An Association between OXPHOS-Related Gene Expression and Malignant Hyperthermia Susceptibility in Human Skeletal Muscle Biopsies
Abstract
:1. Introduction
2. Results
2.1. Transcriptome Analysis
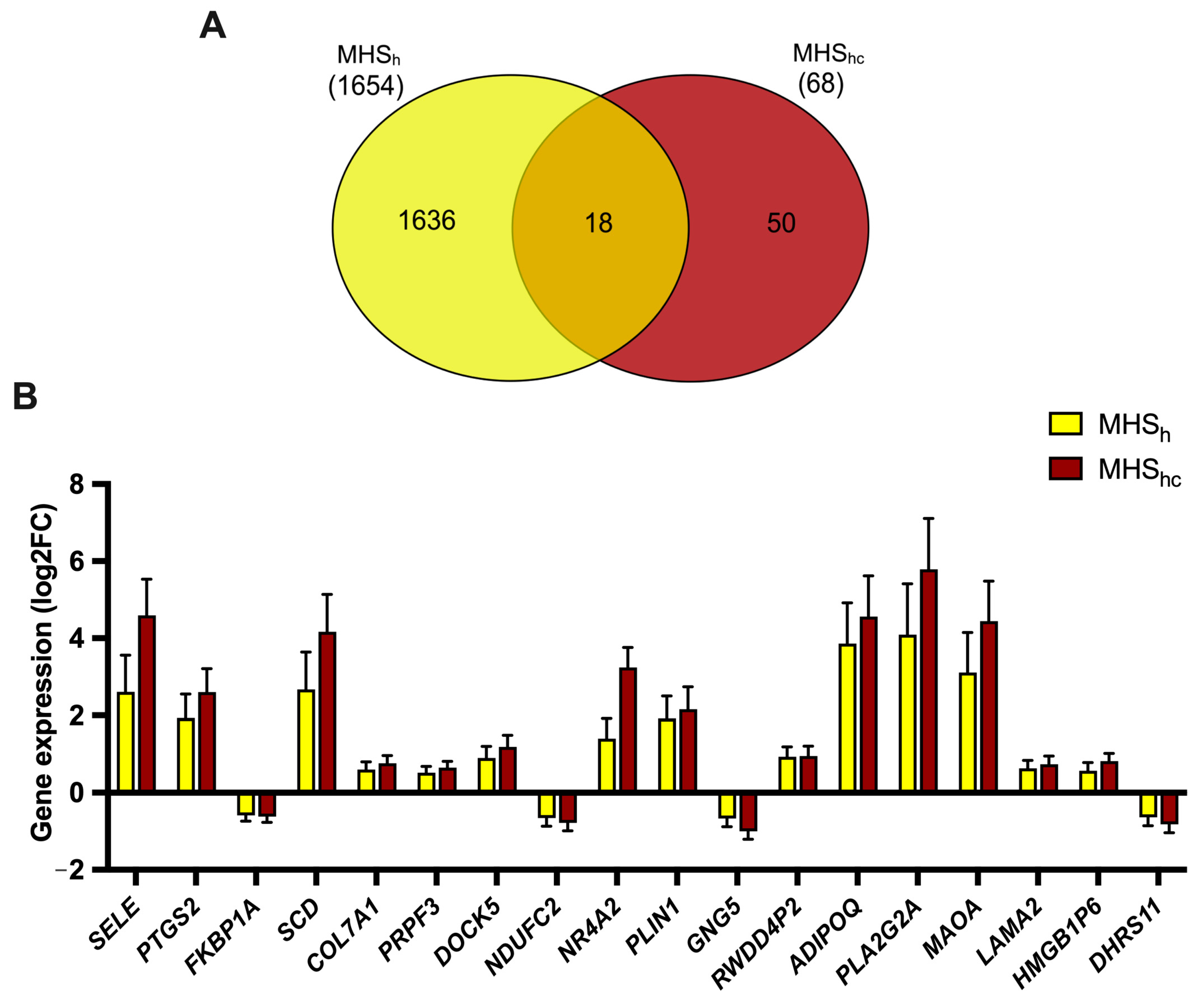
2.2. MHS Sub-Phenotypes
2.3. Validation of OXPHOS-Associated Genes
2.4. Effects of IVCT on Global Gene Expression
3. Discussion
4. Materials and Methods
4.1. Human Muscle Samples
4.2. RNA Sample Preparation
4.3. RNA Sequencing Design
4.4. Library Preparation and Sequencing
4.5. Differential Gene Expression Analysis
4.6. Gene Ontology and Pathway Analysis
4.7. RNAseq Validation
4.8. Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- EMHG. Diagnostic MH Mutations [Online]. 2023. Available online: https://www.emhg.org/diagnostic-mutations (accessed on 25 May 2023).
- Miller, D.; Daly, C.; Aboelsaod, E.; Gardner, L.; Hobson, S.; Riasat, K.; Shepherd, S.; Robinson, R.; Bilmen, J.; Gupta, P.; et al. Genetic epidemiology of malignant hyperthermia in the UK. Br. J. Anaesth. 2018, 121, 944–952. [Google Scholar] [CrossRef]
- Hopkins, P.; Rüffert, H.; Snoeck, M.; Girard, T.; Glahn, K.; Ellis, F.; Müller, C.; Urwyler, A.; European Malignant Hyperthermia Group; Bandschapp, O.; et al. European Malignant Hyperthermia Group guidelines for investigation of malignant hyperthermia susceptibility. Br. J. Anaesth. 2015, 115, 531–539. [Google Scholar] [CrossRef] [PubMed]
- Fiege, M.; Wappler, F.; Weisshorn, R.; Gerbershagen, M.U.; Steinfath, M.; Esch, J.S.A. Results of contracture tests with halothane, caffeine, and ryanodine depend on different malignant hyperthermia-associated ryanodine receptor gene mutations. Anesthesiology 2002, 97, 345–350. [Google Scholar] [CrossRef] [PubMed]
- Carpenter, D.; Robinson, R.L.; Quinnell, R.J.; Ringrose, C.; Hogg, M.; Casson, F.; Booms, P.; Iles, D.E.; Halsall, P.J.; Steele, D.S.; et al. Genetic variation in RYR1 and malignant hyperthermia phenotypes. Br. J. Anaesth. 2009, 103, 538–548. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Riehl, J.; Esteve, E.; Matthaei, K.I.; Goth, S.; Allen, P.D.; Pessah, I.N.; Lopez, J.R. Pharmacologic and functional characterization of malignant hyperthermia in the R163C RyR1 knock-in mouse. Anesthesiology 2006, 105, 1164–1175. [Google Scholar] [CrossRef] [PubMed]
- Yuen, B.; Boncompagni, S.; Feng, W.; Yang, T.; Lopez, J.R.; Matthaei, K.I.; Goth, S.R.; Protasi, F.; Allen, P.D.; Pessah, I.N.; et al. Mice expressing T4826I-RYR1 are viable but exhibit sex- and genotype-dependent susceptibility to malignant hyperthermia and muscle damage. FASEB J. 2012, 26, 1311–1322. [Google Scholar] [CrossRef] [PubMed]
- Chelu, M.G.; Goonasekera, S.A.; Durham, W.J.; Tang, W.; Lueck, J.D.; Riehl, J.; Pessah, I.N.; Zhang, P.; Bhattacharjee, M.B.; Dirksen, R.T.; et al. Heat- and anesthesia-induced malignant hyperthermia in an RyR1 knock-in mouse. FASEB J. 2006, 20, 329–330. [Google Scholar] [CrossRef] [PubMed]
- Lopez, J.; Kaura, V.; Diggle, C.; Hopkins, P.; Allen, P. Malignant hyperthermia, environmental heat stress, and intracellular calcium dysregulation in a mouse model expressing the p.G2435R variant of RYR1. Br. J. Anaesth. 2018, 121, 953–961. [Google Scholar] [CrossRef] [PubMed]
- Durham, W.J.; Aracena-Parks, P.; Long, C.; Rossi, A.E.; Goonasekera, S.A.; Boncompagni, S.; Galvan, D.L.; Gilman, C.P.; Baker, M.R.; Shirokova, N.; et al. RyR1 S-nitrosylation underlies environmental heat stroke and sudden death in Y522S RyR1 knockin mice. Cell 2008, 133, 53–65. [Google Scholar] [CrossRef]
- Michelucci, A.; De Marco, A.; Guarnier, F.A.; Protasi, F.; Boncompagni, S. Antioxidant Treatment Reduces Formation of Structural Cores and Improves Muscle Function in RYR1(Y522S/WT) Mice. Oxid. Med. Cell. Longev. 2017, 2017, 6792694. [Google Scholar] [CrossRef]
- Boncompagni, S.; Rossi, A.E.; Micaroni, M.; Beznoussenko, G.V.; Polishchuk, R.S.; Dirksen, R.T.; Protasi, F. Mitochondria are linked to calcium stores in striated muscle by developmentally regulated tethering structures. Mol. Biol. Cell 2009, 20, 1058–1067. [Google Scholar] [CrossRef]
- Giulivi, C.; Ross-Inta, C.; Omanska-Klusek, A.; Napoli, E.; Sakaguchi, D.; Barrientos, G.; Allen, P.D.; Pessah, I.N. Basal bioenergetic abnormalities in skeletal muscle from ryanodine receptor malignant hyperthermia-susceptible R163C knock-in mice. J. Biol. Chem. 2011, 286, 99–113. [Google Scholar] [CrossRef]
- Chang, L.; Liu, X.; Diggle, C.P.; Boyle, J.P.; Hopkins, P.M.; Shaw, M.-A.; Allen, P.D. Bioenergetic defects in muscle fibers of RYR1 mutant knock-in mice associated with malignant hyperthermia. J. Biol. Chem. 2020, 295, 15226–15235. [Google Scholar] [CrossRef]
- Lavorato, M.; Gupta, P.K.; Hopkins, P.M.; Franzini-Armstrong, C. Skeletal Muscle Microalterations in Patients Carrying Malignant Hyperthermia-Related Mutations of the e-c Coupling Machinery. Eur. J. Transl. Myol. 2016, 26, 6105. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Daly, C.; Miller, D.M.; Allen, P.D.; Boyle, J.P.; Hopkins, P.M.; Shaw, M.-A. Permeabilised skeletal muscle reveals mitochondrial deficiency in malignant hyperthermia-susceptible individuals. Br. J. Anaesth. 2019, 122, 613–621. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Gardner, L.; House, C.; Daly, C.; Allsopp, A.; de Sa, D.R.; Shaw, M.-A.; Hopkins, P.M. Comparison of Transcriptomic Changes in Survivors of Exertional Heat Illness with Malignant Hyperthermia Susceptible Patients. Int. J. Mol. Sci. 2023, 24, 16124. [Google Scholar] [CrossRef] [PubMed]
- Ling, C.; Poulsen, P.; Simonsson, S.; Rönn, T.; Holmkvist, J.; Almgren, P.; Hagert, P.; Nilsson, E.; Mabey, A.G.; Nilsson, P.; et al. Genetic and epigenetic factors are associated with expression of respiratory chain component NDUFB6 in human skeletal muscle. J. Clin. Investig. 2007, 117, 3427–3435. [Google Scholar] [CrossRef] [PubMed]
- Mootha, V.K.; Lindgren, C.M.; Eriksson, K.F.; Subramanian, A.; Sihag, S.; Lehar, J.; Puigserver, P.; Carlsson, E.; Ridderstrale, M.; Laurila, E.; et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003, 34, 267–273. [Google Scholar] [CrossRef]
- Sreekumar, R.; Halvatsiotis, P.; Schimke, J.C.; Nair, K.S. Gene expression profile in skeletal muscle of type 2 diabetes and the effect of insulin treatment. Diabetes 2002, 51, 1913–1920. [Google Scholar] [CrossRef] [PubMed]
- Patti, M.E.; Butte, A.J.; Crunkhorn, S.; Cusi, K.; Berria, R.; Kashyap, S.; Miyazaki, Y.; Kohane, I.; Costello, M.; Saccone, R.; et al. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: Potential role of PGC1 and NRF1. Proc. Natl. Acad. Sci. USA 2003, 100, 8466–8471. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Caballero, L.; Guerrero-Castillo, S.; Nijtmans, L. Unraveling the complexity of mitochondrial complex I assembly: A dynamic process. Biochim. Biophys. Acta 2016, 1857, 980–990. [Google Scholar] [CrossRef]
- Schaefer, A.M.; McFarland, R.; Blakely, E.L.; He, L.; Whittaker, R.G.; Taylor, R.W.; Chinnery, P.F.; Turnbull, D.M. Prevalence of mitochondrial DNA disease in adults. Ann. Neurol. 2008, 63, 35–39. [Google Scholar] [CrossRef]
- Rodenburg, R.J. Mitochondrial complex I-linked disease. Biochim. Biophys. Acta 2016, 1857, 938–945. [Google Scholar] [CrossRef]
- Liu, X.; Trakooljul, N.; Muráni, E.; Krischek, C.; Schellander, K.; Wicke, M.; Wimmers, K.; Ponsuksili, S. Molecular changes in mitochondrial respiratory activity and metabolic enzyme activity in muscle of four pig breeds with distinct metabolic types. J. Bioenerg. Biomembr. 2016, 48, 55–65. [Google Scholar] [CrossRef]
- Borchardt, L.A.; Scharenbrock, A.R.; Olufs, Z.P.G.; Wassarman, D.A.; Perouansky, M. Mutations in Complex I of the Mitochondrial Electron-Transport Chain Sensitize the Fruit Fly (Drosophila melanogaster) to Ether and Non-Ether Volatile Anesthetics. Int. J. Mol. Sci. 2023, 24, 1843. [Google Scholar] [CrossRef]
- Canato, M.; Capitanio, P.; Cancellara, L.; Leanza, L.; Raffaello, A.; Reane, D.V.; Marcucci, L.; Michelucci, A.; Protasi, F.; Reggiani, C. Excessive Accumulation of Ca2 + in Mitochondria of Y522S-RYR1 Knock-in Mice: A Link Between Leak From the Sarcoplasmic Reticulum and Altered Redox State. Front. Physiol. 2019, 10, 1142. [Google Scholar] [CrossRef] [PubMed]
- Görlach, A.; Bertram, K.; Hudecova, S.; Krizanova, O. Calcium and ROS: A mutual interplay. Redox Biol. 2015, 6, 260–271. [Google Scholar] [CrossRef]
- Thompson, S.J.; Riazi, S.; Kraeva, N.; Noseworthy, M.D.; Rayner, T.E.; Schneiderman, J.E.; Cifra, B.; Wells, G.D. Skeletal Muscle Metabolic Dysfunction in Patients With Malignant Hyperthermia Susceptibility. Anesth. Analg. 2017, 125, 434–441. [Google Scholar] [CrossRef] [PubMed]
- Litman, R.S.; Griggs, S.M.; Dowling, J.J.; Riazi, S. Malignant Hyperthermia Susceptibility and Related Diseases. Anesthesiology 2018, 128, 159–167. [Google Scholar] [CrossRef] [PubMed]
- Timmins, M.A.; Rosenberg, H.; Larach, M.G.; Sterling, C.; Kraeva, N.; Riazi, S. Malignant hyperthermia testing in probands without adverse anesthetic reaction. Anesthesiology 2015, 123, 548–556. [Google Scholar] [CrossRef] [PubMed]
- Wappler, F.; Fiege, M.; Steinfath, M.; Agarwal, K.; Scholz, J.; Singh, S.; Matschke, J.; Esch, J.S.A. Evidence for susceptibility to malignant hyperthermia in patients with exercise-induced rhabdomyolysis. Anesthesiology 2001, 94, 95–100. [Google Scholar] [CrossRef]
- Meyer, B.; Wittig, I.; Trifilieff, E.; Karas, M.; Schägger, H. Identification of two proteins associated with mammalian ATP synthase. Mol. Cell Proteom. 2007, 6, 1690–1699. [Google Scholar] [CrossRef]
- Ohsakaya, S.; Fujikawa, M.; Hisabori, T.; Yoshida, M. Knockdown of DAPIT (diabetes-associated protein in insulin-sensitive tissue) results in loss of ATP synthase in mitochondria. J. Biol. Chem. 2011, 286, 20292–20296. [Google Scholar] [CrossRef]
- Päivärinne, H.; Kainulainen, H. DAPIT, a novel protein down-regulated in insulin-sensitive tissues in streptozotocin-induced diabetes. Acta Diabetol. 2001, 38, 83–86. [Google Scholar] [CrossRef] [PubMed]
- Kontro, H.; Hulmi, J.J.; Rahkila, P.; Kainulainen, H. Cellular and tissue expression of DAPIT, a phylogenetically conserved peptide. Eur. J. Histochem. 2012, 56, e18. [Google Scholar] [CrossRef]
- Quinzii, C.M.; Hirano, M. Coenzyme Q and mitochondrial disease. Dev. Disabil. Res. Rev. 2010, 16, 183–188. [Google Scholar] [CrossRef] [PubMed]
- O’reilly, J.; Chintapalli, S.V.; Rutkowsky, J.M.; Tolentino, T.; Lloyd, K.C.K.; Olfert, I.M.; Adams, S.H.; Ono-Moore, K.D. Sex differences in skeletal muscle revealed through fiber type, capillarity, and transcriptomics profiling in mice. Physiol. Rep. 2021, 9, e15031. [Google Scholar] [CrossRef] [PubMed]
- Zahn, J.M.; Sonu, R.; Vogel, H.; Crane, E.; Mazan-Mamczarz, K.; Rabkin, R.; Davis, R.W.; Becker, K.G.; Owen, A.B.; Kim, S.K. Transcriptional profiling of aging in human muscle reveals a common aging signature. PLoS Genet. 2006, 2, e115. [Google Scholar] [CrossRef]
- Bua, E.; Johnson, J.; Herbst, A.; Delong, B.; McKenzie, D.; Salamat, S.; Aiken, J.M. Mitochondrial DNA-deletion mutations accumulate intracellularly to detrimental levels in aged human skeletal muscle fibers. Am. J. Hum. Genet. 2006, 79, 469–480. [Google Scholar] [CrossRef] [PubMed]
- De Magalhaes, J.P.; Curado, J.; Church, G.M. Meta-analysis of age-related gene expression profiles identifies common signatures of aging. Bioinformatics 2009, 25, 875–881. [Google Scholar] [CrossRef]
- de Jong, J.C.; Attema, B.J.; van der Hoek, M.D.; Verschuren, L.; Caspers, M.P.; Kleemann, R.; van der Leij, F.R.; van den Hoek, A.M.; Nieuwenhuizen, A.G.; Keijer, J. Sex differences in skeletal muscle-aging trajectory: Same processes, but with a different ranking. Geroscience 2023, 45, 2367–2386. [Google Scholar] [CrossRef]
- Robinson, R.L.; Brooks, C.; Brown, S.L.; Ellis, F.R.; Halsall, P.J.; Quinnell, R.J.; Shaw, M.-A.; Hopkins, P.M. RYR1 mutations causing central core disease are associated with more severe malignant hyperthermia in vitro contracture test phenotypes. Hum. Mutat. 2002, 20, 88–97. [Google Scholar] [CrossRef]
- Staron, R.S.; Hagerman, F.C.; Hikida, R.S.; Murray, T.F.; Hostler, D.P.; Crill, M.T.; Ragg, K.E.; Toma, K. Fiber type composition of the vastus lateralis muscle of young men and women. J. Histochem. Cytochem. 2000, 48, 623–629. [Google Scholar] [CrossRef]
- Wilson, J.M.; Loenneke, J.P.; Jo, E.; Wilson, G.J.; Zourdos, M.C.; Kim, J.-S. The effects of endurance, strength, and power training on muscle fiber type shifting. J. Strength Cond. Res. 2012, 26, 1724–1729. [Google Scholar] [CrossRef]
- Kelahmetoglu, Y.; Jannig, P.R.; Cervenka, I.; Koch, L.G.; Britton, S.L.; Zhou, J.; Wang, H.; Robinson, M.M.; Nair, K.S.; Ruas, J.L. Comparative Analysis of Skeletal Muscle Transcriptional Signatures Associated With Aerobic Exercise Capacity or Response to Training in Humans and Rats. Front. Endocrinol. 2020, 11, 591476. [Google Scholar] [CrossRef]
- Cheah, A.M. Effect of long chain unsaturated fatty acids on the calcium transport of sarcoplasmic reticulum. Biochim. Biophys. Acta (BBA)—Biomembr. 1981, 648, 113–119. [Google Scholar] [CrossRef]
- Cheah, K.; Cheah, A. Mitochondrial calcium transport and calcium-activated phospholipase in porcine malignant hyperthermia. Biochim. Biophys. Acta (BBA)—Bioenerg. 1981, 634, 70–84. [Google Scholar] [CrossRef]
- Cheah, K.; Cheah, A.M. Skeletal muscle mitochondrial phospholipase A2 and the interaction of mitochondria and sarcoplasmic reticulum in porcine malignant hyperthermia. Biochim. Biophys. Acta (BBA)—Bioenerg. 1981, 638, 40–49. [Google Scholar] [CrossRef]
- Fletcher, J.E.; Mayerberger, S.; Tripolitis, L.; Yudkowsky, M.; Rosenberg, H. Fatty acids markedly lower the threshold for halothane-induced calcium release from the terminal cisternae in human and porcine normal and malignant hyperthermia susceptible skeletal muscle. Life Sci. 1991, 49, 1651–1657. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, J.E.; Rosenberg, H. In vitro muscle contractures induced by halothane and suxamethonium. II: Human skeletal muscle from normal and malignant hyperthermia susceptible patients. Br. J. Anaesth. 1986, 58, 1433–1439. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, J.E.; Rosenberg, H.; Beech, J. Interactions of fatty acids with the calcium release channel in malignant hyperthermia. Adv. Exp. Med. Biol. 1991, 301, 57–69. [Google Scholar] [PubMed]
- Bojko, B.; Vasiljevic, T.; Boyaci, E.; Roszkowska, A.; Kraeva, N.; Moreno, C.A.I.; Koivu, A.; Wąsowicz, M.; Hanna, A.; Hamilton, S.; et al. Untargeted metabolomics profiling of skeletal muscle samples from malignant hyperthermia susceptible patients. Can. J. Anaesth. 2021, 68, 761–772. [Google Scholar] [CrossRef] [PubMed]
- Taylor, R.W.; Pyle, A.; Griffin, H.; Blakely, E.L.; Duff, J.; He, L.; Smertenko, T.; Alston, C.L.; Neeve, V.C.; Best, A.; et al. Use of whole-exome sequencing to determine the genetic basis of multiple mitochondrial respiratory chain complex deficiencies. JAMA 2014, 312, 68–77. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.D.; Holzinger, E.R.; Li, R.; Pendergrass, S.A.; Kim, D. Methods of integrating data to uncover genotype-phenotype interactions. Nat. Rev. Genet. 2015, 16, 85–97. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Kong, C.; Liu, P.; Geng, W.; Tang, H. Identification of Potential Biomarkers for Ryanodine Receptor 1 (RYR1) Mutation-Associated Myopathies Using Bioinformatics Approach. Dis. Markers 2022, 2022, 8787782. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 3. [Google Scholar] [CrossRef]
- Cock, P.J.A.; Fields, C.J.; Goto, N.; Heuer, M.L.; Rice, P.M. The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res. 2010, 38, 1767–1771. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- RStudio Team. RStudio: Integrated Development for R; RStudio, Inc.: Boston, MA, USA, 2015; Available online: http://www.rstudio.com/ (accessed on 10 January 2024).
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’Ayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef] [PubMed]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, RESEARCH0034. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
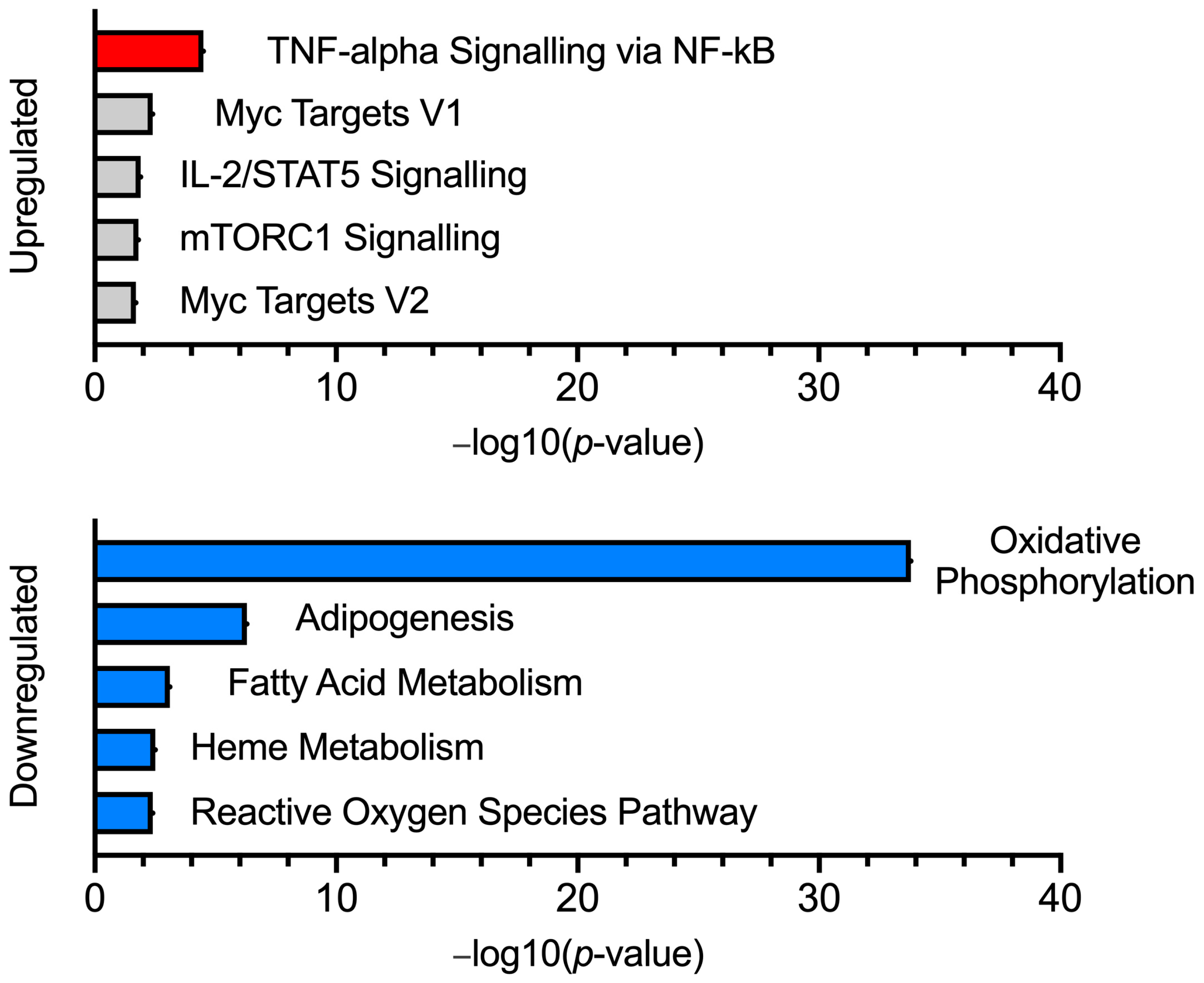


| MHN vs. MHS (Downregulated Genes) Gene Ontology Results | ||||
|---|---|---|---|---|
| Biological Processes | p-Value | Adjusted p-Value | Odds Ratio | Combined Score |
| Aerobic electron transport chain (GO:0019646) | 3.03 × 10−22 | 5.64 × 10−19 | 20.33 | 1007.15 |
| Mitochondrial ATP synthesis coupled electron transport (GO:0042775) | 4.66 × 10−22 | 5.64 × 10−19 | 19.87 | 976.20 |
| Mitochondrial respiratory chain complex I assembly (GO:0032981) | 3.23 × 10−14 | 1.96 × 10−11 | 15.27 | 474.21 |
| NADH dehydrogenase complex assembly (GO:0010257) | 3.23 × 10−14 | 1.96 × 10−11 | 15.27 | 474.21 |
| Mitochondrial respiratory chain complex assembly (GO:0033108) | 1.27 × 10−13 | 6.17 × 10−11 | 10.36 | 307.73 |
| Molecular function | p-value | Adjusted p-value | Odds Ratio | Combined score |
| Oxidoreduction-driven active transmembrane transporter activity (GO:0015453) | 1.35 × 10−20 | 6.05 × 10−18 | 22.50 | 1029.19 |
| Proton transmembrane transporter activity (GO:0015078) | 6.38 × 10−11 | 1.43 × 10−8 | 16.20 | 380.39 |
| NADH dehydrogenase (quinone) activity (GO:0050136) | 1.71 × 10−10 | 1.91 × 10−8 | 17.53 | 394.25 |
| NADH dehydrogenase (ubiquinone) activity (GO:0008137) | 1.71 × 10−10 | 1.91 × 10−8 | 17.53 | 394.25 |
| Active ion transmembrane transporter activity (GO:0022853) | 1.88 × 10−9 | 1.68 × 10−7 | 13.44 | 269.96 |
| Cellular component | p-value | Adjusted p-value | Odds Ratio | Combined score |
| Mitochondrial inner membrane (GO:0005743) | 4.55 × 10−42 | 1.06 × 10−39 | 10.42 | 991.95 |
| Mitochondrial membrane (GO:0031966) | 8.20 × 10−42 | 1.06 × 10−39 | 8.22 | 777.65 |
| Organelle inner membrane (GO:0019866) | 1.97 × 10−38 | 1.70 × 10−36 | 9.35 | 811.84 |
| Mitochondrial matrix (GO:0005759) | 2.17 × 10−18 | 1.40 × 10−16 | 5.51 | 223.92 |
| Mitochondrial respiratory chain complex I (GO:0005747) | 4.31 × 10−13 | 1.86 × 10−11 | 18.76 | 534.17 |
| Patient | Gender | Age at Time of Biopsy | MH Status | Contracture at 2% Halothane | Contracture at 2 mM Caffeine | RYR1 Family Variant |
|---|---|---|---|---|---|---|
| 1 | M | 13 | MHN | 0 | 0 | Negative |
| 2 | M | 11 | MHN | 0 | 0 | Negative |
| 3 | M | 19 | MHN | 0 | 0 | Negative |
| 4 | M | 19 | MHN | 0 | 0 | Negative |
| 5 | M | 12 | MHSh | 1.15 g | 0 | c.7007G>A |
| 6 | M | 39 | MHSh | 0.25 g | 0 | Unknown |
| 7 | M | 30 | MHSh | 0.75 g | 0 | c.4293G>A, c.7879G>A |
| 8 | M | 12 | MHSh | 0.25 g | 0 | c.5183C>T |
| 9 | M | 18 | MHShc | 0.65 g | 2.1 g | c.12700G>C |
| 10 | M | 11 | MHShc | 1.8 g | 0.6 g | c.7292A>T |
| 11 | M | 16 | MHShc | 1.6 g | 0.4 g | c.14201G>A |
| 12 | M | 14 | MHShc | 0.9 g | 0.6 g | c.6617C>T |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, L.; Motley, R.; Daly, C.L.; Diggle, C.P.; Hopkins, P.M.; Shaw, M.-A. An Association between OXPHOS-Related Gene Expression and Malignant Hyperthermia Susceptibility in Human Skeletal Muscle Biopsies. Int. J. Mol. Sci. 2024, 25, 3489. https://doi.org/10.3390/ijms25063489
Chang L, Motley R, Daly CL, Diggle CP, Hopkins PM, Shaw M-A. An Association between OXPHOS-Related Gene Expression and Malignant Hyperthermia Susceptibility in Human Skeletal Muscle Biopsies. International Journal of Molecular Sciences. 2024; 25(6):3489. https://doi.org/10.3390/ijms25063489
Chicago/Turabian StyleChang, Leon, Rebecca Motley, Catherine L. Daly, Christine P. Diggle, Philip M. Hopkins, and Marie-Anne Shaw. 2024. "An Association between OXPHOS-Related Gene Expression and Malignant Hyperthermia Susceptibility in Human Skeletal Muscle Biopsies" International Journal of Molecular Sciences 25, no. 6: 3489. https://doi.org/10.3390/ijms25063489






