-Stranded DNA-Binding Protein with Quercetin and the Structural Basis of Flavonol Inhibition Specificity. Int. J. Mol. Sci. 2022, 23, 588. [Google Scholar] [CrossRef]
- Lin, E.S.; Huang, Y.H.; Luo, R.H.; Basharat, Z.; Huang, C.Y. Crystal Structure of an SSB Protein from Salmonella enterica and Its Inhibition by Flavanonol Taxifolin. Int. J. Mol. Sci. 2022, 23, 4399. [Google Scholar] [CrossRef]
- Huang, C.Y. Crystal structure of SSB complexed with inhibitor myricetin. Biochem. Biophys. Res. Commun. 2018, 504, 704–708. [Google Scholar] [CrossRef]
- Huang, Y.H.; Huang, C.Y. Comparing SSB-PriA Functional and Physical Interactions in Gram-Positive and -Negative Bacteria. Methods Mol. Biol. 2021, 2281, 67–80. [Google Scholar]
- Huang, Y.H.; Guan, H.H.; Chen, C.J.; Huang, C.Y. Staphylococcus aureus single-stranded DNA-binding protein SsbA can bind but cannot stimulate PriA helicase. PLoS ONE 2017, 12, e0182060. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.L.; Cheng, J.H.; Lin, C.Y.; Huang, Y.H.; Huang, C.Y. Characterization of single-stranded DNA-binding protein SsbB from Staphylococcus aureus: SsbB cannot stimulate PriA helicase. RSC Adv. 2018, 8, 28367–28375. [Google Scholar] [CrossRef]
- Huang, Y.H.; Huang, C.Y. SAAV2152 is a single-stranded DNA binding protein: The third SSB in Staphylococcus aureus. Oncotarget 2018, 9, 20239–20254. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Paradzik, T.; Ivic, N.; Filic, Z.; Manjasetty, B.A.; Herron, P.; Luic, M.; Vujaklija, D. Structure-function relationships of two paral- ogous single-stranded DNA-binding proteins from Streptomyces coelicolor: Implication of SsbB in chromosome segregation during sporulation. Nucleic Acids Res. 2013, 41, 3659–3672. [Google Scholar] [CrossRef]
- Yadav, T.; Carrasco, B.; Myers, A.R.; George, N.P.; Keck, J.L.; Alonso, J.C. Genetic recombination in Bacillus subtilis: A division of labor between two single-strand DNA-binding proteins. Nucleic Acids Res. 2012, 40, 5546–5559. [Google Scholar] [CrossRef]
- Su, H.H.; Huang, Y.H.; Lien, Y.; Yang, P.C.; Huang, C.Y. Crystal Structure of DNA Replication Protein SsbA Complexed with the Anticancer Drug 5-Fluorouracil. Int. J. Mol. Sci. 2023, 24, 14899. [Google Scholar] [CrossRef] [PubMed]
- Lin, E.S.; Huang, C.Y. Crystal structure of the single-stranded DNA-binding protein SsbB in complex with the anticancer drug 5-fluorouracil: Extension of the 5-fluorouracil interactome to include the oligonucleotide/oligosaccharide-binding fold protein. Biochem. Biophys. Res. Commun. 2021, 534, 41–46. [Google Scholar] [CrossRef]
- Seibert, C.M.; Raushel, F.M. Structural and catalytic diversity within the amidohydrolase superfamily. Biochemistry 2005, 44, 6383–6391. [Google Scholar] [CrossRef]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef]
- Connors, T.A. The choice of prodrugs for gene directed enzyme prodrug therapy of cancer. Gene Ther. 1995, 2, 702–709. [Google Scholar]
- Vermes, A.; Guchelaar, H.J.; Dankert, J. Flucytosine: A review of its pharmacology, clinical indications, pharmacokinetics, toxicity and drug interactions. J. Antimicrob. Chemother. 2000, 46, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Kazlauskas, A.; Darinskas, A.; Meškys, R.; Tamašauskas, A.; Urbonavičius, J. Isocytosine deaminase Vcz as a novel tool for the prodrug cancer therapy. BMC Cancer 2019, 19, 197. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Li, X.; Fan, J.; Li, H.; Wen, Y.; Meng, C.; Chen, H.; Zhao, Z.; Zhang, Y.; Du, Y.; et al. Structural characterization of an isocytosine-specific deaminase VCZ reveals its application potential in the anti-cancer therapy. iScience 2023, 26, 107672. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Kale, V.; Chen, M. Gene-directed enzyme prodrug therapy. AAPS J. 2015, 17, 102–110. [Google Scholar] [CrossRef]
- Curreri, A.R.; Ansfield, F.J.; Mc, I.F.; Waisman, H.A.; Heidelberger, C. Clinical studies with 5-fluorouracil. Cancer Res. 1958, 18, 478–484. [Google Scholar]
- Eidinoff, M.L.; Knoll, J.E.; Klein, D. Effect of 5-fluorouracil on the incorporation of precursors into nucleic acid pyrimidines. Arch. Biochem. Biophys. 1957, 71, 274–275. [Google Scholar] [CrossRef]


| PDB ID | Organism | Crystal Structure | Unique Ligand |
|---|---|---|---|
| 1H7X | Sus scrofa | Dihydropyrimidine dehydrogenase (DPD) from pig, ternary complex of a mutant enzyme (C671A), NADPH and 5-fluorouracil | FAD, FMN, NDP, SF4, URF |
| 1RXC | Escherichia coli (strain K12) | E. coli uridine phosphorylase: 5-fluorouracil ribose-1-phosphate complex | 5UD, K, PO4, R1P, URF |
| 1UPF | Toxoplasma gondii | Structure of the uracil phosphoribosyltransferase, mutant C128V bound to the drug 5-fluorouracil | SO4, URF |
| 3KVR | Bos taurus | Trapping of an oxocarbenium ion intermediate in UP crystals | R2G, SO4, URF |
| 3KVV | Escherichia coli (strain K12) | Trapping of an oxocarbenium ion intermediate in UP crystals | R2B, SO4, URF |
| 3NAI | Murine norovirus 1 | Crystal structures and functional analysis of murine norovirus RNA-dependent RNA polymerase | GOL, MG, MN3, SO4, URF |
| 3NBQ | Homo sapiens | Human uridine phosphorylase 1 (hUPP1) with 5-fluorouracil | URF |
| 4E1V | Salmonella typhimurium | X-RAY structure of the uridine phosphorylase from Salmonella typhimurium in complex with 5-fluorouracil at 2.15 Å resolution | EDO, GOL, K, URF |
| 4O0O | Momordica balsamina | Crystal structure of the complex of type 1 ribosome inactivating protein from Momordica balsamina with 5-fluorouracil at 2.59 Å resolution | GOL, NAG, URF |
| 4TXN | Schistosoma mansoni | Crystal structure of uridine phosphorylase from Schistosoma mansoni in complex with 5-fluorouracil | SO4, URF |
| 4WRY | Mycobacterium tuberculosis | Crystal structure of Mycobacterium tuberculosis uracil-DNA glycosylase in complex with 5-fluorouracil (B), Form I | CIT, CL, URF |
| 4WRZ | Mycobacterium tuberculosis | Crystal structure of Mycobacterium tuberculosis uracil-DNA glycosylase in complex with 5-fluorouracil (AB), Form I | CIT, CL, IPA, URF |
| 4WS0 | Mycobacterium tuberculosis | Crystal structure of Mycobacterium tuberculosis uracil-DNA glycosylase in complex with 5-fluorouracil (A), Form II | CL, EDO, URF |
| 4WS1 | Mycobacterium tuberculosis | Crystal structure of Mycobacterium tuberculosis uracil-DNA glycosylase in complex with 5-fluorouracil (AB), Form II | CL, EDO, URF |
| 5IAO | Mycobacterium tuberculosis | Structure and mapping of spontaneous mutational sites of PyrR from Mycobacterium tuberculosis | URF |
| 5SXC | Homo sapiens | Crystal structure of PI3Kalpha in complex with fragment 8 | SEP, URF |
| 6KLK | Pseudomonas aeruginosa | Crystal structure of the Pseudomonas aeruginosa dihydropyrimidinase complexed with 5-FU | KCX, URF, ZN |
| 6L0B | Saccharomyces cerevisiae | Crystal structure of dihydroorotase in complex with fluorouracil from Saccharomyces cerevisiae | KCX, URF, ZN |
| 7D8J | Staphylococcus aureus | S. aureus SsbB with 5-FU | URF |
| 7DEP | Staphylococcus aureus | S. aureus SsbB with 5-FU | URF |
| 7YM1 | Staphylococcus aureus | Structure of SsbA protein in complex with the anticancer drug 5-fluorouracil | GOL, URF |
| 8GVZ | Homo sapiens | Crystal structure of the human dihydroorotase domain in complex with the anticancer drug 5-fluorouracil | KCX, URF, ZN |
| 8IS4 | Obesumbacterium proteus | Structure of an isocytosine specific deaminase VCZ in complex with 5-FU | GOL, TRS, URF, ZN |
| Molecule | Type of Binding Site | Area * | Volume * | Dimensions (x, y) of the Cavity (Å) # |
|---|---|---|---|---|
| Dihydropyrimidine dehydrogenase | 59.2 | 23.12 | 4.8, 7.1 | |
| Uridine phosphorylase | 138.5 | 83.4 | 2.8, 6.7 | |
| Uracil phosphoribosyltransferase | 566 | 710 | 11.5, 8.7 | |
| RNA-dependent RNA polymerase | 2976 | 4400 | 17.3, 5.3 | |
| PyrR | Surface | ND | ND | ND |
| Phosphatidylinositol 4,5-bisphosphate 3-kinase | Surface | ND | ND | ND |
| rRNA N-glycosidase | 82.6 | 39.4 | 11.7, 4.7 | |
| Uracil-DNA glycosylase | 278.9 | 169.9 | 3.7, 4.3 | |
| Dihydropyrimidinase | 132.6 | 45.9 | 4.1, 4.3 | |
| Dihydroorotase (yeast) | 144.0 | 44.8 | 6.4, 5.4 | |
| SsbB | 31.1 | 8.8 | 5.3, 8.6 | |
| SsbA | 369 | 452 | 7.1, 5.3 | |
| Dihydroorotase (human) | 289.7 | 213.4 | 6.9, 6.3 | |
| Hydroxydechloroatrazine ethylaminohydrolase | 544 | 437 | 8.7, 11.4 |
| Hydrogen Bond | The Contact Distance within <4 Å | ||||
|---|---|---|---|---|---|
| 5-FU | Dist. [Å] | Residue | 5-FU | Dist. [Å] | Residue |
| Type 1 (P-R type) | |||||
| 1.1 Uridine phosphorylase from Homo sapiens (3NBQ) | |||||
| 5FU [N1] | 4.1 | T141 [OG1] # | 5FU [O2] | 3.6 | E248 [CA] |
| 5FU [N1]/W1 | 3.8/4.0 | R94 [NH2] # | 5FU [O2] | 3.6 | M249 [CG] |
| 5FU [O2] | 2.9 | Q217 [NE2] | 5FU [C2] | 3.8 | I247 [O] |
| 5FU [N3] | 2.7 | Q217 [OE1] | 5FU [N3] | 3.4 | F231 [CE1] |
| 5FU [N3] | 4.0 | R219 [NH1] | 5FU [F5] | 3.7 | L272 [CD1] |
| 5FU [O4] | 2.9 | R219 [NH2] | 5FU [F5] | 3.5 | L273 [CD2] |
| 5FU [O4] | 3.2 | R219 [NH1] * | 5FU [F5] | 3.5 | I281 [CD1] |
| 5FU [F5] | 3.8 | G143 [N] # | |||
| 1.2 rRNA N-glycosidase from Momordica balsamina (4O0O) | |||||
| 5FU [O2] | 3.6 | Y111 [N] | 5FU [O2] | 4.0 | N110 [O2] |
| 5FU [N3] | 3.1 | G109 [O] | |||
| 5FU [F5] | 3.9 | V69 [O] | |||
| 5FU [F5] | 3.8 | R163 [NH1] * | |||
| 5FU [F5] | 3.5 | R163 [NH2] * | |||
| 1.3 PyrR from Mycobacterium tuberculosis (5IAO) | |||||
| 5FU [O2] | 2.7 | H177 [ND1] | 5FU [O4] | 4.0 | D120 [O4] |
| 5FU [O2] | 3.4 | R179 [NH2] # | |||
| 5FU [N3] | 3.0 | H177 [ND1] # | |||
| 5FU [O4] | 3.0 | R58 [NH1] | |||
| 5FU [O4]/W4 | 2.3/3.8 | V176 [O] # | |||
| 5FU [O4]/W4 | 2.3/2.8 | V178 [N] # | |||
| 5FU [F5] | 3.2 | R58 [NH1] * | |||
| 1.4 Dihydropyrimidinase from Pseudomonas aeruginosa (6KLK) | |||||
| 5FU [N1] | 3.1 | S289 [O] # | 5FU [N1] | 2.8 | Y155 [OH] |
| 5FU [O2] | 3.0 | S289 [N] * | 5FU [N1] | 4.0 | H183 [CE1] |
| 5FU [N3] | 3.2 | N337 [O] # | 5FU [O2] | 3.5 | M166 [CB] |
| 5FU [O4] | 2.9 | C318 [SG] * | 5FU [O2] | 3.9 | G338 [N] |
| 5FU [F5] | 3.4 | Kcx150 [OQ2] * | 5FU [C2] | 3.8 | D316 [OD2] |
| 5FU [O2] | 3.5 | H61 [CD2] | |||
| 5FU [O2] | 3.5 | L64 [CD2] | |||
| 5FU [C4] | 3.8 | F66 [CE2] | |||
| 5FU [F5] | 3.5 | F152 [CE1] | |||
| 1.5 Dihydroorotase from Saccharomyces cerevisiae (6L0B) | |||||
| 5FU [N1] | 3.3 | T105 [OG1] # | 5FU [O2] | 3.7 | Kcx98 [OQ2] |
| 5FU [N3] | 3.2 | H16 [ND1] | 5FU [O2] | 3.8 | H137 [ND1] |
| 5FU [O4] | 2.7 | R18 [NH1] | 5FU [O2] | 3.9 | K230 [O] |
| 5FU [O4] | 2.8 | N43 [ND2] | 5FU [O2] | 3.7 | D258 [OD2] |
| 5FU [F5] | 2.8 | R18 [NH2] | 5FU [F5] | 2.7 | T106 [OG] |
| 5FU [F5] | 2.9 | A275 [O] * | 5FU [F5] | 2.6 | H162 [NE2] |
| 5FU [C6] | 3.5 | A260 [CB] | |||
| 5FU [C6] | 3.8 | G276 [CA] | |||
| 1.6 SsbB from Staphylococcus aureus (7DEP) | |||||
| 5FU [N3] | 3.9 | T12 [OG1] # | 5FU [O2] | 3.9 | F48 [CD2] |
| 5FU [O4] | 3.9 | T12 [OG1] # | |||
| 5FU [O4] | 3.5 | K13 [NZ] | |||
| 5FU [O4] | 3.2 | T30 [OG1] * | |||
| 5FU [F5] | 3.3 | N50 [ND2] * | |||
| 1.7 SsbA from Staphylococcus aureus (7YM1) | |||||
| 5FU [O2] | 3.5 | R18 [NH1] # | 5FU [N1] | 3.5 | V52 [CG1] |
| 5FU [O2] | 3.7 | R80 [NH2] # | 5FU [N1] | 4.0 | V96 [CG1] |
| 5FU [O2] | 3.3 | R80 [NE] | 5FU [O2] | 2.7 | E94 [OE1] |
| 5FU [N3] | 2.5 | R18 [NH2] | 5FU [O4] | 4.0 | P21 [CD] |
| 5FU [F5] | 3.7 | F54 [CZ] | |||
| 5FU [F5] | 4.0 | Q78 [NE2] | |||
| 1.8 Dihydroorotase from Homo sapiens (8GVZ) | |||||
| 5FU [N1] | 2.9 | T1562 [OG1] # | 5FU [O2] | 3.8 | K1556 [OQ1] |
| 5FU [N1]/W1 | 3.9/2.8 | T1562 [OG1] # | 5FU [O2] | 4.1 | R1661 [O] |
| 5FU [O2] | 3.9 | H1590 [ND1] | 5FU [O2] | 3.7 | D1686 [OD2] |
| 5FU [O2]/W1 | 3.9/4.0 | T1562 [OG1] # | 5FU [C4] | 4.0 | A1688 |
| 5FU [N3] | 3.9 | H1473 [ND1] # | 5FU [F5] | 3.0 | F1563 [CD2] |
| 5FU [O4] | 2.6 | R1475 [NH1] | 5FU [F5] | 2.6 | H1690 [CE1] |
| 5FU [O4] | 3.4 | R1475 [NH2] # | 5FU [C6] | 2.8 | P1702 [O] |
| 5FU [O4] | 2.8 | N1505 [ND2] | |||
| 5FU [F5] | 2.6 | R1475 [NH2] * | |||
| Type 2 (P type) | |||||
| 2.1 RNA-dependent RNA polymerase from murine norovirus 1 (3NAI) | |||||
| 5FU [N3] | 2.4 | D346 [OD2] * | 5FU [O2] | 3.2 | D245 [OD11] |
| 5FU [O4] | 4.0 | R185 [NH1] # | 5FU [O2] | 2.6 | D347 [OD2] |
| 5FU [F5] | 3.7 | R185 [NH1] * | |||
| 5FU [F5] | 3.6 | R185 [NH2] | |||
| 5FU [F5] | 3.4 | R395 [NH2] * | |||
| 2.2 Phosphatidylinositol 4,5-bisphosphate 3-kinase from Homo sapiens (5SXC) | |||||
| 5FU [O4] | 3.6 | K656 [NZ] | 5FU [N1] | 4.0 | E620 [O] |
| 5FU [F5] | 3.7 | K656 [NZ] * | 5FU [O2] | 3.0 | K621 [O] |
| Type 3 (R type) | |||||
| 3.1 Dihydropyrimidine dehydrogenase from Sus scrofa (1H7X) | |||||
| 5FU [N1] | 2.9 | N609 [OD1] | 5FU [F5] | 3.6 | L162 [C] |
| 5FU [O2] | 2.9 | N609 [ND2] | 5FU [F5] | 3.8 | I163 [CG2] |
| 5FU [O2] | 2.9 | T737 [OG1] | 5FU [C6] | 3.0 | E611 [O] |
| 5FU [O2]/W2 | 3.5/3.2 | G764 [N] # | |||
| 5FU [N3] | 3.0 | N736 [OD1] | |||
| 5FU [O4] | 3.1 | N668 [ND2] | |||
| 5FU [O4] | 3.3 | S670 [OG] | |||
| 5FU [O4] | 3.0 | N736 [ND2] | |||
| 3.2 Uracil phosphoribosyltransferase from Toxoplasma gondii (1UPF) | |||||
| 5FU [O2] | 3.6 | F236 [N] | 5FU [O2] | 3.2 | D235 [OD1] |
| 5FU [N3] | 3.2 | G234 [O] | 5FU [N3] | 3.8 | M166 [CG] |
| 5FU [O4] | 3.4 | I229 [N] | 5FU [C4] | 3.8 | T228 [CD2] |
| 5FU [F5] | 3.8 | I229 [N] * | 5FU [F5] | 3.1 | A168 [CB] |
| 5FU [F5] | 3.0 | Y227 [O] | |||
| 3.3 Uracil-DNA glycosylase from Mycobacterium tuberculosis (4WRY) | |||||
| 5FU [N1] | 3.6 | Y70 [N] | 5FU [O2] | 3.3 | S80 [CA] |
| 5FU [O2] | 2.9 | N127 [ND2] | |||
| 5FU [O2] | 2.8 | F81 [N] | |||
| 5FU [O2]/W2 | 2.6/4.1 | L79 [O] # | |||
| 5FU [N3] | 2.8 | N127 [OD1] | |||
| 5FU [O4] | 2.8 | Q67 [N] | |||
| 5FU [O4] | 3.8 | D68 [N] | |||
| 5FU [O4] | 2.8 | H191 [NE2] * | |||
| 5FU [F5] | 3.9 | S93 [OG] * | |||
| 3.4 Hydroxydechloroatrazine ethylaminohydrolase from Obesumbacterium proteus (8IS4) | |||||
| 5FU [N1] | 3.4 | E237 [OE1] | 5FU [O2] | 3.0 | L300 [CD1] |
| 5FU [N3]/W3 | 3.1/4.0 | D322 [OG1] # | 5FU [O4] | 3.8 | W90 [CH2] |
| 5FU [N3]/W3 | 3.1/2.7 | S326 [OG] # | 5FU [O4] | 3.3 | H70 [NE2] |
| 5FU [O4] | 3.3 | Q73 [NE2] | |||
| 5FU [O4] | 2.7 | Y130 [OH] | |||
| 5FU [F5] | 3.1 | Y130 [OH] | |||
| 5FU [F5] | 3.8 | C207 [SG] * | |||
| PDB ID | Organism | Length | Identities (%) | Positives (%) |
|---|---|---|---|---|
| 3NBQ | Homo sapiens | 310 | 100 | 100 |
| 4TXN | Schistosoma mansoni | 296 | 44 | 64 |
| 1RXC | Escherichia coli (strain K12) | 253 | 32 | 46 |
| 4ETV | Salmonella typhimurium | 253 | 27 | 41 |
| Binding Type | Number | Frequency (%) | F5 | N1 or N3 | N3 | O4 | N1, N3, or F5 | N3, O4, or F5 | Binding Pocket | One Dimension < 11 Å | One Dimension < 12 Å |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Type 1 (P-R type) | 8 | 8/14 (57.1%) | 6/8 (75%) | 6/8 (75%) | 5/8 (35.7%) | 6/8 (75%) | 7/8 (87.5%) | 8/8 (100%) | 7/8 (87.5%) | 7/8 (87.5%) | 8/8 (100%) |
| Type 2 (P type) | 2 | 2/14 (14.3%) | 2/2 (100%) | 0/2 (0%) | 0/2 (0%) | 1/2 (50%) | 2/2 (100%) | 2/2 (100%) | 1/2 (50%) | 1/2 (50%) | 1/2 (50%) |
| Type 3 (R type) | 4 | 4/14 (28.6%) | 3/4 (75%) | 1/4 (25%) | 0/4 (0%) | 2/4 (50%) | 4/4 (100%) | 4/4 (100%) | 4/4 (100%) | 2/4 (50%) | 4/4 (100%) |
| Total | 14 | 14/14 (100%) | 11/14 (78.6%) | 7/14 (50%) | 5/14 (35.7%) | 9/14 (64.3%) | 13/14 (92.9%) | 14/14 (100%) | 12/14 (85.7%) | 9/14 (64.3%) | 13/14 (92.9%) |
| Type | PDB ID | Metal Ion | Interaction |
|---|---|---|---|
| 1 | 6KLK | Zn | ZNα-F5 (3.6 Å) |
| 1 | 6L0B | Zn | Znα-O2 (3.0 Å), Znβ-O2 (2.4 Å) |
| 1 | 8GVZ | Zn | Znα-O2 (3.2 Å), Znβ-O2 (2.7 Å) |
| 2 | 3NAI | Mg | Mg-N3 (3.0 Å) |
| 3 | 8IS4 | Zn | Znα-N3 (3.0 Å) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, E.-S.; Huang, C.-Y. Binding Pattern and Structural Interactome of the Anticancer Drug 5-Fluorouracil: A Critical Review. Int. J. Mol. Sci. 2024, 25, 3404. https://doi.org/10.3390/ijms25063404
Lin E-S, Huang C-Y. Binding Pattern and Structural Interactome of the Anticancer Drug 5-Fluorouracil: A Critical Review. International Journal of Molecular Sciences. 2024; 25(6):3404. https://doi.org/10.3390/ijms25063404
Chicago/Turabian StyleLin, En-Shyh, and Cheng-Yang Huang. 2024. "Binding Pattern and Structural Interactome of the Anticancer Drug 5-Fluorouracil: A Critical Review" International Journal of Molecular Sciences 25, no. 6: 3404. https://doi.org/10.3390/ijms25063404
APA StyleLin, E.-S., & Huang, C.-Y. (2024). Binding Pattern and Structural Interactome of the Anticancer Drug 5-Fluorouracil: A Critical Review. International Journal of Molecular Sciences, 25(6), 3404. https://doi.org/10.3390/ijms25063404






