Electrophilic Compounds in the Human Diet and Their Role in the Induction of the Transcription Factor NRF2
Abstract
1. Introduction
- Some ECs have antioxidant properties, which help neutralize free radicals and protect against oxidative damage to cells [8];
- ECs can modulate the inflammatory response by interacting with proteins involved in inflammation. This may help to reduce chronic inflammation and improve health [9];
- Some ECs have been shown to have anti-tumor effects by targeting specific proteins in cancer cells. These compounds may help prevent the growth and spread of cancer [10];
- ECs can induce the expression of phase II enzymes. These enzymes help detoxify the body by removing harmful substances [11];
- ECs found in olive oil and garlic exert protective effects on the cardiovascular system by improving endothelial function and reducing inflammation [12].
2. NRF2 and Keap1
- The glutathione S-transferase (GST) family, which includes cytosolic, mitochondrial, and microsomal enzymes that catalyze the conjugation of GSH to endogenous electrophiles and xenobiotics [62]. After detoxification by the GST-catalyzed glutathione (GSH) conjugation, the body can eliminate potentially harmful and toxic compounds. GSTs are induced by NRF2 activation and represent an important detoxification pathway [63];
- The UDP-glucuronosyltransferase (UGT) family, which catalyzes the conjugation of a glucuronic acid moiety to a variety of endogenous and exogenous substances, making them more water soluble and easily excreted. Important substrates for glucuronidation include bilirubin and acetaminophen [64]. NRF2 has been shown to induce UGT1A1 and UGT1A6 [65];
- N-acetyltransferase (NAT), acetylate aromatic amines, and hydrazines participate in the metabolism of drugs, carcinogens, and arylamines. Genetic variations in NATs can affect drug efficacy and toxicity [66];
- Sulfotransferases (SULTs) transfer sulfate groups to substrates, enhancing their water solubility. These enzymes are crucial for detoxifying phenolic compounds, drugs, and hormones [67];
- Epoxide hydrolase (EH), although traditionally considered a phase I enzyme, also plays a role during phase II. It detoxifies epoxides formed during phase I reactions, preventing their harmful effects [68];
- Heme oxygenase-1 (HO-1) degrades heme into biliverdin, carbon monoxide, and iron. It has antioxidant and anti-inflammatory properties, protecting cells from oxidative stress. HO-1 protects against a variety of pathologies, including sepsis, hypertension, atherosclerosis, acute lung injury, kidney injury, and pain [69];
- NQO1 reduces quinones, preventing their conversion into reactive oxygen species. It contributes to the cellular defense against oxidative damage [70];
- Glutamate cysteine ligase (GCL), although not directly involved in biotransformation, is critical for GSH biosynthesis, a potent antioxidant that protects cells from oxidative stress [71];
- Ferritin is not an enzyme but is the main intracellular iron storage protein. It helps regulate iron levels and prevents iron-induced oxidative damage [72].
3. Role of NRF2 in Senescence
- Telomeres are protective structures located at the ends of chromosomes. Their length decreases with each successive cell cycle. Senescence happens when the length of the telomeres becomes critically short [78];
- Significant DNA damage that stops the replication of damaged cellular material [79];
- Oxidative stress and restricted nutrition can contribute to cell senescence [80].
4. Electrophilic Compounds Can Interfere with the Keap1 Protein
4.1. Electrophilic Dietary Compounds that Are Not Michael Acceptors
- -
- Isothiocyanates (ITCs) are produced by many plants belonging to the Brassicaceae, Capparaceae, and Caricaceae families. Sulforaphane, sinigrin, allyl isothiocyanate, and methyl isothiocyanate are all types of ITCs [100];
- -
- The compounds obtained from garlic. Allium sativum, a culinary plant, is known for its strong odor and unique taste. The main compounds extracted from garlic are allicin, which is formed when allicin is degraded by crushing or chopping enzymes, allyl sulfides, which are formed from the decomposition of allicin, ajoene, a more stable derivative of allicin formed by chemical reactions, and s-allylcysteine (SAC), which is formed from allicin [101].
4.2. Electrophilic Dietary Compounds That Are Michael Acceptors
- -
- Dithiolethiones are sulfur-containing pentacyclic compounds that exhibit anti-inflammatory, antithrombotic, antioxidant, and chemotherapeutic properties. Researchers are investigating their potential as cancer therapies to prevent cancer in humans, both in the laboratory and in clinical settings [107];
- -
- Curcumin is the major compound in the curcuminoids extracted from turmeric and is a natural polyphenolic molecule. Curcumin contains two α,β-unsaturated residues attached to a carbonyl, so it is a Michael acceptor. Curcumin exists in two tautomeric forms (Keto and Enol). Keto is a solid, but Enol is a liquid [108];
- -
- Chalcones are aromatic ketones that serve as building blocks for several important biological compounds known as bioactive substances. They are present in a variety of foods, such as vegetables, fruits, and teas, as well as in fluorescent materials and chemical intermediates [109]. Chalcones can occur in two forms (cis and trans isomers). Chalcones have a simple chemical compound structure. The trans isomer is thermodynamically more stable. The presence of the Michael acceptor, an α,β-unsaturated carbonyl system, is a key factor in the observed biological activity. They can easily obtain the Michael adduct due to the facile formation of covalent bonds with nucleophiles such as the sulfhydryl group of cysteine residues [110]. Chalcones are predominantly soft electrophiles and soft nucleophiles that have an affinity for thiol moieties. Chalcones are used in medicinal chemistry for many purposes, e.g., as antioxidants, anticancer drugs, antidiabetics, antiviral agents, and antimalarials [111];
- -
- Quinones are a group of compounds present in various bacteria, fungi, and plants. Quinones are ECs that act as Michael acceptors and are stabilized by conjugation. They also act as oxidizing agents and their effect can sometimes be reversed [112];
- -
- Coumarins are phenolic compounds derived from cinnamic acid and are present in several plant species, including edible, medicinal, and aromatic plants from different botanical families, as well as in fungi and bacteria. Coumarins belong to the class of benzopyrans and are present in a variety of medicinal plants [113,114,115,116]. They exhibit a broad spectrum of pharmacological effects, such as anti-inflammatory, anticoagulant, anticancer, antibacterial, antimalarial, antifungal, antiviral, ulcerogenic, and antihypertensive effects. They are present in various parts of plants, such as roots, seeds, nuts, flowers, and fruits, either as heterosides or in their free form. Coumarins are classified as Michael acceptors as they contain an α,β-unsaturated carbonyl [117];
- -
- Terpenoids are the most abundant category of phytochemicals. They can be present in different plant species and play various biological and metabolic roles in living organisms. Green plants, especially those with flowers, have a significantly large number of terpenoid compounds compared with other living organisms [118]. Terpenoids ingested with food have a greater influence on the modulation of the Keap1–NRF2–ARE signaling pathway [119]. Zerumbone is a monocyclic sesquiterpene compound extracted from the rhizomes of Zingiber zerumbet Smith. The compound has three double bonds, two of which are conjugated and one that is isolated, as well as a conjugated carbonyl group. It is structured in an 11-membered ring configuration [120].
5. Role of Hydrogen Sulfide
- Enzymatic synthesis in tissues is carried out by certain enzymes, such as cystathionine-γ-lyase (CSE) and 3-mercaptopyruvate sulfurtransferase (3MST), that convert cysteine or other sulfur-containing molecules into H2S. Organs and tissues such as the brain, heart, kidneys, and lungs contain these enzymes [125];
- Bacteria in the gut play an important role in the production of much of the H2S in the body through their metabolism in the digestive system. These bacteria, such as Escherichia coli and Bacteroides, use sulfur-containing amino acids to produce H2S, which then enters the bloodstream and is distributed throughout the body [126].
5.1. Availability of H2S in Age-Related Diseases and Aging
5.2. Can H2S Donors Induce the H2S-Producing Enzymes CBS, CSE, and 3-MST?
5.3. Electrophilic Sulfur Compounds in the Human Diet
5.4. Dithiolethiones Are Sulfur Compounds
5.5. Sulfurous Waters in Spas
6. Role of Michael Acceptors
6.1. Polyphenols
- The o-hydroxylation of monophenols to o-diphenols (catechols), also known as monophenolase activity;
- The oxidation of o-diphenols to o-quinones, also known as diphenolase activity.
6.2. Quinones
6.3. Chalcones
6.4. Curcuminoids
6.5. Coumarins
6.6. Terpenoids
6.7. Steroids
6.8. Nitro Fatty Acids
6.9. Unsaturated Aldehydes (Cinnamaldehyde and Its Derivatives)
7. Role of Electrophilic Compounds in Diseases
- Researchers are actively investigating how ECs interact with biological molecules. Unraveling these mechanisms will enhance our understanding of their effects on cellular processes and disease pathways [292];
- Targeted therapies. The development of ECs that selectively target specific proteins or pathways holds promise. By designing molecules that interact with specific cellular components, we can create more effective and safer drugs [287];
- Precision medicine. The tailoring of electrophilic therapies to individual patients based on their genetic makeup and disease profile is an exciting avenue. Personalized treatments could optimize efficacy while minimizing side effects [293];
- Some electrophiles exhibit antioxidant properties by activating cellular defense mechanisms. Exploring their potential in conditions like neurodegenerative diseases and cancer is an ongoing area of research [294];
- Innovations in drug delivery can enhance the bioavailability and tissue specificity of ECs. Nanoparticles, liposomes, and other carriers can improve their therapeutic impact [295];
- Combination therapies. The integration of ECs with existing drugs or other treatment modalities could lead to synergistic effects. Combinations may enhance efficacy and reduce resistance [296];
- Safety profiling. Addressing the promiscuity issue (where electrophiles bond with unintended targets) requires rigorous safety profiling. Predictive models and screening assays can help identify potential adverse effects [297];
- Neuroprotection. ECs may play a role in preserving neuronal health. Investigating their impact on neuroinflammation, oxidative stress, and neurodegenerative disorders is crucial [298];
- Metabolic disorders. Exploring electrophiles as regulators of metabolic pathways (e.g., glucose metabolism and lipid homeostasis) could yield novel therapeutic strategies for conditions like diabetes and obesity [299];
- Environmental exposure. Investigating the impact of ECs from environmental sources (e.g., air pollution and dietary components) on human health is an emerging field [300].
8. Bioavailability and Metabolism of Electrophilic Compounds
- -
- Interactions with food components. ECs can interact with other components in the food matrix (such as fibers, proteins, and lipids), affecting their absorption. In the food matrix, ECs can be linked to carbohydrates, organic acids, hemicellulose, and cellulose [305];
- -
- Phase I and II metabolism in the liver. These processes can alter the chemical structure of ECs. Phase I reactions of drug metabolism involve the oxidation, reduction, or hydrolysis of the parent drug, resulting in its conversion to a more polar molecule. This phase yields a polar, water-soluble metabolite that is often still active. Many of the products in this phase can also become substrates for phase II reactions. Phase II reactions involve conjugation by the coupling of the drug or its metabolites to another molecule, such as a glucuronidation, acylation, sulfate, or glycine molecule. This phase yields a large polar metabolite by the addition of endogenous hydrophilic groups to form inactive water-soluble compounds that can be excreted by the body [305];
- -
- Absorption in the small intestine, where most ECs are absorbed. However, their uptake can vary based on their chemical form;
- -
- The gut microbiota can further metabolize ECs, impacting their bioavailability [306].
9. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Cardenas, D. Let not thy food be confused with thy medicine: The Hippocratic misquotation. e-SPEN J. 2013, 8, e260–e262. [Google Scholar] [CrossRef]
- Who, J.; Consultation, F.E. Diet, nutrition and the prevention of chronic diseases. World Health Organ. Tech. Rep. Ser. 2003, 916, 1–149. [Google Scholar]
- Diab, A.; Dastmalchi, L.N.; Gulati, M.; Michos, E.D. A Heart-Healthy Diet for Cardiovascular Disease Prevention: Where Are We Now? Vasc. Health Risk Manag. 2023, 19, 237–253. [Google Scholar] [CrossRef] [PubMed]
- Tuso, P.J.; Ismail, M.H.; Ha, B.P.; Bartolotto, C. Nutritional update for physicians: Plant-based diets. Perm. J. 2013, 17, 61–66. [Google Scholar] [CrossRef] [PubMed]
- McMacken, M.; Shah, S. A plant-based diet for the prevention and treatment of type 2 diabetes. J. Geriatr. Cardiol. 2017, 14, 342–354. [Google Scholar] [CrossRef]
- Fardet, A.; Rock, E. Toward a new philosophy of preventive nutrition: From a reductionist to a holistic paradigm to improve nutritional recommendations. Adv. Nutr. 2014, 5, 430–446. [Google Scholar] [CrossRef] [PubMed]
- Sakanyan, V. Reactive Chemicals and Electrophilic Stress in Cancer: A Minireview. High Throughput 2018, 7, 12. [Google Scholar] [CrossRef] [PubMed]
- Zaky, A.A.; Simal-Gandara, J.; Eun, J.-B.; Shim, J.-H.; Abd El-Aty, A.M. Bioactivities, Applications, Safety, and Health Benefits of Bioactive Peptides From Food and By-Products: A Review. Front. Nutr. 2022, 8, 815640. [Google Scholar] [CrossRef]
- Gersch, M.; Kreuzer, J.; Sieber, S.A. Electrophilic natural products and their biological targets. Nat. Prod. Rep. 2012, 29, 659–682. [Google Scholar] [CrossRef]
- Yoo, M.H.; Xu, X.M.; Carlson, B.A.; Patterson, A.D.; Gladyshev, V.N.; Hatfield, D.L. Targeting thioredoxin reductase 1 reduction in cancer cells inhibits self-sufficient growth and DNA replication. PLoS ONE 2007, 2, e1112. [Google Scholar] [CrossRef]
- Iqbal, I.; Wilairatana, P.; Saqib, F.; Nasir, B.; Wahid, M.; Latif, M.F.; Iqbal, A.; Naz, R.; Mubarak, M.S. Plant Polyphenols and Their Potential Benefits on Cardiovascular Health: A Review. Molecules 2023, 28, 6403. [Google Scholar] [CrossRef]
- Summerhill, V.; Karagodin, V.; Grechko, A.; Myasoedova, V.; Orekhov, A. Vasculoprotective Role of Olive Oil Compounds via Modulation of Oxidative Stress in Atherosclerosis. Front. Cardiovasc. Med. 2018, 5, 188. [Google Scholar] [CrossRef] [PubMed]
- Moi, P.; Chan, K.; Asunis, I.; Cao, A.; Kan, Y.W. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the beta-globin locus control region. Proc. Natl. Acad. Sci. USA 1994, 91, 9926–9930. [Google Scholar] [CrossRef]
- Li, W.; Kong, A.N. Molecular mechanisms of Nrf2-mediated antioxidant response. Mol. Carcinog. 2009, 48, 91–104. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Buttari, B.; Panieri, E.; Profumo, E.; Saso, L. An Overview of Nrf2 Signaling Pathway and Its Role in Inflammation. Molecules 2020, 25, 5474. [Google Scholar] [CrossRef] [PubMed]
- Ngo, V.; Duennwald, M.L. Nrf2 and Oxidative Stress: A General Overview of Mechanisms and Implications in Human Disease. Antioxidants 2022, 11, 2345. [Google Scholar] [CrossRef]
- Taguchi, K.; Yamamoto, M. The KEAP1-NRF2 System as a Molecular Target of Cancer Treatment. Cancers 2020, 13, 46. [Google Scholar] [CrossRef]
- Dodson, M.; de la Vega, M.R.; Cholanians, A.B.; Schmidlin, C.J.; Chapman, E.; Zhang, D.D. Modulating NRF2 in Disease: Timing Is Everything. Annu. Rev. Pharmacol. Toxicol. 2019, 59, 555–575. [Google Scholar] [CrossRef]
- Suzen, S.; Tucci, P.; Profumo, E.; Buttari, B.; Saso, L. A Pivotal Role of Nrf2 in Neurodegenerative Disorders: A New Way for Therapeutic Strategies. Pharmaceuticals 2022, 15, 692. [Google Scholar] [CrossRef]
- Smith, R.E.; Tran, K.; Smith, C.C.; McDonald, M.; Shejwalkar, P.; Hara, K. The Role of the Nrf2/ARE Antioxidant System in Preventing Cardiovascular Diseases. Diseases 2016, 4, 34. [Google Scholar] [CrossRef]
- Lewis, K.N.; Wason, E.; Edrey, Y.H.; Kristan, D.M.; Nevo, E.; Buffenstein, R. Regulation of Nrf2 signaling and longevity in naturally long-lived rodents. Proc. Natl. Acad. Sci. USA 2015, 112, 3722–3727. [Google Scholar] [CrossRef]
- Inouye, S.; Hatori, Y.; Kubo, T.; Saito, S.; Kitamura, H.; Akagi, R. NRF2 and HSF1 coordinately regulate heme oxygenase-1 expression. Biochem. Biophys. Res. Commun. 2018, 506, 7–11. [Google Scholar] [CrossRef]
- Zhang, M.; An, C.; Gao, Y.; Leak, R.K.; Chen, J.; Zhang, F. Emerging roles of Nrf2 and phase II antioxidant enzymes in neuroprotection. Prog. Neurobiol. 2013, 100, 30–47. [Google Scholar] [CrossRef]
- Hou, Y.; Wang, Y.; He, Q.; Li, L.; Xie, H.; Zhao, Y.; Zhao, J. Nrf2 inhibits NLRP3 inflammasome activation through regulating Trx1/TXNIP complex in cerebral ischemia reperfusion injury. Behav. Brain Res. 2018, 336, 32–39. [Google Scholar] [CrossRef]
- Kelley, N.; Jeltema, D.; Duan, Y.; He, Y. The NLRP3 Inflammasome: An Overview of Mechanisms of Activation and Regulation. Int. J. Mol. Sci. 2019, 20, 3328. [Google Scholar] [CrossRef] [PubMed]
- Ryter, S.W.; Ma, K.C.; Choi, A.M.K. Carbon monoxide in lung cell physiology and disease. Am. J. Physiol. Cell Physiol. 2018, 314, C211–C227. [Google Scholar] [CrossRef] [PubMed]
- Motterlini, R.; Otterbein, L.E. The therapeutic potential of carbon monoxide. Nat. Rev. Drug Discov. 2010, 9, 728–743. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.; Nioi, P.; Pickett, C.B. The Nrf2-antioxidant response element signaling pathway and its activation by oxidative stress. J. Biol. Chem. 2009, 284, 13291–13295. [Google Scholar] [CrossRef] [PubMed]
- Villeneuve, N.F.; Lau, A.; Zhang, D.D. Regulation of the Nrf2-Keap1 antioxidant response by the ubiquitin proteasome system: An insight into cullin-ring ubiquitin ligases. Antioxid. Redox Signal. 2010, 13, 1699–1712. [Google Scholar] [CrossRef] [PubMed]
- Joshi, G.; Johnson, J.A. The Nrf2-ARE pathway: A valuable therapeutic target for the treatment of neurodegenerative diseases. Recent Pat. CNS Drug Discov. 2012, 7, 218–229. [Google Scholar] [CrossRef] [PubMed]
- McCord, J.M.; Gao, B.; Hybertson, B.M. The Complex Genetic and Epigenetic Regulation of the Nrf2 Pathways: A Review. Antioxidants 2023, 12, 366. [Google Scholar] [CrossRef] [PubMed]
- Kopacz, A.; Kloska, D.; Forman, H.J.; Jozkowicz, A.; Grochot-Przeczek, A. Beyond repression of Nrf2: An update on Keap1. Free Radic. Biol. Med. 2020, 157, 63–74. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Ru, X.; Wen, T. NRF2, a Transcription Factor for Stress Response and Beyond. Int. J. Mol. Sci. 2020, 21, 4777. [Google Scholar] [CrossRef] [PubMed]
- Jaganjac, M.; Milkovic, L.; Sunjic, S.B.; Zarkovic, N. The NRF2, Thioredoxin, and Glutathione System in Tumorigenesis and Anticancer Therapies. Antioxidants 2020, 9, 1151. [Google Scholar] [CrossRef] [PubMed]
- Sodani, K.; Patel, A.; Kathawala, R.J.; Chen, Z.S. Multidrug resistance associated proteins in multidrug resistance. Chin. J. Cancer 2012, 31, 58–72. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q. Role of nrf2 in oxidative stress and toxicity. Annu. Rev. Pharmacol. Toxicol. 2013, 53, 401–426. [Google Scholar] [CrossRef] [PubMed]
- Baird, L.; Yamamoto, M. The Molecular Mechanisms Regulating the KEAP1-NRF2 Pathway. Mol. Cell. Biol. 2020, 40, e00099-20. [Google Scholar] [CrossRef]
- Kobayashi, M.; Yamamoto, M. Nrf2-Keap1 regulation of cellular defense mechanisms against electrophiles and reactive oxygen species. Adv. Enzym. Regul. 2006, 46, 113–140. [Google Scholar] [CrossRef]
- Ogura, T.; Tong, K.I.; Mio, K.; Maruyama, Y.; Kurokawa, H.; Sato, C.; Yamamoto, M. Keap1 is a forked-stem dimer structure with two large spheres enclosing the intervening, double glycine repeat, and C-terminal domains. Proc. Natl. Acad. Sci. USA 2010, 107, 2842–2847. [Google Scholar] [CrossRef]
- Yamamoto, T.; Suzuki, T.; Kobayashi, A.; Wakabayashi, J.; Maher, J.; Motohashi, H.; Yamamoto, M. Physiological significance of reactive cysteine residues of Keap1 in determining Nrf2 activity. Mol. Cell. Biol. 2008, 28, 2758–2770. [Google Scholar] [CrossRef]
- Fukutomi, T.; Takagi, K.; Mizushima, T.; Ohuchi, N.; Yamamoto, M. Kinetic, thermodynamic, and structural characterizations of the association between Nrf2-DLGex degron and Keap1. Mol. Cell. Biol. 2014, 34, 832–846. [Google Scholar] [CrossRef]
- Itoh, K.; Wakabayashi, N.; Katoh, Y.; Ishii, T.; Igarashi, K.; Engel, J.D.; Yamamoto, M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes. Dev. 1999, 13, 76–86. [Google Scholar] [CrossRef] [PubMed]
- Eggler, A.L.; Liu, G.; Pezzuto, J.M.; van Breemen, R.B.; Mesecar, A.D. Modifying specific cysteines of the electrophile-sensing human Keap1 protein is insufficient to disrupt binding to the Nrf2 domain Neh2. Proc. Natl. Acad. Sci. USA 2005, 102, 10070–10075. [Google Scholar] [CrossRef] [PubMed]
- Turpaev, K.T. Keap1-Nrf2 signaling pathway: Mechanisms of regulation and role in protection of cells against toxicity caused by xenobiotics and electrophiles. Biochemistry 2013, 78, 111–126. [Google Scholar] [CrossRef] [PubMed]
- Kopacz, A.; Rojo, A.I.; Patibandla, C.; Lastra-Martínez, D.; Piechota-Polanczyk, A.; Kloska, D.; Jozkowicz, A.; Sutherland, C.; Cuadrado, A.; Grochot-Przeczek, A. Overlooked and valuable facts to know in the NRF2/KEAP1 field. Free Radic. Biol. Med. 2022, 192, 37–49. [Google Scholar] [CrossRef]
- Baird, L.; Swift, S.; Llères, D.; Dinkova-Kostova, A.T. Monitoring Keap1-Nrf2 interactions in single live cells. Biotechnol. Adv. 2014, 32, 1133–1144. [Google Scholar] [CrossRef]
- Walters, T.S.; McIntosh, D.J.; Ingram, S.M.; Tillery, L.; Motley, E.D.; Arinze, I.J.; Misra, S. SUMO-Modification of Human Nrf2 at K(110) and K(533) Regulates Its Nucleocytoplasmic Localization, Stability and Transcriptional Activity. Cell. Physiol. Biochem. 2021, 55, 141–159. [Google Scholar] [CrossRef]
- Katoh, Y.; Itoh, K.; Yoshida, E.; Miyagishi, M.; Fukamizu, A.; Yamamoto, M. Two domains of Nrf2 cooperatively bind CBP, a CREB binding protein, and synergistically activate transcription. Genes Cells 2001, 6, 857–868. [Google Scholar] [CrossRef]
- Yang, X.; Park, S.H.; Chang, H.C.; Shapiro, J.S.; Vassilopoulos, A.; Sawicki, K.T.; Chen, C.; Shang, M.; Burridge, P.W.; Epting, C.L.; et al. Sirtuin 2 regulates cellular iron homeostasis via deacetylation of transcription factor NRF2. J. Clin. Investig. 2017, 127, 1505–1516. [Google Scholar] [CrossRef]
- Apopa, P.L.; He, X.; Ma, Q. Phosphorylation of Nrf2 in the transcription activation domain by casein kinase 2 (CK2) is critical for the nuclear translocation and transcription activation function of Nrf2 in IMR-32 neuroblastoma cells. J. Biochem. Mol. Toxicol. 2008, 22, 63–76. [Google Scholar] [CrossRef]
- Sun, Z.; Huang, Z.; Zhang, D.D. Phosphorylation of Nrf2 at multiple sites by MAP kinases has a limited contribution in modulating the Nrf2-dependent antioxidant response. PLoS ONE 2009, 4, e6588. [Google Scholar] [CrossRef]
- Alam, J.; Igarashi, K.; Immenschuh, S.; Shibahara, S.; Tyrrell, R.M. Regulation of heme oxygenase-1 gene transcription: Recent advances and highlights from the International Conference (Uppsala, 2003) on Heme Oxygenase. Antioxid. Redox Signal. 2004, 6, 924–933. [Google Scholar] [CrossRef] [PubMed]
- Reichard, J.F.; Motz, G.T.; Puga, A. Heme oxygenase-1 induction by NRF2 requires inactivation of the transcriptional repressor BACH1. Nucleic Acids Res. 2007, 35, 7074–7086. [Google Scholar] [CrossRef]
- Andrés, C.M.C.; Pérez de la Lastra, J.M.; Juan, C.A.; Plou, F.J.; Pérez-Lebeña, E. Polyphenols as Antioxidant/Pro-Oxidant Compounds and Donors of Reducing Species: Relationship with Human Antioxidant Metabolism. Processes 2023, 11, 2771. [Google Scholar] [CrossRef]
- Tsuneyoshi, T. BACH1 mediates the antioxidant properties of aged garlic extract. Exp. Ther. Med. 2020, 19, 1500–1503. [Google Scholar] [CrossRef] [PubMed]
- Dhakshinamoorthy, S.; Jain, A.K.; Bloom, D.A.; Jaiswal, A.K. Bach1 competes with Nrf2 leading to negative regulation of the antioxidant response element (ARE)-mediated NAD(P)H:quinone oxidoreductase 1 gene expression and induction in response to antioxidants. J. Biol. Chem. 2005, 280, 16891–16900. [Google Scholar] [CrossRef] [PubMed]
- Fischhuber, K.; Matzinger, M.; Heiss, E.H. AMPK Enhances Transcription of Selected Nrf2 Target Genes via Negative Regulation of Bach1. Front. Cell Dev. Biol. 2020, 8, 628. [Google Scholar] [CrossRef]
- Davies, K.J.A.; Forman, H.J. Does Bach1 & c-Myc dependent redox dysregulation of Nrf2 & adaptive homeostasis decrease cancer risk in ageing? Free Radic. Biol. Med. 2019, 134, 708–714. [Google Scholar] [CrossRef]
- Li, D.; Sun, D.; Zhu, Y. Expression of nuclear factor erythroid-2-related factor 2, broad complex-tramtrack-bric a brac and Cap’n’collar homology 1 and γ-glutamic acid cysteine synthase in peripheral blood of patients with chronic obstructive pulmonary disease and its clinical significance. Exp. Ther. Med. 2021, 21, 516. [Google Scholar] [CrossRef]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.C. NF-κB signaling in inflammation. Signal Transduct. Target. Ther. 2017, 2, 17023. [Google Scholar] [CrossRef] [PubMed]
- Krajka-Kuźniak, V.; Baer-Dubowska, W. Modulation of Nrf2 and NF-κB Signaling Pathways by Naturally Occurring Compounds in Relation to Cancer Prevention and Therapy. Are Combinations Better Than Single Compounds? Int. J. Mol. Sci. 2021, 22, 8223. [Google Scholar] [CrossRef]
- Allocati, N.; Masulli, M.; Di Ilio, C.; Federici, L. Glutathione transferases: Substrates, inihibitors and pro-drugs in cancer and neurodegenerative diseases. Oncogenesis 2018, 7, 8. [Google Scholar] [CrossRef] [PubMed]
- Hine, C.M.; Mitchell, J.R. NRF2 and the Phase II Response in Acute Stress Resistance Induced by Dietary Restriction. J. Clin. Exp. Pathol. 2012, S4, 7329. [Google Scholar] [CrossRef] [PubMed]
- Rowland, A.; Miners, J.O.; Mackenzie, P.I. The UDP-glucuronosyltransferases: Their role in drug metabolism and detoxification. Int. J. Biochem. Cell Biol. 2013, 45, 1121–1132. [Google Scholar] [CrossRef] [PubMed]
- Buckley, D.B.; Klaassen, C.D. Induction of mouse UDP-glucuronosyltransferase mRNA expression in liver and intestine by activators of aryl-hydrocarbon receptor, constitutive androstane receptor, pregnane X receptor, peroxisome proliferator-activated receptor alpha, and nuclear factor erythroid 2-related factor 2. Drug Metab. Dispos. 2009, 37, 847–856. [Google Scholar] [CrossRef] [PubMed]
- Sim, E.; Abuhammad, A.; Ryan, A. Arylamine N-acetyltransferases: From drug metabolism and pharmacogenetics to drug discovery. Br. J. Pharmacol. 2014, 171, 2705–2725. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, L.C.; Yi, M.; Pedersen, L.G.; Kaminski, A.M. From Steroid and Drug Metabolism to Glycobiology, Using Sulfotransferase Structures to Understand and Tailor Function. Drug Metab. Dispos. 2022, 50, 1027–1041. [Google Scholar] [CrossRef] [PubMed]
- Gautheron, J.; Jéru, I. The Multifaceted Role of Epoxide Hydrolases in Human Health and Disease. Int. J. Mol. Sci. 2020, 22, 13. [Google Scholar] [CrossRef]
- Vilander, L.M.; Vaara, S.T.; Donner, K.M.; Lakkisto, P.; Kaunisto, M.A.; Pettilä, V. Heme oxygenase-1 repeat polymorphism in septic acute kidney injury. PLoS ONE 2019, 14, e0217291. [Google Scholar] [CrossRef]
- Ross, D.; Siegel, D. The diverse functionality of NQO1 and its roles in redox control. Redox Biol. 2021, 41, 101950. [Google Scholar] [CrossRef]
- Lu, S.C. Regulation of glutathione synthesis. Mol. Aspects Med. 2009, 30, 42–59. [Google Scholar] [CrossRef]
- Knovich, M.A.; Storey, J.A.; Coffman, L.G.; Torti, S.V.; Torti, F.M. Ferritin for the clinician. Blood Rev. 2009, 23, 95–104. [Google Scholar] [CrossRef]
- Zhang, Y. Phase II Enzymes. In Encyclopedia of Cancer; Schwab, M., Ed.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 2853–2855. [Google Scholar]
- Childs, B.G.; Durik, M.; Baker, D.J.; van Deursen, J.M. Cellular senescence in aging and age-related disease: From mechanisms to therapy. Nat. Med. 2015, 21, 1424–1435. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Sharpless, N.E. Senescence in Health and Disease. Cell 2017, 169, 1000–1011. [Google Scholar] [CrossRef] [PubMed]
- Juan, C.A.; Pérez de la Lastra, J.M.; Plou, F.J.; Pérez-Lebeña, E. The Chemistry of Reactive Oxygen Species (ROS) Revisited: Outlining Their Role in Biological Macromolecules (DNA, Lipids and Proteins) and Induced Pathologies. Int. J. Mol. Sci. 2021, 22, 4642. [Google Scholar] [CrossRef] [PubMed]
- Roger, L.; Tomas, F.; Gire, V. Mechanisms and Regulation of Cellular Senescence. Int. J. Mol. Sci. 2021, 22, 3173. [Google Scholar] [CrossRef] [PubMed]
- Victorelli, S.; Passos, J.F. Telomeres and Cell Senescence–Size Matters Not. EBioMedicine 2017, 21, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Di Micco, R.; Krizhanovsky, V.; Baker, D.; d’Adda di Fagagna, F. Cellular senescence in ageing: From mechanisms to therapeutic opportunities. Nat. Rev. Mol. Cell Biol. 2021, 22, 75–95. [Google Scholar] [CrossRef] [PubMed]
- Nousis, L.; Kanavaros, P.; Barbouti, A. Oxidative Stress-Induced Cellular Senescence: Is Labile Iron the Connecting Link? Antioxidants 2023, 12, 1250. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Xu, Y.; Luo, Y.; Wang, N.X.; Xiao, J.H. Role of Nrf2 in cell senescence regulation. Mol. Cell Biochem. 2021, 476, 247–259. [Google Scholar] [CrossRef]
- Curieses Andrés, C.M.; Pérez de la Lastra, J.M.; Andrés Juan, C.; Plou, F.J.; Pérez-Lebeña, E. From reactive species to disease development: Effect of oxidants and antioxidants on the cellular biomarkers. J. Biochem. Mol. Toxicol. 2023, 37, e23455. [Google Scholar] [CrossRef]
- Parzych, K.R.; Klionsky, D.J. An overview of autophagy: Morphology, mechanism, and regulation. Antioxid. Redox Signal. 2014, 20, 460–473. [Google Scholar] [CrossRef] [PubMed]
- Bellezza, I.; Giambanco, I.; Minelli, A.; Donato, R. Nrf2-Keap1 signaling in oxidative and reductive stress. Biochim. Biophys. Acta Mol. Cell Res. 2018, 1865, 721–733. [Google Scholar] [CrossRef] [PubMed]
- Fu, W.; Liu, Y.; Yin, H. Mitochondrial Dynamics: Biogenesis, Fission, Fusion, and Mitophagy in the Regulation of Stem Cell Behaviors. Stem Cells Int. 2019, 2019, 9757201. [Google Scholar] [CrossRef] [PubMed]
- Lastra, D.; Escoll, M.; Cuadrado, A. Transcription Factor NRF2 Participates in Cell Cycle Progression at the Level of G1/S and Mitotic Checkpoints. Antioxidants 2022, 11, 946. [Google Scholar] [CrossRef] [PubMed]
- Murata, H.; Takamatsu, H.; Liu, S.; Kataoka, K.; Huh, N.H.; Sakaguchi, M. NRF2 Regulates PINK1 Expression under Oxidative Stress Conditions. PLoS ONE 2015, 10, e0142438. [Google Scholar] [CrossRef]
- Chan, N.C.; Salazar, A.M.; Pham, A.H.; Sweredoski, M.J.; Kolawa, N.J.; Graham, R.L.; Hess, S.; Chan, D.C. Broad activation of the ubiquitin-proteasome system by Parkin is critical for mitophagy. Hum. Mol. Genet. 2011, 20, 1726–1737. [Google Scholar] [CrossRef]
- Villavicencio Tejo, F.; Quintanilla, R.A. Contribution of the Nrf2 Pathway on Oxidative Damage and Mitochondrial Failure in Parkinson and Alzheimer’s Disease. Antioxidants 2021, 10, 1069. [Google Scholar] [CrossRef] [PubMed]
- Kang, L.; Liu, S.; Li, J.; Tian, Y.; Xue, Y.; Liu, X. Parkin and Nrf2 prevent oxidative stress-induced apoptosis in intervertebral endplate chondrocytes via inducing mitophagy and anti-oxidant defenses. Life Sci. 2020, 243, 117244. [Google Scholar] [CrossRef]
- Mi, L.; Hu, J.; Li, N.; Gao, J.; Huo, R.; Peng, X.; Zhang, N.; Liu, Y.; Zhao, H.; Liu, R.; et al. The Mechanism of Stem Cell Aging. Stem Cell Rev. Rep. 2022, 18, 1281–1293. [Google Scholar] [CrossRef]
- Liu, L.; Rando, T.A. Manifestations and mechanisms of stem cell aging. J. Cell Biol. 2011, 193, 257–266. [Google Scholar] [CrossRef]
- Schmidlin, C.J.; Dodson, M.B.; Madhavan, L.; Zhang, D.D. Redox regulation by NRF2 in aging and disease. Free Radic. Biol. Med. 2019, 134, 702–707. [Google Scholar] [CrossRef]
- Ray, S.; Corenblum, M.J.; Anandhan, A.; Reed, A.; Ortiz, F.O.; Zhang, D.D.; Barnes, C.A.; Madhavan, L. A Role for Nrf2 Expression in Defining the Aging of Hippocampal Neural Stem Cells. Cell Transplant. 2018, 27, 589–606. [Google Scholar] [CrossRef] [PubMed]
- Andrés, C.M.C.; Pérez de la Lastra, J.M.; Juan, C.A.; Plou, F.J.; Pérez-Lebeña, E. Antioxidant Metabolism Pathways in Vitamins, Polyphenols, and Selenium: Parallels and Divergences. Int. J. Mol. Sci. 2024, 25, 2600. [Google Scholar] [CrossRef] [PubMed]
- Stefanson, A.L.; Bakovic, M. Dietary regulation of Keap1/Nrf2/ARE pathway: Focus on plant-derived compounds and trace minerals. Nutrients 2014, 6, 3777–3801. [Google Scholar] [CrossRef]
- Luo, Y.; Eggler, A.L.; Liu, D.; Liu, G.; Mesecar, A.D.; van Breemen, R.B. Sites of alkylation of human Keap1 by natural chemoprevention agents. J. Am. Soc. Mass. Spectrom. 2007, 18, 2226–2232. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.D.; Hannink, M. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol. Cell. Biol. 2003, 23, 8137–8151. [Google Scholar] [CrossRef]
- Ohnuma, T.; Nakayama, S.; Anan, E.; Nishiyama, T.; Ogura, K.; Hiratsuka, A. Activation of the Nrf2/ARE pathway via S-alkylation of cysteine 151 in the chemopreventive agent-sensor Keap1 protein by falcarindiol, a conjugated diacetylene compound. Toxicol. Appl. Pharmacol. 2010, 244, 27–36. [Google Scholar] [CrossRef]
- Abdel-Massih, R.M.; Debs, E.; Othman, L.; Attieh, J.; Cabrerizo, F.M. Glucosinolates, a natural chemical arsenal: More to tell than the myrosinase story. Front. Microbiol. 2023, 14, 1130208. [Google Scholar] [CrossRef]
- Bhatwalkar, S.B.; Mondal, R.; Krishna, S.B.N.; Adam, J.K.; Govender, P.; Anupam, R. Antibacterial Properties of Organosulfur Compounds of Garlic (Allium sativum). Front. Microbiol. 2021, 12, 613077. [Google Scholar] [CrossRef]
- Schultz, T.W.; Yarbrough, J.W.; Hunter, R.S.; Aptula, A.O. Verification of the structural alerts for Michael acceptors. Chem. Res. Toxicol. 2007, 20, 1359–1363. [Google Scholar] [CrossRef]
- Little, R.D.; Masjedizadeh, M.R.; Wallquist, O.; Mcloughlin, J.I. The Intramolecular Michael Reaction. Org. React. 2004, 47, 315–552. [Google Scholar] [CrossRef]
- Jackson, P.A.; Widen, J.C.; Harki, D.A.; Brummond, K.M. Covalent Modifiers: A Chemical Perspective on the Reactivity of α,β-Unsaturated Carbonyls with Thiols via Hetero-Michael Addition Reactions. J. Med. Chem. 2017, 60, 839–885. [Google Scholar] [CrossRef]
- de Freitas Silva, M.; Pruccoli, L.; Morroni, F.; Sita, G.; Seghetti, F.; Viegas, C.; Tarozzi, A. The Keap1/Nrf2-ARE Pathway as a Pharmacological Target for Chalcones. Molecules 2018, 23, 1803. [Google Scholar] [CrossRef]
- Chu, H.W.; Sethy, B.; Hsieh, P.W.; Horng, J.T. Identification of Potential Drug Targets of Broad-Spectrum Inhibitors with a Michael Acceptor Moiety Using Shotgun Proteomics. Viruses 2021, 13, 1756. [Google Scholar] [CrossRef]
- Ansari, M.I.; Khan, M.M.; Saquib, M.; Khatoon, S.; Hussain, M.K. Dithiolethiones: A privileged pharmacophore for anticancer therapy and chemoprevention. Future Med. Chem. 2018, 10, 1241–1260. [Google Scholar] [CrossRef]
- Amalraj, A.; Pius, A.; Gopi, S.; Gopi, S. Biological activities of curcuminoids, other biomolecules from turmeric and their derivatives—A review. J. Tradit. Complement. Med. 2017, 7, 205–233. [Google Scholar] [CrossRef]
- Rammohan, A.; Reddy, J.S.; Sravya, G.; Rao, C.N.; Zyryanov, G.V. Chalcone synthesis, properties and medicinal applications: A review. Environ. Chem. Lett. 2020, 18, 433–458. [Google Scholar] [CrossRef]
- Zhuang, C.; Zhang, W.; Sheng, C.; Zhang, W.; Xing, C.; Miao, Z. Chalcone: A Privileged Structure in Medicinal Chemistry. Chem. Rev. 2017, 117, 7762–7810. [Google Scholar] [CrossRef]
- Rajendran, G.; Bhanu, D.; Aruchamy, B.; Ramani, P.; Pandurangan, N.; Bobba, K.N.; Oh, E.J.; Chung, H.Y.; Gangadaran, P.; Ahn, B.C. Chalcone: A Promising Bioactive Scaffold in Medicinal Chemistry. Pharmaceuticals 2022, 15, 1250. [Google Scholar] [CrossRef]
- Bolton, J.L.; Dunlap, T. Formation and Biological Targets of Quinones: Cytotoxic versus Cytoprotective Effects. Chem. Res. Toxicol. 2017, 30, 13–37. [Google Scholar] [CrossRef]
- Linder, M.C.; Hazegh-Azam, M. Copper biochemistry and molecular biology. Am. J. Clin. Nutr. 1996, 63, S797–S811. [Google Scholar]
- Wang, X.J.; Hayes, J.D.; Higgins, L.G.; Wolf, C.R.; Dinkova-Kostova, A.T. Activation of the NRF2 signaling pathway by copper-mediated redox cycling of para- and ortho-hydroquinones. Chem. Biol. 2010, 17, 75–85. [Google Scholar] [CrossRef]
- Satoh, T.; Saitoh, S.; Hosaka, M.; Kosaka, K. Simple ortho- and para-hydroquinones as compounds neuroprotective against oxidative stress in a manner associated with specific transcriptional activation. Biochem. Biophys. Res. Commun. 2009, 379, 537–541. [Google Scholar] [CrossRef]
- Bensasson, R.V.; Zoete, V.; Dinkova-Kostova, A.T.; Talalay, P. Two-step mechanism of induction of the gene expression of a prototypic cancer-protective enzyme by diphenols. Chem. Res. Toxicol. 2008, 21, 805–812. [Google Scholar] [CrossRef]
- Sharifi-Rad, J.; Cruz-Martins, N.; López-Jornet, P.; Lopez, E.P.; Harun, N.; Yeskaliyeva, B.; Beyatli, A.; Sytar, O.; Shaheen, S.; Sharopov, F.; et al. Natural Coumarins: Exploring the Pharmacological Complexity and Underlying Molecular Mechanisms. Oxid. Med. Cell Longev. 2021, 2021, 6492346. [Google Scholar] [CrossRef]
- Singh, B.; Sharma, R.A. Plant terpenes: Defense responses, phylogenetic analysis, regulation and clinical applications. 3 Biotech 2015, 5, 129–151. [Google Scholar] [CrossRef]
- Siraj, M.A.; Islam, M.A.; Al Fahad, M.A.; Kheya, H.R.; Xiao, J.; Simal-Gandara, J. Cancer Chemopreventive Role of Dietary Terpenoids by Modulating Keap1-Nrf2-ARE Signaling System—A Comprehensive Update. Appl. Sci. 2021, 11, 10806. [Google Scholar] [CrossRef]
- Kalantari, K.; Moniri, M.; Boroumand Moghaddam, A.; Abdul Rahim, R.; Bin Ariff, A.; Izadiyan, Z.; Mohamad, R. A Review of the Biomedical Applications of Zerumbone and the Techniques for Its Extraction from Ginger Rhizomes. Molecules 2017, 22, 1645. [Google Scholar] [CrossRef]
- Liang, S.-T.; Chen, C.; Chen, R.-X.; Li, R.; Chen, W.-L.; Jiang, G.-H.; Du, L.-L. Michael acceptor molecules in natural products and their mechanism of action. Front. Pharmacol. 2022, 13, 1033003. [Google Scholar] [CrossRef]
- Andrés, C.M.C.; Pérez de la Lastra, J.M.; Andrés Juan, C.; Plou, F.J.; Pérez-Lebeña, E. Chemistry of Hydrogen Sulfide-Pathological and Physiological Functions in Mammalian Cells. Cells 2023, 12, 2684. [Google Scholar] [CrossRef]
- Munteanu, C.; Rotariu, M.; Turnea, M.; Dogaru, G.; Popescu, C.; Spînu, A.; Andone, I.; Postoiu, R.; Ionescu, E.V.; Oprea, C.; et al. Recent Advances in Molecular Research on Hydrogen Sulfide (H(2)S) Role in Diabetes Mellitus (DM)-A Systematic Review. Int. J. Mol. Sci. 2022, 23, 6720. [Google Scholar] [CrossRef]
- Kimura, H. The physiological role of hydrogen sulfide and beyond. Nitric Oxide 2014, 41, 4–10. [Google Scholar] [CrossRef]
- Sen, U.; Sathnur, P.B.; Kundu, S.; Givvimani, S.; Coley, D.M.; Mishra, P.K.; Qipshidze, N.; Tyagi, N.; Metreveli, N.; Tyagi, S.C. Increased endogenous H2S generation by CBS, CSE, and 3MST gene therapy improves ex vivo renovascular relaxation in hyperhomocysteinemia. Am. J. Physiol. Cell Physiol. 2012, 303, C41–C51. [Google Scholar] [CrossRef]
- Dordević, D.; Jančíková, S.; Vítězová, M.; Kushkevych, I. Hydrogen sulfide toxicity in the gut environment: Meta-analysis of sulfate-reducing and lactic acid bacteria in inflammatory processes. J. Adv. Res. 2021, 27, 55–69. [Google Scholar] [CrossRef]
- Jandhyala, S.M.; Talukdar, R.; Subramanyam, C.; Vuyyuru, H.; Sasikala, M.; Nageshwar Reddy, D. Role of the normal gut microbiota. World J. Gastroenterol. 2015, 21, 8787–8803. [Google Scholar] [CrossRef]
- Abe, K.; Kimura, H. The possible role of hydrogen sulfide as an endogenous neuromodulator. J. Neurosci. 1996, 16, 1066–1071. [Google Scholar] [CrossRef]
- Hosoki, R.; Matsuki, N.; Kimura, H. The possible role of hydrogen sulfide as an endogenous smooth muscle relaxant in synergy with nitric oxide. Biochem. Biophys. Res. Commun. 1997, 237, 527–531. [Google Scholar] [CrossRef]
- Shibuya, N.; Mikami, Y.; Kimura, Y.; Nagahara, N.; Kimura, H. Vascular endothelium expresses 3-mercaptopyruvate sulfurtransferase and produces hydrogen sulfide. J. Biochem. 2009, 146, 623–626. [Google Scholar] [CrossRef]
- Wang, R. Physiological implications of hydrogen sulfide: A whiff exploration that blossomed. Physiol. Rev. 2012, 92, 791–896. [Google Scholar] [CrossRef]
- Buret, A.G.; Allain, T.; Motta, J.P.; Wallace, J.L. Effects of Hydrogen Sulfide on the Microbiome: From Toxicity to Therapy. Antioxid. Redox Signal. 2022, 36, 211–219. [Google Scholar] [CrossRef]
- Kolluru, G.K.; Shen, X.; Bir, S.C.; Kevil, C.G. Hydrogen sulfide chemical biology: Pathophysiological roles and detection. Nitric Oxide 2013, 35, 5–20. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Q.; Ying, J.; Xiang, L.; Zhang, C. The biologic effect of hydrogen sulfide and its function in various diseases. Medicine 2018, 97, e13065. [Google Scholar] [CrossRef] [PubMed]
- Lobo, V.; Patil, A.; Phatak, A.; Chandra, N. Free radicals, antioxidants and functional foods: Impact on human health. Pharmacogn. Rev. 2010, 4, 118–126. [Google Scholar] [CrossRef] [PubMed]
- Olson, K.R. Is hydrogen sulfide a circulating "gasotransmitter" in vertebrate blood? Biochim. Biophys. Acta 2009, 1787, 856–863. [Google Scholar] [CrossRef]
- Kimura, H.; Shibuya, N.; Kimura, Y. Hydrogen sulfide is a signaling molecule and a cytoprotectant. Antioxid. Redox Signal. 2012, 17, 45–57. [Google Scholar] [CrossRef]
- Ju, Y.; Fu, M.; Stokes, E.; Wu, L.; Yang, G. H₂S-Mediated Protein S-Sulfhydration: A Prediction for Its Formation and Regulation. Molecules 2017, 22, 1334. [Google Scholar] [CrossRef]
- Aroca, Á.; Serna, A.; Gotor, C.; Romero, L.C. S-sulfhydration: A cysteine posttranslational modification in plant systems. Plant Physiol. 2015, 168, 334–342. [Google Scholar] [CrossRef]
- Andrés, C.M.C.; Pérez de la Lastra, J.M.; Andrés Juan, C.; Plou, F.J.; Pérez-Lebeña, E. Impact of Reactive Species on Amino Acids;Biological Relevance in Proteins and Induced Pathologies. Int. J. Mol. Sci. 2022, 23, 14049. [Google Scholar] [CrossRef]
- England, K.; Cotter, T.G. Direct oxidative modifications of signalling proteins in mammalian cells and their effects on apoptosis. Redox Rep. 2005, 10, 237–245. [Google Scholar] [CrossRef]
- Yang, G.; Zhao, K.; Ju, Y.; Mani, S.; Cao, Q.; Puukila, S.; Khaper, N.; Wu, L.; Wang, R. Hydrogen sulfide protects against cellular senescence via S-sulfhydration of Keap1 and activation of Nrf2. Antioxid. Redox Signal. 2013, 18, 1906–1919. [Google Scholar] [CrossRef] [PubMed]
- Lv, B.; Chen, S.; Tang, C.; Jin, H.; Du, J.; Huang, Y. Hydrogen sulfide and vascular regulation–An update. J. Adv. Res. 2021, 27, 85–97. [Google Scholar] [CrossRef]
- Shen, Y.; Shen, Z.; Luo, S.; Guo, W.; Zhu, Y.Z. The Cardioprotective Effects of Hydrogen Sulfide in Heart Diseases: From Molecular Mechanisms to Therapeutic Potential. Oxid. Med. Cell Longev. 2015, 2015, 925167. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.L.; Qin, M.; Liu, X.H.; Zhu, Y.Z. The Role of Hydrogen Sulfide on Cardiovascular Homeostasis: An Overview with Update on Immunomodulation. Front. Pharmacol. 2017, 8, 686. [Google Scholar] [CrossRef] [PubMed]
- Kolluru, G.K.; Shackelford, R.E.; Shen, X.; Dominic, P.; Kevil, C.G. Sulfide regulation of cardiovascular function in health and disease. Nat. Rev. Cardiol. 2023, 20, 109–125. [Google Scholar] [CrossRef] [PubMed]
- Meng, G.; Ma, Y.; Xie, L.; Ferro, A.; Ji, Y. Emerging role of hydrogen sulfide in hypertension and related cardiovascular diseases. Br. J. Pharmacol. 2015, 172, 5501–5511. [Google Scholar] [CrossRef]
- Hu, Q.; Lukesh, J.C., 3rd. H(2)S Donors with Cytoprotective Effects in Models of MI/R Injury and Chemotherapy-Induced Cardiotoxicity. Antioxidants 2023, 12, 650. [Google Scholar] [CrossRef]
- Deng, N.H.; Luo, W.; Gui, D.D.; Yan, B.J.; Zhou, K.; Tian, K.J.; Ren, Z.; Xiong, W.H.; Jiang, Z.S. Hydrogen sulfide plays a potential alternative for the treatment of metabolic disorders of diabetic cardiomyopathy. Mol. Cell. Biochem. 2022, 477, 255–265. [Google Scholar] [CrossRef]
- Piragine, E.; Malanima, M.A.; Lucenteforte, E.; Martelli, A.; Calderone, V. Circulating Levels of Hydrogen Sulfide (H(2)S) in Patients with Age-Related Diseases: A Systematic Review and Meta-Analysis. Biomolecules 2023, 13, 1023. [Google Scholar] [CrossRef]
- Felipe Salech, M.; Rafael Jara, L.; Luis Michea, A. Cambios fisiológicos asociados al envejecimiento. Rev. Médica Clínica Las Condes 2012, 23, 19–29. [Google Scholar] [CrossRef]
- Qabazard, B.; Stürzenbaum, S.R. H2S: A New Approach to Lifespan Enhancement and Healthy Ageing? Handb. Exp. Pharmacol. 2015, 230, 269–287. [Google Scholar] [CrossRef] [PubMed]
- Huerta de la Cruz, S.; Rodríguez-Palma, E.J.; Santiago-Castañeda, C.L.; Beltrán-Ornelas, J.H.; Sánchez-López, A.; Rocha, L.; Centurión, D. Exogenous hydrogen sulfide restores CSE and CBS but no 3-MST protein expression in the hypothalamus and brainstem after severe traumatic brain injury. Metab. Brain Dis. 2022, 37, 1863–1874. [Google Scholar] [CrossRef]
- Münke, M.; Kraus, J.P.; Ohura, T.; Francke, U. The gene for cystathionine beta-synthase (CBS) maps to the subtelomeric region on human chromosome 21q and to proximal mouse chromosome 17. Am. J. Hum. Genet. 1988, 42, 550–559. [Google Scholar]
- Yin, P.; Zhao, C.; Li, Z.; Mei, C.; Yao, W.; Liu, Y.; Li, N.; Qi, J.; Wang, L.; Shi, Y.; et al. Sp1 is involved in regulation of cystathionine γ-lyase gene expression and biological function by PI3K/Akt pathway in human hepatocellular carcinoma cell lines. Cell. Signal. 2012, 24, 1229–1240. [Google Scholar] [CrossRef]
- Rodrigues, C.; Percival, S.S. Immunomodulatory Effects of Glutathione, Garlic Derivatives, and Hydrogen Sulfide. Nutrients 2019, 11, 295. [Google Scholar] [CrossRef]
- Shang, A.; Cao, S.Y.; Xu, X.Y.; Gan, R.Y.; Tang, G.Y.; Corke, H.; Mavumengwana, V.; Li, H.B. Bioactive Compounds and Biological Functions of Garlic (Allium sativum L.). Foods 2019, 8, 246. [Google Scholar] [CrossRef]
- Najman, K.; Sadowska, A.; Hallmann, E. Evaluation of Bioactive and Physicochemical Properties of White and Black Garlic (Allium sativum L.) from Conventional and Organic Cultivation. Appl. Sci. 2021, 11, 874. [Google Scholar] [CrossRef]
- Fimognari, C.; Turrini, E.; Ferruzzi, L.; Lenzi, M.; Hrelia, P. Natural isothiocyanates: Genotoxic potential versus chemoprevention. Mutat. Res. 2012, 750, 107–131. [Google Scholar] [CrossRef]
- Zhang, Y. The 1,2-benzenedithiole-based cyclocondensation assay: A valuable tool for the measurement of chemopreventive isothiocyanates. Crit. Rev. Food Sci. Nutr. 2012, 52, 525–532. [Google Scholar] [CrossRef]
- Li, Z.; Liu, Y.; Fang, Z.; Yang, L.; Zhuang, M.; Zhang, Y.; Zhao, W.; Sun, P. Variation of Sulforaphane Levels in Broccoli (Brassica oleracea var. Italica) during Flower Development and the Role of Gene AOP2. J. Liq. Chromatogr. Relat. Technol. 2014, 37, 1199–1211. [Google Scholar] [CrossRef]
- Chiu, Y.C.; Matak, K.; Ku, K.M. Methyl jasmonate treated broccoli: Impact on the production of glucosinolates and consumer preferences. Food Chem. 2019, 299, 125099. [Google Scholar] [CrossRef]
- Yadav, K.; Dhankhar, J. Isothiocyanates—A Review of their Health Benefits and Potential Food Applications. Curr. Res. Nutr. Food Sci. 2022, 10, 476. [Google Scholar] [CrossRef]
- Maruthupandy, M.; Seo, J. Allyl isothiocyanate encapsulated halloysite covered with polyacrylate as a potential antibacterial agent against food spoilage bacteria. Mater. Sci. Eng. C Mater. Biol. Appl. 2019, 105, 110016. [Google Scholar] [CrossRef]
- Castellano, R.; Perruchot, M.H.; Tesseraud, S.; Métayer-Coustard, S.; Baeza, E.; Mercier, Y.; Gondret, F. Methionine and cysteine deficiencies altered proliferation rate and time-course differentiation of porcine preadipose cells. Amino Acids 2017, 49, 355–366. [Google Scholar] [CrossRef]
- Krishnaswamy, K.; Rao, S.B. Failure to produce atherosclerosis in Macaca radiata on a high-methionine, high-fat, pyridoxine-deficient diet. Atherosclerosis 1977, 27, 253–258. [Google Scholar] [CrossRef]
- Mikkelsen, M.D.; Petersen, B.; Olsen, C.; Halkier, B. Biosynthesis and metabolic engineering of glucosinolates. Amino Acids 2002, 22, 279–295. [Google Scholar] [CrossRef] [PubMed]
- Chan, Y.A.; Podevels, A.M.; Kevany, B.M.; Thomas, M.G. Biosynthesis of polyketide synthase extender units. Nat. Prod. Rep. 2009, 26, 90–114. [Google Scholar] [CrossRef] [PubMed]
- Kachungwa Lugata, J.; Ortega, A.D.S.V.; Szabó, C. The Role of Methionine Supplementation on Oxidative Stress and Antioxidant Status of Poultry-A Review. Agriculture 2022, 12, 1701. [Google Scholar] [CrossRef]
- Liu, X.Y.; Li, C.Y.; Bu, H.; Li, Z.; Li, B.; Sun, M.M.; Guo, Y.S.; Zhang, L.; Ren, W.B.; Fan, Z.L.; et al. The neuroprotective potential of phase II enzyme inducer on motor neuron survival in traumatic spinal cord injury in vitro. Cell. Mol. Neurobiol. 2008, 28, 769–779. [Google Scholar] [CrossRef]
- Brown, D.A.; Betharia, S.; Yen, J.H.; Tran, Q.; Mistry, H.; Smith, K. Synthesis and structure-activity relationships study of dithiolethiones as inducers of glutathione in the SH-SY5Y neuroblastoma cell line. Bioorg. Med. Chem. Lett. 2014, 24, 5829–5831. [Google Scholar] [CrossRef]
- Begleiter, A.; Leith, M.K.; Curphey, T.J.; Doherty, G.P. Induction of DT-diaphorase in cancer chemoprevention and chemotherapy. Oncol. Res. 1997, 9, 371–382. [Google Scholar] [PubMed]
- Zipper, L.M.; Mulcahy, R.T. The Keap1 BTB/POZ dimerization function is required to sequester Nrf2 in cytoplasm. J. Biol. Chem. 2002, 277, 36544–36552. [Google Scholar] [CrossRef] [PubMed]
- Sekhar, K.R.; Spitz, D.R.; Harris, S.; Nguyen, T.T.; Meredith, M.J.; Holt, J.T.; Gius, D.; Marnett, L.J.; Summar, M.L.; Freeman, M.L. Redox-sensitive interaction between KIAA0132 and Nrf2 mediates indomethacin-induced expression of gamma-glutamylcysteine synthetase. Free Radic. Biol. Med. 2002, 32, 650–662. [Google Scholar] [CrossRef]
- Li, K.R.; Yang, S.Q.; Gong, Y.Q.; Yang, H.; Li, X.M.; Zhao, Y.X.; Yao, J.; Jiang, Q.; Cao, C. 3H-1,2-dithiole-3-thione protects retinal pigment epithelium cells against Ultra-violet radiation via activation of Akt-mTORC1-dependent Nrf2-HO-1 signaling. Sci. Rep. 2016, 6, 25525. [Google Scholar] [CrossRef] [PubMed]
- Kuo, P.C.; Brown, D.A.; Scofield, B.A.; Yu, I.C.; Chang, F.L.; Wang, P.Y.; Yen, J.H. 3H-1,2-dithiole-3-thione as a novel therapeutic agent for the treatment of experimental autoimmune encephalomyelitis. Brain Behav. Immun. 2016, 57, 173–186. [Google Scholar] [CrossRef] [PubMed]
- Karuri, A.R.; Huang, Y.; Bodreddigari, S.; Sutter, C.H.; Roebuck, B.D.; Kensler, T.W.; Sutter, T.R. 3H-1,2-dithiole-3-thione targets nuclear factor kappaB to block expression of inducible nitric-oxide synthase, prevents hypotension, and improves survival in endotoxemic rats. J. Pharmacol. Exp. Ther. 2006, 317, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Kuo, P.-C.; Yu, I.C.; Scofield, B.A.; Brown, D.A.; Curfman, E.T.; Paraiso, H.C.; Chang, F.-L.; Yen, J.-H. 3H-1,2-Dithiole-3-thione as a novel therapeutic agent for the treatment of ischemic stroke through Nrf2 defense pathway. Brain Behav. Immun. 2017, 62, 180–192. [Google Scholar] [CrossRef] [PubMed]
- Rakitin, O.A. Synthesis and Reactivity of 3H-1,2-dithiole-3-thiones. Molecules 2021, 26, 3595. [Google Scholar] [CrossRef]
- Huang, A.; Seité, S.; Adar, T. The use of balneotherapy in dermatology. Clin. Dermatol. 2018, 36, 363–368. [Google Scholar] [CrossRef]
- Nasermoaddeli, A.; Kagamimori, S. Balneotherapy in medicine: A review. Environ. Health Prev. Med. 2005, 10, 171–179. [Google Scholar] [CrossRef]
- Maraver, F.; Armijo, F.; Fernandez-Toran, M.A.; Armijo, O.; Ejeda, J.M.; Vazquez, I.; Corvillo, I.; Torres-Piles, S. Peloids as Thermotherapeutic Agents. Int. J. Environ. Res. Public Health 2021, 18, 1965. [Google Scholar] [CrossRef] [PubMed]
- Gálvez, I.; Torres-Piles, S.; Ortega-Rincón, E. Balneotherapy, Immune System, and Stress Response: A Hormetic Strategy? Int. J. Mol. Sci. 2018, 19, 1687. [Google Scholar] [CrossRef] [PubMed]
- Kovács, C.; Pecze, M.; Tihanyi, Á.; Kovács, L.; Balogh, S.; Bender, T. The effect of sulphurous water in patients with osteoarthritis of hand. Double-blind, randomized, controlled follow-up study. Clin. Rheumatol. 2012, 31, 1437–1442. [Google Scholar] [CrossRef] [PubMed]
- Vaamonde-García, C.; Vela-Anero, Á.; Hermida-Gómez, T.; Fernández-Burguera, E.; Filgueira-Fernández, P.; Goyanes, N.; Blanco, F.J.; Meijide-Faílde, R. Effect of balneotherapy in sulfurous water on an in vivo murine model of osteoarthritis. Int. J. Biometeorol. 2020, 64, 307–318. [Google Scholar] [CrossRef] [PubMed]
- Protano, C.; Fontana, M.; De Giorgi, A.; Marotta, D.; Cocomello, N.; Crucianelli, S.; Del Cimmuto, A.; Vitali, M. Balneotherapy for osteoarthritis: A systematic review. Rheumatol. Int. 2023, 43, 1597–1610. [Google Scholar] [CrossRef] [PubMed]
- Brglez Mojzer, E.; Knez Hrnčič, M.; Škerget, M.; Knez, Ž.; Bren, U. Polyphenols: Extraction Methods, Antioxidative Action, Bioavailability and Anticarcinogenic Effects. Molecules 2016, 21, 901. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.M.; Rahaman, M.S.; Islam, M.R.; Rahman, F.; Mithi, F.M.; Alqahtani, T.; Almikhlafi, M.A.; Alghamdi, S.Q.; Alruwaili, A.S.; Hossain, M.S.; et al. Role of Phenolic Compounds in Human Disease: Current Knowledge and Future Prospects. Molecules 2022, 27, 233. [Google Scholar] [CrossRef] [PubMed]
- Andrés, C.M.C.; Pérez de la Lastra, J.M.; Andrés Juan, C.; Plou, F.J.; Pérez-Lebeña, E. Superoxide Anion Chemistry;Its Role at the Core of the Innate Immunity. Int. J. Mol. Sci. 2023, 24, 1841. [Google Scholar] [CrossRef]
- Hamad, H.A.M. Phenolic Compounds: Classification, Chemistry, and Updated Techniques of Analysis and Synthesis. In Phenolic Compounds; Farid, A.B., Ed.; IntechOpen: Rijeka, Croatia, 2021; Chapter 4. [Google Scholar]
- Costa, M.; Sezgin-Bayindir, Z.; Losada-Barreiro, S.; Paiva-Martins, F.; Saso, L.; Bravo-Díaz, C. Polyphenols as Antioxidants for Extending Food Shelf-Life and in the Prevention of Health Diseases: Encapsulation and Interfacial Phenomena. Biomedicines 2021, 9, 1909. [Google Scholar] [CrossRef]
- Amankulova, D.; Berganayeva, G.; Kudaibergenova, B.; Zhetpisbay, D.; Sharipova, A.; Dyusebaeva, M. Recent Advances in the Synthesis and Applications of m-Aryloxy Phenols. Molecules 2023, 28, 2657. [Google Scholar] [CrossRef]
- Qiu, Z.; Li, C.-J. Transformations of less-activated phenols and phenol derivatives via C–O cleavage. Chem. Rev. 2020, 120, 10454–10515. [Google Scholar] [CrossRef]
- Floris, B.; Galloni, P.; Conte, V.; Sabuzi, F. Tailored Functionalization of Natural Phenols to Improve Biological Activity. Biomolecules 2021, 11, 1325. [Google Scholar] [CrossRef]
- Tang, J.; Zhang, S.; Zhou, B.-W.; Wang, W.; Zhao, L. Hyperconjugative Aromaticity-Based Circularly Polarized Luminescence Enhancement in Polyaurated Heterocycles. J. Am. Chem. Soc. 2023, 145, 23442–23451. [Google Scholar] [CrossRef]
- Taranto, F.; Pasqualone, A.; Mangini, G.; Tripodi, P.; Miazzi, M.M.; Pavan, S.; Montemurro, C. Polyphenol Oxidases in Crops: Biochemical, Physiological and Genetic Aspects. Int. J. Mol. Sci. 2017, 18, 377. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Were, L. Cysteine’s effects on chlorogenic acid quinone induced greening and browning: Mechanism and effect on antioxidant reducing capacity. Food Chem. 2020, 309, 125697. [Google Scholar] [CrossRef]
- Araji, S.; Grammer, T.A.; Gertzen, R.; Anderson, S.D.; Mikulic-Petkovsek, M.; Veberic, R.; Phu, M.L.; Solar, A.; Leslie, C.A.; Dandekar, A.M.; et al. Novel Roles for the Polyphenol Oxidase Enzyme in Secondary Metabolism and the Regulation of Cell Death in Walnut. Plant Physiol. 2014, 164, 1191–1203. [Google Scholar] [CrossRef] [PubMed]
- Bolton, J.L.; Trush, M.A.; Penning, T.M.; Dryhurst, G.; Monks, T.J. Role of Quinones in Toxicology. Chem. Res. Toxicol. 2000, 13, 135–160. [Google Scholar] [CrossRef]
- Unoki, T.; Akiyama, M.; Kumagai, Y. Nrf2 Activation and Its Coordination with the Protective Defense Systems in Response to Electrophilic Stress. Int. J. Mol. Sci. 2020, 21, 545. [Google Scholar] [CrossRef] [PubMed]
- Nogales-Delgado, S. Polyphenoloxidase (PPO): Effect, Current Determination and Inhibition Treatments in Fresh-Cut Produce. Appl. Sci. 2021, 11, 7813. [Google Scholar] [CrossRef]
- Dulo, B.; Phan, K.; Githaiga, J.; Raes, K.; De Meester, S. Natural Quinone Dyes: A Review on Structure, Extraction Techniques, Analysis and Application Potential. Waste Biomass Valorization 2021, 12, 6339–6374. [Google Scholar] [CrossRef]
- Varela-López, A.; Giampieri, F.; Battino, M.; Quiles, J.L. Coenzyme Q and Its Role in the Dietary Therapy against Aging. Molecules 2016, 21, 373. [Google Scholar] [CrossRef]
- Enriquez, J.A.; Lenaz, G. Coenzyme q and the respiratory chain: Coenzyme q pool and mitochondrial supercomplexes. Mol. Syndromol. 2014, 5, 119–140. [Google Scholar] [CrossRef] [PubMed]
- Nardini, M. Phenolic Compounds in Food: Characterization and Health Benefits. Molecules 2022, 27, 783. [Google Scholar] [CrossRef] [PubMed]
- Soliman, I.A.; Hasanien, Y.A.; Zaki, A.G.; Shawky, H.A.; Nassrallah, A.A. Irradiation impact on biological activities of Anthraquinone pigment produced from Talaromyces purpureogenus and its evaluation, characterization and application in beef burger as natural preservative. BMC Microbiol. 2022, 22, 325. [Google Scholar] [CrossRef] [PubMed]
- Malik, E.M.; Müller, C.E. Anthraquinones As Pharmacological Tools and Drugs. Med. Res. Rev. 2016, 36, 705–748. [Google Scholar] [CrossRef] [PubMed]
- Siva, R.; Mayes, S.; Behera, S.K.; Rajasekaran, C. Anthraquinones dye production using root cultures of Oldenlandia umbellata L. Ind. Crops Prod. 2012, 37, 415–419. [Google Scholar] [CrossRef]
- Hart, P.W.; Rudie, A.W. Anthraquinone a review of the rise and fall of a pulping catalyst. Tappi J. 2014, 13, 23–31. [Google Scholar] [CrossRef]
- Goor, G.; Glenneberg, J.; Jacobi, S. Hydrogen Peroxide. In Ullmann’s Encyclopedia of Industrial Chemistry; Wiley: Hoboken, NJ, USA, 2007. [Google Scholar] [CrossRef]
- Avery, M.L.; Humphrey, J.S.; Primus, T.M.; Decker, D.G.; McGrane, A.P. Anthraquinone protects rice seed from birds. Crop Prot. 1998, 17, 225–230. [Google Scholar] [CrossRef]
- Wu, M.; Jing, Y.; Wong, A.A.; Fell, E.M.; Jin, S.; Tang, Z.; Gordon, R.G.; Aziz, M.J. Extremely Stable Anthraquinone Negolytes Synthesized from Common Precursors. Chem 2020, 6, 1432–1442. [Google Scholar] [CrossRef]
- Locatelli, M.; Epifano, F.; Genovese, S.; Carlucci, G.; Koncić, M.Z.; Kosalec, I.; Kremer, D. Anthraquinone profile, antioxidant and antimicrobial properties of bark extracts of Rhamnus catharticus and R. orbiculatus. Nat. Prod. Commun. 2011, 6, 1275–1280. [Google Scholar] [CrossRef]
- Hu, B.; Zhang, H.; Meng, X.; Wang, F.; Wang, P. Aloe-emodin from rhubarb (Rheum rhabarbarum) inhibits lipopolysaccharide-induced inflammatory responses in RAW264.7 macrophages. J. Ethnopharmacol. 2014, 153, 846–853. [Google Scholar] [CrossRef]
- Sanders, B.; Ray, A.M.; Goldberg, S.; Clark, T.; McDaniel, H.R.; Atlas, S.E.; Farooqi, A.; Konefal, J.; Lages, L.C.; Lopez, J.; et al. Anti-cancer effects of aloe-emodin: A systematic review. J. Clin. Transl. Res. 2018, 3, 283–296. [Google Scholar]
- Sun, W.; Wang, Z.; Sun, M.; Huang, W.; Wang, Y.; Wang, Y. Aloin antagonizes stimulated ischemia/reperfusion-induced damage and inflammatory response in cardiomyocytes by activating the Nrf2/HO-1 defense pathway. Cell Tissue Res. 2021, 384, 735–744. [Google Scholar] [CrossRef]
- Paudel, P.; Jung, H.A.; Choi, J.S. Anthraquinone and naphthopyrone glycosides from Cassia obtusifolia seeds mediate hepatoprotection via Nrf2-mediated HO-1 activation and MAPK modulation. Arch. Pharm. Res. 2018, 41, 677–689. [Google Scholar] [CrossRef]
- Su, C.; Liu, Z.; Wang, Y.; Wang, Y.; Song, E.; Song, Y. The electrophilic character of quinones is essential for the suppression of Bach1. Toxicology 2017, 387, 17–26. [Google Scholar] [CrossRef]
- Gaonkar, S.L.; Vignesh, U.N. Synthesis and pharmacological properties of chalcones: A review. Res. Chem. Intermed. 2017, 43, 6043–6077. [Google Scholar] [CrossRef]
- Karthikeyan, C.; Moorthy, N.S.; Ramasamy, S.; Vanam, U.; Manivannan, E.; Karunagaran, D.; Trivedi, P. Advances in chalcones with anticancer activities. Recent Pat. Anticancer Drug Discov. 2015, 10, 97–115. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Anand, A.; Kumar, V. Recent developments in biological activities of chalcones: A mini review. Eur. J. Med. Chem. 2014, 85, 758–777. [Google Scholar] [CrossRef] [PubMed]
- Sahu, N.K.; Balbhadra, S.S.; Choudhary, J.; Kohli, D.V. Exploring pharmacological significance of chalcone scaffold: A review. Curr. Med. Chem. 2012, 19, 209–225. [Google Scholar] [CrossRef]
- Batovska, D.I.; Todorova, I.T. Trends in utilization of the pharmacological potential of chalcones. Curr. Clin. Pharmacol. 2010, 5, 1–29. [Google Scholar] [CrossRef] [PubMed]
- López, S.N.; Castelli, M.V.; Zacchino, S.A.; Domínguez, J.N.; Lobo, G.; Charris-Charris, J.; Cortés, J.C.; Ribas, J.C.; Devia, C.; Rodríguez, A.M.; et al. In vitro antifungal evaluation and structure-activity relationships of a new series of chalcone derivatives and synthetic analogues, with inhibitory properties against polymers of the fungal cell wall. Bioorg. Med. Chem. 2001, 9, 1999–2013. [Google Scholar] [CrossRef] [PubMed]
- Rastelli, G.; Antolini, L.; Benvenuti, S.; Costantino, L. Structural bases for the inhibition of aldose reductase by phenolic compounds. Bioorg. Med. Chem. 2000, 8, 1151–1158. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-J.; Li, L.-Y.; Wang, S.-S.; Que, S.; Yang, W.-Z.; Zhang, F.-Y.; Gong, N.-B.; Cheng, W.; Liang, H.; Ye, M.; et al. Oxyfadichalcones A–C: Three chalcone dimers fused through a cyclobutane ring from Tibetan medicine Oxytropis falcata Bunge. Tetrahedron 2013, 69, 11074–11079. [Google Scholar] [CrossRef]
- Raj, L.; Ide, T.; Gurkar, A.U.; Foley, M.; Schenone, M.; Li, X.; Tolliday, N.J.; Golub, T.R.; Carr, S.A.; Shamji, A.F.; et al. Selective killing of cancer cells by a small molecule targeting the stress response to ROS. Nature 2011, 475, 231–234. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Curtis-Long, M.J.; Lee, B.W.; Yuk, H.J.; Kim, D.W.; Tan, X.F.; Park, K.H. Inhibition of tyrosinase activity by polyphenol compounds from Flemingia philippinensis roots. Bioorg. Med. Chem. 2014, 22, 1115–1120. [Google Scholar] [CrossRef] [PubMed]
- Mdee, L.K.; Yeboah, S.O.; Abegaz, B.M. Rhuschalcones II-VI, five new bichalcones from the root bark of Rhus pyroides. J. Nat. Prod. 2003, 66, 599–604. [Google Scholar] [CrossRef] [PubMed]
- Pereira, R.; Silva, A.M.S.; Ribeiro, D.; Silva, V.L.M.; Fernandes, E. Bis-chalcones: A review of synthetic methodologies and anti-inflammatory effects. Eur. J. Med. Chem. 2023, 252, 115280. [Google Scholar] [CrossRef]
- Ni, L.; Meng, C.Q.; Sikorski, J.A. Recent advances in therapeutic chalcones. Expert Opin. Ther. Pat. 2004, 14, 1669–1691. [Google Scholar] [CrossRef]
- Zhou, B.; Xing, C. Diverse Molecular Targets for Chalcones with Varied Bioactivities. Med. Chem. 2015, 5, 388–404. [Google Scholar] [CrossRef]
- Egbujor, M.C.; Buttari, B.; Profumo, E.; Telkoparan-Akillilar, P.; Saso, L. An Overview of NRF2-Activating Compounds Bearing α,β-Unsaturated Moiety and Their Antioxidant Effects. Int. J. Mol. Sci. 2022, 23, 8466. [Google Scholar] [CrossRef]
- Sharifi-Rad, J.; Rayess, Y.E.; Rizk, A.A.; Sadaka, C.; Zgheib, R.; Zam, W.; Sestito, S.; Rapposelli, S.; Neffe-Skocińska, K.; Zielińska, D.; et al. Turmeric and Its Major Compound Curcumin on Health: Bioactive Effects and Safety Profiles for Food, Pharmaceutical, Biotechnological and Medicinal Applications. Front. Pharmacol. 2020, 11, 01021. [Google Scholar] [CrossRef]
- Fadus, M.C.; Lau, C.; Bikhchandani, J.; Lynch, H.T. Curcumin: An age-old anti-inflammatory and anti-neoplastic agent. J. Tradit. Complement. Med. 2017, 7, 339–346. [Google Scholar] [CrossRef]
- Benameur, T.; Soleti, R.; Panaro, M.A.; La Torre, M.E.; Monda, V.; Messina, G.; Porro, C. Curcumin as Prospective Anti-Aging Natural Compound: Focus on Brain. Molecules 2021, 26, 4794. [Google Scholar] [CrossRef]
- Nebrisi, E.E. Neuroprotective Activities of Curcumin in Parkinson’s Disease: A Review of the Literature. Int. J. Mol. Sci. 2021, 22, 1248. [Google Scholar] [CrossRef]
- Peng, Y.; Ao, M.; Dong, B.; Jiang, Y.; Yu, L.; Chen, Z.; Hu, C.; Xu, R. Anti-Inflammatory Effects of Curcumin in the Inflammatory Diseases: Status, Limitations and Countermeasures. Drug Des. Devel Ther. 2021, 15, 4503–4525. [Google Scholar] [CrossRef]
- Mansouri, K.; Rasoulpoor, S.; Daneshkhah, A.; Abolfathi, S.; Salari, N.; Mohammadi, M.; Rasoulpoor, S.; Shabani, S. Clinical effects of curcumin in enhancing cancer therapy: A systematic review. BMC Cancer 2020, 20, 791. [Google Scholar] [CrossRef]
- Zoi, V.; Galani, V.; Tsekeris, P.; Kyritsis, A.P.; Alexiou, G.A. Radiosensitization and Radioprotection by Curcumin in Glioblastoma and Other Cancers. Biomedicines 2022, 10, 312. [Google Scholar] [CrossRef]
- Kou, H.; Huang, L.; Jin, M.; He, Q.; Zhang, R.; Ma, J. Effect of curcumin on rheumatoid arthritis: A systematic review and meta-analysis. Front. Immunol. 2023, 14, 1121655. [Google Scholar] [CrossRef]
- Shin, J.W.; Chun, K.S.; Kim, D.H.; Kim, S.J.; Kim, S.H.; Cho, N.C.; Na, H.K.; Surh, Y.J. Curcumin induces stabilization of Nrf2 protein through Keap1 cysteine modification. Biochem. Pharmacol. 2020, 173, 113820. [Google Scholar] [CrossRef]
- Cao, D.; Liu, Z.; Verwilst, P.; Koo, S.; Jangjili, P.; Kim, J.S.; Lin, W. Coumarin-Based Small-Molecule Fluorescent Chemosensors. Chem. Rev. 2019, 119, 10403–10519. [Google Scholar] [CrossRef]
- Küpeli Akkol, E.; Genç, Y.; Karpuz, B.; Sobarzo-Sánchez, E.; Capasso, R. Coumarins and Coumarin-Related Compounds in Pharmacotherapy of Cancer. Cancers 2020, 12, 1959. [Google Scholar] [CrossRef]
- Dennis, R.A.M.; Priscilla, F.; Jennifer, P.; Meryll, D. Phenolic Compounds and Antioxidant Activities of Eight Species of Fabaceae That Are Commonly Used in Traditional Medical Practices in the Republic of Suriname. In Medicinal Plants; Sanjeet, K., Ed.; IntechOpen: Rijeka, Croatia, 2022; Chapter 15. [Google Scholar]
- Al-Khayri, J.M.; Rashmi, R.; Toppo, V.; Chole, P.B.; Banadka, A.; Sudheer, W.N.; Nagella, P.; Shehata, W.F.; Al-Mssallem, M.Q.; Alessa, F.M.; et al. Plant Secondary Metabolites: The Weapons for Biotic Stress Management. Metabolites 2023, 13, 716. [Google Scholar] [CrossRef]
- Hassanein, E.H.M.; Sayed, A.M.; Hussein, O.E.; Mahmoud, A.M. Coumarins as Modulators of the Keap1/Nrf2/ARE Signaling Pathway. Oxid. Med. Cell Longev. 2020, 2020, 1675957. [Google Scholar] [CrossRef] [PubMed]
- Al-Warhi, T.; Sabt, A.; Elkaeed, E.B.; Eldehna, W.M. Recent advancements of coumarin-based anticancer agents: An up-to-date review. Bioorg. Chem. 2020, 103, 104163. [Google Scholar] [CrossRef]
- Garg, S.S.; Gupta, J.; Sharma, S.; Sahu, D. An insight into the therapeutic applications of coumarin compounds and their mechanisms of action. Eur. J. Pharm. Sci. 2020, 152, 105424. [Google Scholar] [CrossRef] [PubMed]
- Jameel, E.; Umar, T.; Kumar, J.; Hoda, N. Coumarin: A Privileged Scaffold for the Design and Development of Antineurodegenerative Agents. Chem. Biol. Drug Des. 2016, 87, 21–38. [Google Scholar] [CrossRef]
- Supuran, C.T. Coumarin carbonic anhydrase inhibitors from natural sources. J. Enzym. Inhib. Med. Chem. 2020, 35, 1462–1470. [Google Scholar] [CrossRef]
- Carneiro, A.; Matos, M.J.; Uriarte, E.; Santana, L. Trending Topics on Coumarin and Its Derivatives in 2020. Molecules 2021, 26, 501. [Google Scholar] [CrossRef]
- Zhang, L.; Xu, Z. Coumarin-containing hybrids and their anticancer activities. Eur. J. Med. Chem. 2019, 181, 111587. [Google Scholar] [CrossRef] [PubMed]
- Reddy, D.S.; Kongot, M.; Kumar, A. Coumarin hybrid derivatives as promising leads to treat tuberculosis: Recent developments and critical aspects of structural design to exhibit anti-tubercular activity. Tuberculosis 2021, 127, 102050. [Google Scholar] [CrossRef]
- Xu, Z.; Chen, Q.; Zhang, Y.; Liang, C. Coumarin-based derivatives with potential anti-HIV activity. Fitoterapia 2021, 150, 104863. [Google Scholar] [CrossRef]
- Feng, D.; Zhang, A.; Yang, Y.; Yang, P. Coumarin-containing hybrids and their antibacterial activities. Arch. Pharm. 2020, 353, e1900380. [Google Scholar] [CrossRef]
- Song, X.F.; Fan, J.; Liu, L.; Liu, X.F.; Gao, F. Coumarin derivatives with anticancer activities: An update. Arch. Pharm. 2020, 353, e2000025. [Google Scholar] [CrossRef]
- Annunziata, F.; Pinna, C.; Dallavalle, S.; Tamborini, L.; Pinto, A. An Overview of Coumarin as a Versatile and Readily Accessible Scaffold with Broad-Ranging Biological Activities. Int. J. Mol. Sci. 2020, 21, 4618. [Google Scholar] [CrossRef] [PubMed]
- Harborne, J.B. The Natural Coumarins: Occurrence, Chemistry and Biochemistry (Book). Plant Cell Environ. 1982, 5, 435–436. [Google Scholar] [CrossRef]
- Di Stasi, L.C. Natural Coumarin Derivatives Activating Nrf2 Signaling Pathway as Lead Compounds for the Design and Synthesis of Intestinal Anti-Inflammatory Drugs. Pharmaceuticals 2023, 16, 511. [Google Scholar] [CrossRef] [PubMed]
- Dinkova-Kostova, A.T.; Liby, K.T.; Stephenson, K.K.; Holtzclaw, W.D.; Gao, X.; Suh, N.; Williams, C.; Risingsong, R.; Honda, T.; Gribble, G.W.; et al. Extremely potent triterpenoid inducers of the phase 2 response: Correlations of protection against oxidant and inflammatory stress. Proc. Natl. Acad. Sci. USA 2005, 102, 4584–4589. [Google Scholar] [CrossRef]
- Djedjibegovic, J.; Marjanovic, A.; Panieri, E.; Saso, L. Ellagic Acid-Derived Urolithins as Modulators of Oxidative Stress. Oxid. Med. Cell Longev. 2020, 2020, 5194508. [Google Scholar] [CrossRef] [PubMed]
- Bae, G.S.; Kim, D.G.; Jo, I.J.; Choi, S.B.; Kim, M.J.; Shin, J.Y.; Kim, D.U.; Song, H.J.; Joo, M.; Park, S.J. Heme oxygenase-1 induced by desoxo-narchinol-A attenuated the severity of acute pancreatitis via blockade of neutrophil infiltration. Int. Immunopharmacol. 2019, 69, 225–234. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.W.; Yoon, C.S.; Kim, Y.C.; Oh, H. Desoxo-narchinol A and Narchinol B Isolated from Nardostachys jatamansi Exert Anti-neuroinflammatory Effects by Up-regulating of Nuclear Transcription Factor Erythroid-2-Related Factor 2/Heme Oxygenase-1 Signaling. Neurotox. Res. 2019, 35, 230–243. [Google Scholar] [CrossRef]
- Tsoyi, K.; Jang, H.J.; Lee, Y.S.; Kim, Y.M.; Kim, H.J.; Seo, H.G.; Lee, J.H.; Kwak, J.H.; Lee, D.U.; Chang, K.C. (+)-Nootkatone and (+)-valencene from rhizomes of Cyperus rotundus increase survival rates in septic mice due to heme oxygenase-1 induction. J. Ethnopharmacol. 2011, 137, 1311–1317. [Google Scholar] [CrossRef]
- Kim, N.; Hwangbo, C.; Lee, S.; Lee, J.H. Eupatolide inhibits PDGF-induced proliferation and migration of aortic smooth muscle cells through ROS-dependent heme oxygenase-1 induction. Phytother. Res. 2013, 27, 1700–1707. [Google Scholar] [CrossRef]
- Prasannan, R.; Kalesh, K.A.; Shanmugam, M.K.; Nachiyappan, A.; Ramachandran, L.; Nguyen, A.H.; Kumar, A.P.; Lakshmanan, M.; Ahn, K.S.; Sethi, G. Key cell signaling pathways modulated by zerumbone: Role in the prevention and treatment of cancer. Biochem. Pharmacol. 2012, 84, 1268–1276. [Google Scholar] [CrossRef] [PubMed]
- Leung, W.S.; Yang, M.L.; Lee, S.S.; Kuo, C.W.; Ho, Y.C.; Huang-Liu, R.; Lin, H.W.; Kuan, Y.H. Protective effect of zerumbone reduces lipopolysaccharide-induced acute lung injury via antioxidative enzymes and Nrf2/HO-1 pathway. Int. Immunopharmacol. 2017, 46, 194–200. [Google Scholar] [CrossRef] [PubMed]
- Ha, D.T.; Phuong, T.T.; Oh, J.; Bae, K.; Thuan, N.D.; Na, M. Palbinone from Paeonia suffruticosa protects hepatic cells via up-regulation of heme oxygenase-1. Phytother. Res. 2014, 28, 308–311. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, J.; Jia, W.; Zhao, A.; Li, T. Distinct immunosuppressive effect by Isodon serra extracts. Int. Immunopharmacol. 2005, 5, 1957–1965. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.K.; Sangwan, N.S.; Sangwan, R.S. Phcog rev.: Plant review Andrographis paniculata (Kalmegh): A review. Pharmacogn. Rev. 2007, 1, 283–298. [Google Scholar]
- Kim, Y.M.; Kim, H.J.; Chang, K.C. Glycyrrhizin reduces HMGB1 secretion in lipopolysaccharide-activated RAW 264.7 cells and endotoxemic mice by p38/Nrf2-dependent induction of HO-1. Int. Immunopharmacol. 2015, 26, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Mou, K.; Pan, W.; Han, D.; Wen, X.; Cao, F.; Miao, Y.; Li, P. Glycyrrhizin protects human melanocytes from H2O2-induced oxidative damage via the Nrf2-dependent induction of HO-1. Int. J. Mol. Med. 2019, 44, 253–261. [Google Scholar] [CrossRef] [PubMed]
- Heyninck, K.; Sabbe, L.; Chirumamilla, C.S.; Szarc Vel Szic, K.; Vander Veken, P.; Lemmens, K.J.A.; Lahtela-Kakkonen, M.; Naulaerts, S.; Op de Beeck, K.; Laukens, K.; et al. Withaferin A induces heme oxygenase (HO-1) expression in endothelial cells via activation of the Keap1/Nrf2 pathway. Biochem. Pharmacol. 2016, 109, 48–61. [Google Scholar] [CrossRef]
- Chew, L.Y.; Zhang, H.; He, J.; Yu, F. The Nrf2-Keap1 pathway is activated by steroid hormone signaling to govern neuronal remodeling. Cell Rep. 2021, 36, 109466. [Google Scholar] [CrossRef] [PubMed]
- Freeman, B.A.; Baker, P.R.; Schopfer, F.J.; Woodcock, S.R.; Napolitano, A.; d’Ischia, M. Nitro-fatty acid formation and signaling. J. Biol. Chem. 2008, 283, 15515–15519. [Google Scholar] [CrossRef] [PubMed]
- Piesche, M.; Roos, J.; Kühn, B.; Fettel, J.; Hellmuth, N.; Brat, C.; Maucher, I.V.; Awad, O.; Matrone, C.; Comerma Steffensen, S.G.; et al. The Emerging Therapeutic Potential of Nitro Fatty Acids and Other Michael Acceptor-Containing Drugs for the Treatment of Inflammation and Cancer. Front. Pharmacol. 2020, 11, 1297. [Google Scholar] [CrossRef] [PubMed]
- Kansanen, E.; Jyrkkänen, H.-K.; Volger, O.L.; Leinonen, H.; Kivelä, A.M.; Häkkinen, S.-K.; Woodcock, S.R.; Schopfer, F.J.; Horrevoets, A.J.; Ylä-Herttuala, S. Nrf2-dependent and-independent responses to nitro-fatty acids in human endothelial cells. J. Biol. Chem. 2009, 284, 33233–33241. [Google Scholar] [CrossRef] [PubMed]
- Schopfer, F.J.; Vitturi, D.A.; Jorkasky, D.K.; Freeman, B.A. Nitro-fatty acids: New drug candidates for chronic inflammatory and fibrotic diseases. Nitric Oxide 2018, 79, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Rao, P.V.; Gan, S.H. Cinnamon: A multifaceted medicinal plant. Evid. Based Complement. Altern. Med. 2014, 2014, 642942. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.C.; Chung, Y.L.; Wu, M.L.; Chuang, S.M. Cinnamaldehyde enhances Nrf2 nuclear translocation to upregulate phase II detoxifying enzyme expression in HepG2 cells. J. Agric. Food Chem. 2011, 59, 5164–5171. [Google Scholar] [CrossRef]
- O’Brien, J.; Wendell, S.G. Electrophile Modulation of Inflammation: A Two-Hit Approach. Metabolites 2020, 10, 453. [Google Scholar] [CrossRef]
- Gai, C.; Harnor, S.J.; Zhang, S.; Cano, C.; Zhuang, C.; Zhao, Q. Advanced approaches of developing targeted covalent drugs. RSC Med. Chem. 2022, 13, 1460–1475. [Google Scholar] [CrossRef]
- Mons, E.; Jansen, I.D.C.; Loboda, J.; van Doodewaerd, B.R.; Hermans, J.; Verdoes, M.; van Boeckel, C.A.A.; van Veelen, P.A.; Turk, B.; Turk, D.; et al. The Alkyne Moiety as a Latent Electrophile in Irreversible Covalent Small Molecule Inhibitors of Cathepsin K. J. Am. Chem. Soc. 2019, 141, 3507–3514. [Google Scholar] [CrossRef]
- Ghosh, A.K.; Samanta, I.; Mondal, A.; Liu, W.R. Covalent Inhibition in Drug Discovery. ChemMedChem 2019, 14, 889–906. [Google Scholar] [CrossRef]
- Roth, G.J.; Stanford, N.; Majerus, P.W. Acetylation of prostaglandin synthase by aspirin. Proc. Natl. Acad. Sci. USA 1975, 72, 3073–3076. [Google Scholar] [CrossRef] [PubMed]
- Boike, L.; Henning, N.J.; Nomura, D.K. Advances in covalent drug discovery. Nat. Rev. Drug Discov. 2022, 21, 881–898. [Google Scholar] [CrossRef] [PubMed]
- Bauer, R.A. Covalent inhibitors in drug discovery: From accidental discoveries to avoided liabilities and designed therapies. Drug Discov. Today 2015, 20, 1061–1073. [Google Scholar] [CrossRef] [PubMed]
- Selvaraju, K.; Mofers, A.; Pellegrini, P.; Salomonsson, J.; Ahlner, A.; Morad, V.; Hillert, E.K.; Espinosa, B.; Arnér, E.S.J.; Jensen, L.; et al. Cytotoxic unsaturated electrophilic compounds commonly target the ubiquitin proteasome system. Sci. Rep. 2019, 9, 9841. [Google Scholar] [CrossRef] [PubMed]
- Reddi, R.N.; Rogel, A.; Gabizon, R.; Rawale, D.G.; Harish, B.; Marom, S.; Tivon, B.; Arbel, Y.S.; Gurwicz, N.; Oren, R.; et al. Sulfamate Acetamides as Self-Immolative Electrophiles for Covalent Ligand-Directed Release Chemistry. J. Am. Chem. Soc. 2023, 145, 3346–3360. [Google Scholar] [CrossRef] [PubMed]
- LoPachin, R.M.; Gavin, T. Reactions of electrophiles with nucleophilic thiolate sites: Relevance to pathophysiological mechanisms and remediation. Free Radic. Res. 2016, 50, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Baron, G.; Altomare, A.; Della Vedova, L.; Gado, F.; Quagliano, O.; Casati, S.; Tosi, N.; Bresciani, L.; Del Rio, D.; Roda, G.; et al. Unraveling the parahormetic mechanism underlying the health-protecting effects of grapeseed procyanidins. Redox Biol. 2024, 69, 102981. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.C.; Wang, Z. Precision Medicine: Disease Subtyping and Tailored Treatment. Cancers 2023, 15, 3837. [Google Scholar] [CrossRef]
- Satoh, T.; McKercher, S.R.; Lipton, S.A. Nrf2/ARE-mediated antioxidant actions of pro-electrophilic drugs. Free Radic. Biol. Med. 2013, 65, 645–657. [Google Scholar] [CrossRef]
- Lv, Y.; Li, W.; Liao, W.; Jiang, H.; Liu, Y.; Cao, J.; Lu, W.; Feng, Y. Nano-Drug Delivery Systems Based on Natural Products. Int. J. Nanomed. 2024, 19, 541–569. [Google Scholar] [CrossRef]
- Repash, E.M.; Pensabene, K.M.; Palenchar, P.M.; Eggler, A.L. Solving the Problem of Assessing Synergy and Antagonism for Non-Traditional Dosing Curve Compounds Using the DE/ZI Method: Application to Nrf2 Activators. Front. Pharmacol. 2021, 12, 686201. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Kar, S. Application of artificial intelligence and machine learning in early detection of adverse drug reactions (ADRs) and drug-induced toxicity. Artif. Intell. Chem. 2023, 1, 100011. [Google Scholar] [CrossRef]
- Singh, S.; Nagalakshmi, D.; Sharma, K.K.; Ravichandiran, V. Natural antioxidants for neuroinflammatory disorders and possible involvement of Nrf2 pathway: A review. Heliyon 2021, 7, e06216. [Google Scholar] [CrossRef] [PubMed]
- Lillich, F.F.; Imig, J.D.; Proschak, E. Multi-Target Approaches in Metabolic Syndrome. Front. Pharmacol. 2020, 11, 554961. [Google Scholar] [CrossRef] [PubMed]
- Lavezzi, A.M.; Ramos-Molina, B. Environmental Exposure Science and Human Health. Int. J. Environ. Res. Public. Health 2023, 20, 5764. [Google Scholar] [CrossRef] [PubMed]
- Stielow, M.; Witczyńska, A.; Kubryń, N.; Fijałkowski, Ł.; Nowaczyk, J.; Nowaczyk, A. The Bioavailability of Drugs-The Current State of Knowledge. Molecules 2023, 28, 8038. [Google Scholar] [CrossRef] [PubMed]
- Bertelli, A.; Biagi, M.; Corsini, M.; Baini, G.; Cappellucci, G.; Miraldi, E. Polyphenols: From Theory to Practice. Foods 2021, 10, 2595. [Google Scholar] [CrossRef]
- Żyżelewicz, D.; Oracz, J. Bioavailability and Bioactivity of Plant Antioxidants. Antioxidants 2022, 11, 2336. [Google Scholar] [CrossRef]
- Di Lorenzo, C.; Colombo, F.; Biella, S.; Stockley, C.; Restani, P. Polyphenols and Human Health: The Role of Bioavailability. Nutrients 2021, 13, 273. [Google Scholar] [CrossRef] [PubMed]
- Almazroo, O.A.; Miah, M.K.; Venkataramanan, R. Drug Metabolism in the Liver. Clin. Liver Dis. 2017, 21, 1–20. [Google Scholar] [CrossRef]
- Zhang, F.; He, F.; Li, L.; Guo, L.; Zhang, B.; Yu, S.; Zhao, W. Bioavailability Based on the Gut Microbiota: A New Perspective. Microbiol. Mol. Biol. Rev. 2020, 84, 2. [Google Scholar] [CrossRef]
- Orlando, P.; Nartea, A.; Silvestri, S.; Marcheggiani, F.; Cirilli, I.; Dludla, P.V.; Fiorini, R.; Pacetti, D.; Loizzo, M.R.; Lucci, P.; et al. Bioavailability Study of Isothiocyanates and Other Bioactive Compounds of Brassica oleracea L. var. Italica Boiled or Steamed: Functional Food or Dietary Supplement? Antioxidants 2022, 11, 209. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.S. Allicin and Other Functional Active Components in Garlic: Health Benefits and Bioavailability. Int. J. Food Prop. 2007, 10, 245–268. [Google Scholar] [CrossRef]
- Lawson, L.D.; Hunsaker, S.M. Allicin Bioavailability and Bioequivalence from Garlic Supplements and Garlic Foods. Nutrients 2018, 10, 812. [Google Scholar] [CrossRef] [PubMed]
- Chhabria, S.; Desai, K. Purification and characterisation of alliinase produced by Cupriavidus necator and its application for generation of cytotoxic agent: Allicin. Saudi J. Biol. Sci. 2018, 25, 1429–1438. [Google Scholar] [CrossRef] [PubMed]
- Scalbert, A.; Williamson, G. Dietary intake and bioavailability of polyphenols. J. Nutr. 2000, 130, 2073s–2085s. [Google Scholar] [CrossRef]
- Padilla-González, G.F.; Grosskopf, E.; Sadgrove, N.J.; Simmonds, M.S.J. Chemical Diversity of Flavan-3-Ols in Grape Seeds: Modulating Factors and Quality Requirements. Plants 2022, 11, 809. [Google Scholar] [CrossRef]
- Rudrapal, M.; Khairnar, S.J.; Khan, J.; Dukhyil, A.B.; Ansari, M.A.; Alomary, M.N.; Alshabrmi, F.M.; Palai, S.; Deb, P.K.; Devi, R. Dietary polyphenols and their role in oxidative stress-induced human diseases: Insights into protective effects, antioxidant potentials and mechanism (s) of action. Front. Pharmacol. 2022, 13, 283. [Google Scholar] [CrossRef]
- Nakamura, Y.; Miyoshi, N. Electrophiles in foods: The current status of isothiocyanates and their chemical biology. Biosci. Biotechnol. Biochem. 2010, 74, 242–255. [Google Scholar] [CrossRef]
- de Araújo, F.F.; de Paulo Farias, D.; Neri-Numa, I.A.; Pastore, G.M. Polyphenols and their applications: An approach in food chemistry and innovation potential. Food Chem. 2021, 338, 127535. [Google Scholar] [CrossRef]
- Fraga, C.G.; Croft, K.D.; Kennedy, D.O.; Tomás-Barberán, F.A. The effects of polyphenols and other bioactives on human health. Food Funct. 2019, 10, 514–528. [Google Scholar] [CrossRef]
- Velderrain-Rodríguez, G.; Palafox-Carlos, H.; Wall-Medrano, A.; Ayala-Zavala, J.; Chen, C.O.; Robles-Sánchez, M.; Astiazaran-García, H.; Alvarez-Parrilla, E.; González-Aguilar, G. Phenolic compounds: Their journey after intake. Food Funct. 2014, 5, 189–197. [Google Scholar] [CrossRef]
- Teng, H.; Chen, L. Polyphenols and bioavailability: An update. Crit. Rev. Food Sci. Nutr. 2019, 59, 2040–2051. [Google Scholar] [CrossRef]
- Williamson, G.; Clifford, M.N. Role of the small intestine, colon and microbiota in determining the metabolic fate of polyphenols. Biochem. Pharmacol. 2017, 139, 24–39. [Google Scholar] [CrossRef]
- Chen, H.; Sang, S. Biotransformation of tea polyphenols by gut microbiota. J. Funct. Foods 2014, 7, 26–42. [Google Scholar] [CrossRef]
- Aravind, S.M.; Wichienchot, S.; Tsao, R.; Ramakrishnan, S.; Chakkaravarthi, S. Role of dietary polyphenols on gut microbiota, their metabolites and health benefits. Food Res. Int. 2021, 142, 110189. [Google Scholar] [CrossRef]
- Aura, A.-M. Microbial metabolism of dietary phenolic compounds in the colon. Phytochem. Rev. 2008, 7, 407–429. [Google Scholar] [CrossRef]
- Bermúdez-Soto, M.-J.; Tomás-Barberán, F.-A.; García-Conesa, M.-T. Stability of polyphenols in chokeberry (Aronia melanocarpa) subjected to in vitro gastric and pancreatic digestion. Food Chem. 2007, 102, 865–874. [Google Scholar] [CrossRef]
- Gabriel, A.; Chinenye, O. Toxicity of Polyphenols Consumed as Food and Nutraceuticals: Remedies through Nanotherapeutic Approaches. In Polyphenols: Food, Nutraceutical, and Nanotherapeutic Applications; Rudrapal, M., Ed.; Wiley: Hoboken, NJ, USA, 2023; pp. 290–311. [Google Scholar]
- Cianciosi, D.; Forbes-Hernández, T.Y.; Regolo, L.; Alvarez-Suarez, J.M.; Navarro-Hortal, M.D.; Xiao, J.; Quiles, J.L.; Battino, M.; Giampieri, F. The reciprocal interaction between polyphenols and other dietary compounds: Impact on bioavailability, antioxidant capacity and other physico-chemical and nutritional parameters. Food Chem. 2022, 375, 131904. [Google Scholar] [CrossRef]
- Dima, C.; Assadpour, E.; Dima, S.; Jafari, S.M. Nutraceutical nanodelivery; an insight into the bioaccessibility/bioavailability of different bioactive compounds loaded within nanocarriers. Crit. Rev. Food Sci. Nutr. 2021, 61, 3031–3065. [Google Scholar] [CrossRef]
- D’Archivio, M.; Filesi, C.; Varì, R.; Scazzocchio, B.; Masella, R. Bioavailability of the polyphenols: Status and controversies. Int. J. Mol. Sci. 2010, 11, 1321–1342. [Google Scholar] [CrossRef]
- Domínguez-Avila, J.A.; Wall-Medrano, A.; Velderrain-Rodríguez, G.R.; Chen, C.-Y.O.; Salazar-López, N.J.; Robles-Sánchez, M.; González-Aguilar, G.A. Gastrointestinal interactions, absorption, splanchnic metabolism and pharmacokinetics of orally ingested phenolic compounds. Food Funct. 2017, 8, 15–38. [Google Scholar] [CrossRef]
- Day, A.J.; Cañada, F.J.; Díaz, J.C.; Kroon, P.A.; Mclauchlan, R.; Faulds, C.B.; Plumb, G.W.; Morgan, M.R.; Williamson, G. Dietary flavonoid and isoflavone glycosides are hydrolysed by the lactase site of lactase phlorizin hydrolase. FEBS Lett. 2000, 468, 166–170. [Google Scholar] [CrossRef]
- Chen, L.; Cao, H.; Xiao, J. Polyphenols: Absorption, bioavailability, and metabolomics. In Polyphenols: Properties, Recovery, and Applications; Elsevier: Amsterdam, The Netherlands, 2018; pp. 45–67. [Google Scholar]
- Majee, C.; Mazumder, R.; Choudhary, A.N. An Insight into the Hepatoprotective Activity and Structure-activity Relationships of Flavonoids. Mini Rev. Med. Chem. 2023, 23, 131–149. [Google Scholar] [CrossRef]
- Rodríguez-Daza, M.C.; Pulido-Mateos, E.C.; Lupien-Meilleur, J.; Guyonnet, D.; Desjardins, Y.; Roy, D. Polyphenol-mediated gut microbiota modulation: Toward prebiotics and further. Front. Nutr. 2021, 8, 689456. [Google Scholar] [CrossRef]
- Sharma, R.; Diwan, B.; Singh, B.P.; Kulshrestha, S. Probiotic fermentation of polyphenols: Potential sources of novel functional foods. Food Prod. Process. Nutr. 2022, 4, 21. [Google Scholar] [CrossRef]
- Rana, A.; Samtiya, M.; Dhewa, T.; Mishra, V.; Aluko, R.E. Health benefits of polyphenols: A concise review. J. Food Biochem. 2022, 46, e14264. [Google Scholar] [CrossRef]
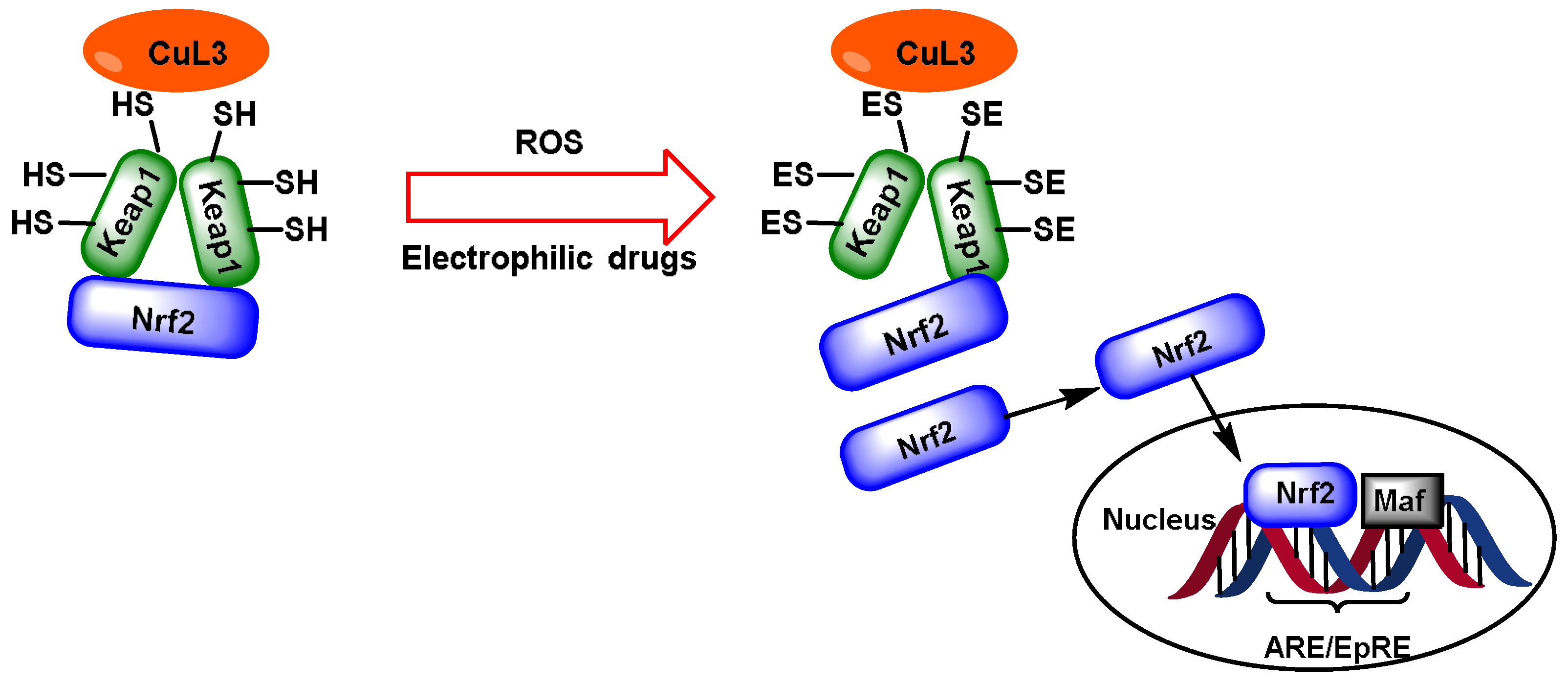
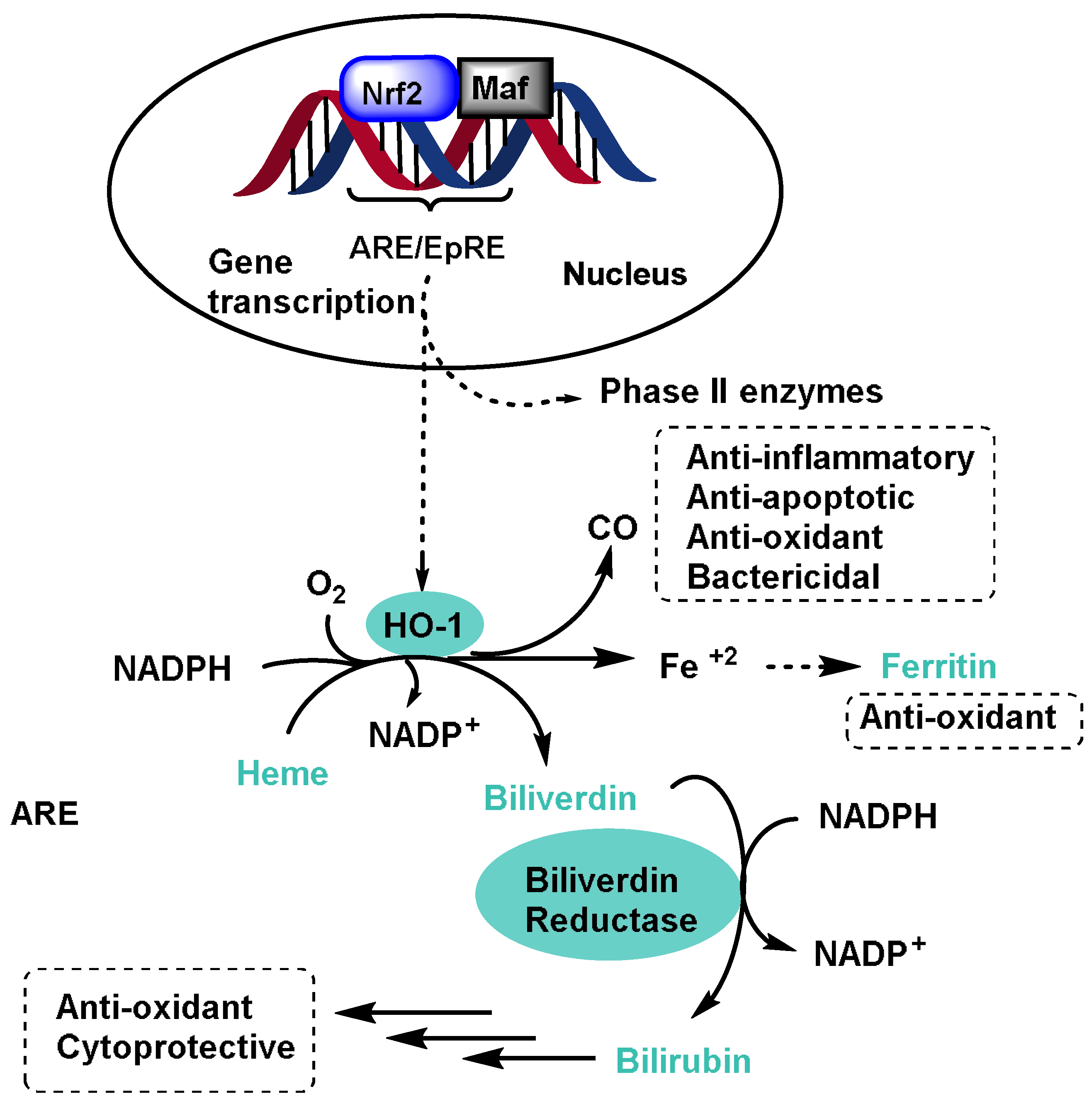
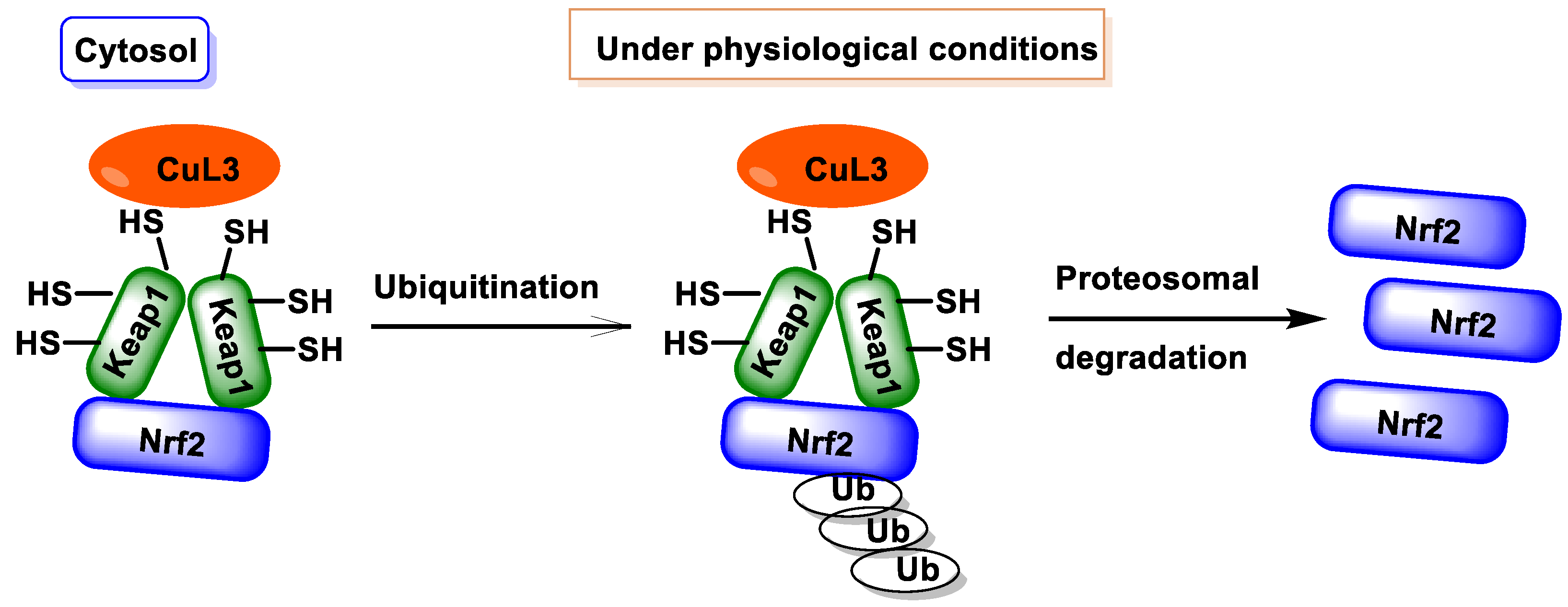
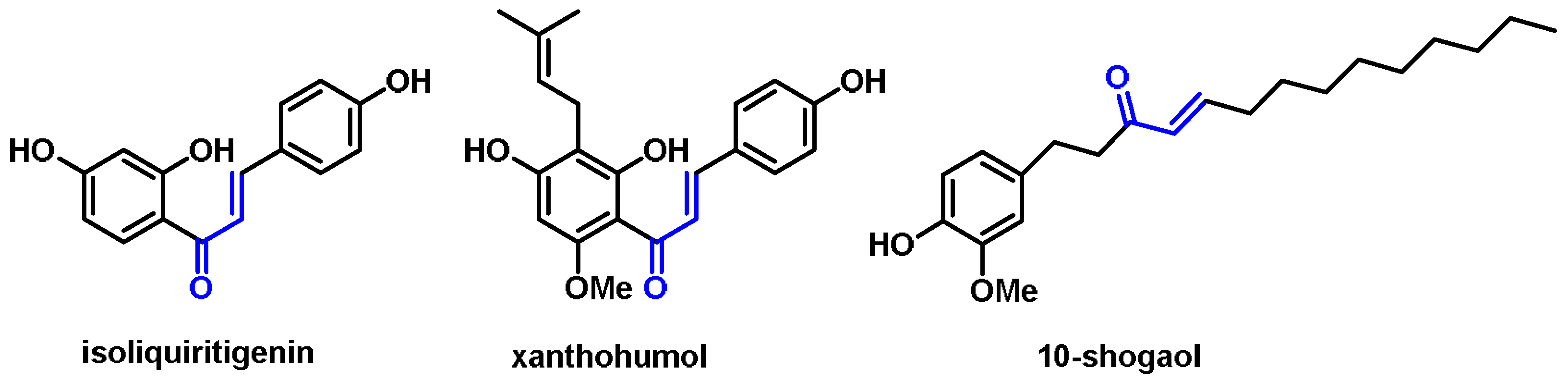

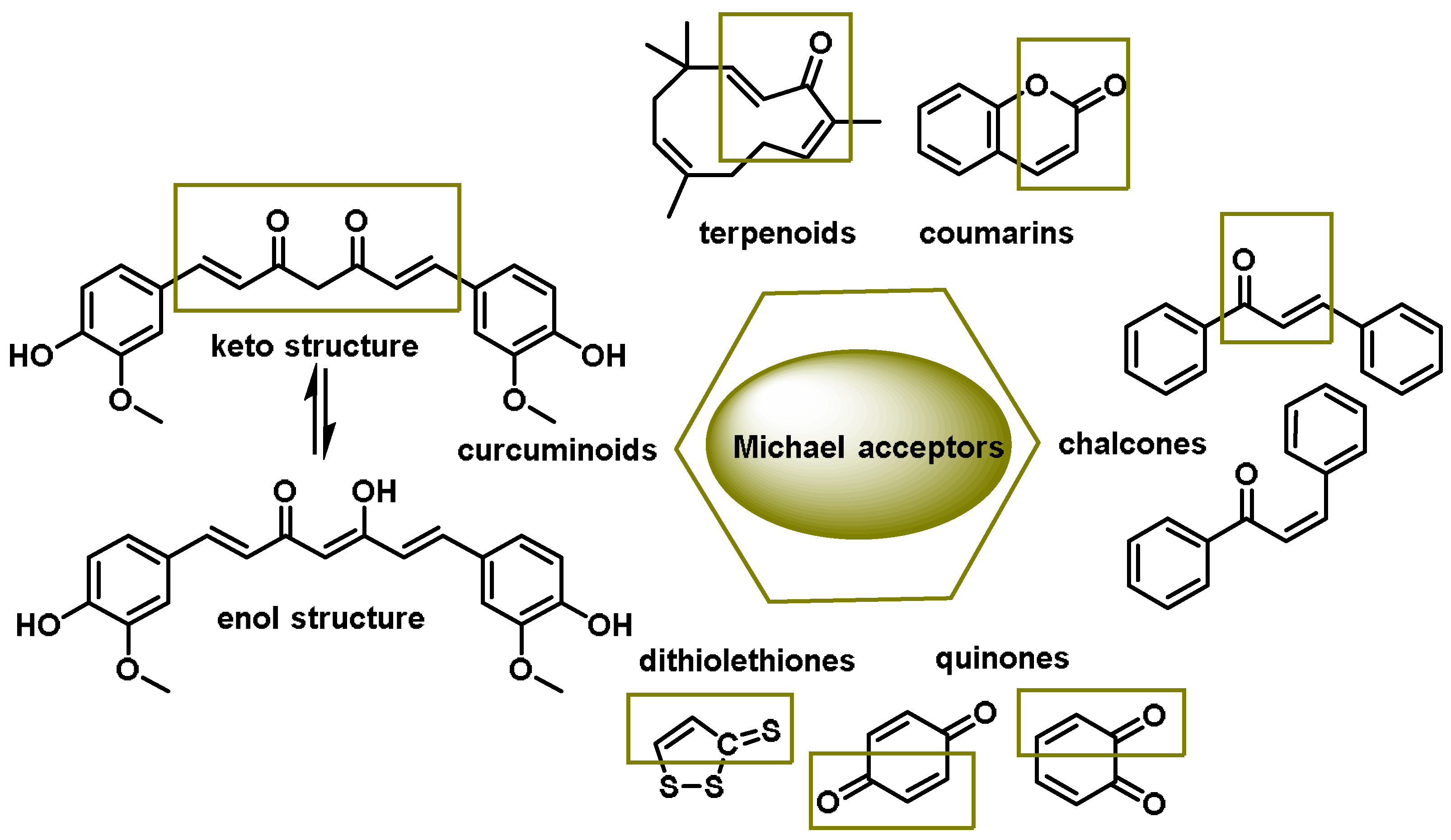
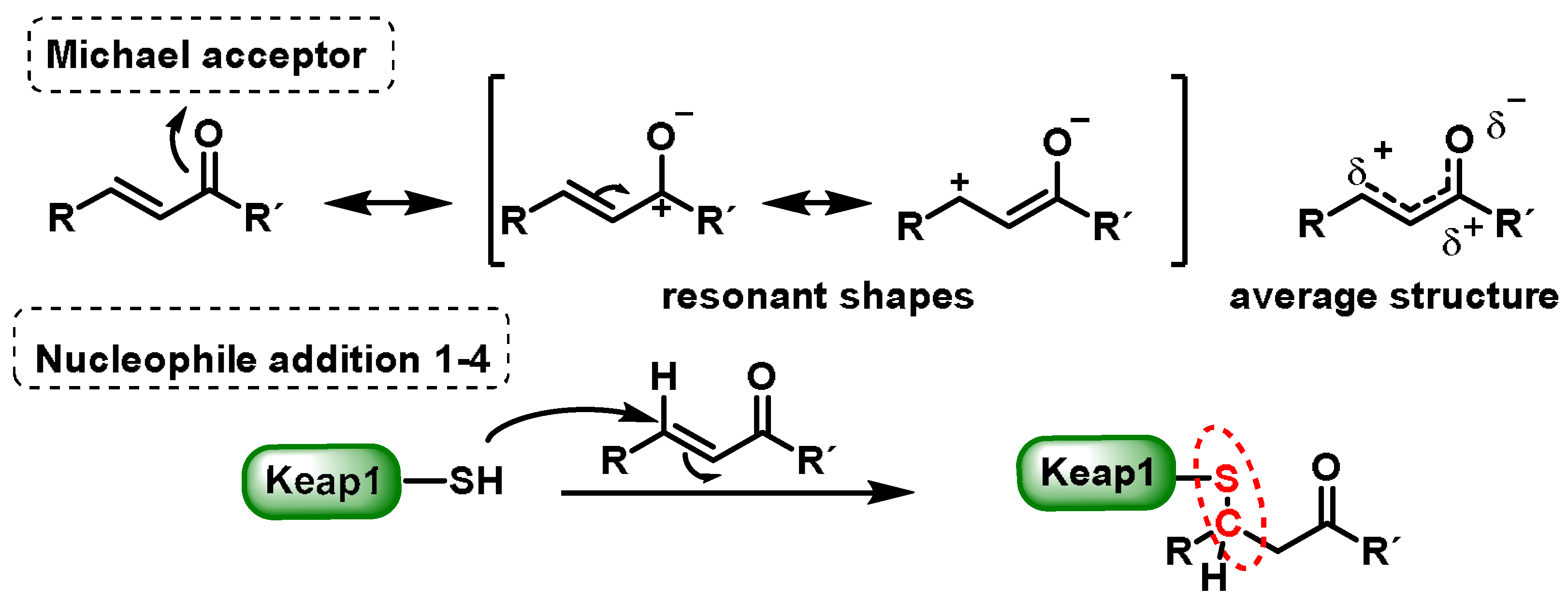


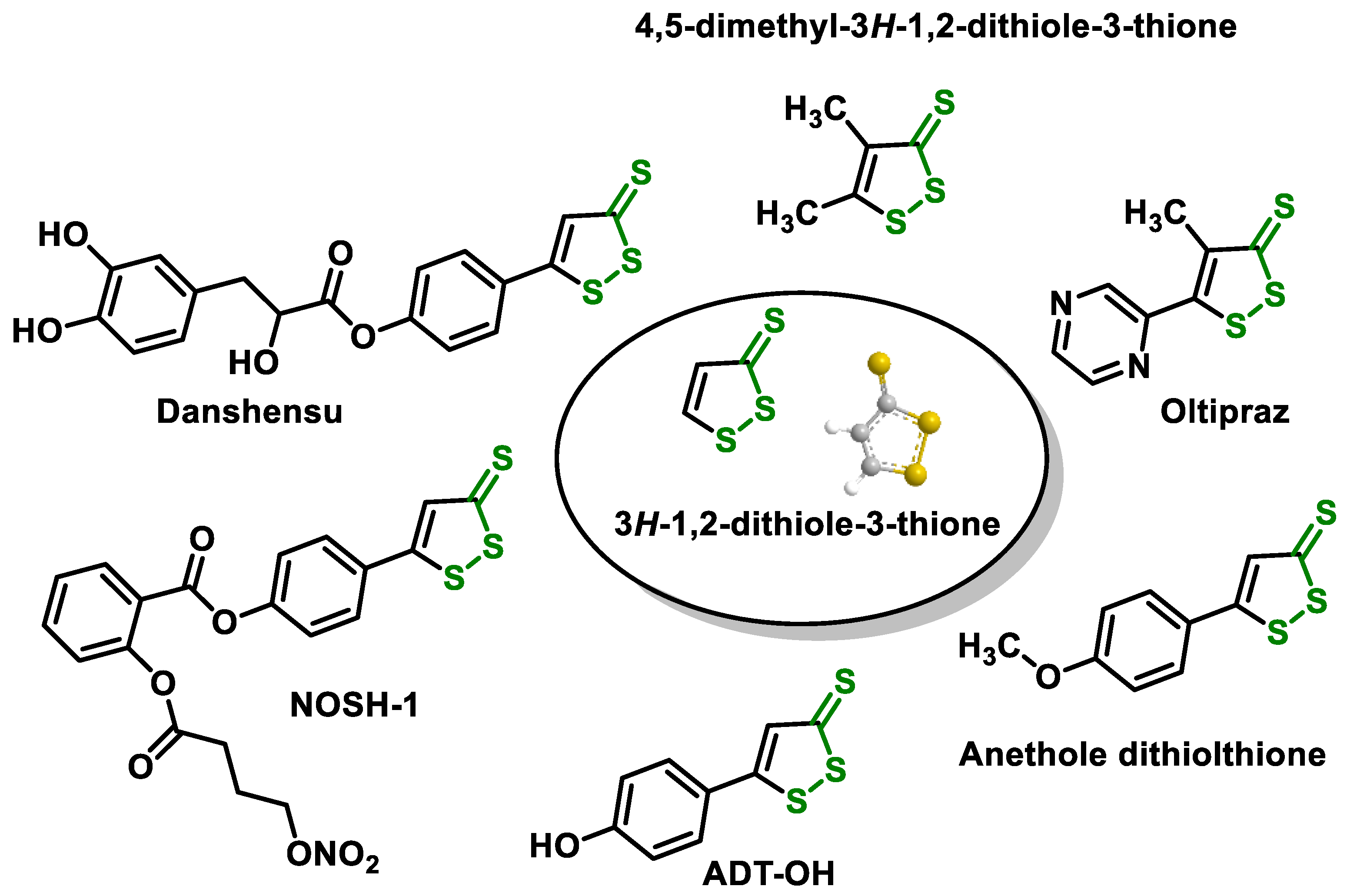
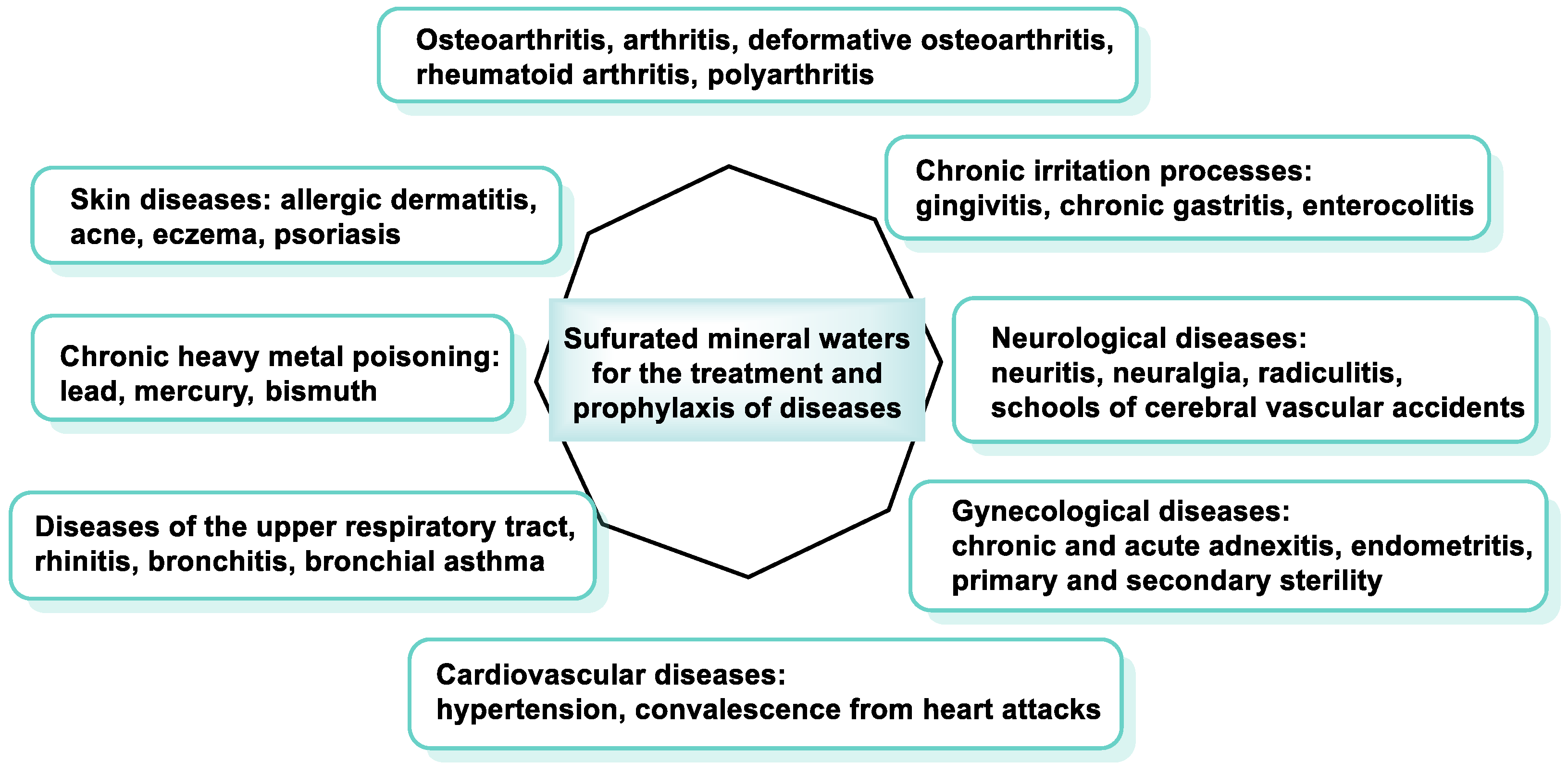
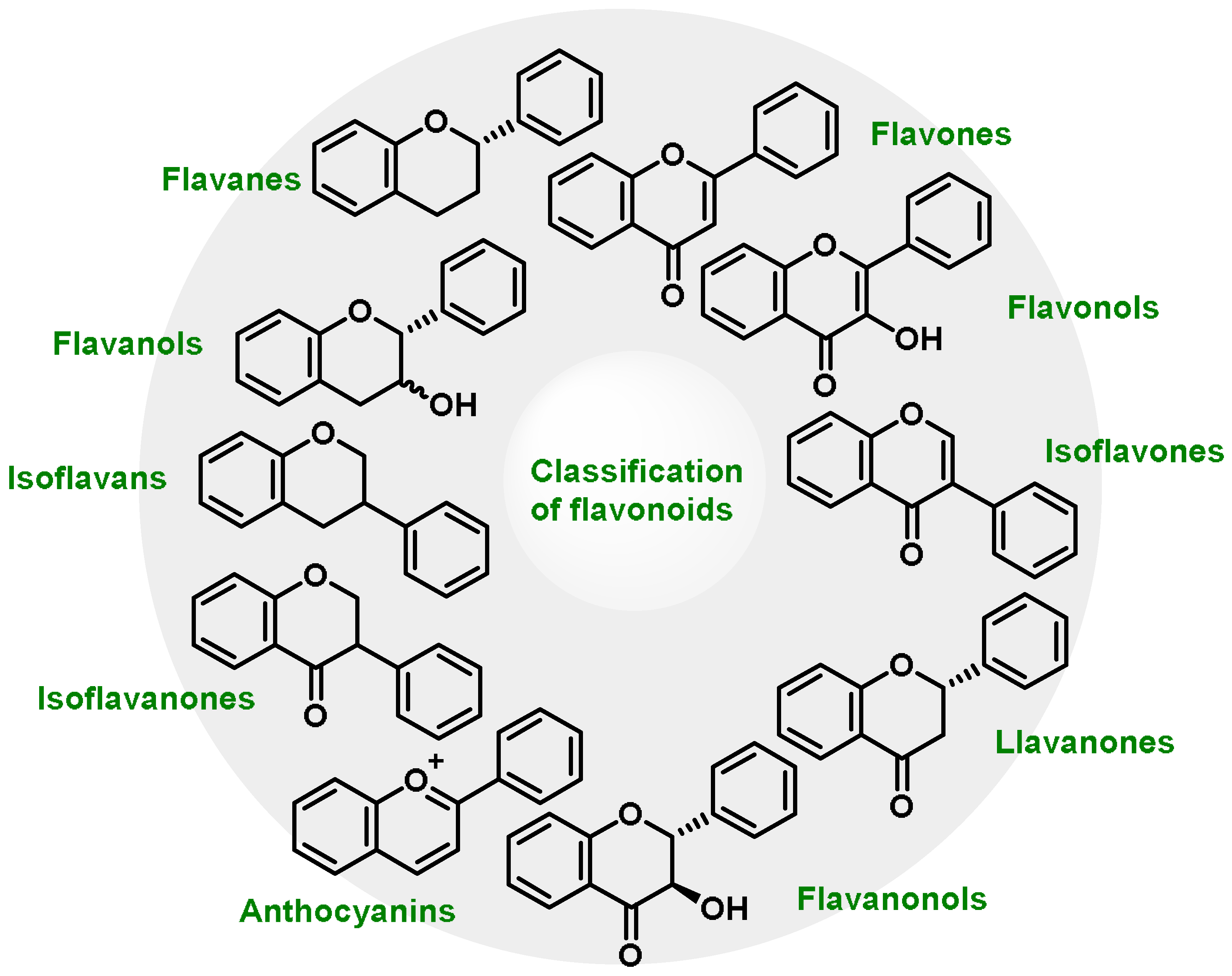
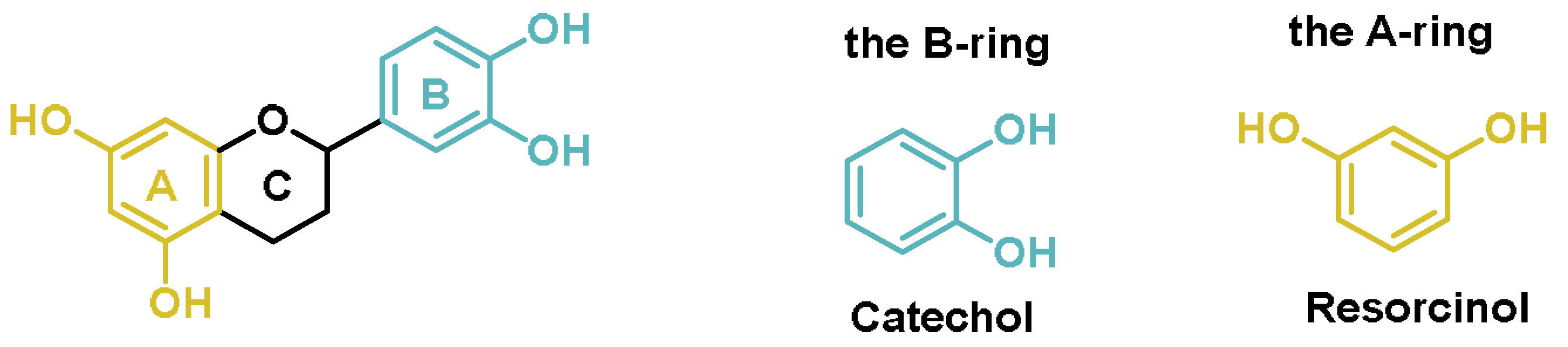
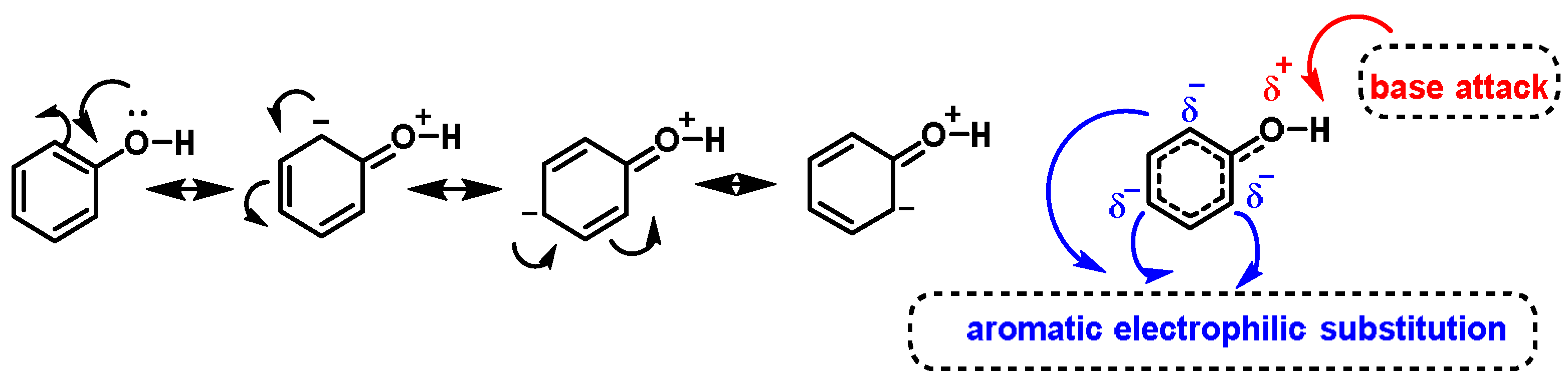
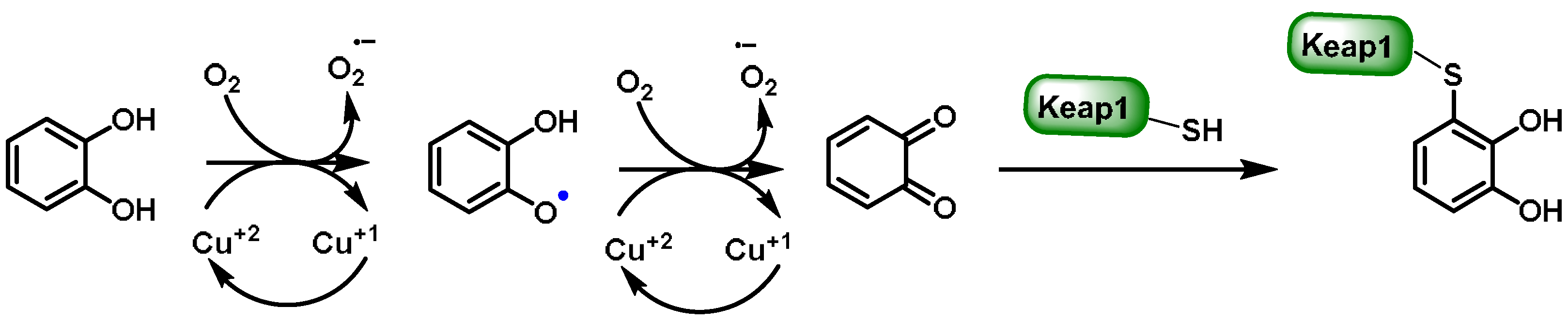
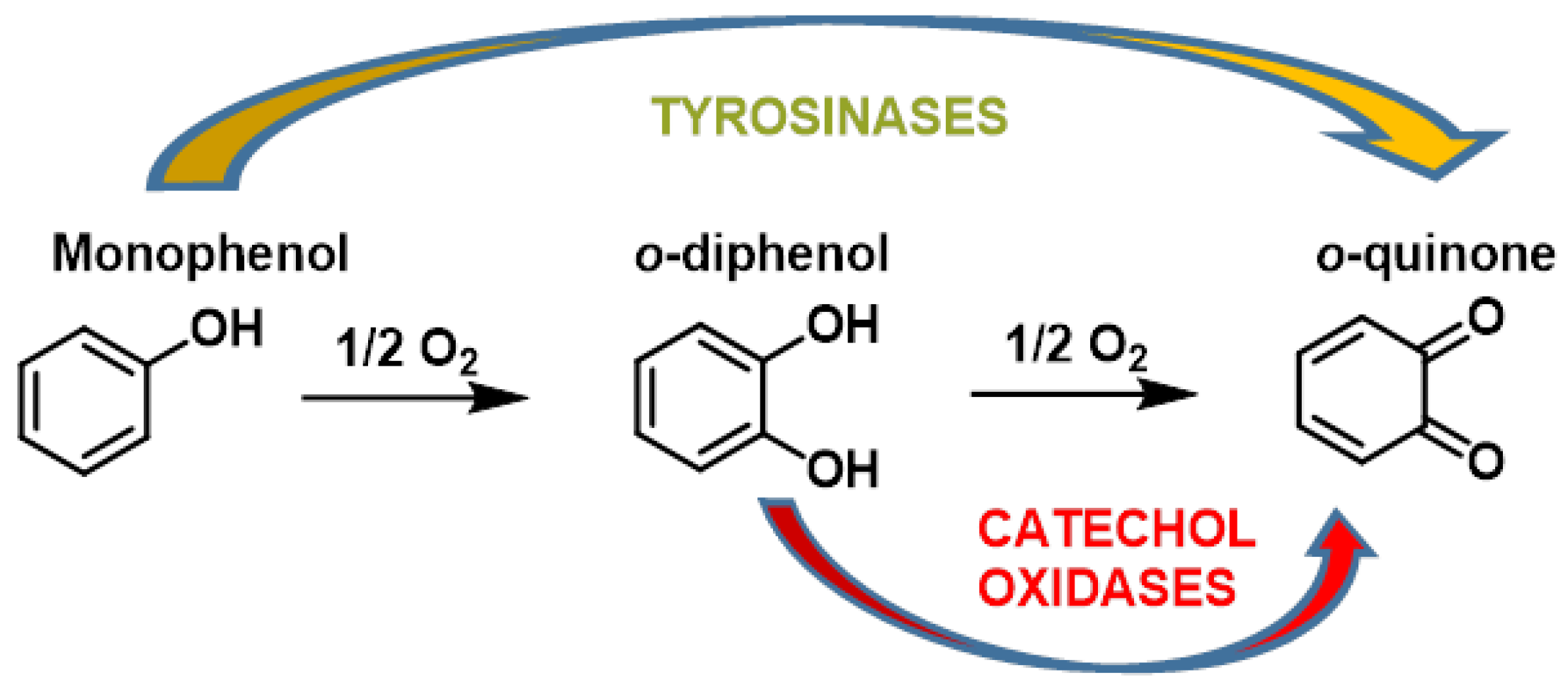
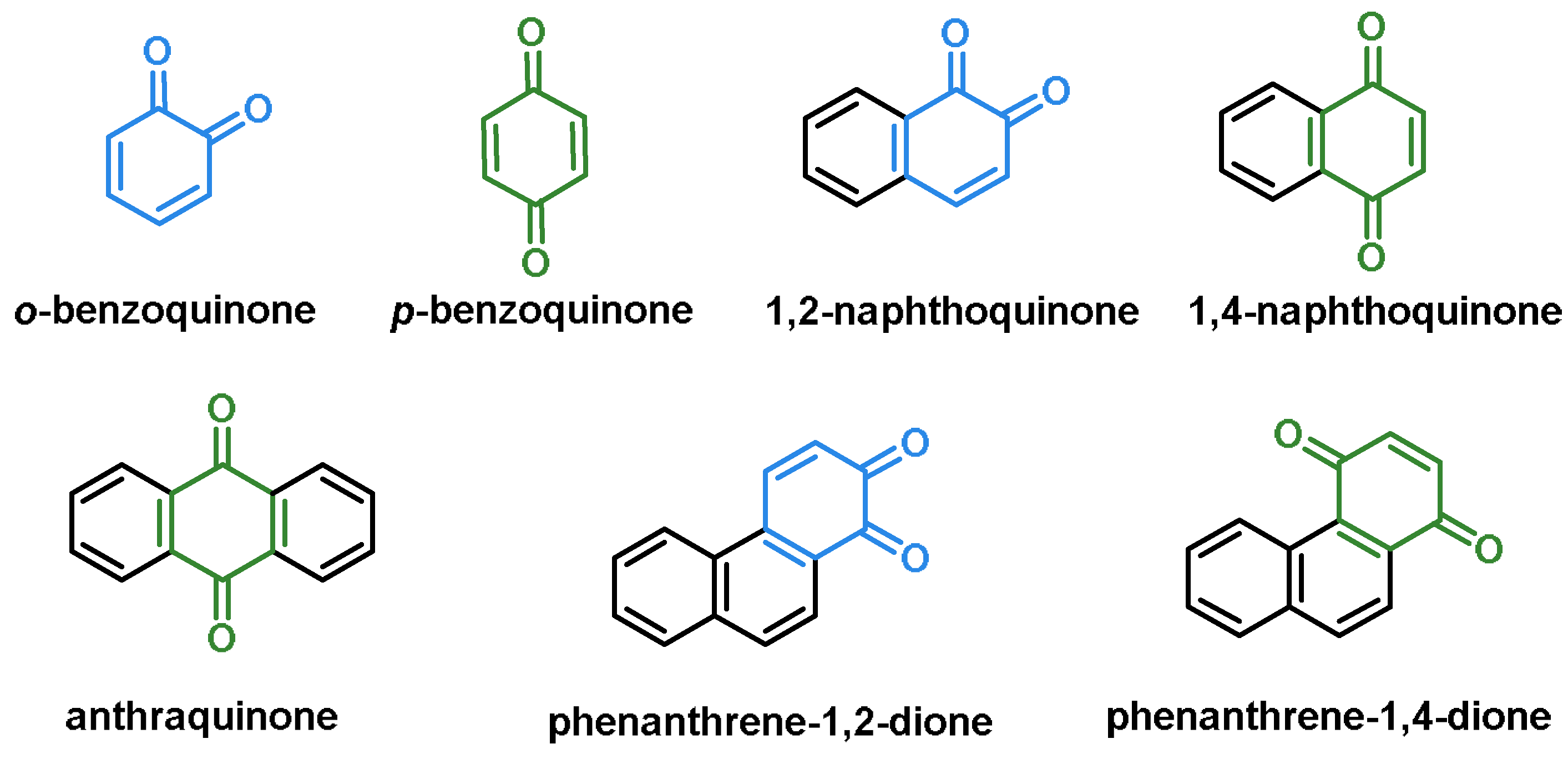

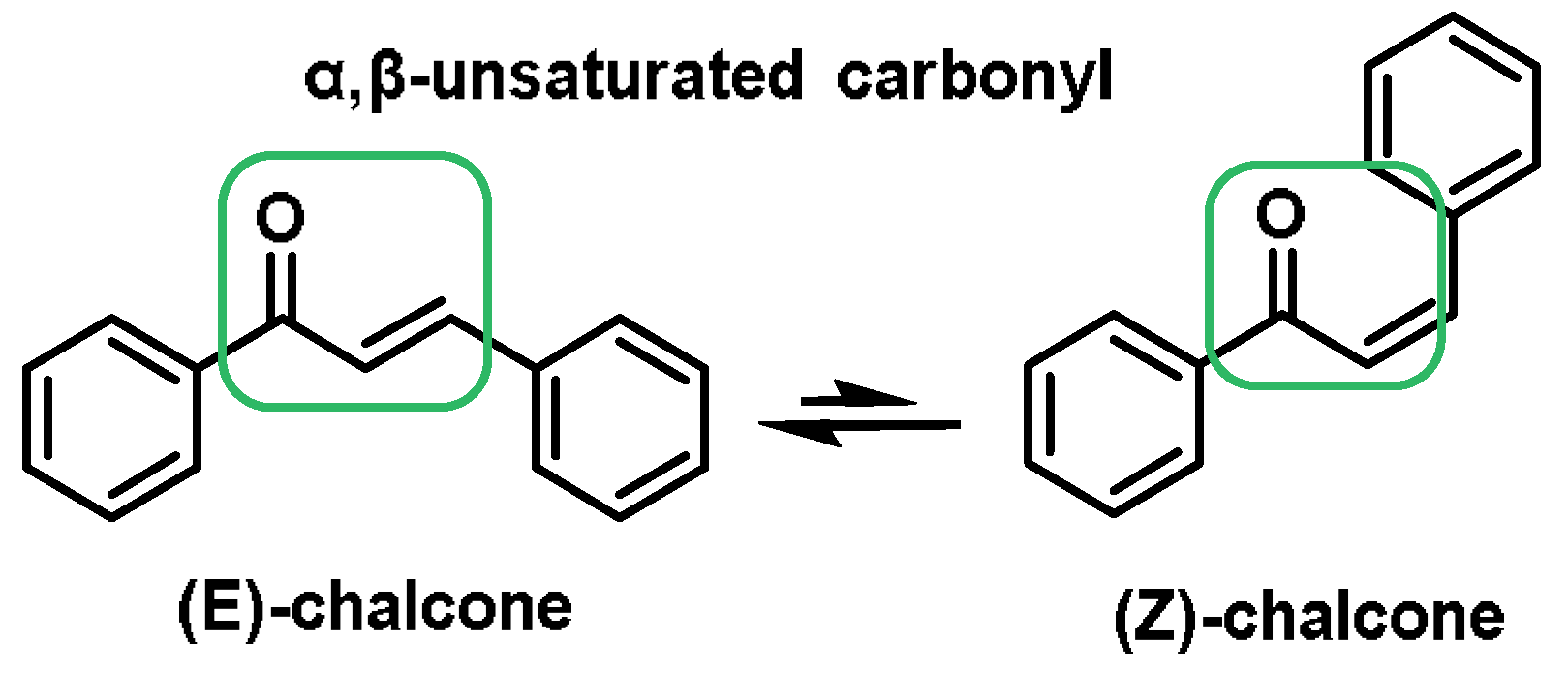


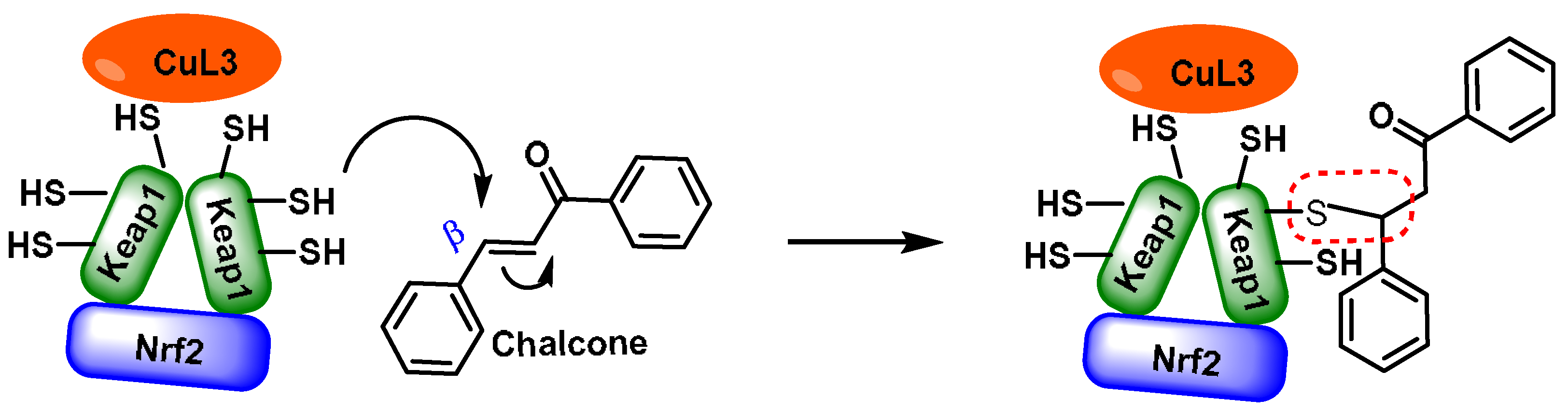
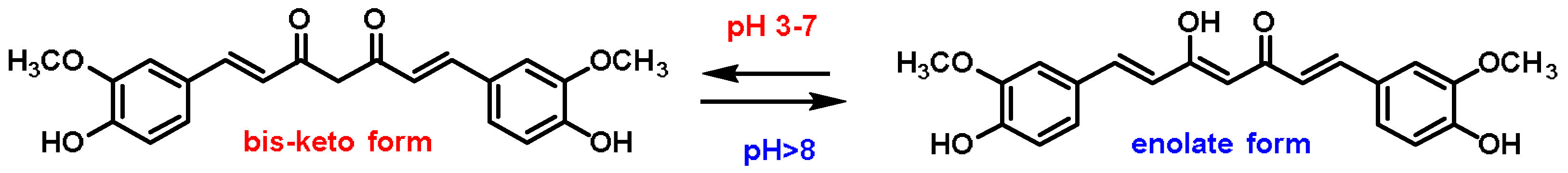
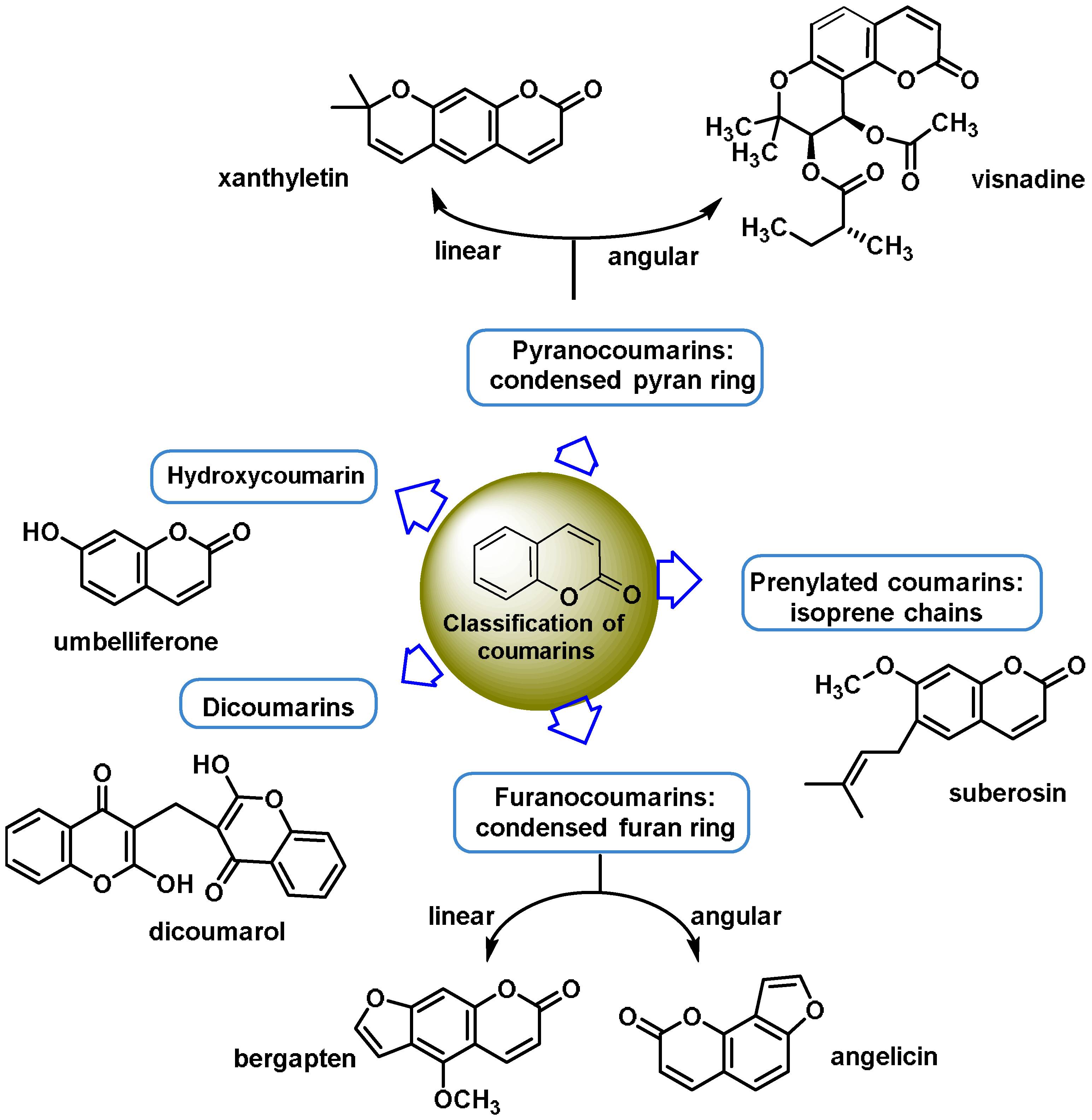
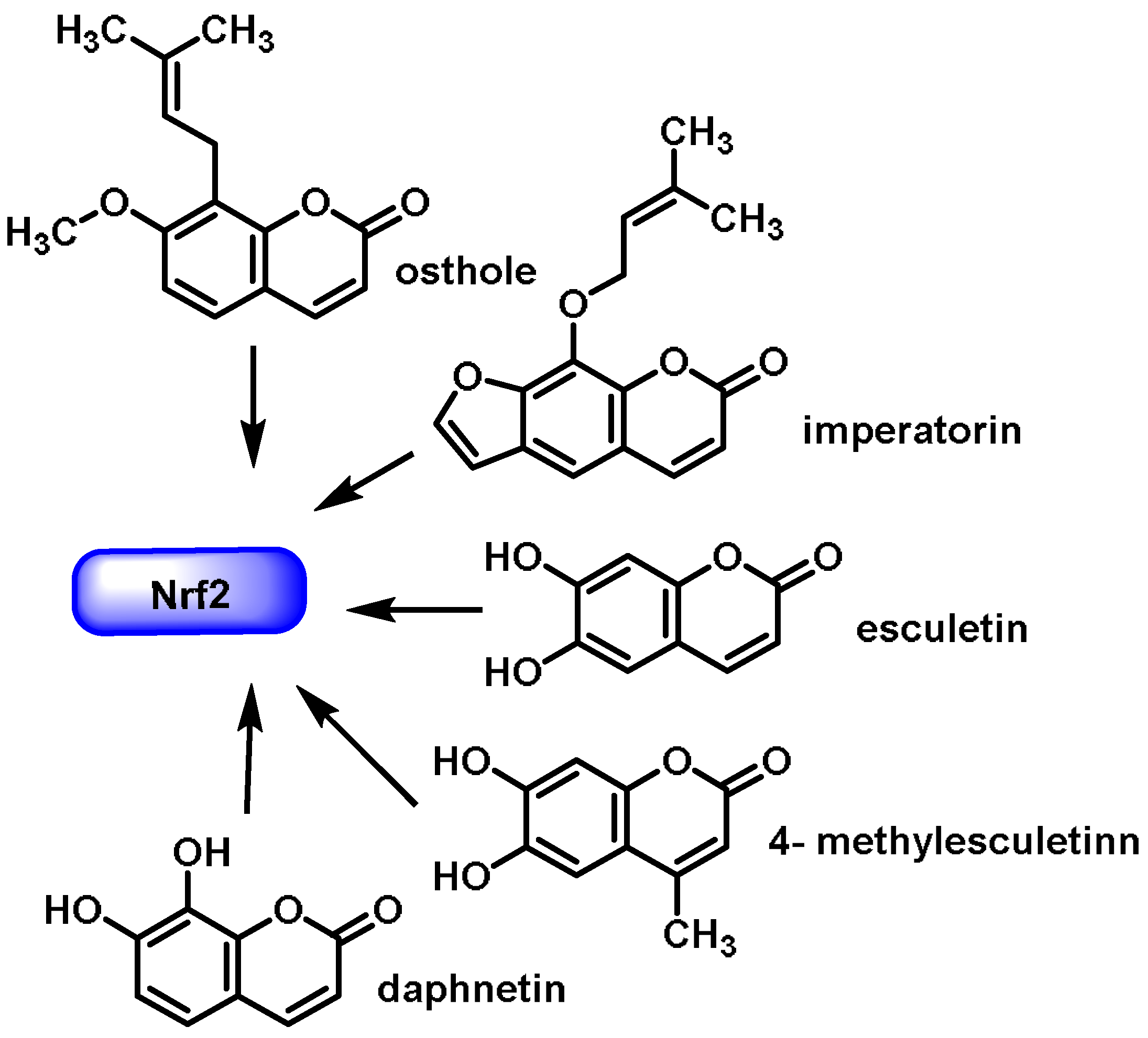
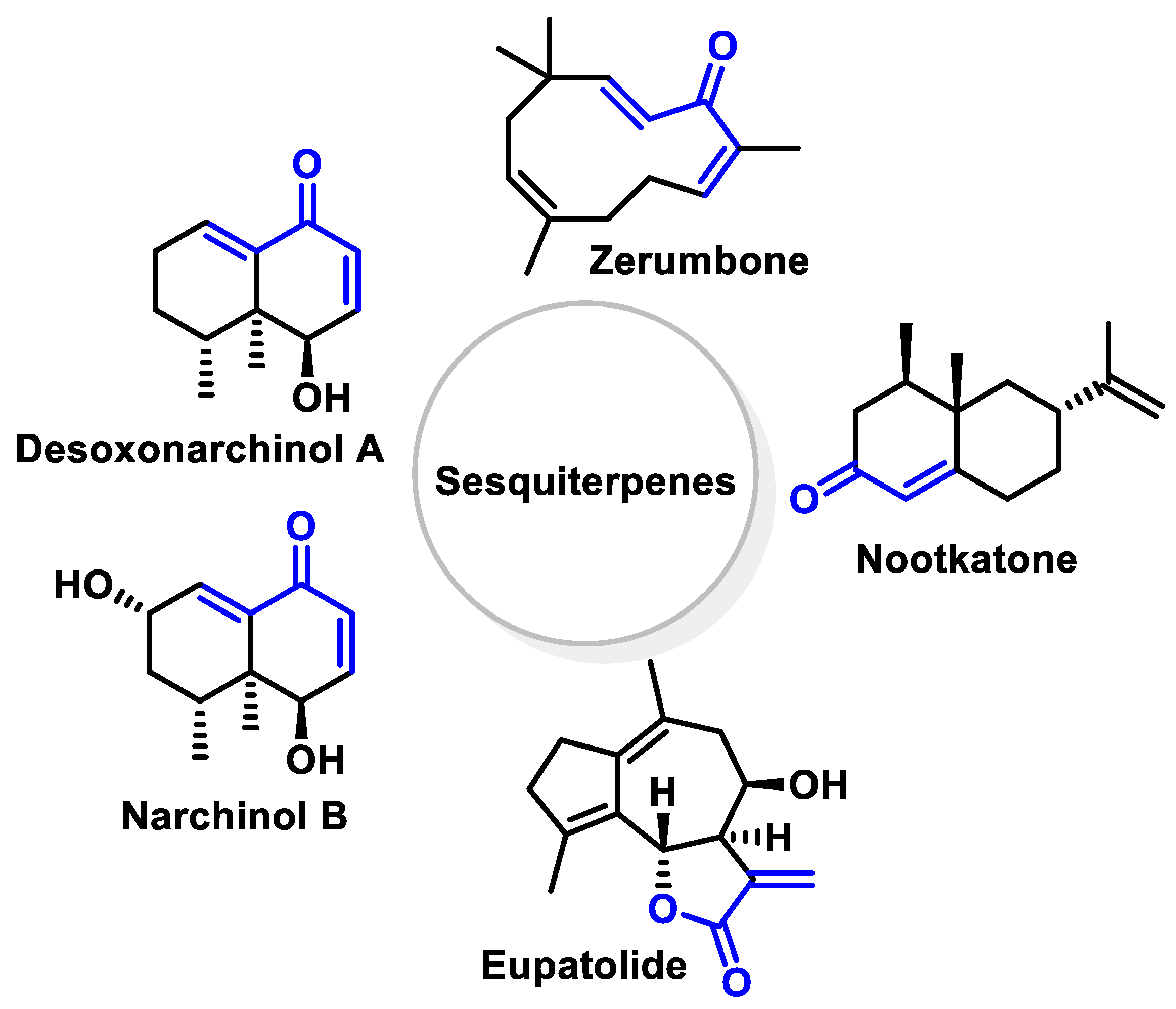
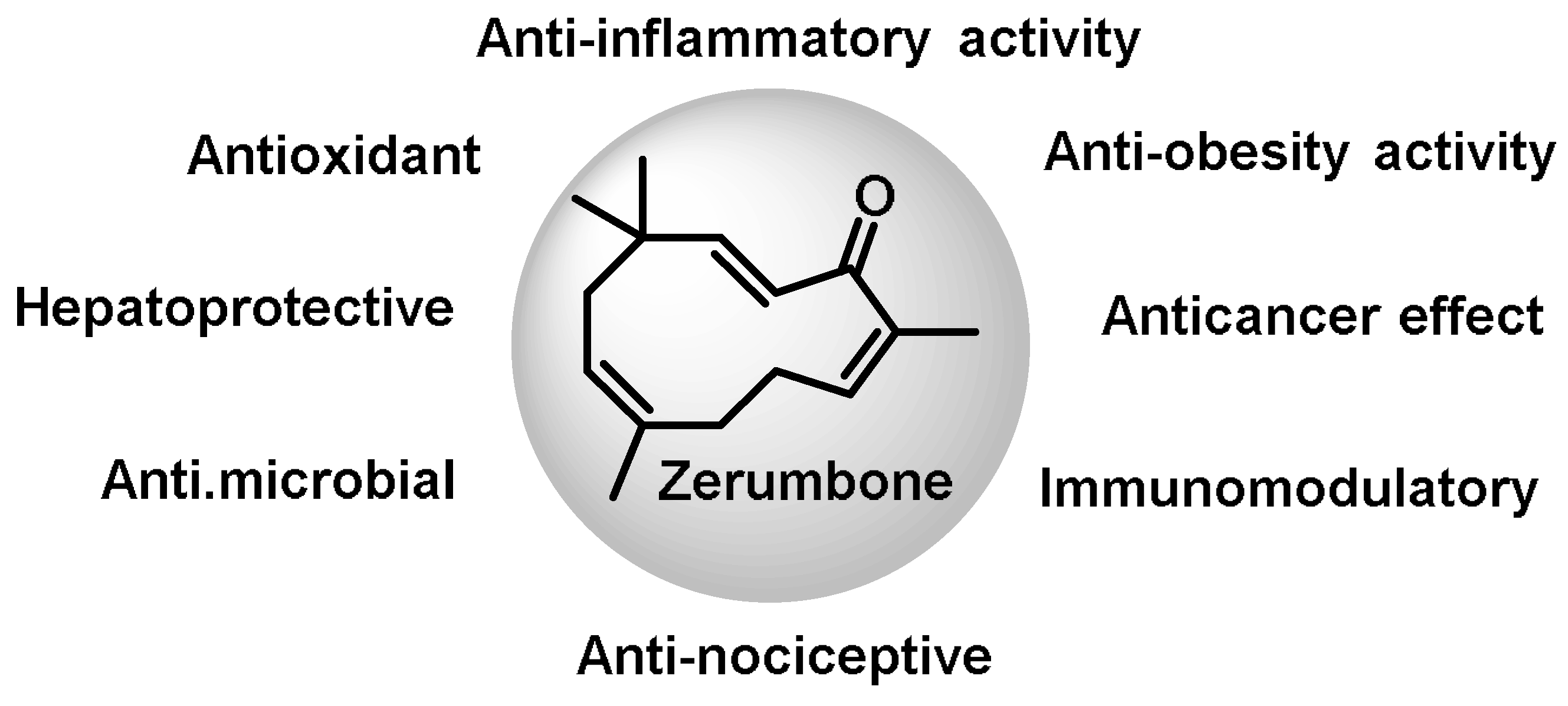
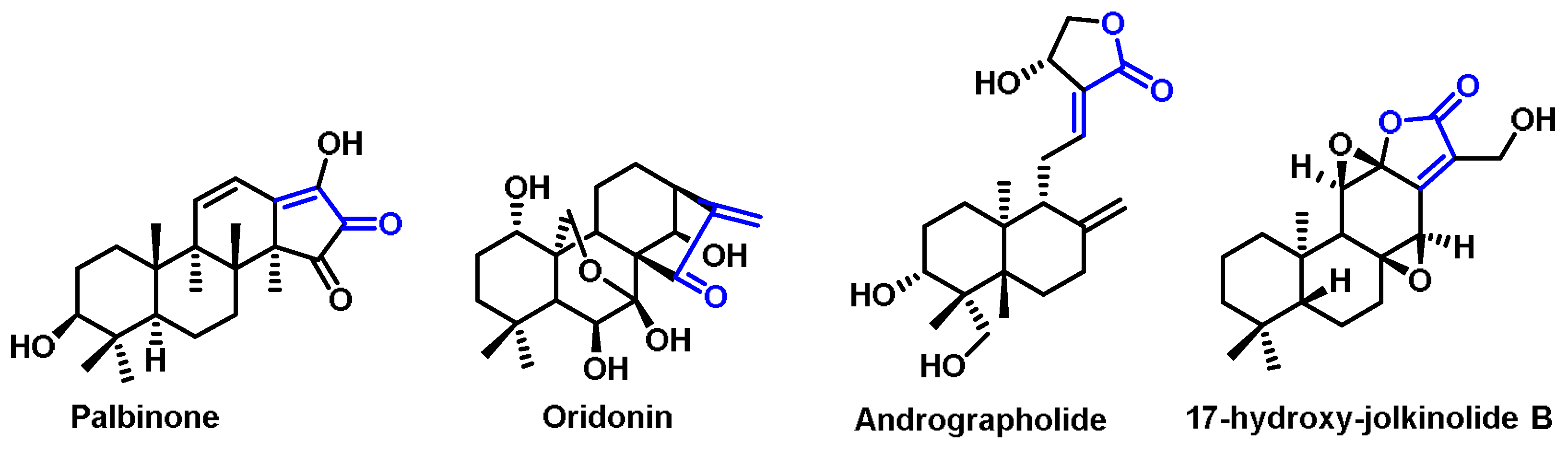
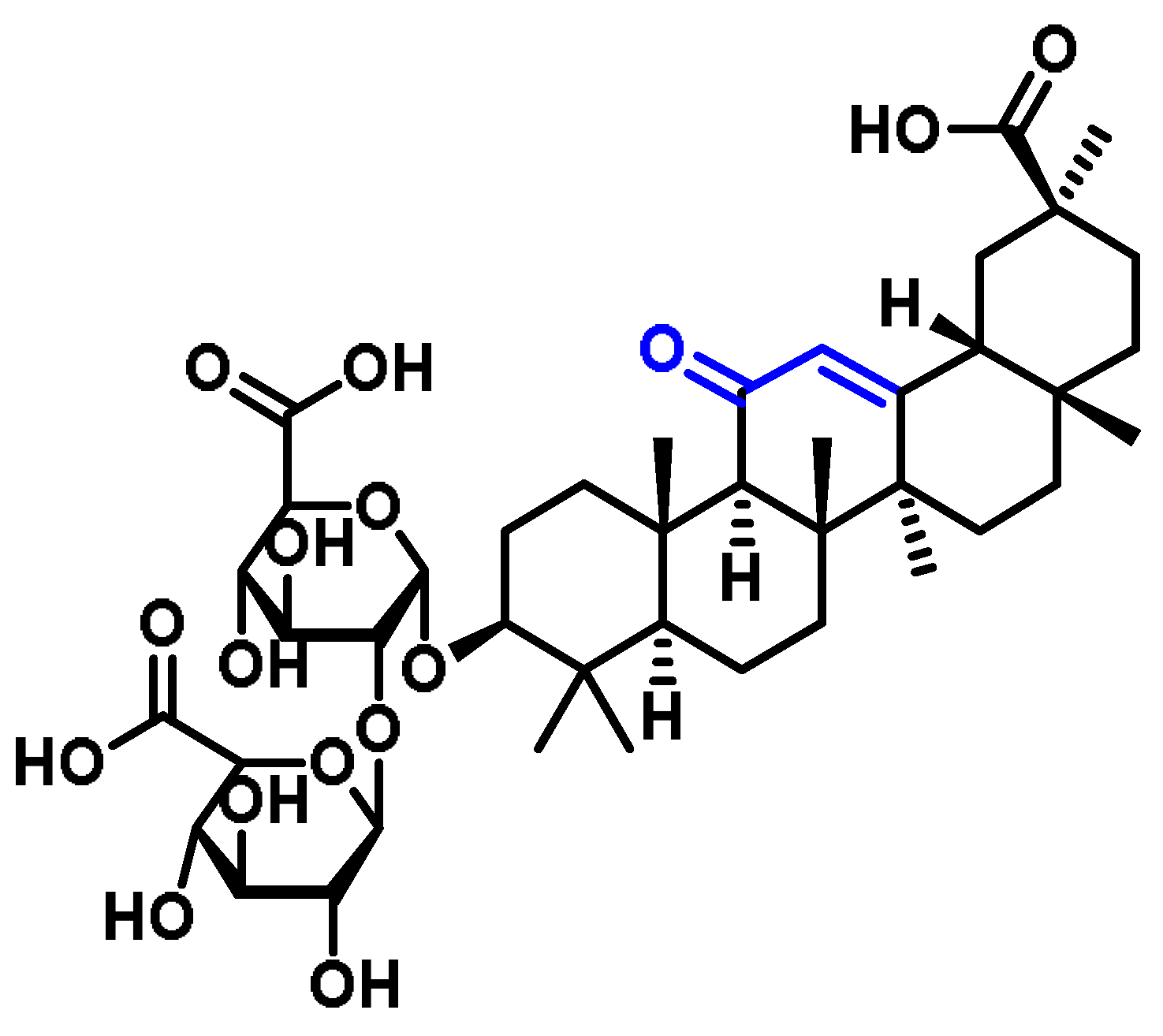
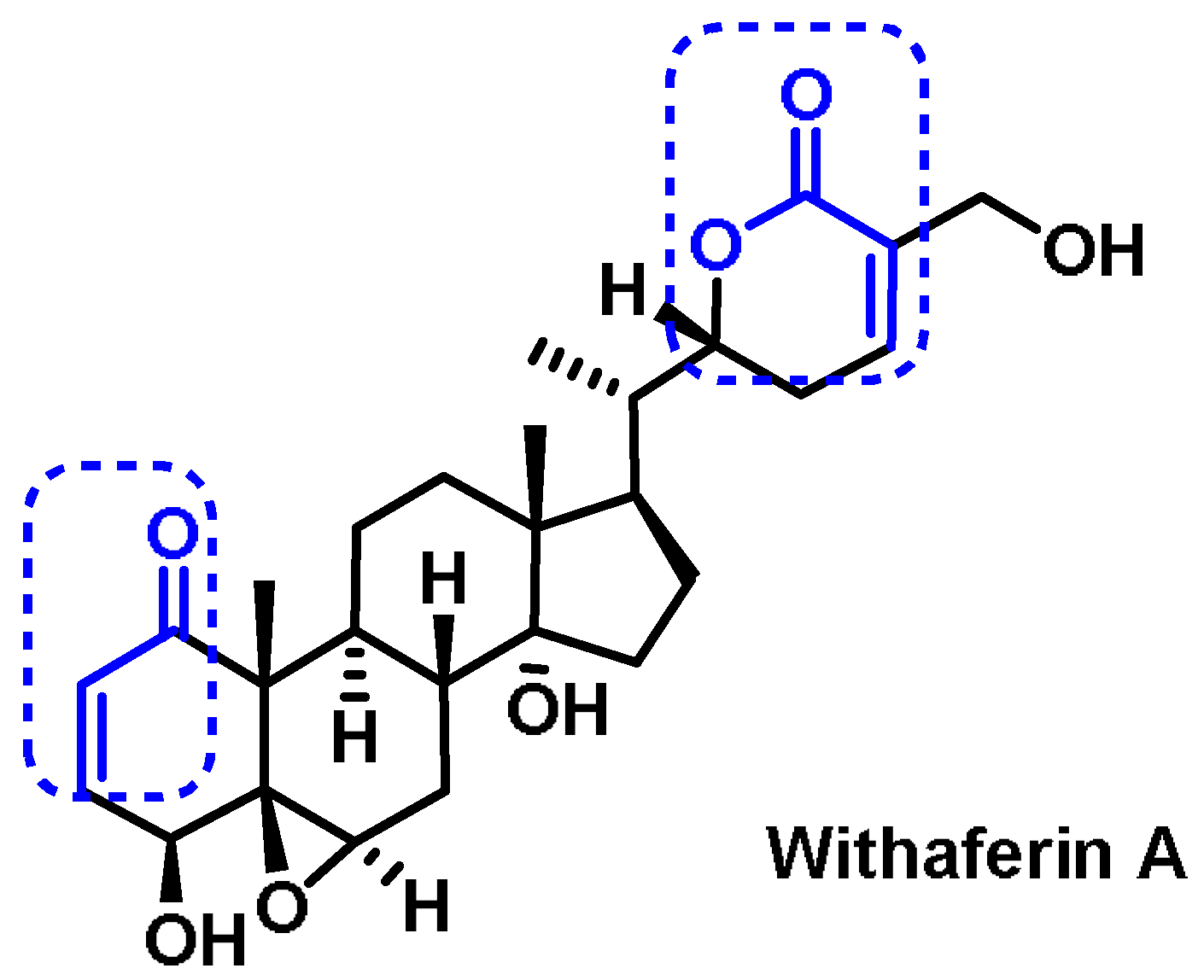
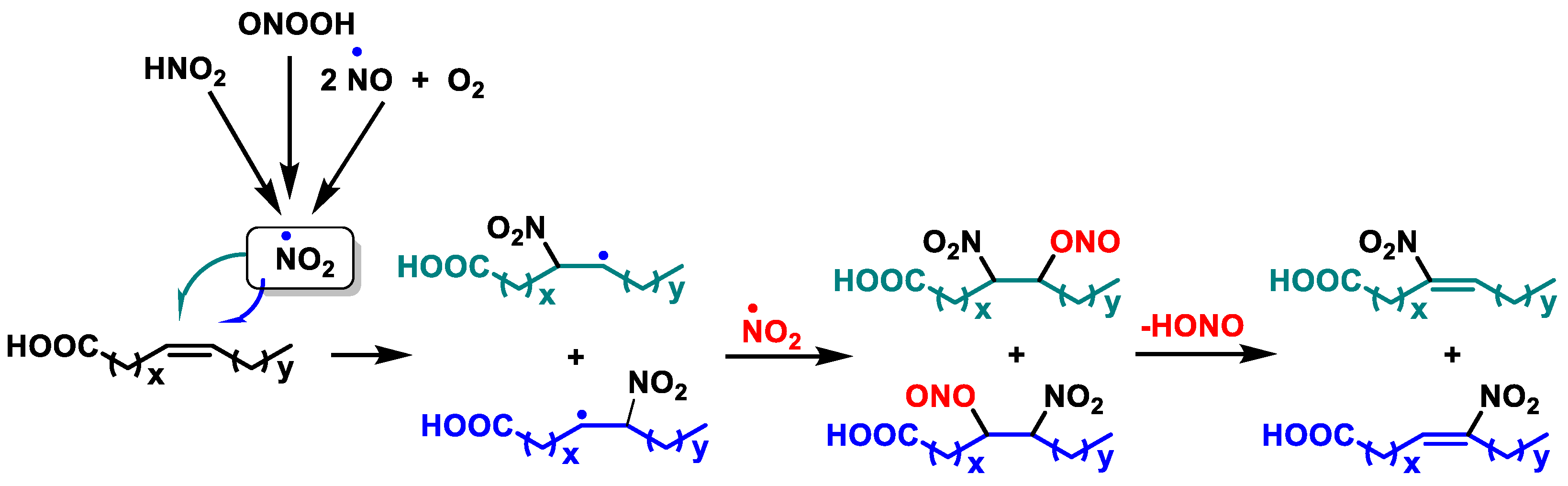

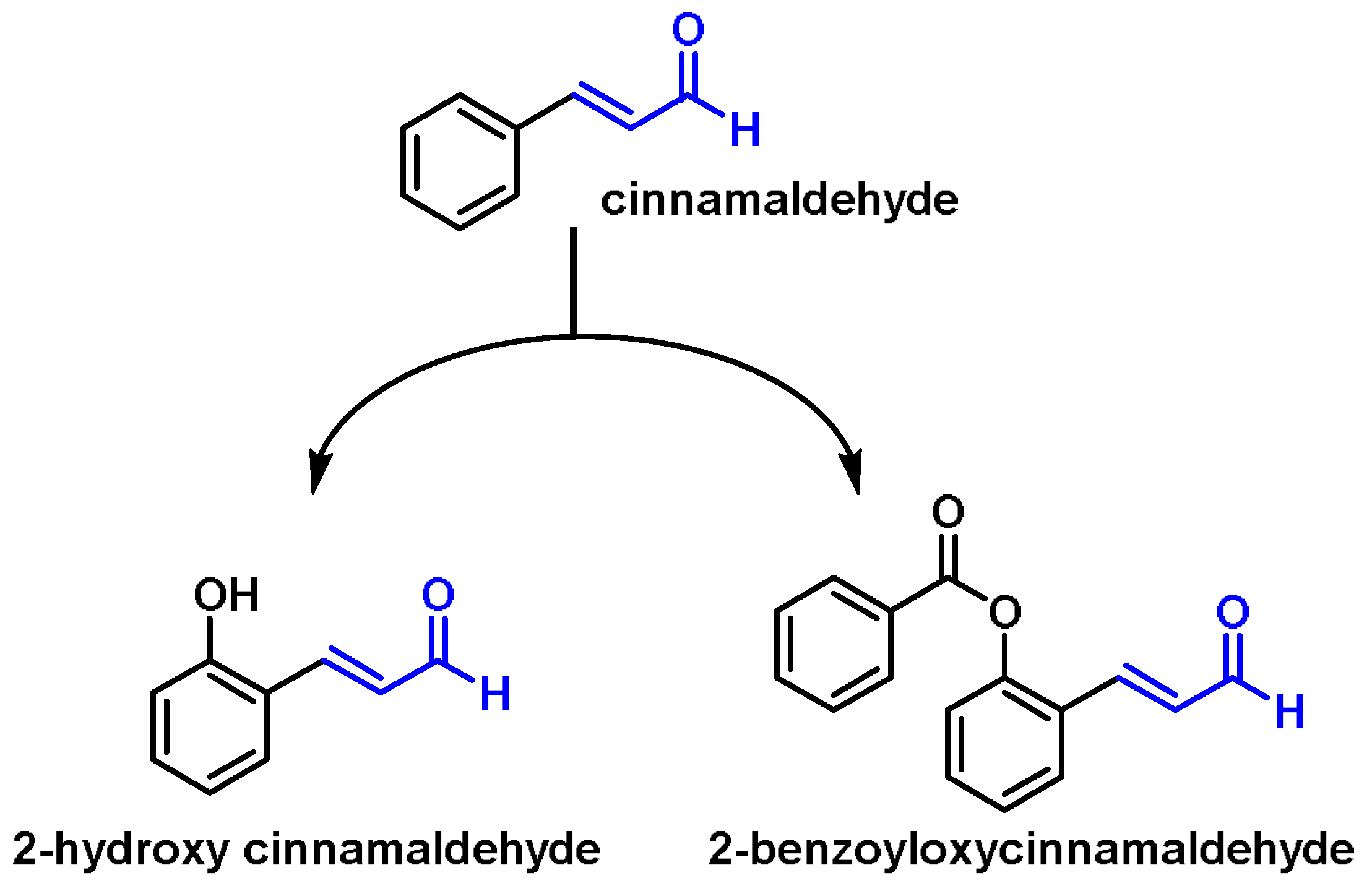
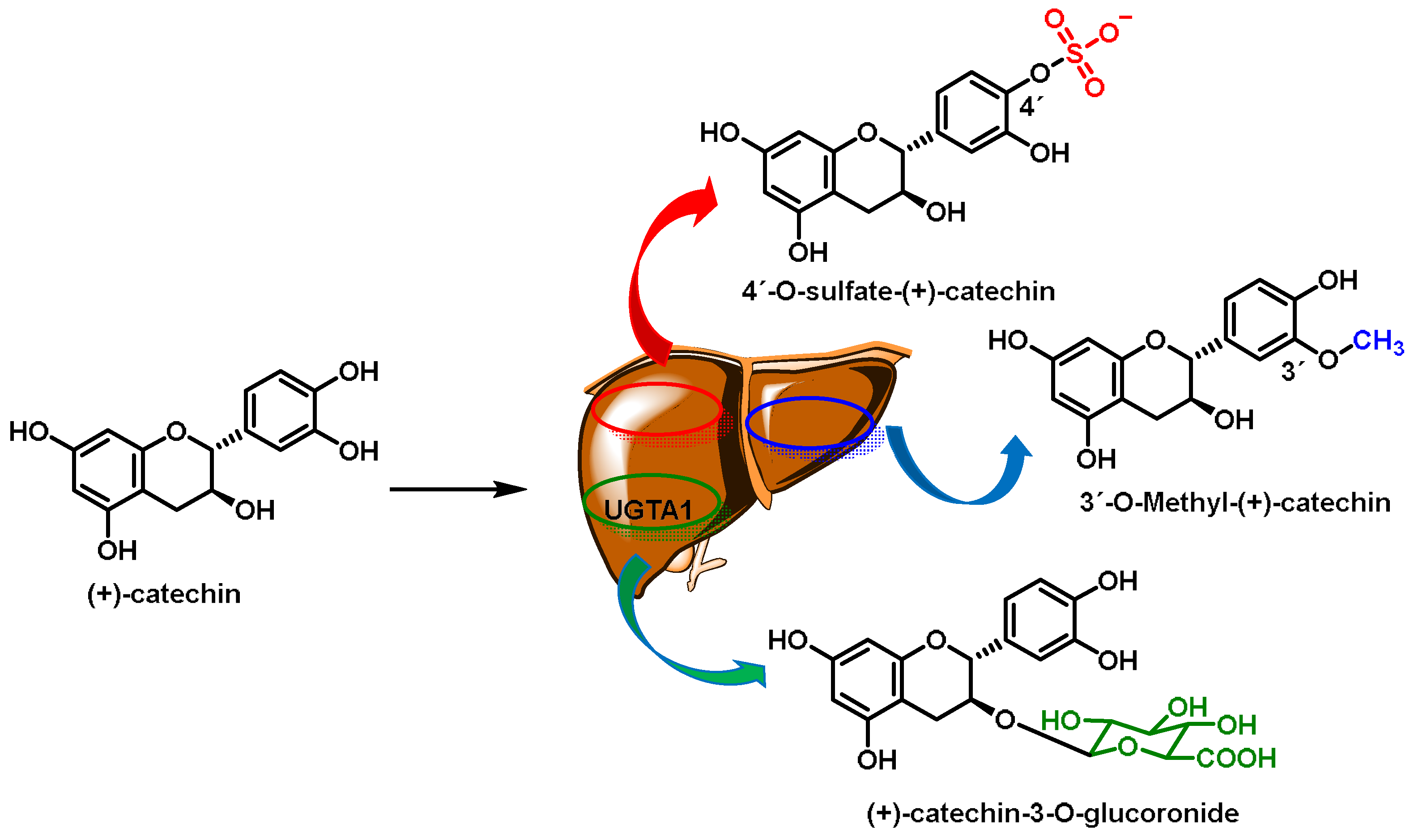
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Andrés, C.M.C.; Pérez de la Lastra, J.M.; Bustamante Munguira, E.; Juan, C.A.; Plou, F.J.; Pérez Lebeña, E. Electrophilic Compounds in the Human Diet and Their Role in the Induction of the Transcription Factor NRF2. Int. J. Mol. Sci. 2024, 25, 3521. https://doi.org/10.3390/ijms25063521
Andrés CMC, Pérez de la Lastra JM, Bustamante Munguira E, Juan CA, Plou FJ, Pérez Lebeña E. Electrophilic Compounds in the Human Diet and Their Role in the Induction of the Transcription Factor NRF2. International Journal of Molecular Sciences. 2024; 25(6):3521. https://doi.org/10.3390/ijms25063521
Chicago/Turabian StyleAndrés, Celia María Curieses, José Manuel Pérez de la Lastra, Elena Bustamante Munguira, Celia Andrés Juan, Francisco J. Plou, and Eduardo Pérez Lebeña. 2024. "Electrophilic Compounds in the Human Diet and Their Role in the Induction of the Transcription Factor NRF2" International Journal of Molecular Sciences 25, no. 6: 3521. https://doi.org/10.3390/ijms25063521
APA StyleAndrés, C. M. C., Pérez de la Lastra, J. M., Bustamante Munguira, E., Juan, C. A., Plou, F. J., & Pérez Lebeña, E. (2024). Electrophilic Compounds in the Human Diet and Their Role in the Induction of the Transcription Factor NRF2. International Journal of Molecular Sciences, 25(6), 3521. https://doi.org/10.3390/ijms25063521






