The MYB Transcription Factor GmMYB78 Negatively Regulates Phytophthora sojae Resistance in Soybean
Abstract
:1. Introduction
2. Results
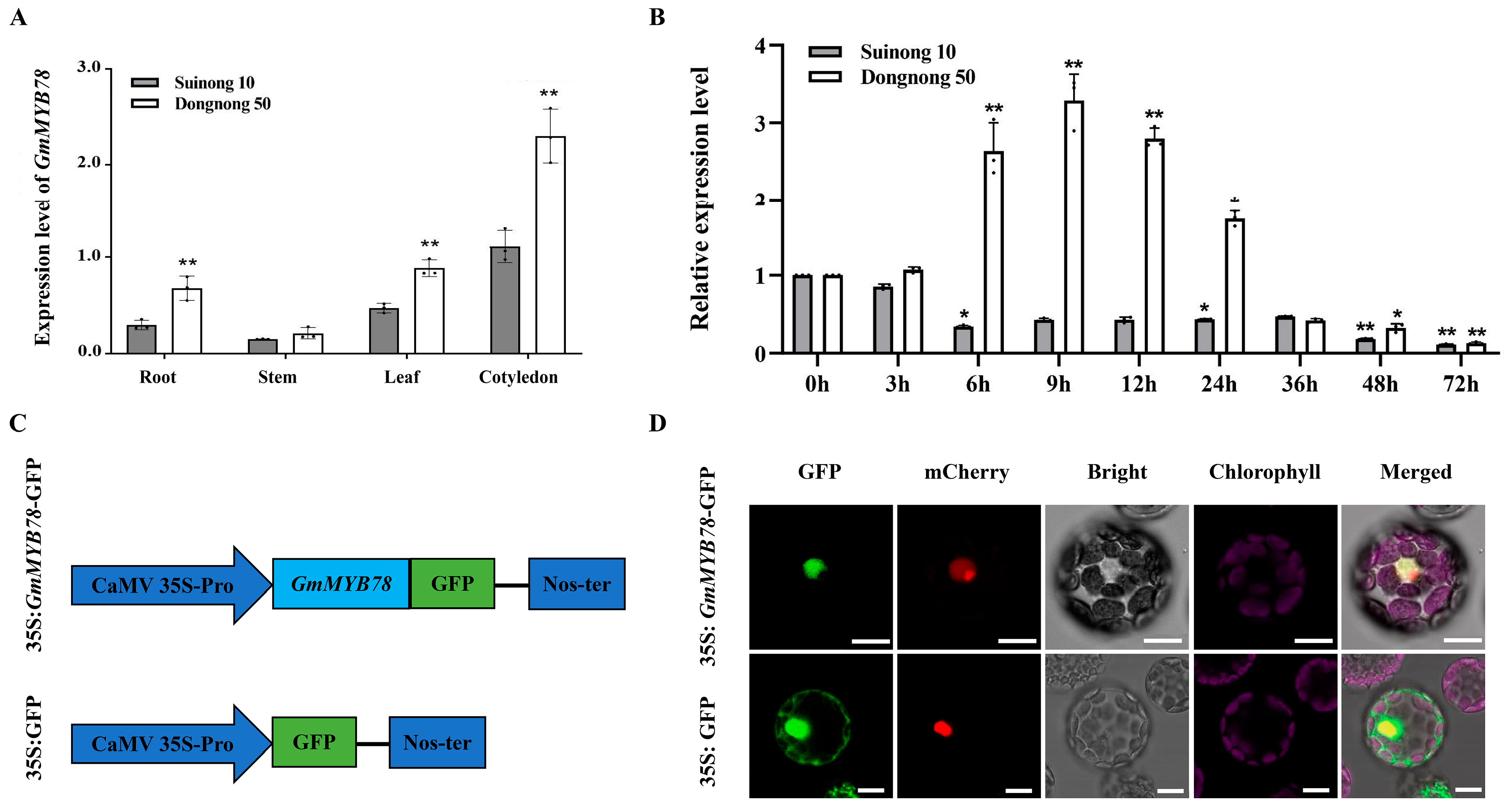
2.1. GmMYB78 Is Induced by P. sojae in Susceptible Soybean Cultivar and Localized in the Nucleus
2.2. GmMYB78 Increases Susceptibility to P. sojae in Transgenic Soybean Hairy Roots
2.3. GmMYB78 Inhibits Pathogenesis-Related (PR) Gene Expression in Response to P. sojae Infection
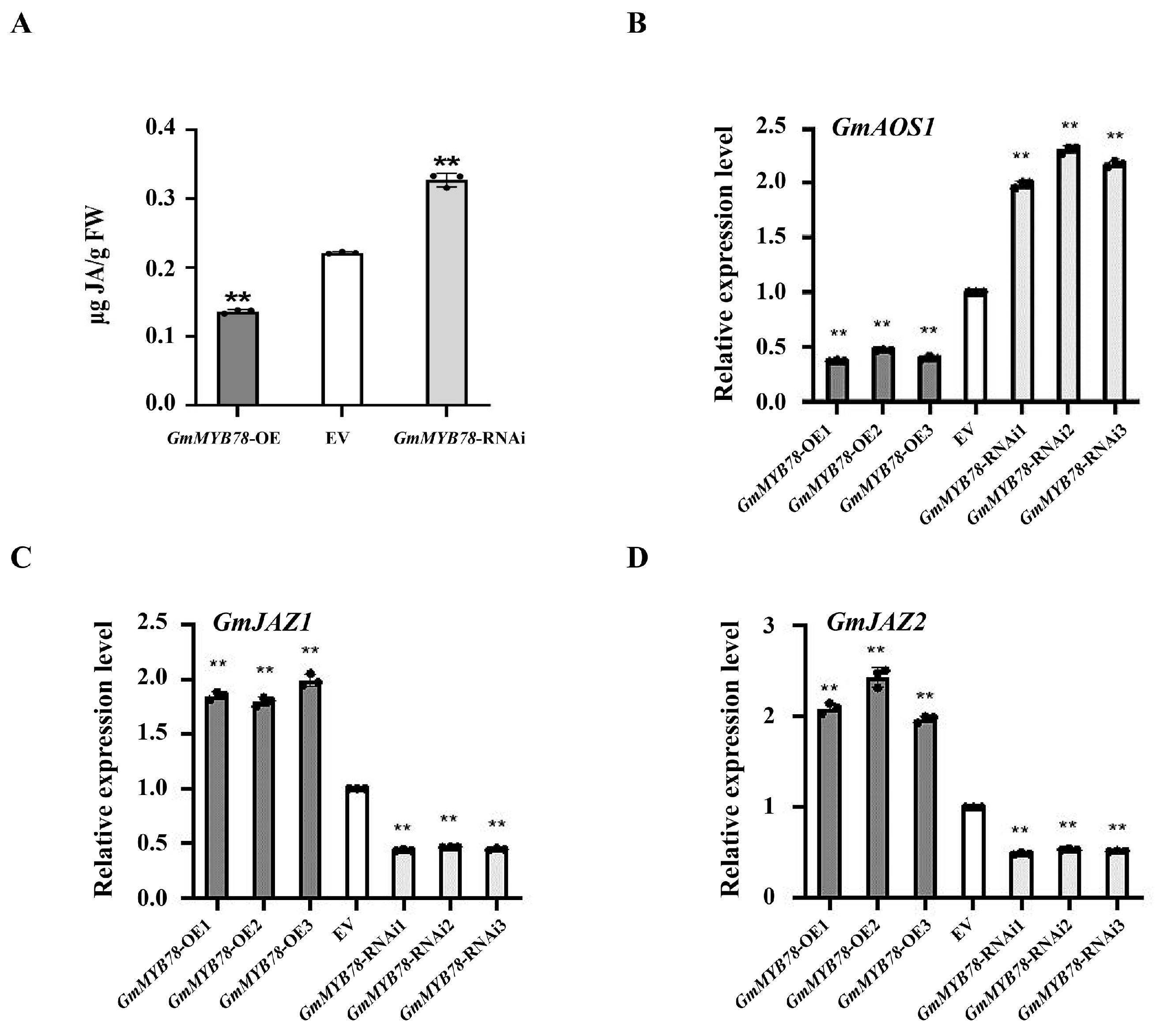
2.4. GmMYB78 Is a Negative Regulator of JA-Dependent Signaling during the Response to P. sojae
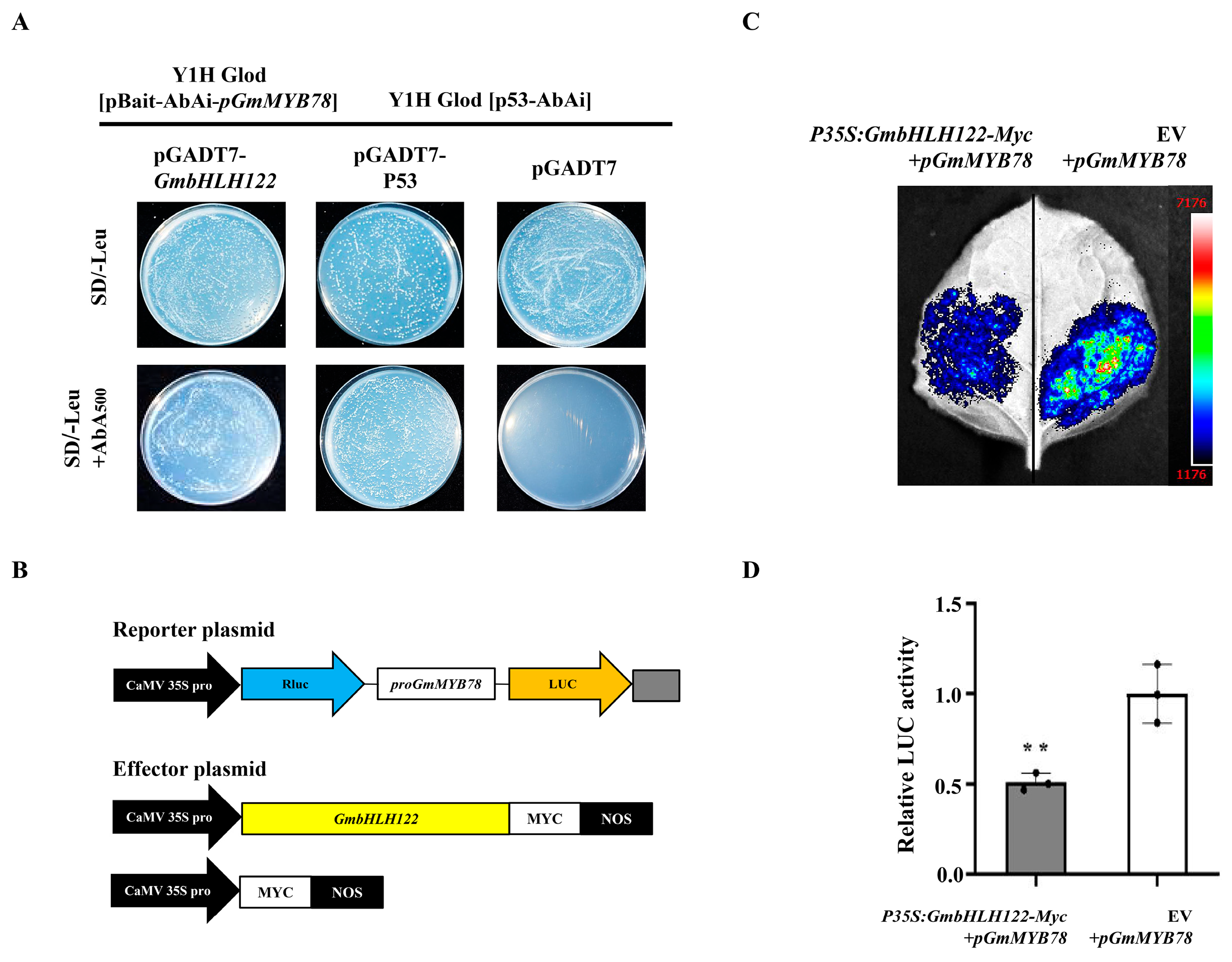
2.5. GmMYB78 Regulates the Transcription of GmbZIP25
2.6. GmbHLH122 Regulates the Transcription of GmMYB78
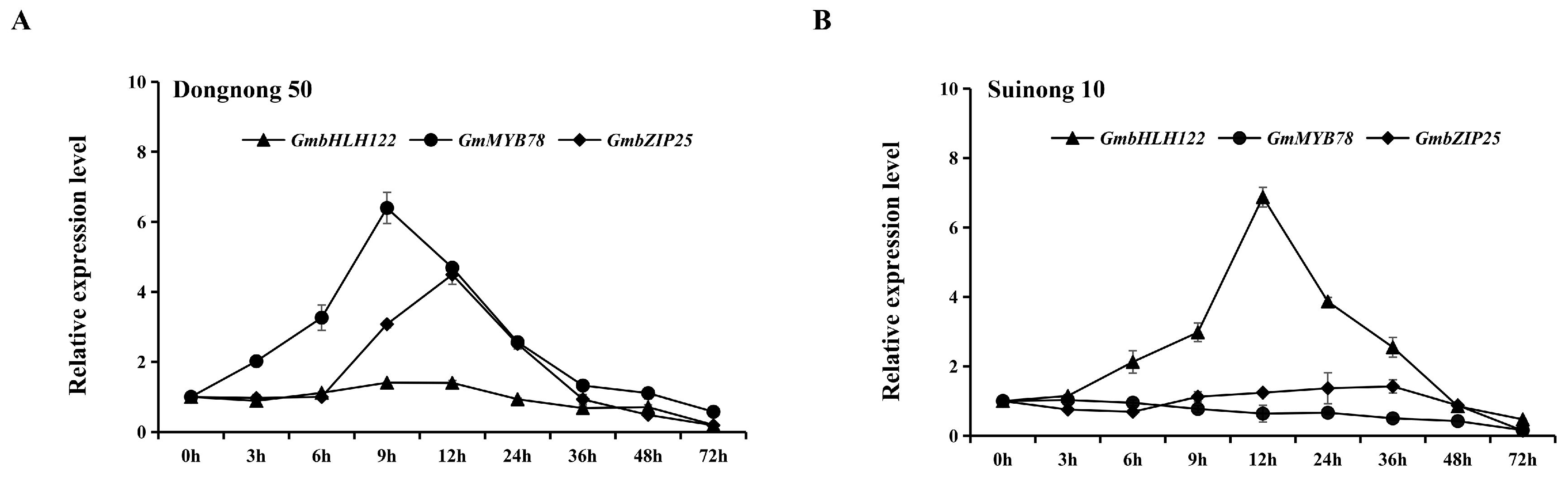
2.7. Expression Analysis of GmbLHL122, GmMYB78, and GmbZIP25 in Response to P. sojae
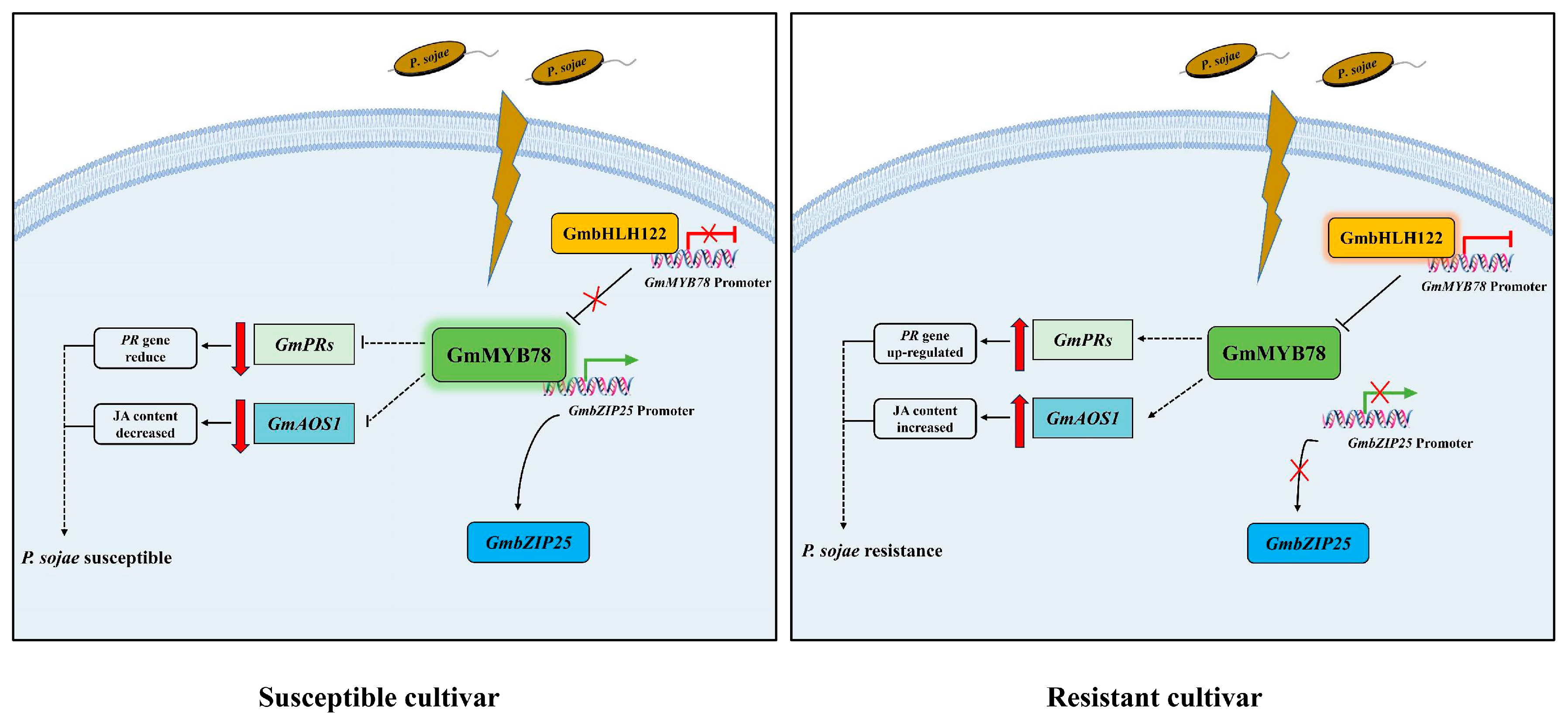
3. Discussion
3.1. GmMYB78 Negatively Regulates P. sojae Infection by Inhibiting the Expression of Pathogenesis-Related Genes
3.2. GmMYB78 Is Involved in Soybean Defense Response to P. sojae through JA Signaling Pathway
3.3. GmbHLH122-GmMYB78-GmbZIP25 Regulatory Module Is Involved in the Response to P. sojae in Soybean
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. RT-qPCR
4.3. Gene Cloning, Sequence Analyses, and Plasmid Construction
4.4. Subcellular Localization Assays of the GmMYB78 Protein
4.5. Agrobacterium Rhizogenes-Mediated Transformation of Soybean Hairy Roots
4.6. Assessment of Soybean Disease Responses
4.7. Determination of JA Levels
4.8. RNA-Seq and Metabolome Analysis
4.9. Yeast One-Hybrid (Y1H) Assays
4.10. Transient Transcription Dual-Luciferase Assays
4.11. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Schmitthenner, A. Problems and progress in control of Phytophthora root rot of soybean. Plant Dis. 1985, 69, 362–368. [Google Scholar] [CrossRef]
- Tyler, B.M. Phytophthora sojae root rot pathogen of soybean and model oomycete. Mol. Plant Pathol. 2007, 8, 1–8. [Google Scholar] [CrossRef]
- Hill, C.B.; Li, Y.; Hartman, G.L. Resistance to the soybean aphid in soybean germplasm. Crop. Sci. 2004, 44, 98–106. [Google Scholar] [CrossRef]
- Rushton, P.J.; Somssich, I.E. Transcriptional control of plant genes responsive to pathogens. Curr. Opin. Plant Biol. 1998, 1, 311–315. [Google Scholar] [CrossRef]
- Ng, D.W.K.; Abeysinghe, J.K.; Kamali, M. Regulating the regulators: The control of transcription factors in plant defense signaling. Int. J. Mol. Sci. 2018, 19, 3737. [Google Scholar] [CrossRef] [PubMed]
- Gu, K.D.; Zhang, Q.Y.; Yu, J.Q.; Wang, J.H.; Zhang, F.J.; Wang, C.K.; Zhao, Y.W.; Sun, C.H.; You, C.X.; Hu, D.G.; et al. R2R3-MYB transcription factor MdMYB73 confers increased resistance to the fungal pathogen Botryosphaeria dothidea in apples via the salicylic acid pathway. J. Agric. Food Chem. 2021, 69, 447–458. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Jiang, L.; Du, B.; Ning, B.; Ding, X.; Zhang, C.; Song, B.; Liu, S.; Zhao, M.; Zhao, Y.; et al. GmMKK4-activated GmMPK6 stimulates GmERF113 to trigger resistance to Phytophthora sojae in soybean. Plant J. 2022, 111, 473–495. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Q.; Dong, L.; Gao, T.; Liu, T.; Li, N.; Wang, L.; Chang, X.; Wu, J.; Xu, P.; Zhang, S. The bHLH transcription factor GmPIB1 facilitates resistance to Phytophthora sojae in Glycine max. J. Exp. Bot. 2018, 69, 2527–2541. [Google Scholar] [CrossRef]
- Dong, L.; Cheng, Y.; Wu, J.; Cheng, Q.; Li, W.; Fan, S.; Jiang, L.; Xu, Z.; Kong, F.; Zhang, D.; et al. Overexpression of GmERF5, a new member of the soybean EAR motif-containing ERF transcription factor, enhances resistance to Phytophthora sojae in soybean. J. Exp. Bot. 2015, 66, 2635–2647. [Google Scholar] [CrossRef]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef]
- Ramalingam, A.; Kudapa, H.; Pazhamala, L.T.; Garg, V.; Varshney, R.K. Gene expression and yeast two-hybrid studies of 1R-MYB transcription factor mediating drought stress response in Chickpea (Cicer arietinum L.). Front. Plant Sci. 2015, 6, 1117. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.F.; Yin, X.R.; Allan, A.C.; Lin-Wang, K.; Shi, Y.N.; Huang, Y.J.; Ferguson, I.B.; Xu, C.J.; Chen, K.S. The role of MrbHLH1 and MrMYB1 in regulating anthocyanin biosynthetic genes in tobacco and Chinese bayberry (Myrica rubra) during anthocyanin biosynthesis. Plant Cell Tissue Organ Cult. 2013, 115, 285–298. [Google Scholar] [CrossRef]
- Shen, X.J.; Wang, Y.Y.; Zhang, Y.X.; Guo, W.; Jiao, Y.Q.; Zhou, X.A. Overexpression of the wild soybean R2R3-MYB transcription factor GsMYB15 enhances resistance to salt stress and Helicoverpa armigera in transgenic Arabidopsis. Int. J. Mol. Sci. 2018, 19, 3958. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.Y.; Song, J.; Ferguson, A.C.; Klisch, D.; Simpson, K.; Mo, R.; Taylor, B.; Mitsuda, N.; Wilson, Z.A. Transcription factor MYB26 is key to spatial specificity in anther secondary thickening formation. Plant Physiol. 2017, 175, 333–350. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.C.; Gao, X.K.; Shi, M.; Sun, M.H.; Li, K.L.; Cai, Y.; Chen, C.A.; Wang, C.; Maoz, I.; Guo, X.H.; et al. Jasmonic acid regulates the biosynthesis of medicinal metabolites via the JAZ9-MYB76 complex in Salvia miltiorrhiza. Hortic. Res. 2023, 10, uhad004. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Pan, L.J.; Yang, Z.; Su, M.W.; Xu, J.; Jiang, X.; Yin, X.Z.; Wang, T.; Wan, F.F.; Chi, X.Y. A MYB-related transcription factor from peanut, AhMYB30, improves freezing and salt stress tolerance in transgenic Arabidopsis through both DREB/CBF and ABA-signaling pathways. Front. Plant Sci. 2023, 14, 1136626. [Google Scholar] [CrossRef]
- Hu, Z.W.; Zhong, X.; Zhang, H.R.; Luo, X.C.; Wang, Y.X.; Wang, Y.; Liu, T.; Zhang, Y.; Wang, X.P.; An, H.L.; et al. GhMYB18 confers Aphis gossypii Glover resistance through regulating the synthesis of salicylic acid and flavonoids in cotton plants. Plant Cell Rep. 2023, 42, 355–369. [Google Scholar] [CrossRef] [PubMed]
- Hawku, M.D.; He, F.X.; Bai, X.X.; Islam, M.A.; Huang, X.L.; Kang, Z.S.; Guo, J. A R2R3 MYB transcription factor, TaMYB391, is positively involved in wheat resistance to Puccinia striiformis f. sp. tritici. Int. J. Mol. Sci. 2022, 23, 14070. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.X.; Li, X.; He, Q.; Guo, D.X.; Liu, C.Q.; Cao, J.Y.; Wu, Z.Y.; Kang, Z.S.; Wang, X.J. TaMYB29: A novel R2R3-MYB transcription factor involved in wheat defense against stripe rust. Front. Plant Sci. 2021, 12, 783388. [Google Scholar] [CrossRef] [PubMed]
- Li, D.K.; Li, Y.; Zhang, L.; Wang, X.Y.; Zhao, Z.; Tao, Z.W.; Wang, J.M.; Wang, J.; Lin, M.; Li, X.F.; et al. Arabidopsis ABA receptor RCAR1/PYL9 interacts with an R2R3-type MYB transcription factor, AtMYB44. Int. J. Mol. Sci. 2014, 15, 8473–8490. [Google Scholar] [CrossRef]
- Chen, H.H.; Lai, L.Y.; Li, L.X.; Liu, L.P.; Jakada, B.H.; Huang, Y.M.; He, Q.; Chai, M.N.; Niu, X.P.; Qin, Y. AcoMYB4, an Ananas comosus L. MYB transcription factor, functions in osmotic stress through negative regulation of ABA signaling. Int. J. Mol. Sci. 2020, 21, 5727. [Google Scholar] [CrossRef]
- Zhao, H.X.; Yao, P.F.; Zhao, J.L.; Wu, H.L.; Wang, S.; Chen, Y.; Hu, M.F.; Wang, T.; Li, C.L.; Wu, Q. A novel R2R3-MYB transcription factor FtMYB22 negatively regulates salt and drought stress through ABA-dependent pathway. Int. J. Mol. Sci. 2022, 23, 14549. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Hyun, W.Y.; Nguyen, H.N.; Jeong, C.Y.; Xiong, L.; Hong, S.W.; Lee, H. AtMyb7, a subgroup 4 R2R3 Myb, negatively regulates ABA-induced inhibition of seed germination by blocking the expression of the bZIP transcription factor ABI5. Plant Cell Environ. 2015, 38, 559–571. [Google Scholar] [CrossRef] [PubMed]
- Ku, Y.S.; Ni, M.; Muñoz, N.B.; Xiao, Z.X.; Lo, A.W.Y.; Chen, P.; Li, M.W.; Cheung, M.Y.; Xie, M.; Lam, H.M. ABAS1 from soybean is a 1R-subtype MYB transcriptional repressor that enhances ABA sensitivity. J. Exp. Bot. 2020, 71, 2970–2981. [Google Scholar] [CrossRef]
- Zhang, S.; Zhao, Q.C.; Zeng, D.X.; Xu, J.H.; Zhou, H.G.; Wang, F.L.; Ma, N.; Li, Y.H. RhMYB108, an R2R3-MYB transcription factor, is involved in ethylene- and JA-induced petal senescence in rose plants. Hortic. Res. 2019, 6, 131. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.Y.; He, X.; Tu, L.L.; Zhu, L.F.; Zhu, S.T.; Ge, Z.H.; Zhang, X.L. GhJAZ2 negatively regulates cotton fiber initiation by interacting with the R2R3-MYB transcription factor GhMYB25-like. Plant J. 2016, 88, 921–935. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.L.; Zhang, C.; Zhang, H.; Duan, Y.; Zou, Z.W.; Zhou, L.; Zhu, X.J.; Fang, W.P.; Ma, Y.C. CsMYB transcription factors participate in jasmonic acid signal transduction in response to cold stress in tea plant (Camellia sinensis). Plants 2022, 11, 2869. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Q.; Judelson, H.S. Myb transcription factors in the oomycete Phytophthora with novel diversified DNA-binding domains and developmental stage-specific expression. Gene 2010, 453, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Chen, T.; Kan, J.; Yao, Y.; Guo, D.; Yang, Y.; Ling, X.; Wang, J.; Zhang, B. The GhMYB36 transcription factor confers resistance to biotic and abiotic stress by enhancing PR1 gene expression in plants. Plant Biotechnol. J. 2022, 20, 722–735. [Google Scholar] [CrossRef]
- Zheng, H.Y.; Dong, L.L.; Han, X.Y.; Jin, H.B.; Yin, C.C.; Han, Y.L.; Li, B.; Qin, H.J.; Zhang, J.S.; Shen, Q.H.; et al. The TuMYB46L-TuACO3 module regulates ethylene biosynthesis in einkorn wheat defense to powdery mildew. New Phytol. 2020, 225, 2526–2541. [Google Scholar] [CrossRef]
- Shim, J.S.; Jung, C.; Lee, S.; Min, K.; Lee, Y.-W.; Choi, Y.; Lee, J.S.; Song, J.T.; Kim, J.-K.; Choi, Y.D. AtMYB44 regulates WRKY70 expression and modulates antagonistic interaction between salicylic acid and jasmonic acid signaling. Plant J. 2013, 73, 483–495. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.W.; Qi, M.; Yang, Y. A novel jasmonic acid-inducible rice myb gene associates with fungal infection and host cell death. Mol. Plant-Microbe Interact. MPMI 2001, 14, 527–535. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.L.; Zhang, C.L.; Wang, G.L.; Wang, Y.X.; Qi, C.H.; Zhao, Q.; You, C.X.; Li, Y.Y.; Hao, Y.J. The R2R3 MYB transcription factor MdMYB30 modulates plant resistance against pathogens by regulating cuticular wax biosynthesis. BMC Plant Biol. 2019, 19, 362. [Google Scholar] [CrossRef] [PubMed]
- Shriti, S.; Paul, S.; Das, S. Overexpression of CaMYB78 transcription factor enhances resistance response in chickpea against Fusarium oxysporum and negatively regulates anthocyanin biosynthetic pathway. Protoplasma 2023, 260, 589–605. [Google Scholar] [CrossRef] [PubMed]
- Noman, A.; Hussain, A.; Adnan, M.; Khan, M.I.; Ashraf, M.F.; Zainab, M.; Khan, K.A.; Ghramh, H.A.; He, S.L. A novel MYB transcription CaPHL8 provide clues about evolution of pepper immunity againstsoil borne pathogen. Microb. Pathog. 2019, 137, 103758. [Google Scholar] [CrossRef]
- Bai, Q.X.; Duan, B.B.; Ma, J.C.; Fen, Y.N.; Sun, S.J.; Long, Q.M.; Lv, J.J.; Wan, D.S. Coexpression of PalbHLH1 and PalMYB90 genes from Populus alba enhances pathogen resistance in poplar by increasing the flavonoid content. Front. Plant Sci. 2020, 10, 1772. [Google Scholar] [CrossRef]
- Saxena, S.; Pal, L.; Naik, J.; Singh, Y.; Verma, P.K.; Chattopadhyay, D.; Pandey, A. The R2R3-MYB-SG7 transcription factor CaMYB39 orchestrates surface phenylpropanoid metabolism and pathogen resistance in chickpea. New Phytol. 2023, 238, 798–816. [Google Scholar] [CrossRef]
- Al-Attala, M.; Wang, X.; Abou-Attia, M.; Duan, X.; Kang, Z. A novel TaMYB4 transcription factor involved in the defence response against Puccinia striiformis f. sp. tritici and abiotic stresses. Plant Mol. Biol. 2014, 84, 589–603. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.H.; Guo, D.L.; Li, G.R.; Yang, Y.J.; Zhang, G.H.; Li, S.H.; Liang, Z.C. The grapevine R2R3-type MYB transcription factor VdMYB1 positively regulates defense responses by activating the stilbene synthase gene 2 (VdSTS2). BMC Plant Biol. 2019, 19, 478. [Google Scholar] [CrossRef]
- Qiu, B.L.; Chen, H.J.; Zheng, L.L.; Su, L.L.; Cui, X.M.; Ge, F.; Liu, D.Q. An MYB transcription factor modulates Panax notoginseng resistance against the root rot pathogen Fusarium solani by regulating the jasmonate acid signaling pathway and photosynthesis. Phytopathology 2022, 112, 1323–1334. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Gao, H.; Sun, Y.; Jiang, L.; He, S.; Song, B.; Liu, S.; Zhao, M.; Wang, L.; Liu, Y.; et al. The BTB/POZ domain protein GmBTB/POZ promotes the ubiquitination and degradation of the soybean AP2/ERF-like transcription factor GmAP2 to regulate the defense response to Phytophthora sojae. J. Exp. Bot. 2021, 72, 7891–7908. [Google Scholar] [CrossRef] [PubMed]
- Song, S.S.; Qi, T.C.; Fan, M.; Zhang, X.; Gao, H.; Huang, H.; Wu, D.W.; Guo, H.W.; Xie, D.X. The bHLH subgroup IIId factors negatively regulate jasmonate-mediated plant defense and development. PLoS Genet. 2013, 9, e1003653. [Google Scholar] [CrossRef]
- Zhu, Y.T.; Hu, X.Q.; Wang, P.; Wang, H.W.; Ge, X.Y.; Li, F.G.; Hou, Y.X. GhODO1, an R2R3-type MYB transcription factor, positively regulates cotton resistance to Verticillium dahliae via the lignin biosynthesis and jasmonic acid signaling pathway. Int. J. Biol. Macromol. 2022, 201, 580–591. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Yang, B.; Xu, H.W.; Wu, J.B.; Xu, Z.Y.; Wang, Y.C. The Phytophthora effector Avh94 manipulates host jasmonic acid signaling to promote infection. J. Integr. Plant Biol. 2022, 64, 2199–2210. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.B.; Wei, Y.Y.; Cao, Z.D.; Jiang, S.; Chen, Y.; Shao, X.F. The jasmonic acid signaling pathway is associated with terpinen-4-ol-Induced disease resistance against Botrytis cinerea in strawberry fruit. J. Agric. Food Chem. 2021, 69, 10678–10687. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Ai, G.; Shen, D.Y.; Chai, C.Y.; Jia, Y.L.; Liu, W.J.; Dou, D.L. Bioinformatical analysis and prediction of Nicotiana benthamiana bHLH transcription factors in Phytophthora parasitica resistance. Genomics 2019, 111, 473–482. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.J.; Dong, L.D.; Han, D.; Zhang, F.; Wu, J.J.; Jiang, L.Y.; Cheng, Q.; Li, R.P.; Lu, W.C.; Meng, F.S.; et al. GmWRKY31 and GmHDL56 enhances resistance to Phytophthora sojae by regulating defense-related gene expression in soybean. Front. Plant Sci. 2017, 8, 781. [Google Scholar] [CrossRef]
- Prouse, M.B.; Campbell, M.M. The interaction between MYB proteins and their target DNA binding sites. Biochim. Biophys. Acta-Gene Regul. Mech. 2012, 1819, 67–77. [Google Scholar] [CrossRef]
- Tanikawa, J.; Yasukawa, T.; Enari, M.; Ogata, K.; Nishimura, Y.; Ishii, S.; Sarai, A. Recognition of specific DNA sequences by the c-myb protooncogene product: Role of three repeat units in the DNA-binding domain. Proc. Natl. Acad. Sci. USA 1993, 90, 9320–9324. [Google Scholar] [CrossRef]
- Bilaud, T.; Koering, C.E.; Binet-Brasselet, E.; Ancelin, K.; Pollice, A.; Gasser, S.M.; Gilson, E. The telobox, a Myb-related telomeric DNA binding motif found in proteins from yeast, plants and human. Nucleic Acids Res. 1996, 24, 1294–1303. [Google Scholar] [CrossRef] [PubMed]
- Albert, N.W.; Griffiths, A.G.; Cousins, G.R.; Verry, I.M.; Williams, W.M. Anthocyanin leaf markings are regulated by a family of R2R3-MYB genes in the genus Trifolium. New Phytol. 2015, 205, 882–893. [Google Scholar] [CrossRef]
- Sajeevan, R.S.; Abdelmeguid, I.; Saripella, G.V.; Lenman, M.; Alexandersson, E. Comprehensive transcriptome analysis of different potato cultivars provides insight into early blight disease caused by Alternaria solani. BMC Plant Biol. 2023, 23, 130. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, Z.X.; Luo, R.X.; Sun, Y.; Yang, C.F.; Li, X.; Gao, A.P.; Pu, J.J. Genome-wide characterization, identification and expression profile of MYB transcription factor gene family during abiotic and biotic stresses in mango (Mangifera indica). Plants 2022, 11, 3141. [Google Scholar] [CrossRef] [PubMed]
- Longsaward, R.; Pengnoo, A.; Kongsawadworakul, P.; Viboonjun, U. A novel rubber tree PR-10 protein involved in host-defense response against the white root rot fungus Rigidoporus microporus. BMC Plant Biol. 2023, 23, 157. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.X.; Zhang, X.M.; Zhang, Q.H.; Chai, S.Y.; Yin, W.C.; Gao, M.; Li, Z.; Wang, X.P. The transcription factors VaERF16 and VaMYB306 interact to enhance resistance of grapevine to Botrytis cinerea infection. Mol. Plant Pathol. 2022, 23, 1415–1432. [Google Scholar] [CrossRef]
- Zhang, C.; Gao, H.; Li, R.; Han, D.; Wang, L.; Wu, J.; Xu, P.; Zhang, S. GmBTB/POZ, a novel BTB/POZ domain-containing nuclear protein, positively regulates the response of soybean to Phytophthora sojae infection. Mol. Plant Pathol. 2019, 20, 78–91. [Google Scholar] [CrossRef]
- Dutta, S.; Kumawat, G.; Singh, B.P.; Gupta, D.K.; Singh, S.; Dogra, V.; Gaikwad, K.; Sharma, T.R.; Raje, R.S.; Bandhopadhya, T.K.; et al. Development of genic-SSR markers by deep transcriptome sequencing in pigeonpea Cajanus cajan (L.) Millspaugh. BMC Plant Biol. 2011, 11, 17. [Google Scholar] [CrossRef] [PubMed]
- Yates, S.A.; Swain, M.T.; Hegarty, M.J.; Chernukin, I.; Lowe, M.; Allison, G.G.; Ruttink, T.; Abberton, M.T.; Jenkins, G.; Skot, L. De novo assembly of red clover transcriptome based on RNA-Seq data provides insight into drought response, gene discovery and marker identification. BMC Genom. 2014, 15, 453. [Google Scholar] [CrossRef]
- Gupta, M.; Sharma, G.; Saxena, D.; Budhwar, R.; Vasudevan, M.; Gupta, V.; Gupta, A.; Gupta, R.; Chandran, D. Dual RNA-Seq analysis of Medicago truncatula and the pea powdery mildew Erysiphe pisi uncovers distinct host transcriptional signatures during incompatible and compatible interactions and pathogen effector candidates. Genomics 2020, 112, 2130–2145. [Google Scholar] [CrossRef]
- Nagy, E.D.; Guo, Y.F.; Tang, S.X.; Bowers, J.E.; Okashah, R.A.; Taylor, C.A.; Zhang, D.; Khanal, S.; Heesacker, A.F.; Khalilian, N.; et al. A high-density genetic map of Arachis duranensis, a diploid ancestor of cultivated peanut. BMC Genom. 2012, 13, 469. [Google Scholar] [CrossRef] [PubMed]
- Alexander, D.; Goodman, R.M.; Gut-Rella, M.; Glascock, C.; Weymann, K.; Friedrich, L.; Maddox, D.; Ahl-Goy, P.; Luntz, T.; Ward, E. Increased tolerance to two oomycete pathogens in transgenic tobacco expressing pathogenesis-related protein 1a. Proc. Natl. Acad. Sci. USA 1993, 90, 7327–7331. [Google Scholar] [CrossRef] [PubMed]
- Sessa, G.; Yang, X.Q.; Raz, V.; Eyal, Y.; Fluhr, R. Dark induction and subcellular localization of the pathogenesis-related PRB-1b protein. Plant Mol. Biol. 1995, 28, 537–547. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.F.; Jiang, L.Y.; Wu, J.J.; Li, W.B.; Fan, S.J.; Zhang, S.Z. Isolation and characterization of a pathogenesis-related protein 10 gene (GmPR10) with induced expression in soybean (Glycine max) during infection with Phytophthora sojae. Mol. Biol. Rep. 2014, 41, 4899–4909. [Google Scholar] [CrossRef]
- Breen, S.; Williams, S.J.; Outram, M.; Kobe, B.; Solomon, P.S. Emerging insights into the functions of pathogenesis-related protein 1. Trends Plant Sci. 2017, 22, 871–879. [Google Scholar] [CrossRef]
- van Verk, M.C.; Pappaioannou, D.; Neeleman, L.; Bol, J.F.; Linthorst, H.J.M. A novel WRKY transcription factor is required for induction of PR-1a gene expression by salicylic acid and bacterial elicitors. Plant Physiol. 2008, 146, 1983–1995. [Google Scholar] [CrossRef]
- Zhang, Y.; Fan, W.; Kinkema, M.; Li, X.; Dong, X. Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene. Proc. Natl. Acad. Sci. USA 1999, 96, 6523–6528. [Google Scholar] [CrossRef]
- Zhao, Y.; Chang, X.; Qi, D.; Dong, L.; Wang, G.; Fan, S.; Jiang, L.; Cheng, Q.; Chen, X.; Han, D.; et al. A novel soybean ERF transcription factor, GmERF113, increases resistance to Phytophthora sojae infection in soybean. Front. Plant Sci. 2017, 8, 299. [Google Scholar] [CrossRef]
- He, K.; Gou, X.P.; Yuan, T.; Lin, H.H.; Asami, T.; Yoshida, S.; Russell, S.D.; Li, J. BAK1 and BKK1 regulate Brassinosteroid-dependent growth and BrassinosteroidIndependent cell-death pathways. Curr. Biol. 2007, 17, 1109–1115. [Google Scholar] [CrossRef] [PubMed]
- Ward, E.R.; Uknes, S.J.; Williams, S.C.; Dincher, S.S.; Wiederhold, D.L.; Alexander, D.C.; Ahl-Goy, P.; Metraux, J.P.; Ryals, J.A. Coordinate gene activity in response to agents that Induce systemic acquired resistance. Plant Cell 1991, 3, 1085–1094. [Google Scholar] [CrossRef] [PubMed]
- Du, M.M.; Zhao, J.H.; Tzeng, D.T.W.; Liu, Y.Y.; Deng, L.; Yang, T.X.; Zhai, Q.Z.; Wu, F.M.; Huang, Z.; Zhou, M.; et al. MYC2 orchestrates a hierarchical transcriptional cascade that regulates jasmonate-mediated plant immunity in tomato. Plant Cell 2017, 29, 1883–1906. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.B.; Liu, Q.N.; Cao, Y.R.; Zhang, Y.; Chen, D.B.; Lou, X.Y.; Cheng, S.H.; Cao, L.Y. The OsMPK15 negatively regulates Magnaporthe oryza and Xoo disease resistance via SA and JA signaling pathway in rice. Front. Plant Sci. 2019, 10, 752. [Google Scholar] [CrossRef]
- He, X.; Zhu, L.F.; Wassan, G.M.; Wang, Y.J.; Miao, Y.H.; Shaban, M.; Hu, H.Y.; Sun, H.; Zhang, X.L. GhJAZ2 attenuates cotton resistance to biotic stresses via the inhibition of the transcriptional activity of GhbHLH171. Mol. Plant Pathol. 2018, 19, 896–908. [Google Scholar] [CrossRef]
- Uji, Y.; Taniguchi, S.; Tamaoki, D.; Shishido, H.; Akimitsu, K.; Gomi, K. Overexpression of OsMYC2 results in the up-regulation of early JA-rresponsive genes and bacterial blight resistance in rice. Plant Cell Physiol. 2016, 57, 1814–1827. [Google Scholar] [CrossRef]
- Ji, S.D.; Wang, Z.Y.; Wang, J.J.; Fan, H.J.; Wang, Y.C.; Liu, Z.H. Properties analysis of transcription factor gene TasMYB36 from Trichoderma asperellum CBS433.97 and its heterogeneous transfomation to improve antifungal ability of Populus. Sci. Rep. 2017, 7, 12801. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.J.; Li, M.; Li, T.T.; Chen, Y.J.; Zhang, L.J.; Zhao, G.F.; Zhuang, J.H.; Zhao, W.Y.; Gao, L.P.; Xia, T. Airborne fungus-induced biosynthesis of anthocyanins in Arabidopsis thaliana via jasmonic acid and salicylic acid signaling. Plant Sci. 2020, 300, 110635. [Google Scholar] [CrossRef]
- Ren, H.R.; Bai, M.J.; Sun, J.J.; Liu, J.Y.; Ren, M.; Dong, Y.W.; Wang, N.; Ning, G.G.; Wang, C.Q. RcMYB84 and RcMYB123 mediate jasmonate-induced defense responses against Botrytis cinerea in rose (Rosa chinensis). Plant J. 2020, 103, 1839–1849. [Google Scholar] [CrossRef]
- Sugimoto, K.; Takeda, S.; Hirochika, H. MYB-related transcription factor NtMYB2 induced by wounding and elicitors is a regulator of the tobacco retrotransposon Tto1 and defense-related genes. Plant Cell 2000, 12, 2511–2528. [Google Scholar] [CrossRef] [PubMed]
- Chakravarthy, S.; Tuori, R.P.; D’Ascenzo, M.D.; Fobert, P.R.; Després, C.; Martin, G.B. The tomato transcription factor Pti4 regulates defense-related gene expression via GCC box and non-GCC box cis elements. Plant Cell 2003, 15, 3033–3050. [Google Scholar] [CrossRef]
- Zhu, C.Y.; Li, Z.Y.; Tang, Y.Z.; Zhang, L.Q.; Wen, J.H.; Wang, Z.M.; Su, Y.Y.; Chen, Y.E.; Zhang, H.Y. TaWRKY10 plays a key role in the upstream of circadian gene TaLHY in wheat. Plant Sci. 2021, 310, 110973. [Google Scholar] [CrossRef]
- Gao, Y.; Jia, S.W.; Wang, C.L.; Wang, F.J.; Wang, F.J.; Zhao, K.J. BjMYB1, a transcription factor implicated in plant defence through activating BjCHI1 chitinase expression by binding to a W-box-like element. J. Exp. Bot. 2016, 67, 4647–4658. [Google Scholar] [CrossRef]
- Zhang, M.F.; Wang, J.Q.; Luo, Q.J.; Yang, C.; Yang, H.B.; Cheng, Y.J. CsMYB96 enhances citrus fruit resistance against fungal pathogen by activating salicylic acid biosynthesis and facilitating defense metabolite accumulation. J. Plant Physiol. 2021, 264, 153472. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Cai, H.Y.; Bai, M.Y.; Zhang, M.; Chen, F.Q.; Huang, Y.M.; Priyadarshani, S.; Chai, M.N.; Liu, L.P.; Liu, Y.H.; et al. A soybean bZIP transcription factor GmbZIP19 confers multiple biotic and abiotic stress responses in plant. Int. J. Mol. Sci. 2020, 21, 4701. [Google Scholar] [CrossRef]
- Song, M.; Fang, S.Q.; Li, Z.G.; Li, X.; Liu, W.B.; Zhang, Y.; Lin, C.H.; Miao, W.G. CsAtf1, a bZIP transcription factor, is involved in fludioxonil sensitivity and virulence in the rubber tree anthracnose fungus Colletotrichum siamense. Fungal Genet. Biol. 2022, 158, 103649. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.T.; Lin, R.M.; Li, Y.Y.; Wang, P.; Feng, J.; Chen, W.Q.; Xu, S.C. TabZIP74 acts as a positive regulator in wheat stripe rust resistance and involves root development by mRNA splicing. Front. Plant Sci. 2019, 10, 1551. [Google Scholar] [CrossRef] [PubMed]
- Li, D.D.; Li, X.M.; Liu, X.T.; Zhang, Z. Comprehensive analysis of bZIP gene family and function of RcbZIP17 on Botrytis resistance in rose (Rosa chinensis). Gene 2023, 849, 146867. [Google Scholar] [CrossRef]
- Zhang, S.Z.; Xu, P.F.; Wu, J.J.; Xue, A.G.; Zhang, J.X.; Li, W.B.; Chen, C.; Chen, W.Y.; Lv, H.Y. Races of Phytophthora sojae and their virulences on soybean cultivars in Heilongjiang, China. Plant Dis. 2010, 94, 87–91. [Google Scholar] [CrossRef]
- Ward, E.W.B.; Lazarovits, G.; Unwin, C.H.; Buzzell, R.I. Hypocotyl reactions and glyceollin in soybeans inoculated with zoospores of phytophthora megasperma var sojae. Phytopathology 1979, 69, 951–955. [Google Scholar] [CrossRef]
- Morris, P.F.; Savard, M.E.; Ward, E.W.B.J.P.; Pathology, M.P. Identification and accumulation of isoflavonoids and isoflavone glucosides in soybean leaves and hypocotyls in resistance responses to Phytophthora megasperma f. sp. glycinea. Physiol. Mol. Plant Pathol. 1991, 39, 229–244. [Google Scholar] [CrossRef]
- Fehr, W.; Caviness, C.; Burmood, D.; Pennington, J. Stage of development descriptions for soybeans, Glycine Max (L.) Merrill 1. Crop. Sci. 1971, 11, 929–931. [Google Scholar] [CrossRef]
- Kerschen, A.; Napoli, C.A.; Jorgensen, R.A.; Müller, A.E. Effectiveness of RNA interference in transgenic plants. FEBS Lett. 2004, 566, 223–228. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.D.; Cho, Y.H.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef]
- Graham, T.L.; Graham, M.Y.; Subramanian, S.; Yu, O. RNAi silencing of genes for elicitation or biosynthesis of 5-deoxyisoflavonoids suppresses race-specific resistance and hypersensitive cell death in Phytophthora sojae infected tissues. Plant Physiol. 2007, 144, 728–740. [Google Scholar] [CrossRef] [PubMed]
- Kereszt, A.; Li, D.; Indrasumunar, A.; Nguyen, C.D.; Nontachaiyapoom, S.; Kinkema, M.; Gresshoff, P.M. Agrobacterium rhizogenes-mediated transformation of soybean to study root biology. Nat. Protoc. 2007, 2, 948–952. [Google Scholar] [CrossRef]
- Dou, D.; Wang, B.; Zhu, S.; Tang, Y.; Wang, Z.; Sun, J.; Li, R.; Zhang, Z. Transgenic tobacco with NDR1 gene improved its resistance to two fungal diseases. Sci. Agric. Sin. 2003, 36, 1120–1124. [Google Scholar]
- Zhu, F.; Xi, D.H.; Deng, X.G.; Peng, X.J.; Tang, H.; Chen, Y.J.; Jian, W.; Feng, H.; Lin, H.H. The chilli veinal mottle virus regulates expression of the tobacco mosaic virus resistance gene N and jasmonic acid/ethylene signaling is essential for systemic resistance against chilli veinal mottle virus in tobacco. Plant Mol. Biol. Rep. 2014, 32, 382–394. [Google Scholar] [CrossRef]
- Meng, X.; Xu, J.; He, Y.; Yang, K.Y.; Mordorski, B.; Liu, Y.; Zhang, S. Phosphorylation of an ERF transcription factor by Arabidopsis MPK3/MPK6 regulates plant defense gene induction and fungal resistance. Plant Cell 2013, 25, 1126–1142. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, H.; Ma, J.; Zhao, Y.; Zhang, C.; Zhao, M.; He, S.; Sun, Y.; Fang, X.; Chen, X.; Ma, K.; et al. The MYB Transcription Factor GmMYB78 Negatively Regulates Phytophthora sojae Resistance in Soybean. Int. J. Mol. Sci. 2024, 25, 4247. https://doi.org/10.3390/ijms25084247
Gao H, Ma J, Zhao Y, Zhang C, Zhao M, He S, Sun Y, Fang X, Chen X, Ma K, et al. The MYB Transcription Factor GmMYB78 Negatively Regulates Phytophthora sojae Resistance in Soybean. International Journal of Molecular Sciences. 2024; 25(8):4247. https://doi.org/10.3390/ijms25084247
Chicago/Turabian StyleGao, Hong, Jia Ma, Yuxin Zhao, Chuanzhong Zhang, Ming Zhao, Shengfu He, Yan Sun, Xin Fang, Xiaoyu Chen, Kexin Ma, and et al. 2024. "The MYB Transcription Factor GmMYB78 Negatively Regulates Phytophthora sojae Resistance in Soybean" International Journal of Molecular Sciences 25, no. 8: 4247. https://doi.org/10.3390/ijms25084247



