Abstract
Leaf senescence is the terminal stage of leaf development, and its initiation and progression are closely controlled by the integration of a myriad of endogenous signals and environmental stimuli. It has been documented that WRKY transcription factors (TFs) play essential roles in regulating leaf senescence, yet the molecular mechanism of WRKY-mediated leaf senescence still lacks detailed elucidation in crop plants. In this study, we cloned and identified a tobacco WRKY TF gene, designated NtWRKY70b, acting as a positive regulator of natural leaf senescence. The expression profile analysis showed that NtWRKY70b transcript levels were induced by aging and hydrogen peroxide (H2O2) and downregulated upon hydrogen sulfide (H2S) treatment. The physiological and biochemical assays revealed that overexpression of NtWRKY70b (OE) clearly promoted leaf senescence, triggering increased levels of reactive oxygen species (ROS) and decreased H2S content, while disruption of NtWRKY70b by chimeric repressor silencing technology (SRDX) significantly delayed the onset of leaf senescence, leading to a decreased accumulation of ROS and elevated concentration of H2S. The quantitative real-time PCR analysis showed that the expression levels of various senescence-associated genes and ROS biosynthesis-related genes (NtRbohD and NtRbohE) were upregulated in OE lines, while the expression of H2S biosynthesis-related genes (NtDCD and NtCYSC1) were inhibited in OE lines. Furthermore, the Yeast one-hybrid analysis (Y1H) and dual luciferase assays showed that NtWRKY70b could directly upregulate the expression of an ROS biosynthesis-related gene (NtRbohD) and a chlorophyll degradation-related gene (NtPPH) by binding to their promoter sequences. Accordingly, these results indicated that NtWYKY70b directly activated the transcript levels of NtRbohD and NtPPH and repressed the expression of NtDCD and NtCYCS1, thereby promoting ROS accumulation and impairing the endogenous H2S production, and subsequently accelerating leaf aging. These observations improve our knowledge of the regulatory mechanisms of WRKY TFs controlling leaf senescence and provide a novel method for ensuring high agricultural crop productivity via genetic manipulation of leaf senescence in crops.
1. Introduction
In plants, leaf senescence is a crucial stage of plant development, which is stimulated by integrating multiple internal factors and external environmental cues [1,2]. During this process, plants have evolved highly elaborate and sophisticated senescence-regulating mechanisms consisting of the fine-tuned modulation of the integration of multiple phytohormones, including abscisic acid (ABA), jasmonic acid (JA), and salicylic acid (SA), and the degradation of subcellular organelles (e.g., chloroplast and mitochondria) and metabolic changes like the hydrolysis of chlorophyll and other macromolecules, e.g., proteins, lipids, and nucleic acids, followed by nutrient remobilization and reallocation throughout the plant’s life cycle to ensure the reproductive success [3,4,5,6]. It is well known that the recycling of nutrient from senescing leaves to new organs, including growing leaves and fruits and developing seed, is a crucial process for plant fitness and crop productivity under fluctuating environmental conditions. An increasing number of reports have demonstrated that precocious leaf senescence negatively influences the yield and quality of crops, while prolongation of leaf longevity remarkably improves the plant biomass and crop yields [2,4,5,6,7,8]. Thus, the proper timing of the initiation and progress of leaf senescence is critical for plant development and crops’ productivity, yet the understanding of how plants sense and respond to internal aging signals and external environmental cues and then initiate leaf senescence is still fragmentary.
Emerging evidence has demonstrated that during leaf senescence, an increasing number of transcription factor (TF) genes from multiple families, such as NAC and WRKY, are significantly upregulated [9,10,11], implying that TFs play essential roles in transcriptional regulation of senescence-associated genes (SAG) in this process. WRKY transcription factors (TFs), defined by the WRKY domain consisting of the conserved amino acid sequence WRKYGQK at its N-terminal, comprise a superfamily of regulatory biomolecules in plants which widely participate in a myriad of key biological processes, including multiple developmental and physiological processes and various biotic and abiotic stress responses [12,13,14,15,16,17]. In Arabidopsis, a transcriptome analysis revealed that numerous WRKY genes were dramatically induced in senescing leaves [11], indicating important roles of WRKY TFs in leaf senescence regulation. Recent genetic and molecular biological investigation have provided various evidence to demonstrate that WRKY members play vital roles in the coordinated regulation of leaf senescence. For example, overexpression of several WRKY genes such as AtWRKY75, AtWRKY45, AtWRKY55, and AtWRKY42 positively influences leaf senescence, while the disruption of these TFs genes prolongs leaf longevity [9,18,19,20,21]. Conversely, AtWRKY25 and AtWRKY70 act as negative regulators of modulation of leaf senescence [22,23,24]. Recently, WRKY47 and WRKY70, from Brassica napus, were identified as participating in controlling leaf senescence [17,25].
Studies concerning free radical damage reactions in plant have revealed that reactive oxygen species (ROS) have diverse functions in various developmental and physiological processes, including seed germination, root morphogenesis, stomatal movement, leaf development, and abiotic stress responses [4,18,26,27,28]. It has been reported that respiratory burst oxidase homologs (Rbohs), as homologs of mammalian NADPH oxidases, participate in accumulation of apoplastic ROS (e.g., O2− and H2O2), which are targeted to the plasma membrane [29]. In Arabidopsis, Rbohs genes class consists of 10 members, namely AtRbohA–AtRbohJ [29]. Recent studies have demonstrated that multiple Rbohs genes (e.g., RbohD, RbohE, and RbohF) are involved in various physiological processes and biotic/abiotic stress responses in Arabidopsis, tobacco, rice, and oilseed rape [30,31,32,33,34,35]. Numerous studies have reported that high levels of endogenous ROS are a typical physiological feature in aging leaves, and their burst leads to a disturbance of redox state and dramatic oxidative damage to the cell membrane, ultimately causing leaf senescence and cell death in plants [32,36,37]. Consequently, ROS play a vital function in regulating the onset and progress of leaf aging, yet little is known about the upstream transcriptional network which is involved in controlling the accumulation of ROS during leaf senescence.
Hydrogen sulfide (H2S) is a colorless, highly soluble gas with a foul odor similar to rotten eggs, which has been regarded as a toxic gas for many years [38]. Recently, in plants, it has been understood that H2S, as the third key endogenous gasotransmitter besides nitric oxide (NO) and carbon monoxide (CO), has important functions in a myriad of developmental and physiological processes, including root development, seedlings growth, stomatal movement, flowering time, leaf senescence, fruit ripening, and plant responses to abiotic stresses (e.g., drought, salt, low temperature, and osmotic stresses) [27,39,40,41,42,43,44,45,46,47,48]. Furthermore, emerging studies have demonstrated that H2S influences various signaling pathways via increased levels of the persulfidation of its target proteins. For example, in Arabidopsis, H2S was reported to modulate the ABA signaling network by improving the persulfidation of SnRK2.6 and ABI4, two curial components of the ABA signal transduction, leading to these proteins’ functions changing [43,44]. In heading Chinese cabbage, H2S was found to be involved in controlling flowering by s-sulfhydration of BraFLCs, some key MADs box transcription factors for regulating the timing of flowering, thereby changing their binding ability to downstream promoters, subsequently causing early flowering [45].
Genetic and molecular studies have revealed that WRKY TFs, ROS, and H2S have vital functions in the modulation of leaf senescence [26,46,49]. However, the molecular mechanism driving their relationships in the leaf aging process remains largely unknown. Tobacco is an important economic crop and also a significant model organism for life science research, yet little is understood on the molecular mechanisms of leaf senescence in tobacco. Based our previous transcriptome assay, in this study, we isolated and identified a WRKY TF gene, namely NtWRKY70b, which positively influenced leaf senescence through modified levels of ROS and H2S in tobacco. Our investigations revealed that NtWRKY70b acts as a new positive regulator in regulating leaf senescence through modulating the metabolism of ROS and H2S.
2. Results
2.1. Identification and Sequence Analysis of NtWYKY70b Orthologs
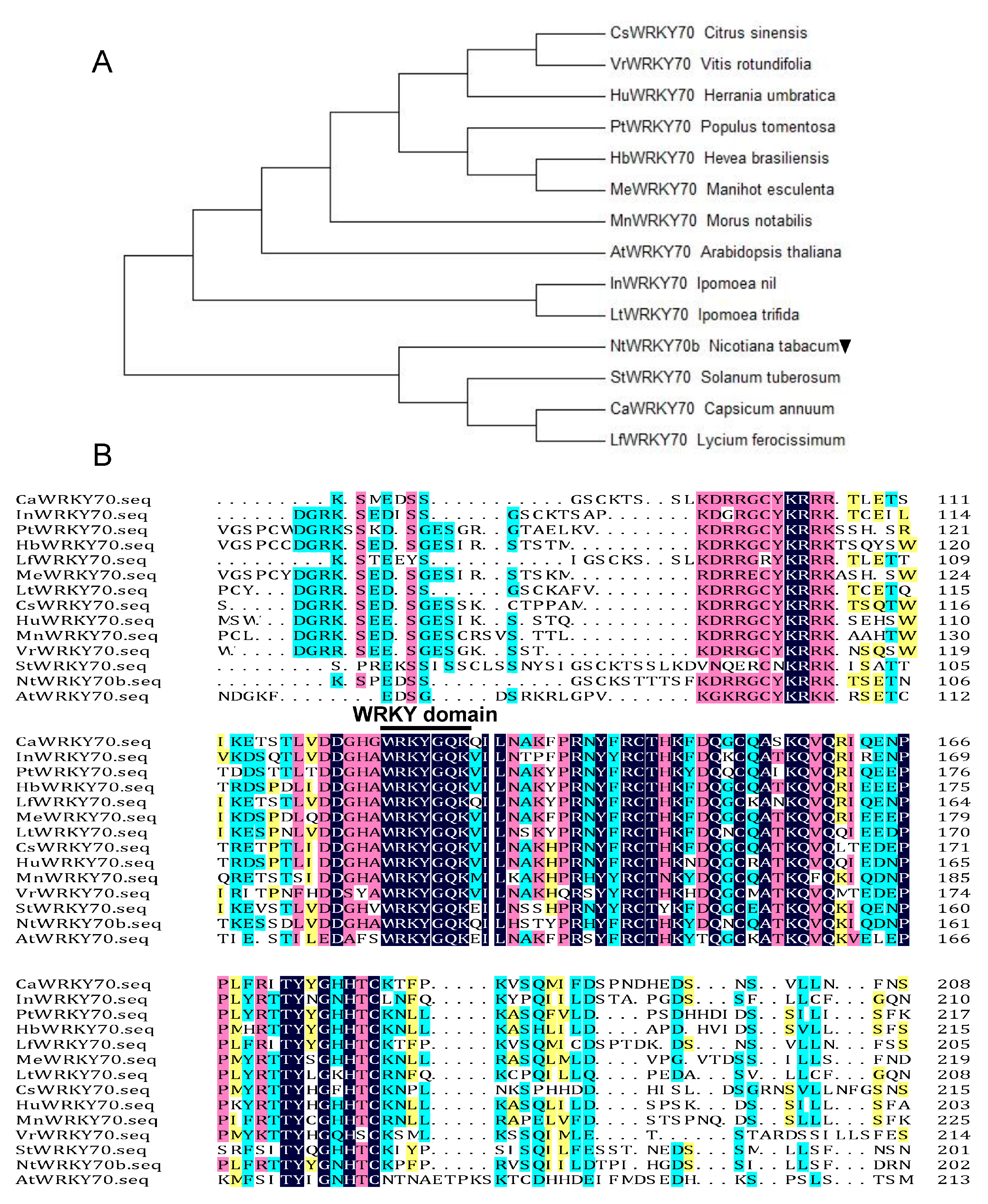
Based on our previous transcriptome data designed to screen senescence-associated genes with senescent leaves from the Nicotiana tabacum “K326” [6], we isolated a senescence-induced transcription factor gene named NtWYKY70b. According to the data of the National Center for Biotechnology Information (NCBI) and the Solanaceae Genomics Network, we searched the homologous genes of NtWRKY in other species and performed a multiple alignment and phylogenetic evolution analysis of these sequences by the DNAMAN and Neighbor-Joining (NJ) method, respectively. The results showed that NtWRKY70b encoded a 300-amino-acid protein containing conserved WRKY domains, including the WRKYGQ/KK core motif (Figure 1A,B), whose amino acid sequence was shown to be highly homologous to AtWRKY70 and LfWRKY70. Thus, the NtWRKY70b could be a senescence-associated WRKY transcription factor.
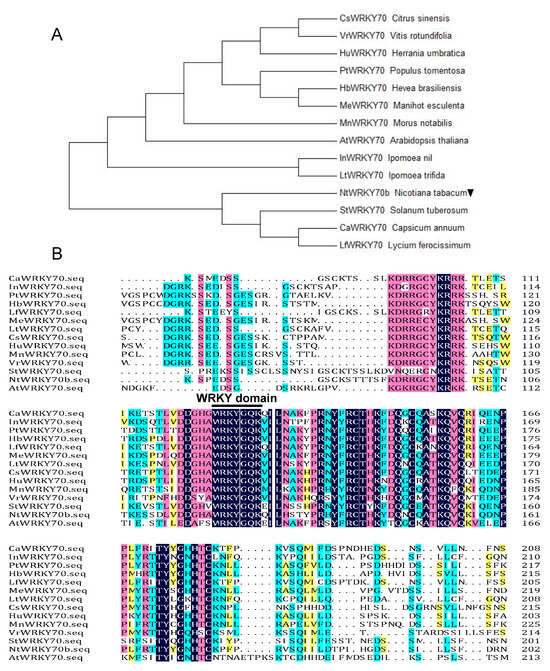
Figure 1.
Bioinformatics analysis and characterization of NtWRKY70b. (A) Phylogenetic analysis of WRKY70 orthologs from different species of plants utilizing the neighbor-joining (NJ) method in MEGA 7.0. NtWRKY70b is marked by a downward black triangle. (B) Sequences alignment of NtWRKY70b with 13 other WRKY70-like homologous protein sequences. The conserved core motif WRKYGQK in the WRKY domain is indicated by a black line. The NCBI accession numbers of these different WRKY homologous proteins are listed in Supplementary Table S1.
2.2. Expression of NtWRKY70b Is Upregulated during Leaf Senescence
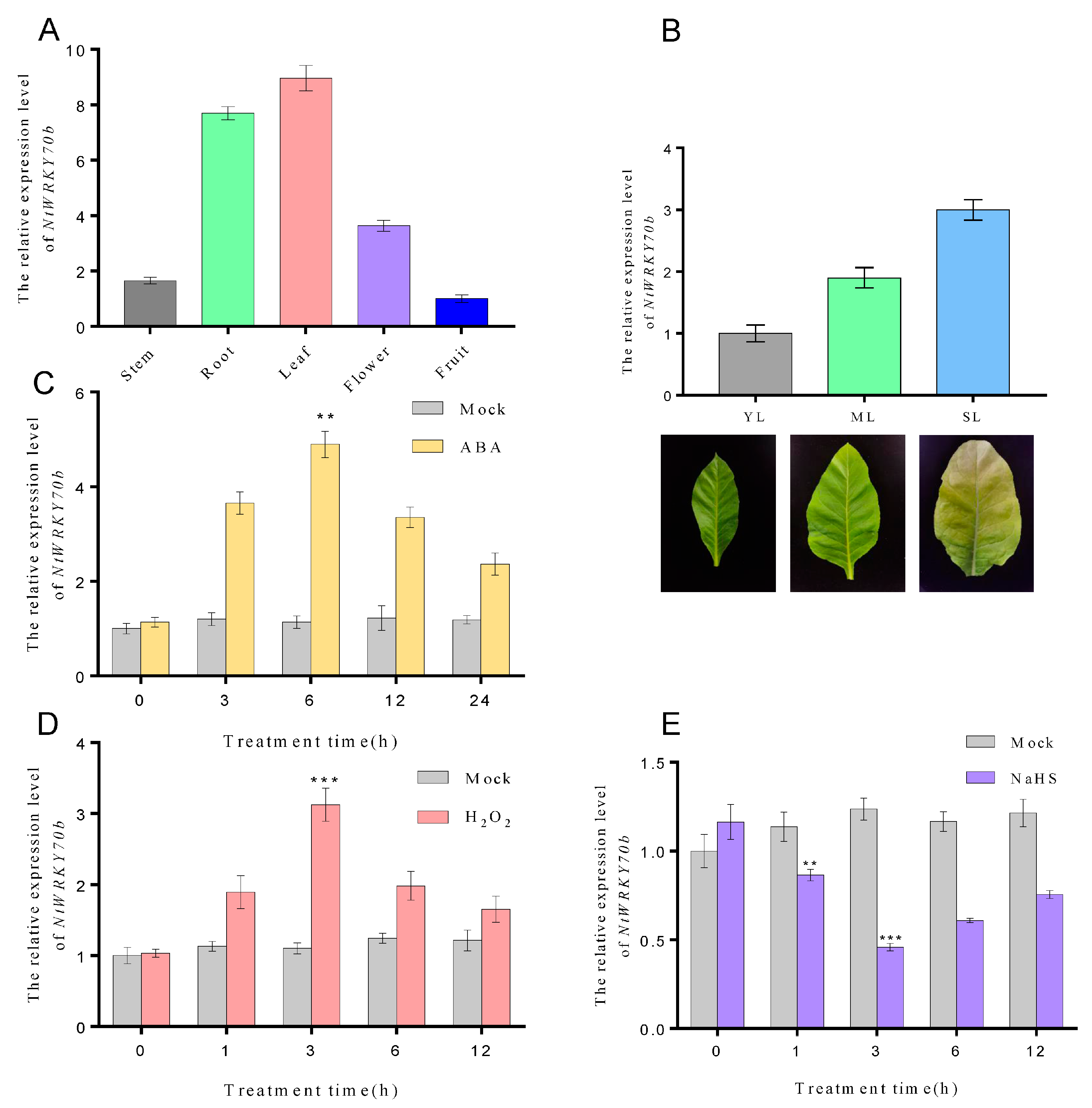
Our previous transcriptome data showed that the expression levels of NtWRKY70b were significantly elevated during natural aging. In order to deeply explore the spatiotemporal expression profiles of NtWRKY70b, we performed quantitative real-time PCR (qRT-PCR) to detect its transcript level in various tissues (e.g., root, stem, flower, fruit, young leaf, mature leaf, and senescent leaf) and under multiple abiotic stress conditions. As the results indicate, NtWRKY70b was expressed ubiquitously in all tissues mentioned above, and its expression was highest in the leaf, followed by root, flower, and stem, with the lowest level in fruit (Figure 2A). Furthermore, we found that the transcript level of NtWRKY70b was gradually increased during leaf aging processes (Figure 2B). Under ABA and H2O2 conditions, the expression of NtWRKY70b was strongly upregulated. As the qRT-PCR analysis showed, the expression levels of NtWRKY70b began to accumulate after 3 h of exposure to ABA treatment and peaked after 6 h of ABA treatment, exhibiting a five-fold increase compared to that in control check (CK) plants, and then declined gradually. Similarly, when plants were exposed to H2O2 treatment, the transcript levels of NtWRKY70b were significantly induced compared to that in CK plants (Figure 2C,D). Conversely, H2S treatment caused a decreased expression level of NtWRKY70b. Upon application of exogenous sodium hydrosulfide (NaHS, a H2S donor), the expression levels of NtWRKY70b started to decline after 1 h of NaHS treatment, and bottomed out after 3 h of exposure to NaHS, showing a 0.4-fold decrease relative to that of the CK group (Figure 2E). In summary, these findings indicated that NtWRKY70b might be involved in regulating leaf senescence and the abiotic stress response in plants through multiple signaling networks (e.g., ABA, ROS, and H2S).
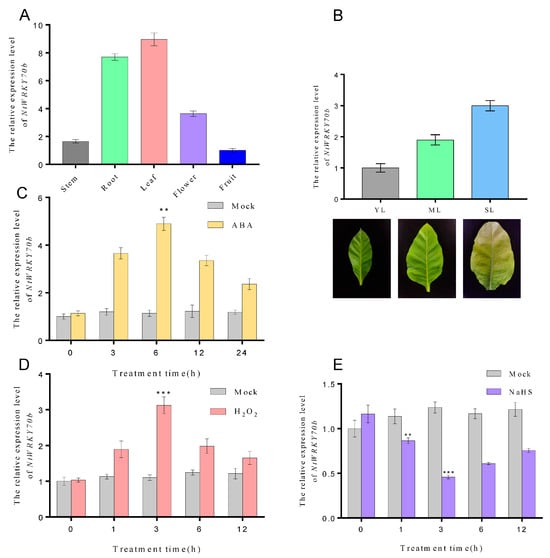
Figure 2.
Transcriptional patterns of NtWRKY70b in tobacco. (A,B) The expression levels of NtWRKY70b in various tissues of tobacco “K326”, such as stem, root, leaf, flower, and fruit (A), and in leaves at different stages of development, including young leaves YL, mature leaves ML, and senescent leaves SL (B). (C–E) The expression patterns of NtWRKY70b in 12-day-old “K326” seedlings under exogenous molecules treatments: ABA (100 μM), NaHS (10 μM), H2O2 (10 mM), and water as control (Mock). The values are represented as means ± SD, n = 3. Asterisks indicate statistically significant differences levels (Student’s t test, ** p < 0.01, and *** p < 0.001) with corresponding controls.
2.3. NtWRKY70b Is Localized in the Nucleus and Acts as a Transcriptional Activator
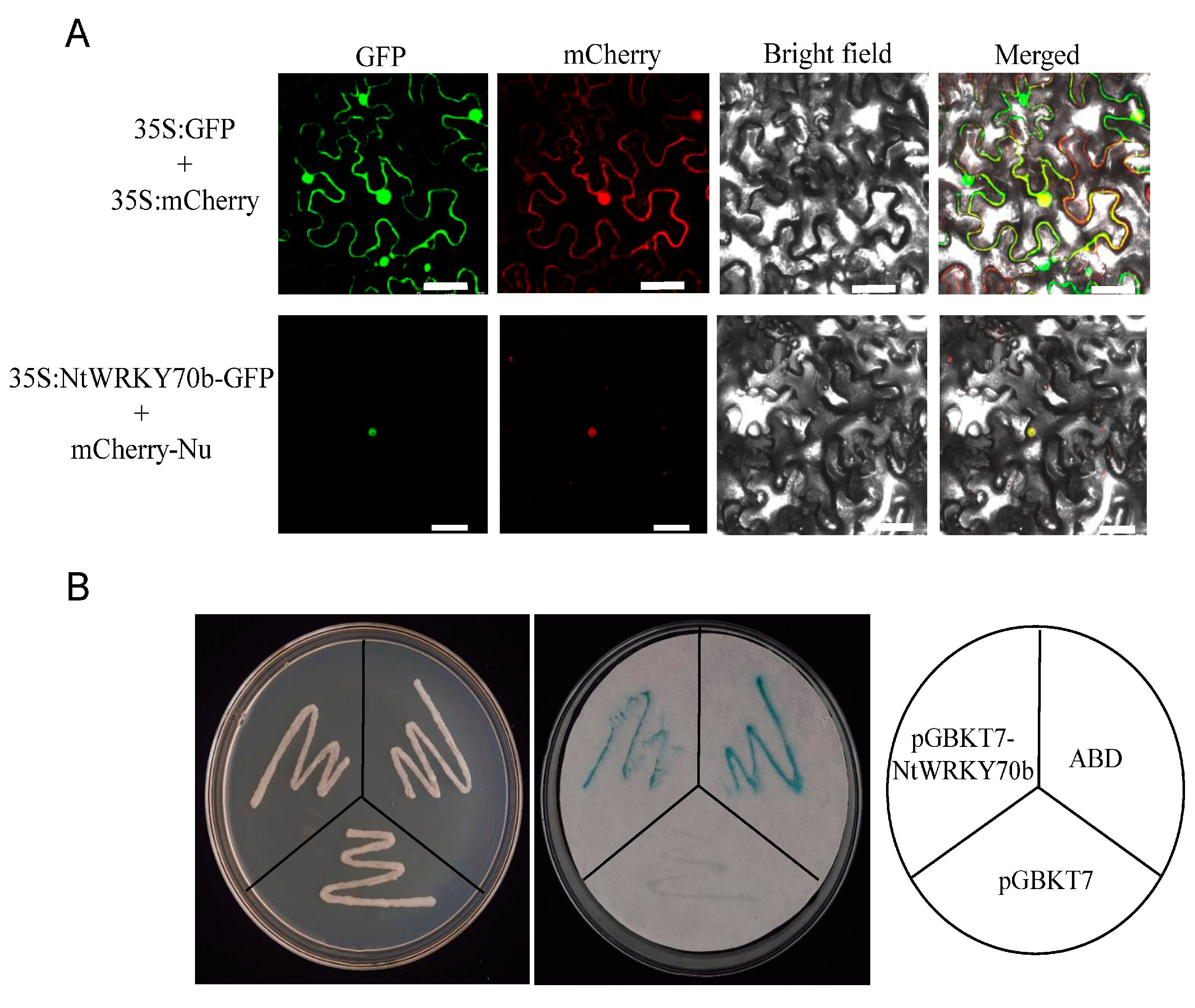
To investigate the subcellular localization of NtWRKY70b protein, we performed a green fluorescent protein (GFP)-labeled assay for the study of subcellular localization. The 35S:NtWRKY70b was transformed into leaves of N. benthamiana by the agrobacterium-mediated method. The fluorescent observation data showed that the NtWRKY70b-GFP signal was positioned in the nucleus, which was co-localized with the known nuclear marker BES1n-mCherry (Figure 3A), while the empty GFP was ubiquitously distributed throughout the cells. These results indicated that the NtWRKY70b was directly targeted to the nucleus of cells.
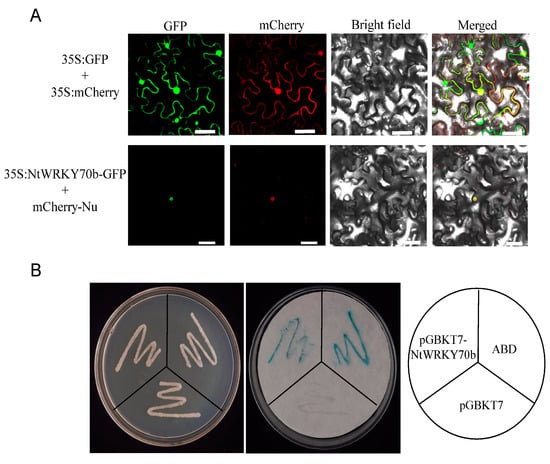
Figure 3.
Transcription factor characterization of NtWRKY70b. (A) NtWRKY70b is targeted in the nucleus in cells. NtWRKY70b-GFP, Empty GFP, and a nuclear marker (BES1n-mCherry) were expressed in the leaves of N. benthamiana (bars for 50 µm). (B) Transcriptional activation assay of NtWRKY70b in the yeast AH109. protein. The pGBKT7-AD performed the role of the positive control, while empty pGBKT7 was the negative control. The plates were incubated for 3 days and then subjected to the galactosidase assay.
To further test whether NtWRKY70b has the ability to activate or repress transcription, we constructed a recombinant yeast expression cassette pGBKT7-NtWRKY70b, then transformed it into the AHA109 yeast strain. As our observations show, all yeast transformants could grow normally on the plate containing SD/-Trp, which showed that these vectors had been successfully transformed into yeast cells. As a positive result of the transactivation assay, β-galactosidase gene (lacZ) is expressed. Yeast colonies that express lacZ turn blue in the presence of the chromogenic substrate X-β-Gal. The galactosidase assays showed that these yeast colonies carrying pGBKT7-NtWRKY70b and pGBKT7-AD displayed a remarkable blue color, while the empty control colonies did not appear blue (Figure 3B). The experiment confirmed that NtWRKY70b acted as a transcriptional activator to induce the expression of the β-gal reporter gene. Thus, the above results suggested that NtWRKY70b is a nuclear-localized transcriptional activator in cells.
2.4. NtWRKY70b Accelerates Dark-Induced Leaf Senescence
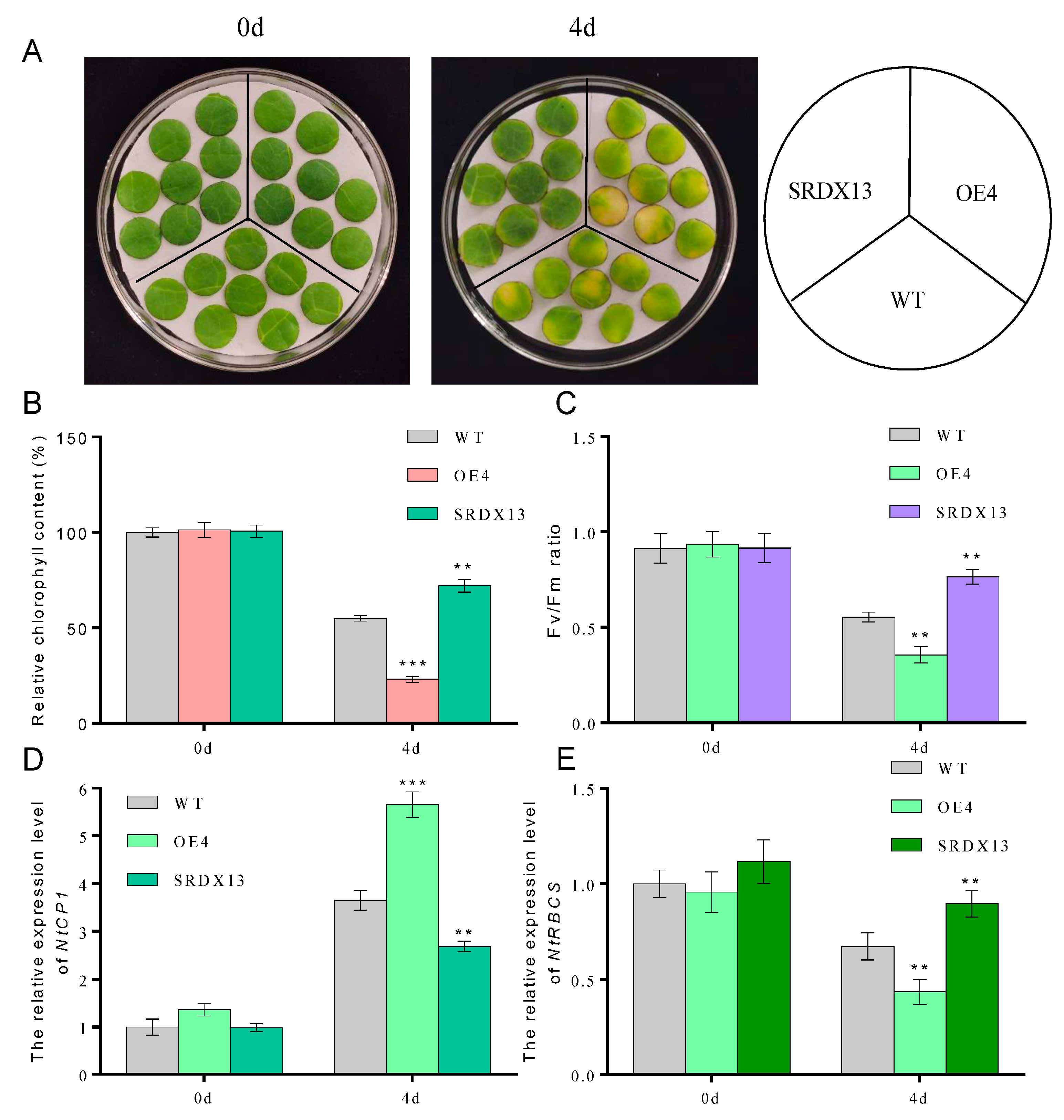
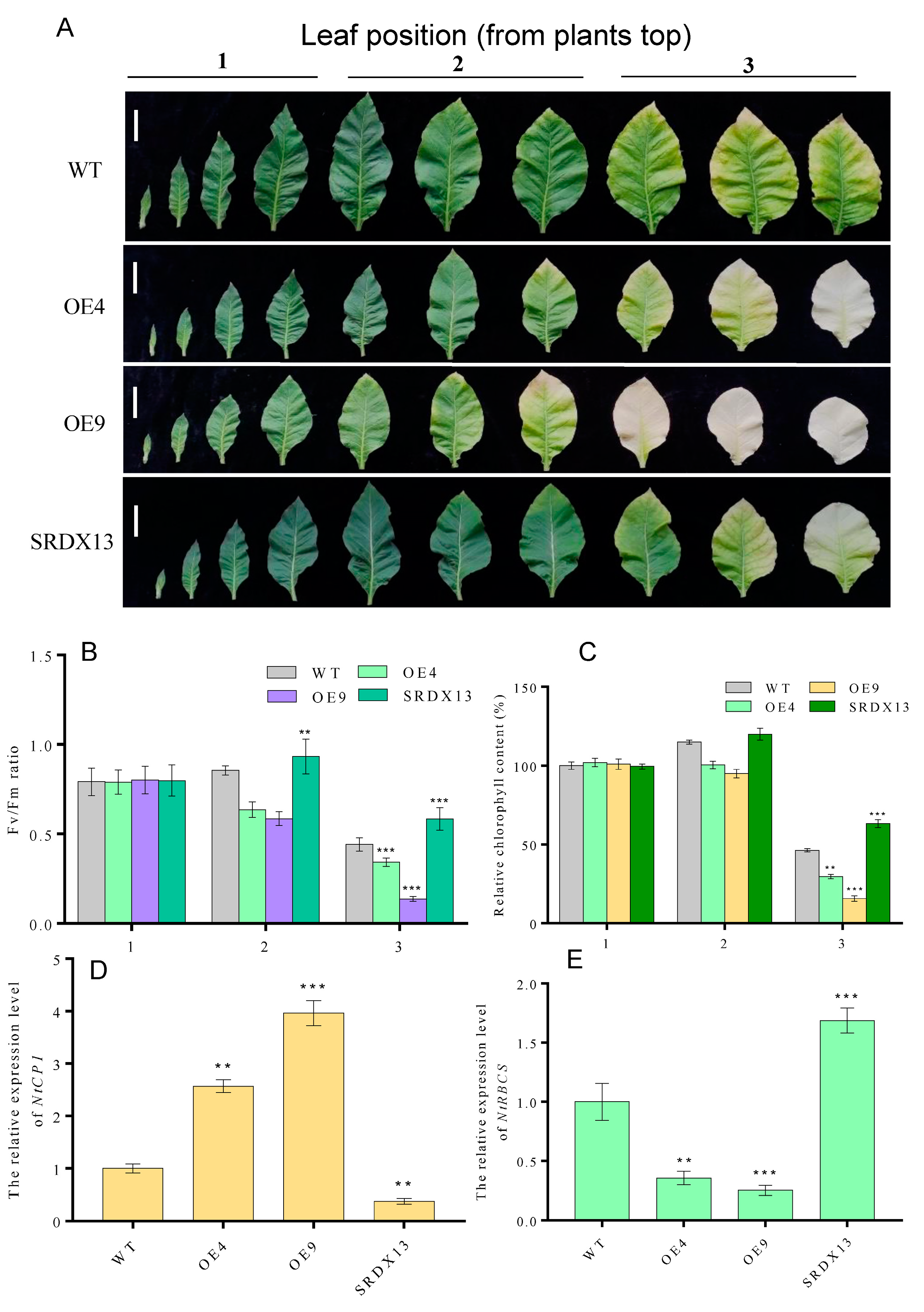
It is well known that dark treatment is often used to effectively accelerate the induction of leaf senescence [4,6]. To elucidate the potential biological roles of NtWRKY70b in senescence processes, we constructed overexpression cassettes 35S:NtWRKY70b (OE) and the dominant-negative vectors 35S:NtWRKY70b-SRDX (SRDX) and then transiently transformed these vectors into the tobacco (N. benthamiana) leaves. The qRT-PCR analysis showed that the transcript level of NtWRKY70b in transgenic leaves was increased approximately 23-fold relative to its level in the leaves containing the empty vector (CK). After the above-mentioned transformed tobacco leaves were subjected to dark treatment for 3 days, it was found that OE leaves showed an early yellowing phenomenon compared to CK (Supplementary Figure S1A,B). Moreover, the relative chlorophyll content and Fv/Fm in OE leaves were dramatically decreased compared with that in CK, and overexpression of NtWRKY70b triggered an elevated level of relative electrolytic leakage. However, the SRDX leaves showed the opposite phenotype (Supplementary Figure S1C–E). In addition, we obtained independent NtWRKY70b-overexpressing plants (12 lines) and NtWRKY70b:SRDX plants (10 lines) based on common tobacco “K326” by the agrobacterium-mediated method. To further test the key function of NtWRKY70b in darkness-induced leaf senescence, two independent transgenic lines (OE4 and SRDX13) were chosen to undergo a similar darkness treatment. In parallel with the findings in those transiently transgenic leaves, after exposure to darkness conditions for 4 days, transgenic line OE4 displayed significantly accelerated leaf aging relative to wild-type plants (WT), while SRDX13 plants regulated leaf senescence negatively. Moreover, the physiological analysis of chlorophyll content and Fv/Fm ratio in those transgenic plants demonstrated that NtWRKY70b plays positive roles in the regulation of dark-triggered senescence (Figure 4A–C). We also detected the transcript levels of CYSTEINE PROTEINASE 1 (NtCP1, SENESCENCE-ASSOCIATED GENE 12/SAG12 homolog in tobacco) and the RIBULOSE BISPHOSPHATE CARBOXYLASE SMALL CHAIN (NtRBCS), two well-known senescence-associated genes [6]. The expression of NtCP1 was remarkably increased in the overexpressing lines OE4 and decreased in the SRDX13 lines compared with that in WT, while the expression trend of NtRBCS in OE4 and SRDX13 lines was opposite to that of NtCP1 (Figure 4D,E). Therefore, these results indicate that NtWRKY70b takes part in promoting dark-induced leaf senescence.
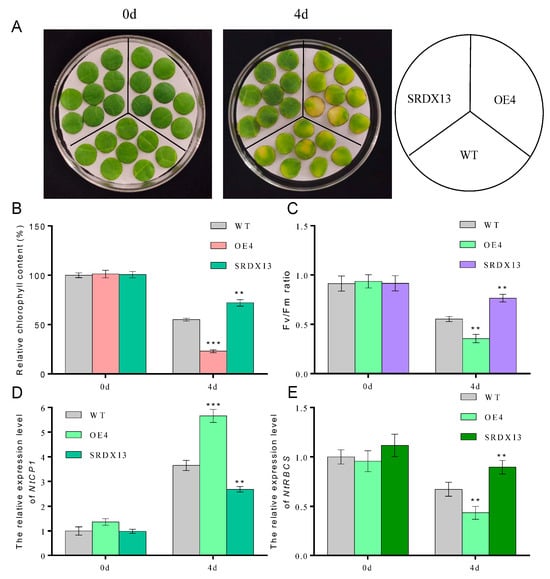
Figure 4.
Overexpression of NtWRKY70b accelerates dark-induced leaf senescence. (A) Phenotype observations of ten-week-old and 8th leaves of NtWRKY70b-OE, NtWRKY70b-SRDX, and WT tobacco plants “K326” for four days under dark conditions. (B,C) Chlorophyll content (A) and Fv/Fm (C) of leaves in (A). (D,E) The expression level of NtCP1 (D) and NtRBCS (E) of these transgenic lines in (A). The values are represented as means ± SD, n = 3. Asterisks indicate statistically significant differences levels (Student’s t test, ** p < 0.01, and *** p < 0.001) with corresponding controls.
2.5. NtWRKY70b Promotes Age-Triggered Leaf Senescence via Modulating Levels of ROS and H2S
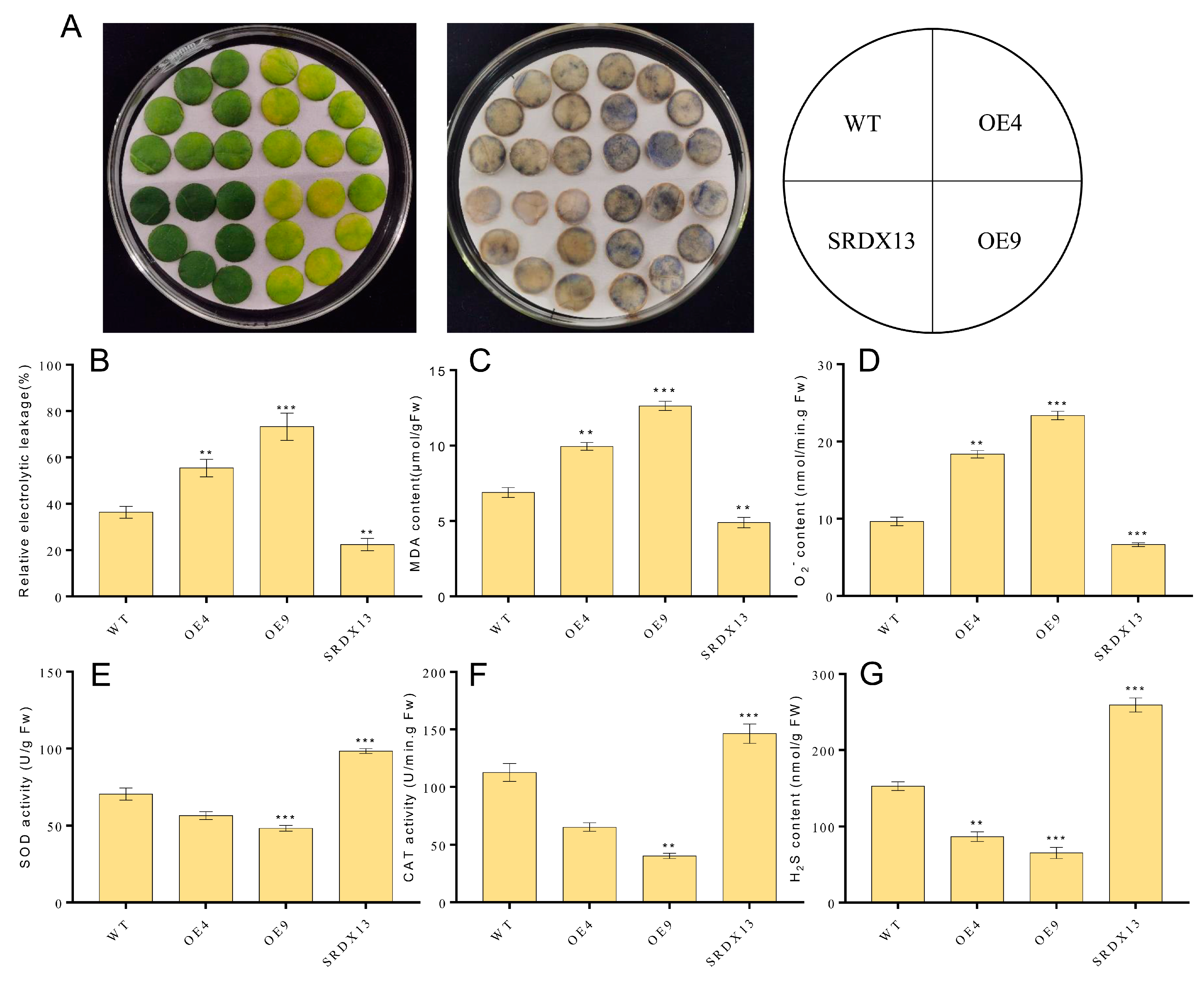
As described above, the expression of NtWRKY70b was upregulated gradually during natural senescence. In order to further explore whether NtWRKY70b is involved in regulating the progress of leaf aging, we first studied the aging phenotype of NtWRKY70b transgenic lines under normal growth conditions. The results showed that compared with WT, the NtWRKY70b-overexpressing lines OE4 and OE9 showed an obviously premature senescence phenotype, while the disruption of NtWRKY70b function caused delayed leaf senescence (Figure 5A). In order to further verify the above aging phenotype from the physiological level, we tested the relative chlorophyll content and Fv/Fm ratio of these transgenic lines. The results showed that the relative chlorophyll content and Fv/Fm ratio of OE4 and OE9 were significantly lower than those of WT, while the relative chlorophyll content and Fv/Fm ratio of SRDX13 plants were increased significantly compared with that of WT (Figure 5B,C). Furthermore, the expression of senescence marker genes NtCP1 and NtRBCS in transgenic lines is shown in Figure 5D,E. In OE4 and OE9 lines, the expression of NtCP1 is higher, while the expression of NtRBCS is lower, compared to WT. Numerous reports have demonstrated that excess ROS induces leaf senescence. To determine whether NtWRKY70b influences ROS accumulation in the leaf senescence process, we detected the ROS metabolism by NBT staining, relative electrolytic leakage, malondialdehyde (MDA), and activity analysis of multiple antioxidases. The observation indicated that overexpressing lines OE4 and OE9 contained a higher accumulation of O2− than WT, while SRDX13 had a lower level of ROS relative to WT. Moreover, the relative electrolytic leakage and malondialdehyde (MDA) were significantly higher in OE4 and OE9 than those in WT (Figure 6A–D). We also tested the activities of some antioxidases, including superoxide dismutase (SOD) and catalase (CAT) (Figure 6E,F). There were decreases in the activities of SOD and CAT in the OE4 and OE9 lines, leading to elevated levels of ROS. According to the NaHS-reduced expression of NtWRKY70b mentioned above, we detected endogenous H2S production in OE, SRDX, and WT plants during leaf senescence. As shown in Figure 6G, the H2S content in NtWRKY70b-OE lines showed a significant decrease compared to that in WT, while there was more H2S production in NtWRKY70b-SRDX lines. It can be postulated that NtWRKY70b plays essential functions in promoting leaf senescence via regulating the accumulation of ROS and H2S scavenging.
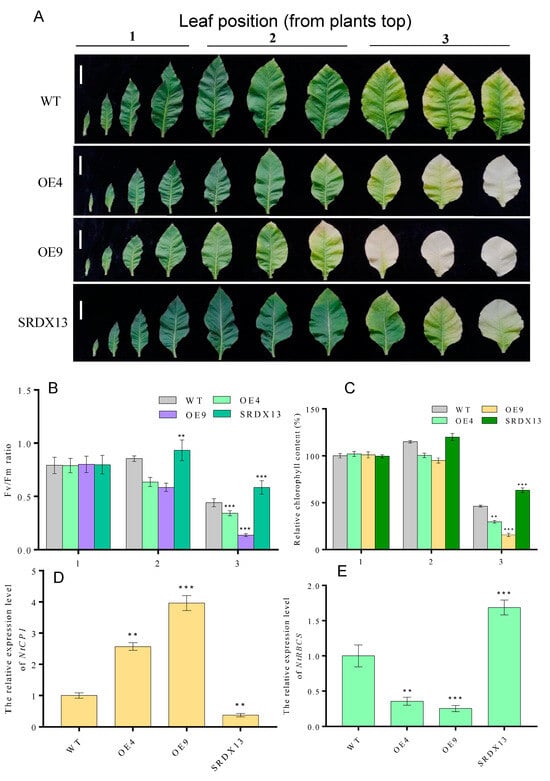
Figure 5.
Overexpression of NtWRKY70b promotes age-dependent leaf senescence. (A) Phenotype of twelve-week-old transgenic plants ((bars for 5 cm)). (B,C) Chlorophyll content (B) and Fv/Fm (C) of leaves in (A). (D,E) The expression level of NtCP1 (D) and NtRBCS (E) of these transgenic lines in (A). (1) The first to fourth leaves as counted from the top, (2) the fifth to seventh leaves from the top, and (3) the eighth to tenth leaves from the top. The values are represented as means ± SD, n = 3. Asterisks indicate statistically significant differences levels (Student’s t-test, ** p < 0.01, and *** p < 0.001) with corresponding controls.
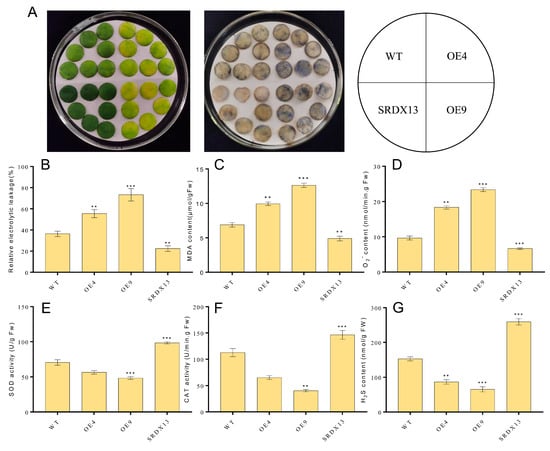
Figure 6.
NtWRKY70b regulates ROS and H2S metabolism during leaf senescence. (A) NBT staining. Three independent experiments were conducted (n ≥ 20), showing similar results. (B) Ions leakage rate. (C) MDA content. (D) O2.− content. (E,F) Activity analysis of antioxidant enzymes SOD € and CAT (F) in transgenic plants and WT. (G) H2S content. The data are means ± SD of three biological replicates. Asterisks indicate statistically significant differences levels (Student’s t test, ** p < 0.01, and *** p < 0.001) with corresponding controls.
2.6. NtWRKY70b Regulates the Expression of Synthesis-Related Genes of ROS and H2S
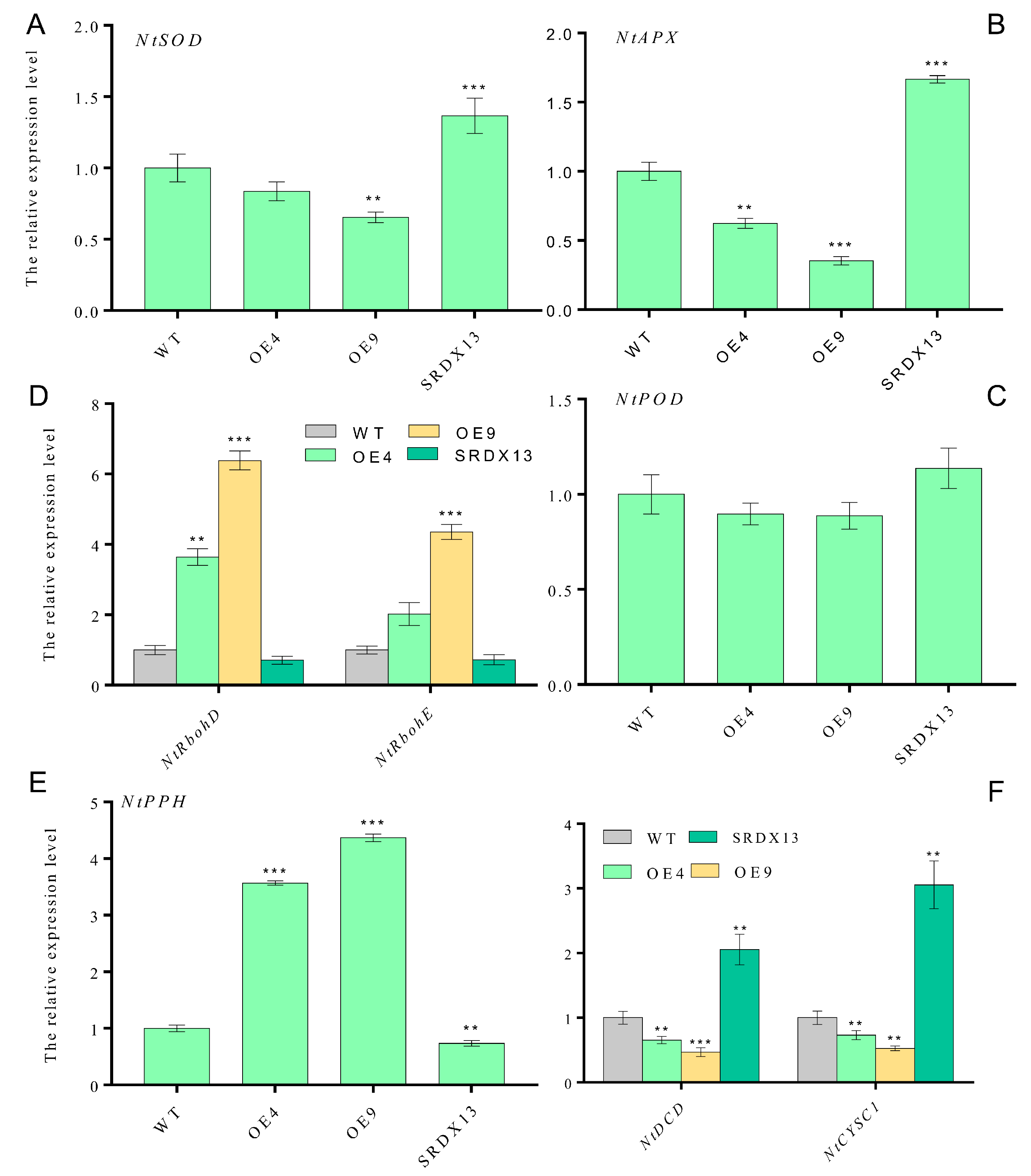
To further decipher the molecular mechanism of NtWRKY70b influencing leaf senescence, through qRT-PCR analysis, we determined the transcriptional level of some ROS-synthesis/scavenging-related genes (e.g., NtRbohD, NtRbohE, NtSOD, NtAPX, and NtPOD), H2S-synthesis-related genes (e.g., NtDCD and NtCYSC1), and chlorophyll-catabolic genes (e.g., NtPPH). These results showed that the expression levels of ROS-scavenging-related genes (NtSOD, NtAPX, and NtPOD) were decreased in overexpressing plants (OE4 and OE9) compared with that in WT (Figure 7A–C), but there was an obvious increase in NtRbohD and NtRbohE transcription in overexpressing plants (Figure 7D), thereby triggering excess ROS production. In addition, we found that the expression level of NtDCD1 and NtCYSC1, two novel H2S biosynthesis genes, were significantly downregulated in NtWRKY70b-OE plants (Figure 7F), implying that NtWRKY70b inhibited the production of H2S through directly or indirectly regulating the expression of NtDCD1 and NtCYSC1. In summary, these results demonstrates that NtWRKY70b regulates the accumulation of ROS and H2S via directly or indirectly regulating the expression of their synthesis-related genes, subsequently promoting natural leaf senescence.
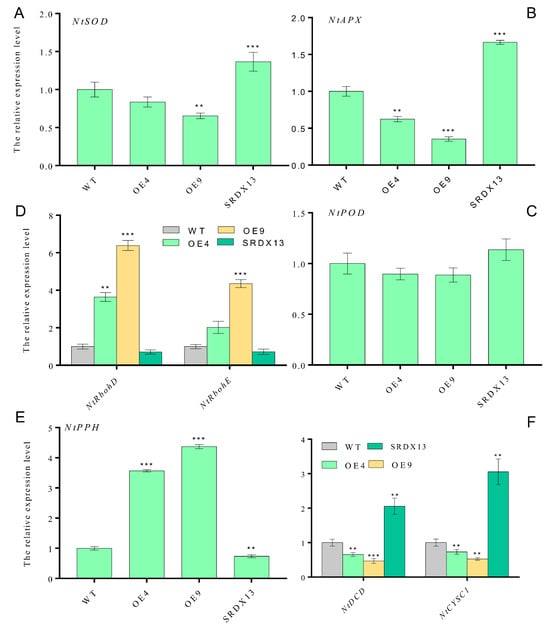
Figure 7.
NtWRKY70b influences the expression levels of some biosynthesis genes of ROS and H2S. (A) NtSOD. (B) NtAPX. (C) NtPOD. (D) NtRbohD and NtRbohE. (E) NtPPH. (F) NtDCD and NtCYSC1. The 8th leaves were harvested from 12-week-old transgenic plants grown under natural long-day conditions in soil and were used for qRT-PCR analysis. The expression levels of these genes were normalized to that of actin, an internal control. The data are means ± SD of three biological replicates. Asterisks indicate statistically significant differences levels (Student’s t test, ** p < 0.01, and *** p < 0.001) with corresponding controls.
2.7. NtWRKY70b Directly Regulates Expression of NtRbohD and NtPPH by Binding to Their Promoters
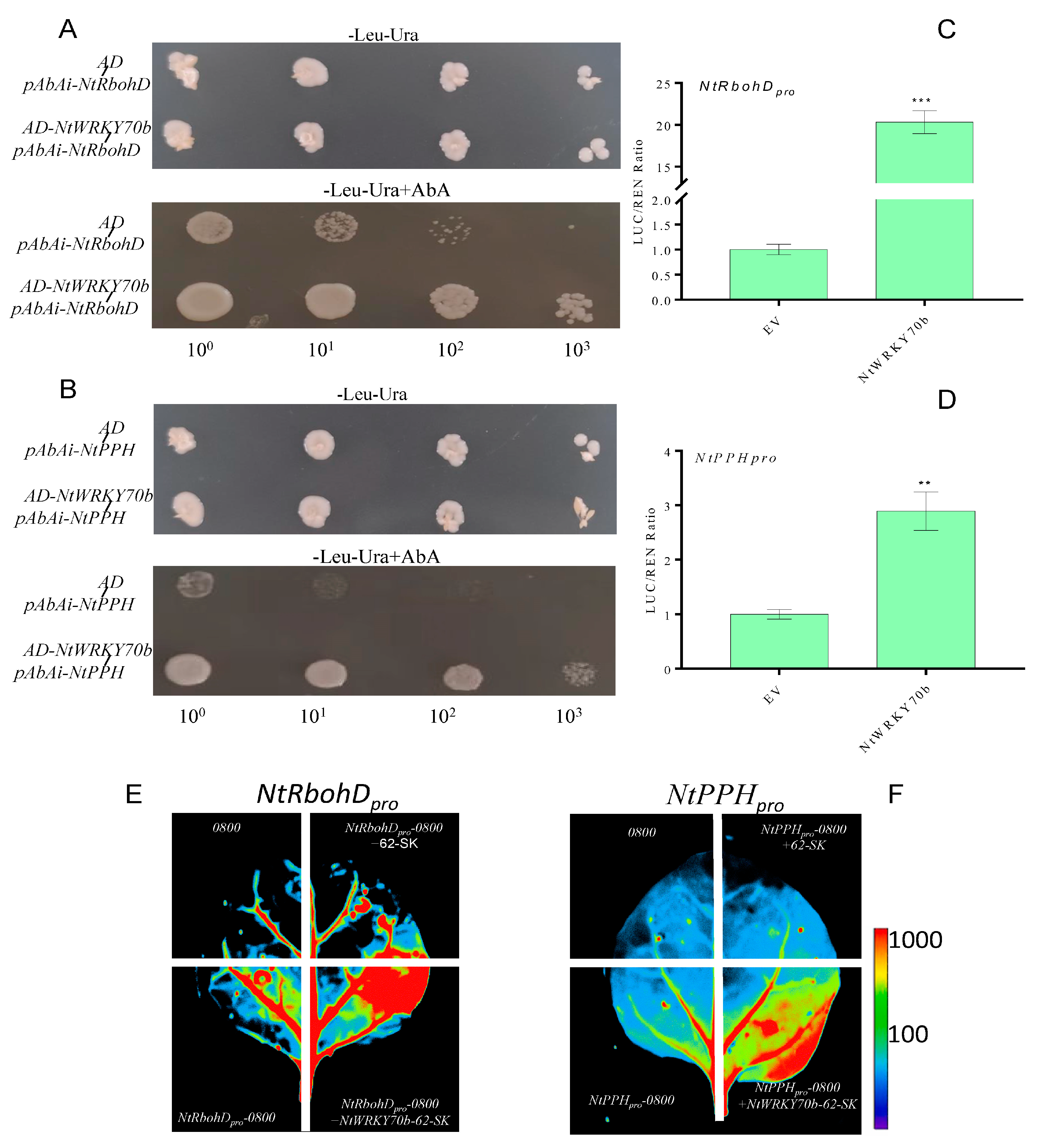
The observations mentioned above revealed that the expression levels of ROS-synthesis/scavenging-related genes are significantly changed in NtWRKY70b-OE lines, and the ROS content is the highest in the overexpression lines. To verify whether NtWRKY70b promotes endogenous ROS accumulation by directly regulating the expression of these genes, we first analyzed the promoter sequences of related genes. We found that the promoter sequences of NtRbohD contains one W-box element, which is the conserved sequence bound by WRKY TFs. Furthermore, in the promoter sequences of NtPPH, there were several W-box elements (Supplementary Figure S2). These findings indicate that NtRbohD and NtPPH could be direct target genes of NtWRKY70b. We used yeast one-hybrid analysis (Y1H) to verify whether NtWRKY70b could directly bind to the promoters of the above two genes. The pGADT7-NtWRKY70b construct was co-transfected with pAbAi-NtRbohD pro and pAbAi-NtPPH pro plasmids into yeast strain Y1HGold, respectively. The experimental results demonstrated that NtWRKY70b directly bound to the promoter sequences of NtRbohD and NtPPH in yeast cells (Figure 8A,B). Furthermore, we performed the dual-luciferase assay with the N. benthamiana leaf system to confirm NtWRKY70b directly binding to the promoter sequences of NtRbohD and NtPPH. The promoters of NtRbohD and NtPPH were inserted into the pGreenII0800 vector, then these recombinant plasmids were transformed into leaves of the N. benthamiana through the agrobacterium-mediated method. The observations from the dual-luciferase reporter system demonstrated that NtWRKY70b directly regulates pGreenII0800-LUC-NtRbohDpro and pGreenII0800-LUC-NtPPHpro in vivo (Figure 8C–F). Overall, these results reveal that NtWRKY70b directly binds to the promoter sequences of NtRbohD and NtPPH and regulates their expression, thereby promoting age-dependent leaf senescence.
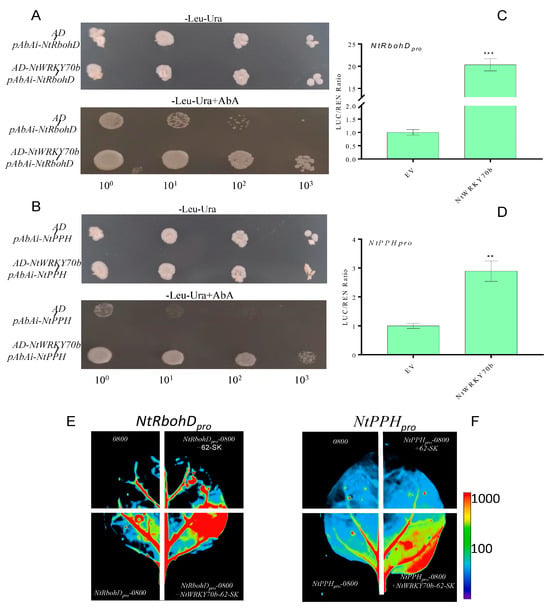
Figure 8.
NtRbohD and NtPPH are the direct target genes of NtWRKY70b. (A,B) NtWRKY70b binds to the promoters of NtRbohD and NtPPH in yeast cells. (C–F) Dual-luciferase reporter assays show that NtWRKY70b directly affects the expression of NtRbohD (C,E) and NtPPH (D,F) in tobacco leaves. Three independent experiments were performed, showing similar results. The data are means ± SD of three biological replicates. Asterisks indicate statistically significant differences levels (Student’s t test, ** p < 0.01, and *** p < 0.001) with corresponding controls.
2.8. H2S Suppresses Early Aging Phenotype of NtWRKY70b-Overexpressing Plants
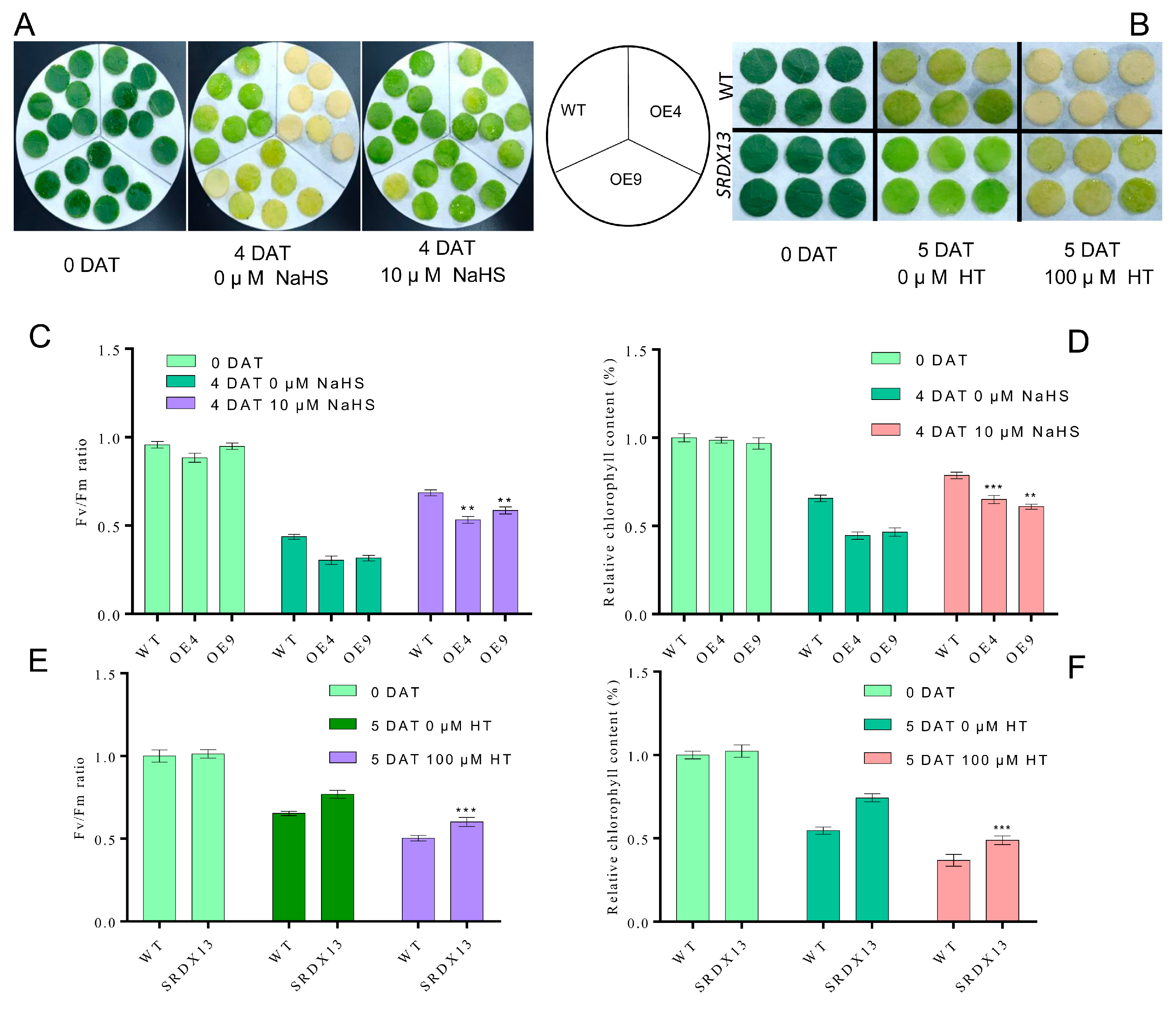
Emerging studies have revealed that H2S plays essential roles in the inhibition of leaf senescence [46,48]. Given our findings that, during leaf senescence, NtWRKY70b suppresses the expression of NtDCD1 and NtCYSC1, two putative H2S biosynthesis genes, thereby inhibiting H2S production, in order to confirm H2S acting as a downstream of NtWRKY70b, we studied whether the modification of H2S content in NtWRKY70b-OE lines could influence the accelerated leaf senescence phenotype. To this end, pharmacological tests were performed to check H2S functions in the leaf senescence process using sodium hydrosulfide (NaHS, a H2S donor) and hypotaurine (HT, a H2S scavenger). Upon application of NaHS treatment, these transgenic lines and WT displayed a delayed senescence phenotype compared to CK (Figure 9A), while HT treatment accelerated the progress of leaf senescence (Figure 9B). We also determined the chlorophyll content and Fv/Fm ratio of these NtWRKY70b-OE, NtWRKY70b-SRDX, and WT plants under NaHS and HT treatments (Figure 9C–F). These findings showed that pretreatment with NaHS increased chlorophyll content and decreased electrolytic leakage in plants compared to CK, yet HT treatment triggered opposite results. Collectively, these results indicate that the decreased H2S content in NtWRKY70b-OE lines serves as a key mechanism underlying its accelerated leaf senescence phenotype.
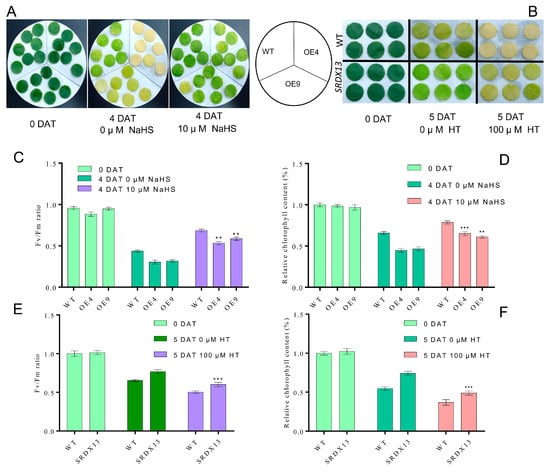
Figure 9.
H2S represses the premature leaf senescence of NtWRKY70b-OE plants. (A) Phenotypic analysis of NtWRKY70b-OE and WT leaves after four days of 0 and 10 μM NaHS treatment under dark conditions. (B) Phenotypic observation of NtWRKY70b-SRDX and WT after five days of 0 and 100 μM HT treatment under dark condition. (C) Fv/Fm ratio of leaves in (A,D) Relative chlorophyll content of leaves in (A,E) Fv/Fm ratio of leaves in (B,F) Relative chlorophyll content of leaves in (B). The data are means ± SD of three biological replicates. Asterisks indicate statistically significant differences levels (Student’s t test, ** p < 0.01, and *** p < 0.001) with corresponding controls.
3. Discussion
Senescence is a tightly programmed process of plant cell death, regulated by integrating external environmental and internal developmental signals. Plants have evolved a myriad of highly complex strategies to adapt to changing environmental conditions, such as leaf senescence, thereby enhancing plant fitness and reproduction [50]. An increasing number of studies have revealed that premature senescence significantly affects crop yield. Some stay-green mutants of crop plants could significantly increase biomass and yield. Therefore, dissecting the molecular mechanisms of leaf senescence can provide a theoretical basis for breeding varieties with higher yields [51,52]. This study aimed to revealed the molecular mechanism of NtWRKY70b regulating leaf senescence in tobacco.
Transcription factors, acting as hubs, participate in the leaf senescence process by regulating genes involved in chlorophyll degradation metabolism, aging-related genes, hormone synthesis genes, etc. They bind to specific cis-acting elements in the target gene promoters to regulate downstream gene expression, leading to the activation or inhibition of target genes during senescence. In Arabidopsis, the genes of 96 transcription factors are upregulated at least threefold in senescent leaves [53]. These transcription factors belong to 20 different families, with the largest families being NAC, WRKY, C2H2-type zinc finger, AP2/EREBP, and MYB proteins. WRKY TFs, as one of the largest transcription factor families in plants, play essential roles in the leaf senescence process. In the past decades, transcriptome analysis has demonstrated that a large number of genes are senescence-associated genes (SAGs), but only a small fraction of genes have been functionally characterized. In this study, we cloned and identified a tobacco WRKY TF, designated NtWRKY70b, acting as a positive regulator of natural leaf senescence. The expression profile analysis showed that NtWRKY70b transcript levels were induced by aging and hydrogen peroxide (H2O2) and downregulated upon hydrogen sulfide (H2S) treatment. Overexpression of NtWRKY70b (OE) clearly promoted leaf senescence, triggering increased levels of reactive oxygen species (ROS) and decreased H2S content, while functional inhibition of NtWRKY70b delays leaf senescence. In summary, our study provides new insights into the connection between transcription factors and leaf senescence.
Previous studies have demonstrated that reactive oxygen species (ROS) serve as important signaling molecules, playing crucial roles in various growth and developmental processes, including plant senescence [35,54]. When ROS accumulate excessively in plant cells, lipid peroxidation is initiated, leading to cell membrane damage and, in severe cases, cell death. As signaling molecules, ROS can regulate plant senescence through complex signaling systems. However, the upstream regulatory mechanism of endogenous ROS biosynthesis in the leaf senescence process is largely unknown. Here, we found that the accumulations of superoxide anions (O2−), ion leakage, and MDA content in NtWRKY70b-OE lines were higher than that in WT, and ROS-scavenging enzyme activity in NtWRKY70b-OE was lower. Yet the results of SRDX13 showed opposite trends. These results indicate that overexpression of NtWRKY70b leads to massive ROS accumulation in senescent leaves. We further explored its molecular mechanisms. In this study, we found that the ROS-synthesis-related genes NtRbohD and NtRbohE were upregulated in NtWRKY70b-OE, while the expression of H2S-synthesis-related genes (e.g., NtDCD1 and NtCYSC1) was downregulated. It is well known that hydrogen sulfide (H2S) is an important gasotransmitter that could enhance the expression of various stress-resistant genes and signal transduction, alleviating oxidative stress damage to plants [55]. Studies have reported that the blocking function in LCD1 reduces H2S production, leading to premature senescence of tomato leaves [46]. In addition, exogenous H2S reduces the production of superoxide anions (O2.−), malondialdehyde (MDA), and H2O2 in tomato fruits [56]. In summary, H2S could negatively regulate tomato leaf senescence, but its regulatory network of regulating leaf senescence in plants remains unclear. In this study, we designed pharmacological experiments involving exogenous application of NaHS and HT. The results indicate that HT can alleviate the stay-green phenotype of NtWRKY70b-SRDX, while exogenous NaHS delays the premature senescence of NtWRKY70b-OE, implying that H2S is a key downstream biomolecule of NtWRKY70b. In Arabidopsis, 10 Rboh genes may function in a tissue- or organ-specific manner [19]. We found that the expression levels of two Rboh putative homologous genes (e.g., NtRbohD and NtRbohE) in tobacco were significantly upregulated during leaf senescence, and NtWRKY70b could bind to their promoter sequences and activate their expressions. It is further elucidated that the molecular mechanism of NtWRKY70b accelerates leaf senescence by regulating the accumulation of ROS. From the above results, it can be found that overexpression of NtWRKY70b can change the expression levels of ROS- and H2S-related genes, and further studies showed that NtWRKY70b can promote the production of reactive oxygen species by directly binding to the promoters of NtRbohD and NtPPH, but it has not been found that NtWRKY70b can directly regulate the expression of other genes (e.g., NtRbohE, NtSOD, NtAPX, NtPOD, NtDCD, or NtCYSC1). So, we can speculate that in addition to NtWRKY70b directly regulating NtRbohD and NtPPH expression, there may be other indirect regulatory pathways and feedback inhibition pathways in the process of NtWRKY70b regulating leaf senescence.
Leaf senescence represents the final stage of leaf growth and development, and its complex regulatory networks have been extensively studied and reported by many researchers. An increasing number of reports have revealed that transcription factors (TFs) act as crucial regulators in regulating leaf senescence. In this study, we identified a novel transcription factor, NtWRKY70b, which played a positive regulatory role in leaf senescence process. Furthermore, these findings indicate that NtWRKY70b directly binds to the promoters of the ROS-synthesis-related gene NtRbohD and the chlorophyll degradation gene NtPPH, thereby activating their expression and subsequently promoting ROS accumulation and accelerating cellular oxidation and chlorophyll degradation processes, ultimately leading to premature leaf senescence. Intriguingly, we found that the overexpression of NtWRKY70b caused decreased expression levels of the H2S synthesis genes (e.g., NtDCD1 and NtCYSC1), inhibiting endogenous H2S synthesis then accelerating leaf senescence. It will be important to study the molecular mechanism of H2S participates in the regulation of leaf senescence. In Arabidopsis, it was reported that AtWRKY70 TF played a negative regulatory role in leaf senescence [24]. The difference with our observations may be due to the functional specificity of WRKY TFs derived from different plant species. So, it will be interesting to further investigate the transcriptional and translational regulatory network of NtWRKY70b taking part in regulating the onset and progress of plant senescence, and whether this mechanism is conserved for other plant species. A deeper understanding of the molecular and physiological mechanisms of H2S-mediated leaf senescence is crucial for advancing agricultural practices, optimizing crop productivity, and managing natural ecosystems in a changing environment.
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
All transgenic plants used in this study were in the Nicotiana tabacum L. Cv. “K326” background. Nicotiana benthamiana was used for transient transformation assays. Those seeds mentioned above were surface-sterilized with 75% ethanol and grown on 1/2 Murashige and Skoog medium containing 20 g/L sucrose, 0.5 g/L MES and 1.1% agar (PH = 5.8) for 10 days. The seedings of those lines were transferred to soil to continue growth under the same growth conditions (28 °C, 16 h light/8 h dark and 55% relative humidity). When cultivating “K326” to 12 true leaves, the 3rd, 6th and 10th leaves from the top were named as young leaves (YL), mature leaves (ML), and senescence leaves (SL), respectively. The 5-week-old Nicotiana benthamiana leaves were selected for subcellular localization and transient transformation experiments.
4.2. RNA Extraction and Quantitative Real-Time PCR Analysis
Total RNA was extracted from “K326” using the M5 Total RNA Extraction Reagent (Mei5bio, Beijing, China) according to the manufacturer’s instructions, and first-strand cDNAs were synthesized by the HiScript III RT SuperMix by qPCR (Vazyme, Nanjing, China). qRT-PCR was performed on Thermal Cycler Dice® Real-Time System III (TaKaRa, Kusatsu, Japan) by using SYBR Green Master Mix as described previously [4]. The NtActin was used as an internal control gene, and the expression levels of genes were calculated with the 2−ΔΔCT method. Primers used for qRT-PCR are listed in the Supplementary Table S2.
4.3. Subcellular Localization of NtWRKY70b Protein
To clarify the subcellular localization of the NtWRKY70b protein, the CDS sequence of the NtWRKY70b gene was connected with linearized 2301-S2-GFP vector to construct 2301-S2-GFP-NtWRKY70b fusion expression vector, which was transformed into the Agrobacterium strain GV3101. 2301-S2-GFP-NtWRKY70b and the nucleus marker were injected into the leaves of 1-month-old Nicotiana benthamiana. After 1 d dark treatment and 2 d long-day condition (16 h light/8 h dark), the fluorescence signals were observed by laser confocal microscope (LEICA TCS SP5II, Wetzlar, Germany).
4.4. Transactivation Activity Assay
To explore transactivation activities of NtWYKY70b protein, the CDS fragment of NtWYKY70b was amplified by PCR and fused in-frame to the BD domain of pGBKT7 vector, yielding the pGBKT7-NtWYKY70b fusion constructs. The recombinant plasmids of pGBKT7-NtWYKY70b, pGBKT7-AD, and the empty vector pGBKT7 were transformed into yeast strain AH109, respectively. Yeast transformants harboring the above plasmids were plated on SD/-Trp and SD/-Trp/-His medium and incubated at 28 °C for 72 h. We attached the colony to the filter paper and then transferred the filter paper to liquid nitrogen and froze it for 20 s. We removed the filter paper and melted it at room temperature. We repeated the freezing and thawing process three times. Transactivation activity of the fused NtWYKY70b protein was assessed by the growth and production of blue pigments.
4.5. Generation of Transgenic Plants
For NtWYKY70b overexpression lines, the CDS fragment of NtWYKY70b was cloned into pCAMBIA2301-S2 vector under the control of cauliflower mosaic virus (CaMV) 35S promoter, constructing the pCAMBIA2301-S2-NtWYKY70b recombinant plasmid. For NtWYKY70b -SRDX lines, the coding region sequence of NtWYKY70b without termination codons was used as a template, and an SRDX inhibitory domain was added at the end of the reverse primer to construct NtWYKY70b-SRDX expression vector. Those above fused plasmids were transformed into Agrobacterium strain EHA105, then introduced into leaves of “K326” through Agrobacterium-mediated transformation. The transgenic lines were screened through planting seeds on MS medium containing 200 mg/L kanamycin.
4.6. Transient Transformation of NtWYKY70b
To study the transient expression, the coding sequence of NtWYKY70b was amplified and a wpCAMBIA2301-S2 vector was inserted, yielding the pCAMBIA2301-S2-NtWRKY70b construct. pCAMBIA2301-S2-NtWRKY70b recombinant plasmid and pCAMBIA2301-S2 empty plasmid (CK) were transformed into Agrobacterium strain GV3101, respectively. Those above strains were re-suspended in solution for injection (10 mmol/L MES, 200 mmol/L AS, 10 mmol/L MgCl2, pH = 5.7), then infiltrated into leaves of 1-month-old Nicotiana benthamiana to explore the transient expression of NtWRKY70b. After dark treatment for 24 h and normal condition for 48 h, the infected leaves were detached for dark-induced assay. Under dark conditions for another 5 d, the phenotypes were observed as described previously [4].
4.7. Leaf Senescence Assay
For phenotypic assay, seeds of WT, NtWRKY70b overexpression, and NtWRKY70b-SRDX lines were surface-sterilized with 75% ethanol and grown on 1/2 MS medium [2.2 g/L MS, 0.5 g/L MES, 20 g/L sucrose, 200 mg/L Kanamycin and 1.1% agar (PH = 5.8)] for 14 d, then transferred to soil to continue growing under the long-day condition at the same time. The leaves from any genotype plants were detached and defined as (1), (2), and (3), respectively. Leaves of 2-month-old different genotypes were harvested for phenotype observation and measurement of physiological values.
4.8. Physiological and Biochemical Assays
Two-month-old leaves of WT and different transgenic lines were detached for physiological and biochemical measurements.
The chlorophyll content was measured as described previously [6]. Briefly, chlorophyll contents were measured by the colorimetric method, and three biological replicates were performed. The leaves of different genotype plants were cut into pieces and flash-frozen using liquid nitrogen. Chlorophyll was extracted from leaf samples with 95% ethanol, and then chlorophyll content was counted according to the absorbance at 649 nm and 665 nm. The Fv/Fm was measured by a Hansatech m-pea fluorescence spectrometer.
For ions leakage rates, leaves were placed in deionized water and placed in a vacuum pump for 30 min, after which the conductivity was measured as E1. Then, the leaves were boiled for 10 min and cooled down to room temperature, and the conductivity was measured as E2. Ions leakage rates (%) = E1/E2 × 100.
The malondialdehyde (MDA) accumulation was measured by the thiobarbituric acid-based method. In brief, the leaves were cut into small pieces, then mixed with 10% TCA and 0.6% TBA. After being boiled for 15 min, the leaves were cooled down to room temperature, and absorbance was measured at 532 nm, 600 nm, and 450 nm by using a spectrophotometer. The MDA content was measured as described [39].
The superoxide anions (O2−) content was measured by superoxide anion assay kit (Nanjing Jiancheng). The superoxide dismutase (SOD) and catalase (CAT) activities were analyzed by SOD Detection Kit (Nanjing Jiancheng) and CAT Detection Kit (Nanjing Jiancheng) according to the operating instructions. The enzyme liquid was extracted for determination of superoxide dismutase (SOD) as well as catalase (CAT) activities according to a previous study [39]. Three replicates were performed in these experiments.
The hydrogen sulfide content was measured as described previously [27].
4.9. Yeast One-Hybrid Analysis
The CDS fragment of NtWYKY70b was cloned and fused to pGADT7 vector, and the NtRbohD (880 bp upstream from ATG) and NtPPH (680 bp upstream from ATG) promoter sequences containing the NtWYKY70b-binding sites were inserted into a pAbAi vector. Those above vectors and empty vector were inserted into yeast strain Y1HGold. The control and experimental groups were grown on SD/-Ura-Leu and SD/-Ura-Leu + AbA (50 ng/mL and 100 ng/mL) medium to evaluate the DNA binding activity of NtWYKY70b by observing the growth status of the yeast cells at 28 °C for 3 d.
4.10. Dual-Luciferase Assay
The promoter sequences of NtRbohD and NtPPH were amplified from “K326” cDNA by PCR, respectively, and then inserted into pGreenII0800-LUC vector as a reporter plasmid. The CDS fragment of NtWYKY70b was fused with the vector pGreenII 62-SK as an effector plasmid [27]. Those above plasmids were infiltrated into leaves of 1-month-old Nicotiana benthamiana through Agrobacterium-mediated transformation. The ratio of LUC/REN was detected by Dual-Luciferase Reporter Assay System Kit (Promega, Madison, WI, USA) following the manufacturer’s instructions.
Supplementary Materials
The supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms25073686/s1.
Author Contributions
X.Z., Y.S., H.W., X.L. and S.L. designed the research; X.Z., Y.S., H.W., Y.Z. and S.L. performed the research; X.Z., Y.S., H.W. and S.L. analyzed the data; and X.Z., Y.S., H.W., X.L. and S.L. wrote the paper. The project administration was carried out by S.L.; funding acquisition by X.L. and S.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by The Talents of High-Level Scientific Research Foundation of Qingdao Agricultural University (6631120074 and 6651118004) and the Foundation of Shandong Province Modern Agricultural Technology System (SDAIT-25-05). The APC was funded by The Talents of High-Level Scientific Research Foundation of Qingdao Agricultural University.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Acknowledgments
We are very grateful to Shengcheng Han (Beijing Normal university) for kindly providing the mCherry plasmid.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Kim, H.J.; Nam, H.G.; Lim, P.O. Regulatory Network of NAC Transcription Factors in Leaf Senescence. Curr. Opin. Plant Biol. 2016, 33, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, S.; Guo, Y. Signal Transduction in Leaf Senescence: Progress and Perspective. Plants 2019, 8, 405. [Google Scholar] [CrossRef] [PubMed]
- Quirino, B.F.; Noh, Y.-S.; Himelblau, E.; Amasino, R.M. Molecular Aspects of Leaf Senescence. Trends Plant Sci. 2000, 5, 278–282. [Google Scholar] [CrossRef] [PubMed]
- Mao, C.; Lu, S.; Lv, B.; Zhang, B.; Shen, J.; He, J.; Luo, L.; Xi, D.; Chen, X.; Ming, F. A Rice Nac Transcription Factor Promotes Leaf Senescence via ABA Biosynthesis. Plant Physiol. 2017, 174, 1747–1763. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Gan, S. Leaf Senescence: Signals, Execution, and Regulation. Curr. Top. Dev. Biol. 2005, 71, 83–112. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Ji, X.; Zhang, X.; Wu, H.; Sun, Y.; Zhu, Y.; Su, S.; Wei, S.; Liu, X. A Cationic Amino Acid Transporter NtCAT1 Promotes Leaf Senescence by the Accumulation of ABA in Nicotiana tabacum. Agronomy 2023, 13, 1691. [Google Scholar] [CrossRef]
- Guo, Y.; Ren, G.; Zhang, K.; Li, Z.; Miao, Y.; Guo, H. Leaf Senescence: Progression, Regulation, and Application. Mol. Hortic. 2021, 1, 5. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Wang, Y.; Zhu, Y.; Tang, J.; Hu, B.; Liu, L.; Ou, S.; Wu, H.; Sun, X.; Chu, J.; et al. OsNAP Connects Abscisic Acid and Leaf Senescence by Fine-Tuning Abscisic Acid Biosynthesis and Directly Targeting Senescence-Associated Genes in Rice. Proc. Natl. Acad. Sci. USA 2014, 111, 10013–10018. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Cui, X.; Yang, B.; Xu, S.; Wei, X.; Zhao, P.; Niu, F.; Sun, M.; Wang, C.; Cheng, H.; et al. WRKY55 Transcription Factor Positively Regulates Leaf Senescence and Defense Response through Modulating the Transcription of Genes Implicated in ROS and SA Biosynthesis in Arabidopsis. Development 2020, 147, dev189647. [Google Scholar] [CrossRef]
- Breeze, E.; Harrison, E.; McHattie, S.; Hughes, L.; Hickman, R.; Hill, C.; Kiddle, S.; Kim, Y.S.; Penfold, C.A.; Jenkins, D.; et al. High-Resolution Temporal Profiling of Transcripts during Arabidopsis Leaf Senescence Reveals a Distinct Chronology of Processes and Regulation. Plant Cell 2011, 23, 873–894. [Google Scholar] [CrossRef]
- Guo, Y.; Cai, Z.; Gan, S. Transcriptome of Arabidopsis Leaf Senescence. Plant Cell Environ. 2004, 27, 521–549. [Google Scholar] [CrossRef]
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKY Superfamily of Plant Transcription Factors. Trends Plant Sci. 2000, 5, 199–206. [Google Scholar] [CrossRef]
- Xu, Y.-H.; Wang, J.-W.; Wang, S.; Wang, J.-Y.; Chen, X.-Y. Characterization of GaWRKY1, a Cotton Transcription Factor That Regulates the Sesquiterpene Synthase Gene (+)- δ -Cadinene Synthase-A. Plant Physiol. 2004, 135, 507–515. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.-L.; Xie, Z.; Zou, X.; Casaretto, J.; Ho, T.D.; Shen, Q.J. A Rice WRKY Gene Encodes a Transcriptional Repressor of the Gibberellin Signaling Pathway in Aleurone Cells. Plant Physiol. 2004, 134, 1500–1513. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Ai, C.; Jing, S.; Yu, D. Research Progress on Functional Analysis of Rice WRKY Genes. Rice Sci. 2010, 17, 60–72. [Google Scholar] [CrossRef]
- Zhou, X.; Jiang, Y.; Yu, D. WRKY22 Transcription Factor Mediates Dark-Induced Leaf Senescence in Arabidopsis. Mol. Cells 2011, 31, 303–314. [Google Scholar] [CrossRef]
- Liu, T.; Li, Y.; Wang, C.; Zhang, D.; Liu, J.; He, M.; Chen, M.; Guo, Y. Brassica napus Transcription Factor Bna.A07.WRKY70 Negatively Regulates Leaf Senescence in Arabidopsis thaliana. Plants 2023, 12, 347. [Google Scholar] [CrossRef]
- Guo, P.; Li, Z.; Huang, P.; Li, B.; Fang, S.; Chu, J.; Guo, H. A Tripartite Amplification Loop Involving the Transcription Factor WRKY75, Salicylic Acid, and Reactive Oxygen Species Accelerates Leaf Senescence. Plant Cell 2017, 29, 2854–2870. [Google Scholar] [CrossRef]
- Niu, F.; Cui, X.; Zhao, P.; Sun, M.; Yang, B.; Deyholos, M.K.; Li, Y.; Zhao, X.; Jiang, Y. WRKY42 Transcription Factor Positively Regulates Leaf Senescence through Modulating SA and ROS Synthesis in Arabidopsis thaliana. Plant J. 2020, 104, 171–184. [Google Scholar] [CrossRef]
- Woo, H.R.; Masclaux-Daubresse, C.; Lim, P.O. Plant Senescence: How Plants Know When and How to Die. J. Exp. Bot. 2018, 69, 715–718. [Google Scholar] [CrossRef]
- Chen, L.; Xiang, S.; Chen, Y.; Li, D.; Yu, D. Arabidopsis WRKY45 Interacts with the DELLA Protein RGL1 to Positively Regulate Age-Triggered Leaf Senescence. Mol. Plant 2017, 10, 1174–1189. [Google Scholar] [CrossRef] [PubMed]
- Doll, J.; Muth, M.; Riester, L.; Nebel, S.; Bresson, J.; Lee, H.-C.; Zentgraf, U. Arabidopsis thaliana WRKY25 Transcription Factor Mediates Oxidative Stress Tolerance and Regulates Senescence in a Redox-Dependent Manner. Front. Plant Sci. 2020, 10, 492676. [Google Scholar] [CrossRef] [PubMed]
- Besseau, S.; Li, J.; Palva, E.T. WRKY54 and WRKY70 Co-Operate as Negative Regulators of Leaf Senescence in Arabidopsis thaliana. J. Exp. Bot. 2012, 63, 2667–2679. [Google Scholar] [CrossRef]
- Ülker, B.; Shahid Mukhtar, M.; Somssich, I.E. The WRKY70 Transcription Factor of Arabidopsis Influences Both the Plant Senescence and Defense Signaling Pathways. Planta 2007, 226, 125–137. [Google Scholar] [CrossRef] [PubMed]
- Cui, R.; Feng, Y.; Yao, J.; Shi, L.; Wang, S.; Xu, F. The Transcription Factor BnaA9.WRKY47 Coordinates Leaf Senescence and Nitrogen Remobilization in Brassica napus. J. Exp. Bot. 2023, 74, 5606–5619. [Google Scholar] [CrossRef] [PubMed]
- Baxter, A.; Mittler, R.; Suzuki, N. ROS as Key Players in Plant Stress Signalling. J. Exp. Bot. 2014, 65, 1229–1240. [Google Scholar] [CrossRef]
- Sun, Y.; Song, K.; Guo, M.; Wu, H.; Ji, X.; Hou, L.; Liu, X.; Lu, S. A NAC Transcription Factor from ‘Sea Rice 86’ Enhances Salt Tolerance by Promoting Hydrogen Sulfide Production in Rice Seedlings. Int. J. Mol. Sci. 2022, 23, 6435. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Zhi, W.; Qiao, H.; Huang, J.; Li, S.; Lu, Q.; Wang, N.; Li, Q.; Zhou, Q.; Sun, J.; et al. H2O2-Dependent Oxidation of the Transcription Factor GmNTL1 Promotes Salt Tolerance in Soybean. Plant Cell 2023, 36, 112–135. [Google Scholar] [CrossRef] [PubMed]
- Torres, M.A.; Dangl, J.L. Functions of the Respiratory Burst Oxidase in Biotic Interactions, Abiotic Stress and Development. Curr. Opin. Plant Biol. 2005, 8, 397–403. [Google Scholar] [CrossRef]
- Liu, D.; Li, Y.-Y.; Zhou, Z.-C.; Xiang, X.; Liu, X.; Wang, J.; Hu, Z.-R.; Xiang, S.-P.; Li, W.; Xiao, Q.-Z.; et al. Tobacco Transcription Factor BHLH123 Improves Salt Tolerance by Activating NADPH Oxidase NtRbohE Expression. Plant Physiol. 2021, 186, 1706–1720. [Google Scholar] [CrossRef]
- Adachi, H.; Nakano, T.; Miyagawa, N.; Ishihama, N.; Yoshioka, M.; Katou, Y.; Yaeno, T.; Shirasu, K.; Yoshioka, H. WRKY Transcription Factors Phosphorylated by MAPK Regulate a Plant Immune NADPH Oxidase in Nicotiana benthamiana. Plant Cell 2015, 27, 2645–2663. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Ye, C.; Zhao, Y.; Cheng, X.; Wang, Y.; Jiang, Y.-Q.; Yang, B. An Oilseed Rape WRKY-Type Transcription Factor Regulates ROS Accumulation and Leaf Senescence in Nicotiana benthamiana and Arabidopsis through Modulating Transcription of RbohD and RbohF. Planta 2018, 247, 1323–1338. [Google Scholar] [CrossRef]
- Mittler, R. ROS Are Good. Trends Plant Sci. 2017, 22, 11–19. [Google Scholar] [CrossRef]
- Marino, D.; Dunand, C.; Puppo, A.; Pauly, N. A Burst of Plant NADPH Oxidases. Trends Plant Sci. 2012, 17, 9–15. [Google Scholar] [CrossRef]
- Waszczak, C.; Carmody, M.; Kangasjärvi, J. Reactive Oxygen Species in Plant Signaling. Annu. Rev. Plant Biol. 2018, 69, 209–236. [Google Scholar] [CrossRef]
- Wu, A.; Allu, A.D.; Garapati, P.; Siddiqui, H.; Dortay, H.; Zanor, M.I.; Asensi-Fabado, M.A.; Munná-Bosch, S.; Antonio, C.; Tohge, T.; et al. JUNGBRUNNEN1, a Reactive Oxygen Species-Responsive NAC Transcription Factor, Regulates Longevity in Arabidopsis. Plant Cell 2012, 24, 482–506. [Google Scholar] [CrossRef]
- Lee, S.; Seo, P.J.; Lee, H.J.; Park, C.M. A NAC Transcription Factor NTL4 Promotes Reactive Oxygen Species Production during Drought-Induced Leaf Senescence in Arabidopsis. Plant J. 2012, 70, 831–844. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Mu, Y.; Hao, X.; Yang, J.; Zhang, D.; Jin, Z.; Pei, Y. H2S Aids Osmotic Stress Resistance by S-Sulfhydration of Melatonin Production-Related Enzymes in Arabidopsis thaliana. Plant Cell Rep. 2022, 41, 365–376. [Google Scholar] [CrossRef]
- Hou, L.; Wang, Z.; Gong, G.; Zhu, Y.; Ye, Q.; Lu, S.; Liu, X. Hydrogen Sulfide Alleviates Manganese Stress in Arabidopsis. Int. J. Mol. Sci. 2022, 23, 5046. [Google Scholar] [CrossRef]
- Sun, C.; Yao, G.; Li, L.; Li, T.; Zhao, Y.; Hu, K.; Zhang, C.; Zhang, H. E3 Ligase BRG3 Persulfidation Delays Tomato Ripening by Reducing Ubiquitination of the Repressor WRKY71. Plant Physiol. 2023, 192, 616–632. [Google Scholar] [CrossRef]
- Zhang, J.; Zhou, M.; Zhou, H.; Zhao, D.; Gotor, C.; Romero, L.C.; Shen, J.; Ge, Z.; Zhang, Z.; Shen, W.; et al. Hydrogen Sulfide, a Signaling Molecule in Plant Stress Responses. J. Integr. Plant Biol. 2021, 63, 146–160. [Google Scholar] [CrossRef]
- Fang, H.; Liu, Z.; Long, Y.; Liang, Y.; Jin, Z.; Zhang, L.; Liu, D.; Li, H.; Zhai, J.; Pei, Y. The Ca2+/Calmodulin2-Binding Transcription Factor TGA3 Elevates LCD Expression and H2S Production to Bolster Cr6+ Tolerance in Arabidopsis. Plant J. 2017, 91, 1038–1050. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Zhang, J.; Zhou, M.; Zhou, H.; Cui, B.; Gotor, C.; Romero, L.C.; Fu, L.; Yang, J.; Foyer, C.H.; et al. Persulfidation-Based Modification of Cysteine Desulfhydrase and the NADPH Oxidase RBOHD Controls Guard Cell Abscisic Acid Signaling. Plant Cell 2020, 32, 1000–1017. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Zhang, J.; Shen, J.; Zhou, H.; Zhao, D.; Gotor, C.; Romero, L.C.; Fu, L.; Li, Z.; Yang, J.; et al. Hydrogen Sulfide-Linked Persulfidation of ABI4 Controls ABA Responses through the Transactivation of MAPKKK18 in Arabidopsis. Mol. Plant 2021, 14, 921–936. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, L.; Pei, Z.; Zhang, L.; Liu, Z.; Liu, D.; Hao, X.; Jin, Z.; Pei, Y. Hydrogen Sulfide Promotes Flowering in Heading Chinese Cabbage by S-Sulfhydration of BraFLCs. Hortic. Res. 2021, 8, 19. [Google Scholar] [CrossRef]
- Hu, K.; Peng, X.; Yao, G.; Zhou, Z.; Yang, F.; Li, W.; Zhao, Y.; Li, Y.; Han, Z.; Chen, X.; et al. Roles of a Cysteine Desulfhydrase Lcd1 in Regulating Leaf Senescence in Tomato. Int. J. Mol. Sci. 2021, 22, 13078. [Google Scholar] [CrossRef] [PubMed]
- Geng, Z.-K.; Ma, L.; Rong, Y.-L.; Li, W.-J.; Yao, G.-F.; Zhang, H.; Hu, K.-D. A Hydrogen-Sulfide-Repressed Methionine Synthase SlMS1 Acts as a Positive Regulator for Fruit Ripening in Tomato. Int. J. Mol. Sci. 2022, 23, 12239. [Google Scholar] [CrossRef]
- Zhao, Y.-Q.; Hu, K.-D.; Yao, G.-F.; Wang, S.-Y.; Peng, X.-J.; Zhang, H. A D-Cysteine Desulfhydrase, SlDCD2, Participates in Tomato Fruit Ripening by Modulating ROS Homoeostasis and Ethylene Biosynthesis. Hortic. Res. 2023, 10, uhad014. [Google Scholar] [CrossRef] [PubMed]
- Woo, H.R.; Kim, H.J.; Lim, P.O.; Nam, H.G. Leaf Senescence: Systems and Dynamics Aspects. Annu. Rev. Plant Biol. 2019, 70, 347–376. [Google Scholar] [CrossRef]
- Munné-Bosch, S.; Alegre, L. Die and Let Live: Leaf Senescence Contributes to Plant Survival under Drought Stress. Funct. Plant Biol. 2004, 31, 203. [Google Scholar] [CrossRef]
- Yoo, S.-C.; Cho, S.-H.; Zhang, H.; Paik, H.-C.; Lee, C.-H.; Li, J.; Yoo, J.-H.; Lee, B.-W.; Koh, H.-J.; Seo, H.S.; et al. Quantitative Trait Loci Associated with Functional Stay-Green SNU-SG1 in Rice. Mol. Cells 2007, 24, 83–94. [Google Scholar] [CrossRef] [PubMed]
- Siqueira, J.A.; da Fonseca-Pereira, P.; Fernie, A.R.; Nunes-Nesi, A.; Araújo, W.L. Recycling Amino Acids Ensures Meiosis and Seed Development. Trends Plant Sci. 2022, 27, 1084–1086. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Balazadeh, S.; Mueller-Roeber, B. Tomato Fruit Ripening Factor NOR Controls Leaf Senescence. J. Exp. Bot. 2019, 70, 2727–2740. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Shabala, S.; Niu, Y.; Chen, Z.H.; Shabala, L.; Meinke, H.; Venkataraman, G.; Pareek, A.; Xu, J.; Zhou, M. Molecular Mechanisms of Salinity Tolerance in Rice. Crop J. 2021, 9, 506–520. [Google Scholar] [CrossRef]
- Li, Y.; Sun, D.; Xu, K.; Jin, L.; Peng, R. Hydrogen Sulfide Enhances Plant Tolerance to Waterlogging Stress. Plants 2021, 10, 1928. [Google Scholar] [CrossRef]
- Yao, G.-F.; Wei, Z.-Z.; Li, T.-T.; Tang, J.; Huang, Z.-Q.; Yang, F.; Li, Y.-H.; Han, Z.; Hu, F.; Hu, L.-Y.; et al. Modulation of Enhanced Antioxidant Activity by Hydrogen Sulfide Antagonization of Ethylene in Tomato Fruit Ripening. J. Agric. Food Chem. 2018, 66, 10380–10387. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).