Gene Silencing of Angiopoietin-like 3 (ANGPTL3) Induced De Novo Lipogenesis and Lipid Accumulation in Huh7 Cell Line
Abstract
:1. Introduction
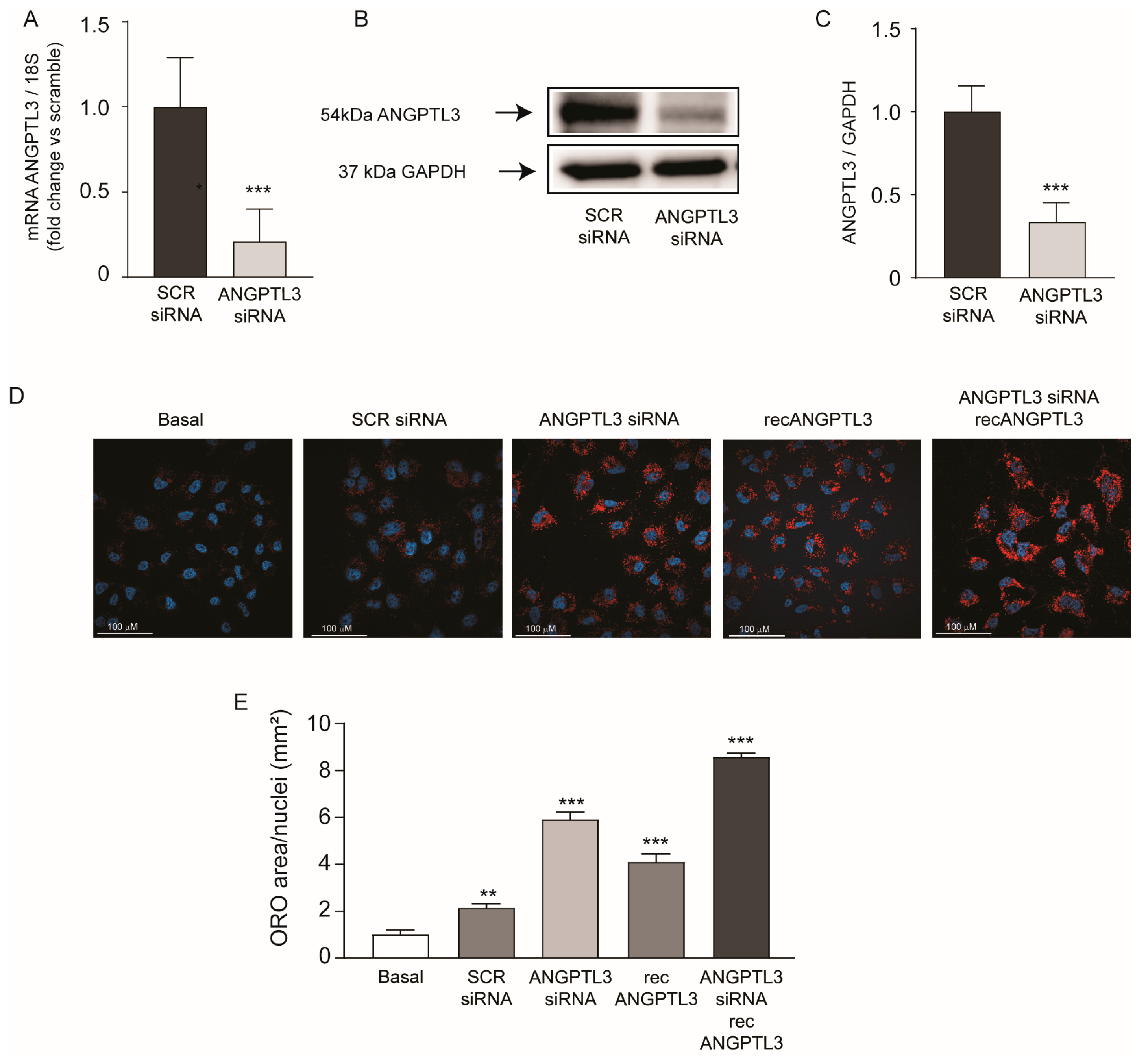
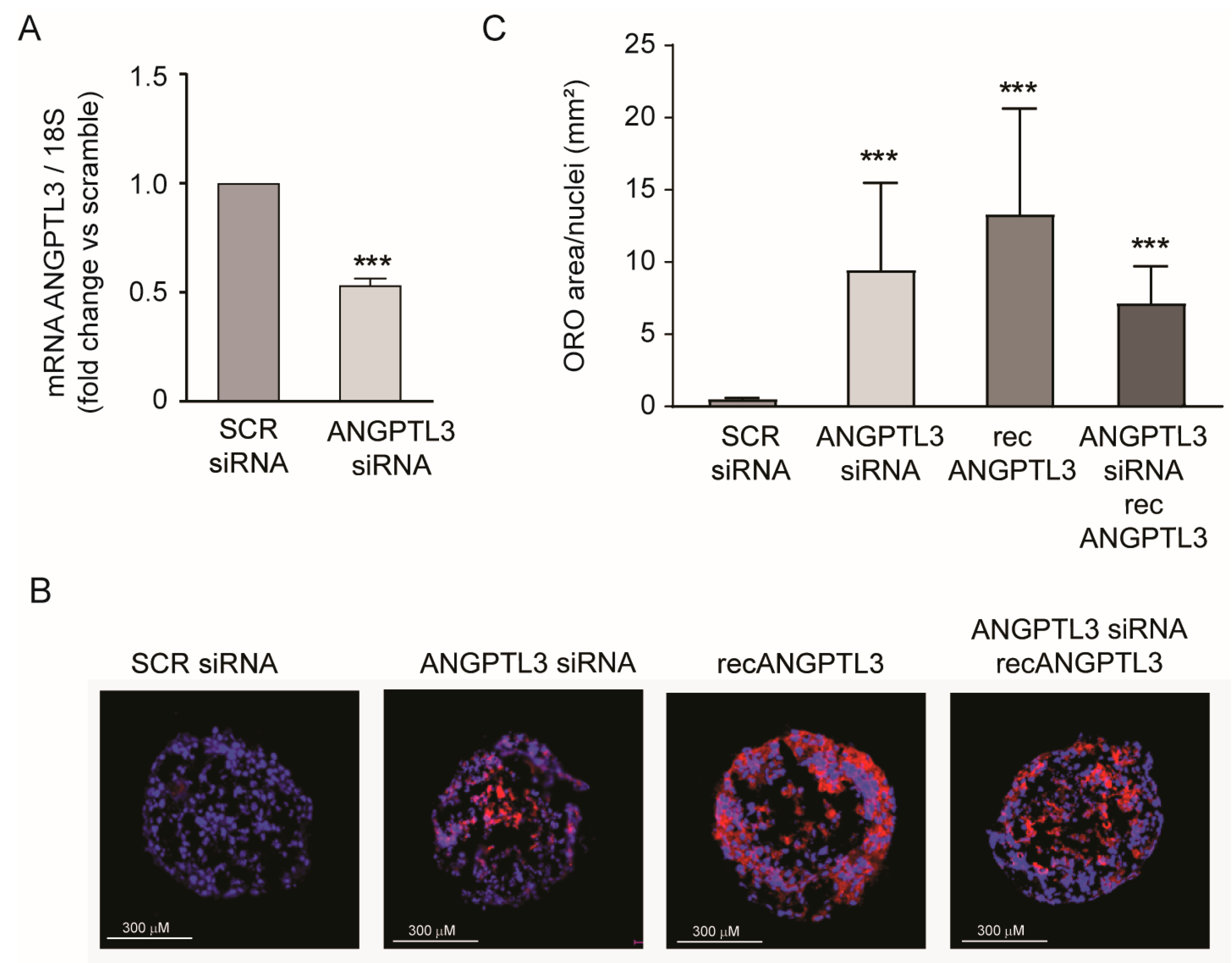
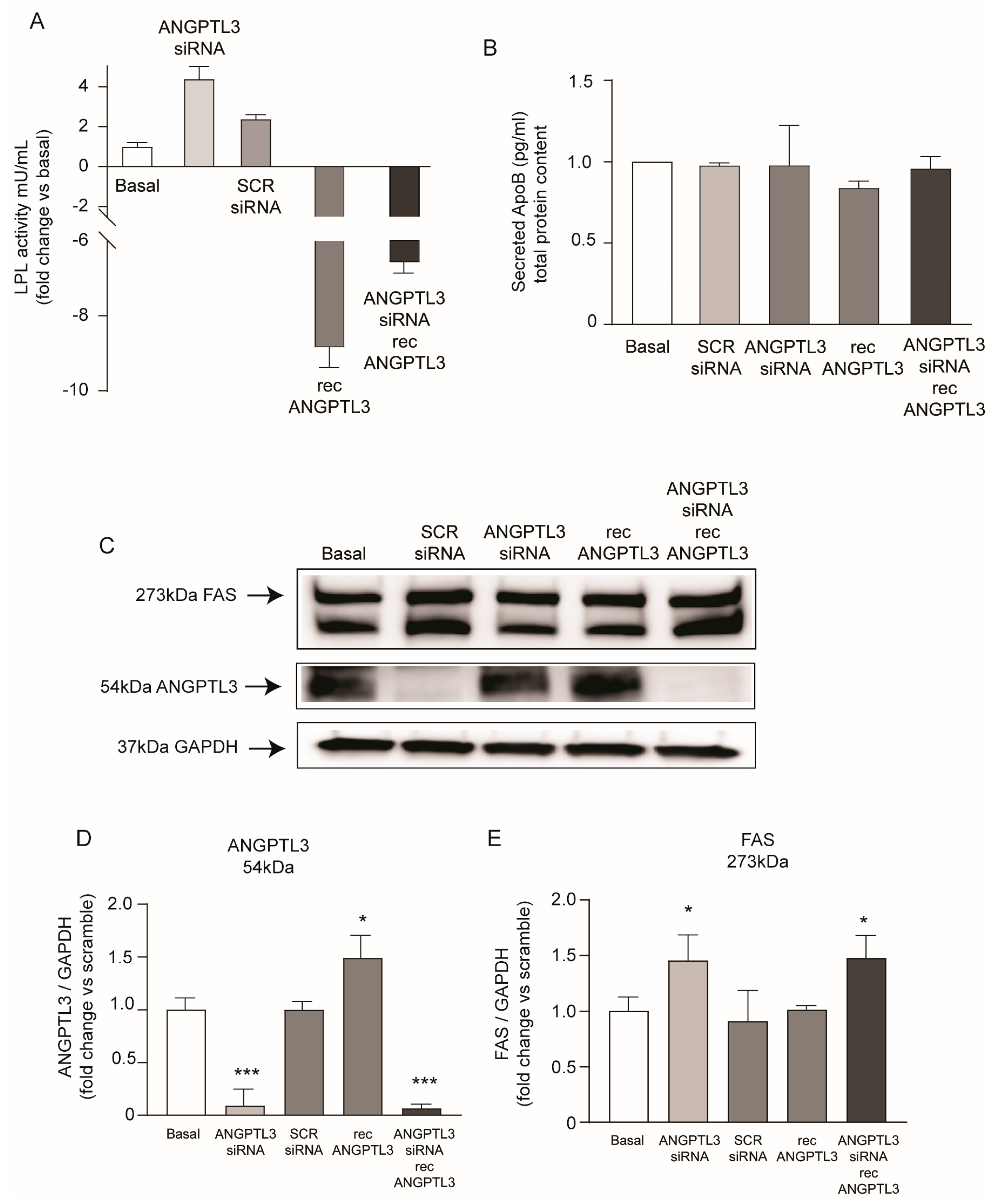
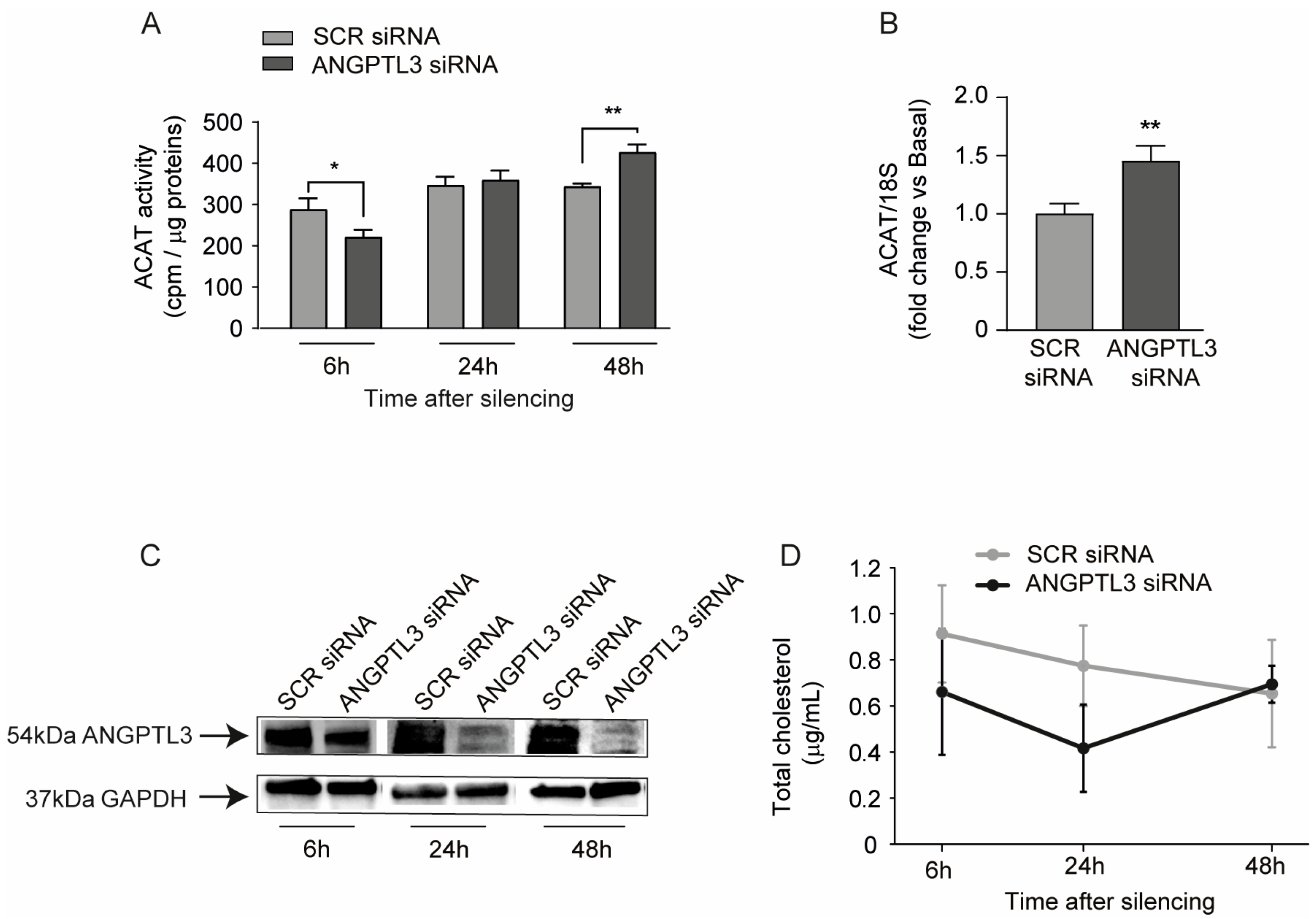
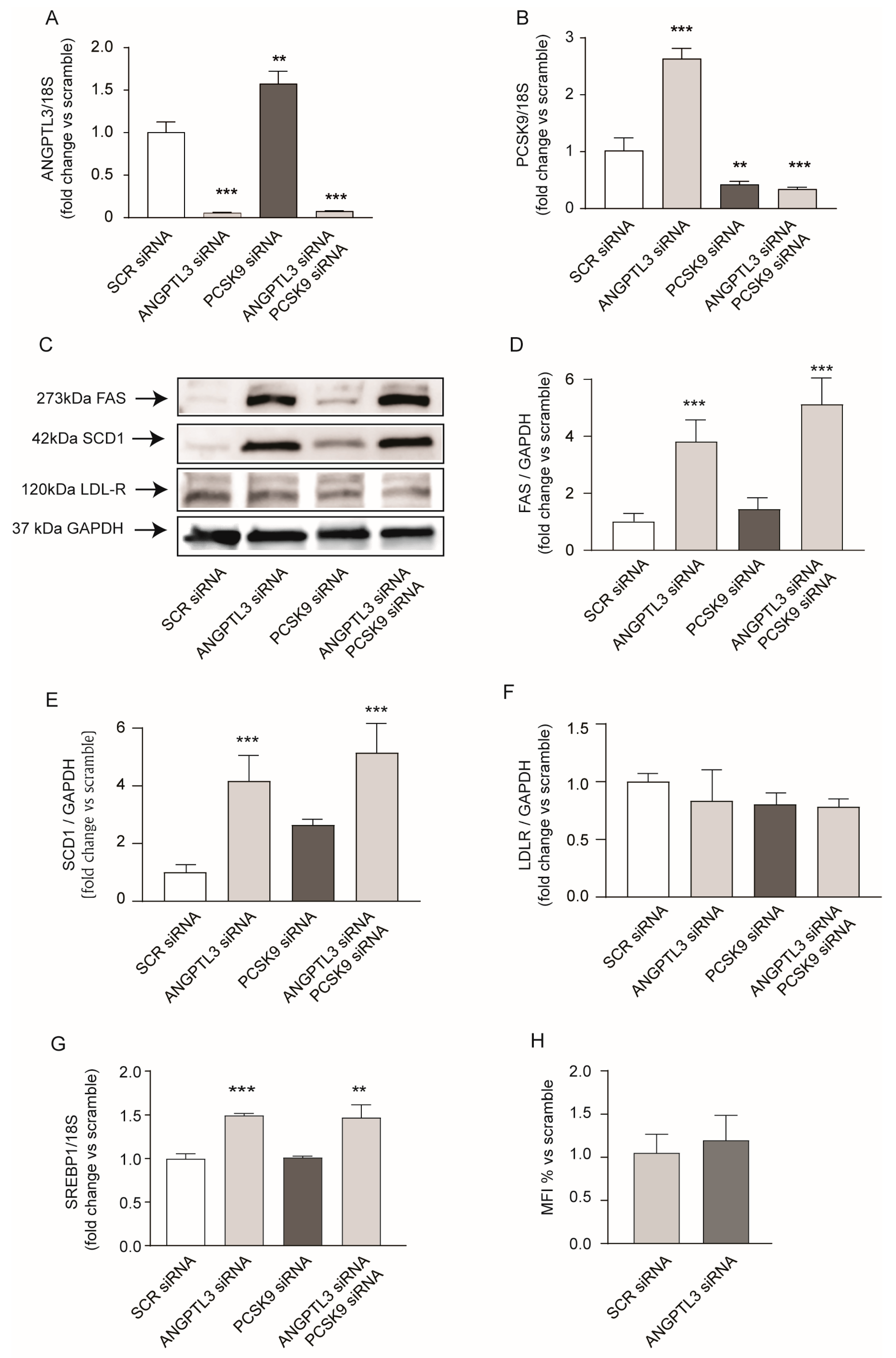
2. Results
3. Discussion
4. Materials and Methods
4.1. Cell Cultures
4.2. ANGPTL3 Silencing and Recombinant Protein Administration
4.3. Western Blotting
4.4. Reverse Transcription and Quantitative PCR (RT-qPCR)
4.5. ELISA Assay for ApoB and PCSK9
4.6. Neutral Lipid Staining with Oil Red-O
4.7. Lipoprotein Lipase Activity Assay
4.8. Cholesterol Esterification Assay (ACAT Activity)
4.9. Cholesterol Determination
4.10. Fluorescent LDL Uptake Cell-Based Assay
4.11. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Conklin, D.; Gilbertson, D.; Taft, D.W.; Maurer, M.F.; Whitmore, T.E.; Smith, D.L.; Walker, K.M.; Chen, L.H.; Wattler, S.; Nehls, M.; et al. Identification of a mammalian angiopoietin-related protein expressed specifically in liver. Genomics 1999, 62, 477–482. [Google Scholar] [CrossRef] [PubMed]
- Koishi, R.; Ando, Y.; Ono, M.; Shimamura, M.; Yasumo, H.; Fujiwara, T.; Horikoshi, H.; Furukawa, H. Angptl3 regulates lipid metabolism in mice. Nat. Genet. 2002, 30, 151–157. [Google Scholar] [CrossRef] [PubMed]
- Shimizugawa, T.; Ono, M.; Shimamura, M.; Yoshida, K.; Ando, Y.; Koishi, R.; Ueda, K.; Inaba, T.; Minekura, H.; Kohama, T.; et al. ANGPTL3 decreases very low density lipoprotein triglyceride clearance by inhibition of lipoprotein lipase. J. Biol. Chem. 2002, 277, 33742–33748. [Google Scholar] [CrossRef] [PubMed]
- Shimamura, M.; Matsuda, M.; Kobayashi, S.; Ando, Y.; Ono, M.; Koishi, R.; Furukawa, H.; Makishima, M.; Shimomura, I. Angiopoietin-like protein 3, a hepatic secretory factor, activates lipolysis in adipocytes. Biochem. Biophys. Res. Commun. 2003, 301, 604–609. [Google Scholar] [CrossRef] [PubMed]
- Shimamura, M.; Matsuda, M.; Yasumo, H.; Okazaki, M.; Fujimoto, K.; Kono, K.; Shimizugawa, T.; Ando, Y.; Koishi, R.; Kohama, T.; et al. Angiopoietin-like protein3 regulates plasma HDL cholesterol through suppression of endothelial lipase. Arterioscler. Thromb. Vasc. Biol. 2007, 27, 366–372. [Google Scholar] [CrossRef]
- McCoy, M.G.; Sun, G.S.; Marchadier, D.; Maugeais, C.; Glick, J.M.; Rader, D.J. Characterization of the lipolytic activity of endothelial lipase. J. Lipid Res. 2002, 43, 921–929. [Google Scholar] [CrossRef]
- Lupo, M.G.; Ferri, N. Angiopoietin-Like 3 (ANGPTL3) and Atherosclerosis: Lipid and Non-Lipid Related Effects. J. Cardiovasc. Dev. Dis. 2018, 5, 39. [Google Scholar] [CrossRef] [PubMed]
- Dewey, F.E.; Gusarova, V.; Dunbar, R.L.; O’Dushlaine, C.; Schurmann, C.; Gottesman, O.; McCarthy, S.; Van Hout, C.V.; Bruse, S.; Dansky, H.M.; et al. Genetic and Pharmacologic Inactivation of ANGPTL3 and Cardiovascular Disease. N. Engl. J. Med. 2017, 377, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Ruscica, M.; Zimetti, F.; Adorni, M.P.; Sirtori, C.R.; Lupo, M.G.; Ferri, N. Pharmacological aspects of ANGPTL3 and ANGPTL4 inhibitors: New therapeutic approaches for the treatment of atherogenic dyslipidemia. Pharmacol. Res. 2020, 153, 104653. [Google Scholar] [CrossRef] [PubMed]
- Gusarova, V.; Alexa, C.A.; Wang, Y.; Rafique, A.; Kim, J.H.; Buckler, D.; Mintah, I.J.; Shihanian, L.M.; Cohen, J.C.; Hobbs, H.H.; et al. ANGPTL3 blockade with a human monoclonal antibody reduces plasma lipids in dyslipidemic mice and monkeys. J. Lipid Res. 2015, 56, 1308–1317. [Google Scholar] [CrossRef] [PubMed]
- Gaudet, D.; Karwatowska-Prokopczuk, E.; Baum, S.J.; Hurh, E.; Kingsbury, J.; Bartlett, V.J.; Figueroa, A.L.; Piscitelli, P.; Singleton, W.; Witztum, J.L.; et al. Vupanorsen, an N-acetyl galactosamine-conjugated antisense drug to ANGPTL3 mRNA, lowers triglycerides and atherogenic lipoproteins in patients with diabetes, hepatic steatosis, and hypertriglyceridaemia. Eur. Heart J. 2020, 41, 3936–3945. [Google Scholar] [CrossRef]
- Graham, M.J.; Lee, R.G.; Brandt, T.A.; Tai, L.J.; Fu, W.; Peralta, R.; Yu, R.; Hurh, E.; Paz, E.; McEvoy, B.W.; et al. Cardiovascular and Metabolic Effects of ANGPTL3 Antisense Oligonucleotides. N. Engl. J. Med. 2017, 377, 222–232. [Google Scholar] [CrossRef] [PubMed]
- Raal, F.J.; Rosenson, R.S.; Reeskamp, L.F.; Hovingh, G.K.; Kastelein, J.J.P.; Rubba, P.; Ali, S.; Banerjee, P.; Chan, K.C.; Gipe, D.A.; et al. Evinacumab for Homozygous Familial Hypercholesterolemia. N. Engl. J. Med. 2020, 383, 711–720. [Google Scholar] [CrossRef]
- Wang, Y.; Gusarova, V.; Banfi, S.; Gromada, J.; Cohen, J.C.; Hobbs, H.H. Inactivation of ANGPTL3 reduces hepatic VLDL-triglyceride secretion. J. Lipid Res. 2015, 56, 1296–1307. [Google Scholar] [CrossRef] [PubMed]
- Luo, F.; Das, A.; Fang, Z. Evinacumab for Homozygous Familial Hypercholesterolemia. N. Engl. J. Med. 2021, 384, e17. [Google Scholar] [CrossRef]
- Ando, Y.; Shimizugawa, T.; Takeshita, S.; Ono, M.; Shimamura, M.; Koishi, R.; Furukawa, H. A decreased expression of angiopoietin-like 3 is protective against atherosclerosis in apoE-deficient mice. J. Lipid Res. 2003, 44, 1216–1223. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.C.; Desai, U.; Gololobov, G.; Hong, S.; Feng, X.; Yu, X.C.; Gay, J.; Wilganowski, N.; Gao, C.; Du, L.L.; et al. Identification of a new functional domain in angiopoietin-like 3 (ANGPTL3) and angiopoietin-like 4 (ANGPTL4) involved in binding and inhibition of lipoprotein lipase (LPL). J. Biol. Chem. 2009, 284, 13735–13745. [Google Scholar] [CrossRef] [PubMed]
- Bergmark, B.A.; Marston, N.A.; Bramson, C.R.; Curto, M.; Ramos, V.; Jevne, A.; Kuder, J.F.; Park, J.G.; Murphy, S.A.; Verma, S.; et al. Effect of Vupanorsen on Non-High-Density Lipoprotein Cholesterol Levels in Statin-Treated Patients with Elevated Cholesterol: TRANSLATE-TIMI 70. Circulation 2022, 145, 1377–1386. [Google Scholar] [CrossRef] [PubMed]
- Stefanutti, C.; Chan, D.C.; Di Giacomo, S.; Morozzi, C.; Watts, G.F. Long-Term Efficacy and Safety of Evinacumab in Patients with Homozygous Familial Hypercholesterolemia: Real-World Clinical Experience. Pharmaceuticals 2022, 15, 1389. [Google Scholar] [CrossRef] [PubMed]
- Rosenson, R.S.; Gaudet, D.; Ballantyne, C.M.; Baum, S.J.; Bergeron, J.; Kershaw, E.E.; Moriarty, P.M.; Rubba, P.; Whitcomb, D.C.; Banerjee, P.; et al. Evinacumab in severe hypertriglyceridemia with or without lipoprotein lipase pathway mutations: A phase 2 randomized trial. Nat. Med. 2023, 29, 729–737. [Google Scholar] [CrossRef]
- Yue, P.; Tanoli, T.; Wilhelm, O.; Patterson, B.; Yablonskiy, D.; Schonfeld, G. Absence of fatty liver in familial hypobetalipoproteinemia linked to chromosome 3p21. Metabolism 2005, 54, 682–688. [Google Scholar] [CrossRef] [PubMed]
- Inaba, T.; Matsuda, M.; Shimamura, M.; Takei, N.; Terasaka, N.; Ando, Y.; Yasumo, H.; Koishi, R.; Makishima, M.; Shimomura, I. Angiopoietin-like protein 3 mediates hypertriglyceridemia induced by the liver X receptor. J. Biol. Chem. 2003, 278, 21344–21351. [Google Scholar] [CrossRef] [PubMed]
- Ruscica, M.; Macchi, C.; Fogacci, F.; Ferri, N.; Grandi, E.; Rizzoli, E.; D’Addato, S.; Borghi, C.; Cicero, A.F.; Brisighella Heart Study, G. Angiopoietin-like 3 and subclinical peripheral arterial disease: Evidence from the Brisighella Heart Study. Eur. J. Prev. Cardiol. 2019, 27, 2251–2254. [Google Scholar] [CrossRef] [PubMed]
- Ferri, N.; Ruscica, M. Proprotein convertase subtilisin/kexin type 9 (PCSK9) and metabolic syndrome: Insights on insulin resistance, inflammation, and atherogenic dyslipidemia. Endocrine 2016, 54, 588–601. [Google Scholar] [CrossRef] [PubMed]
- Schulz, R.; Schluter, K.D.; Laufs, U. Molecular and cellular function of the proprotein convertase subtilisin/kexin type 9 (PCSK9). Basic. Res. Cardiol. 2015, 110, 4. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.S.; Goldstein, J.L. The SREBP pathway: Regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 1997, 89, 331–340. [Google Scholar] [CrossRef] [PubMed]
- Lakoski, S.G.; Lagace, T.A.; Cohen, J.C.; Horton, J.D.; Hobbs, H.H. Genetic and metabolic determinants of plasma PCSK9 levels. J. Clin. Endocrinol. Metab. 2009, 94, 2537–2543. [Google Scholar] [CrossRef] [PubMed]
- Theocharidou, E.; Papademetriou, M.; Reklou, A.; Sachinidis, A.; Boutari, C.; Giouleme, O. The Role of PCSK9 in the Pathogenesis of Non-alcoholic Fatty Liver Disease and the Effect of PCSK9 Inhibitors. Curr. Pharm. Des. 2018, 24, 3654–3657. [Google Scholar] [CrossRef] [PubMed]
- Ruhanen, H.; Haridas, P.A.N.; Jauhiainen, M.; Olkkonen, V.M. Angiopoietin-like protein 3, an emerging cardiometabolic therapy target with systemic and cell-autonomous functions. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2020, 1865, 158791. [Google Scholar] [CrossRef]
- Burks, K.H.; Xie, Y.; Gildea, M.; Jung, I.H.; Mukherjee, S.; Lee, P.; Pudupakkam, U.; Wagoner, R.; Patel, V.; Santana, K.; et al. ANGPTL3 deficiency impairs lipoprotein production and produces adaptive changes in hepatic lipid metabolism. J. Lipid Res. 2024, 65, 100500. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.X.; Redon, V.; Yu, H.; Querbes, W.; Pirruccello, J.; Liebow, A.; Deik, A.; Trindade, K.; Wang, X.; Musunuru, K.; et al. Role of angiopoietin-like 3 (ANGPTL3) in regulating plasma level of low-density lipoprotein cholesterol. Atherosclerosis 2018, 268, 196–206. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.S.; Goldstein, J.L. A proteolytic pathway that controls the cholesterol content of membranes, cells, and blood. Proc. Natl. Acad. Sci. USA 1999, 96, 11041–11048. [Google Scholar] [CrossRef]
- Wang, X.; Sato, R.; Brown, M.S.; Hua, X.; Goldstein, J.L. SREBP-1, a membrane-bound transcription factor released by sterol-regulated proteolysis. Cell 1994, 77, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Ruhanen, H.; Haridas, P.A.N.; Minicocci, I.; Taskinen, J.H.; Palmas, F.; di Costanzo, A.; D’Erasmo, L.; Metso, J.; Partanen, J.; Dalli, J.; et al. ANGPTL3 deficiency alters the lipid profile and metabolism of cultured hepatocytes and human lipoproteins. Biochim. Et Biophys. Acta Mol. Cell Biol. Lipids 2020, 1865, 158679. [Google Scholar] [CrossRef]
- Jeong, H.J.; Lee, H.S.; Kim, K.S.; Kim, Y.K.; Yoon, D.; Park, S.W. Sterol-dependent regulation of proprotein convertase subtilisin/kexin type 9 expression by sterol-regulatory element binding protein-2. J. Lipid Res. 2008, 49, 399–409. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Samarghandi, A.; Zhang, N.; Yao, Z.; Xiong, M.; Teng, B.B. Proprotein convertase subtilisin/kexin type 9 interacts with apolipoprotein B and prevents its intracellular degradation, irrespective of the low-density lipoprotein receptor. Arterioscler. Thromb. Vasc. Biol. 2012, 32, 1585–1595. [Google Scholar] [CrossRef]
- Bini, S.; D’Erasmo, L.; Minicocci, I.; Di Costanzo, A.; Tramontano, D.; Pomanti, G.; Covino, S.; Arca, M.; Pecce, V. ApoB secretion and intracellular lipid content are modulated by ANGPTL3 and PCSK9 in HEPG2 cells. Atherosclerosis 2023, 379, S14. [Google Scholar] [CrossRef]
- Mehlem, A.; Hagberg, C.E.; Muhl, L.; Eriksson, U.; Falkevall, A. Imaging of neutral lipids by oil red O for analyzing the metabolic status in health and disease. Nat. Protoc. 2013, 8, 1149–1154. [Google Scholar] [CrossRef] [PubMed]
- Canavesi, M.; Baldini, N.; Leonardi, A.; Sironi, G.; Bellosta, S.; Bernini, F. In vitro inhibitory effect of lercanidipine on cholesterol accumulation and matrix metalloproteinases secretion by macrophages. J. Cardiovasc. Pharmacol. 2004, 44, 416–422. [Google Scholar] [CrossRef] [PubMed]
- Pinzon Grimaldos, A.; Pacella, I.; Bini, S.; Tucci, G.; Cammarata, I.; Di Costanzo, A.; Minicocci, I.; D’Erasmo, L.; Arca, M.; Piconese, S. ANGPTL3 deficiency associates with the expansion of regulatory T cells with reduced lipid content. Atherosclerosis 2022, 362, 38–46. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rossi, I.; Marodin, G.; Lupo, M.G.; Adorni, M.P.; Papotti, B.; Dall’Acqua, S.; Ferri, N. Gene Silencing of Angiopoietin-like 3 (ANGPTL3) Induced De Novo Lipogenesis and Lipid Accumulation in Huh7 Cell Line. Int. J. Mol. Sci. 2024, 25, 3708. https://doi.org/10.3390/ijms25073708
Rossi I, Marodin G, Lupo MG, Adorni MP, Papotti B, Dall’Acqua S, Ferri N. Gene Silencing of Angiopoietin-like 3 (ANGPTL3) Induced De Novo Lipogenesis and Lipid Accumulation in Huh7 Cell Line. International Journal of Molecular Sciences. 2024; 25(7):3708. https://doi.org/10.3390/ijms25073708
Chicago/Turabian StyleRossi, Ilaria, Giorgia Marodin, Maria Giovanna Lupo, Maria Pia Adorni, Bianca Papotti, Stefano Dall’Acqua, and Nicola Ferri. 2024. "Gene Silencing of Angiopoietin-like 3 (ANGPTL3) Induced De Novo Lipogenesis and Lipid Accumulation in Huh7 Cell Line" International Journal of Molecular Sciences 25, no. 7: 3708. https://doi.org/10.3390/ijms25073708
APA StyleRossi, I., Marodin, G., Lupo, M. G., Adorni, M. P., Papotti, B., Dall’Acqua, S., & Ferri, N. (2024). Gene Silencing of Angiopoietin-like 3 (ANGPTL3) Induced De Novo Lipogenesis and Lipid Accumulation in Huh7 Cell Line. International Journal of Molecular Sciences, 25(7), 3708. https://doi.org/10.3390/ijms25073708






