Abstract
Cancers remain the second leading cause of mortality in the world. Preclinical and clinical studies point an important role of cancer/leukaemia stem cells (CSCs/LSCs) in the colonisation at secondary organ sites upon metastatic spreading, although the precise mechanisms for specific actions are still not fully understood. Reviewing the present knowledge on the crucial role of CSCs/LSCs, their plasticity, and population heterogeneity in treatment failures in cancer patients is timely. Standard chemotherapy, which acts mainly on rapidly dividing cells, is unable to adequately affect CSCs with a low proliferation rate. One of the proposed mechanisms of CSC resistance to anticancer agents is the fact that these cells can easily shift between different phases of the cell cycle in response to typical cell stimuli induced by anticancer drugs. In this work, we reviewed the recent studies on CSC/LSC alterations associated with disease recurrence, and we systematised the functional assays, markers, and novel methods for CSCs screening. This review emphasises CSCs’ involvement in cancer progression and metastasis, as well as CSC/LSC targeting by synthetic and natural compounds aiming at their elimination or modulation of stemness properties.
1. Introduction
Despite extensive research into the nature of cancers, they remain a leading cause of death. Based on the GLOBOCAN 2020 registry with over 10 million fatal events in 2020 alone, including lung cancer (1,796,144), colorectal (935,173), liver (830,180), stomach (768,793), breast (684,996), oesophagus (544,076), pancreas (466,003), prostate (375,304), cervix uteri (341,831), leukaemia (311,594), and so on [1]. Thus, many questions are still to be answered in this regard: What else do we need to know to tame cancers? Have we reached the glass ceiling in this regard? How long is this path? Or perhaps, is this exploration a never ending story?
Many authors have defined cancerous process and its sounds like a mantra. The phenomenon has been explained by accumulation of genetic and epigenetic alterations, which enhance cell transformation into a specific (cancerous) phenotype, i.e., limited apoptosis, infinite replicative capacity, increased motility, and pro-angiogenic ability [2,3]. It is also important to mention the altered energetic metabolism (the Warburg effect) and facility to convert into endothelial-like cells in order to maintain metabolic balance in tumour-dependent hypoxic areas (vascular mimicry), and the potential ability to enter and exit a quiescent state and immune evasion by cancer cells, which are also fundamental features of cancer transformation [4,5]. Delving deeper into the topic, we need to account for the role of cancer stem cells (CSCs), which constitute a specific tumour cell population. These cells show particular characteristics like localisation within the tumour, promotion tumour initiation, a high capacity to create colonies, pro-metastatic, pro-recurrence, and, last but not least, low drug sensitivity. According to the literature, CSCs exhibit great pro-neoplastic potential by maintaining pre-neoplastic foci, i.e., ideal tumour-initiating environments [2,6,7]. Furthermore, CSCs possess the potential to differentiate into multiple cell lineages, including pericytes, endothelial cells, or cancer-associated fibroblasts, and they are able to remodel their microenvironment, which enables the recruitment of other cells; consequently, they participate in the tumour growth and spreading [7,8].
In spite of great advances, modern chemotherapeutic agents and immunotherapies have not eliminated the severe worldwide cancer mortality [9,10]. Standard anticancer agents do not distinguish normal cells from cancer cells; thus, the chemo-related side effects are common and cause serious discomfort among oncology patients [11]. The individual/intrinsic profile of each patient must be taken into consideration, since therapeutic failures might be associated with de novo lower sensitivity to drugs or the acquisition of treatment resistance as a result of the therapy used [12]. The insufficient treatment response may be due to the heterogeneity of cancers, which is also associated with CSC biology [2,13]. Thus, the main purpose of stratified medicine is to translate the molecular status of tumour cells into predictive and prognostic indexes that can be applied to personalise treatments leading to longer survival and reduced toxicity [10,14]. In this line, patients who are stratified as high risk for relapse could be treated with adjuvant mode, while patients without detectable CSCs after neoadjuvant treatment and surgery might be adequate for less intensive follow-up procedures. Well-defined risk factors that are related to shorter survival rate in oncology patients include advanced age, unfavourable genetic profile, associated comorbidities, as well as overtreatment and treatment-related toxicity. The ideal balance between a patient’s risk and favourable outcomes are of utmost relevance to providing a therapy decision [9,10,11].
In this review, we discuss the general concept, characteristics, and detection technologies of CSCs and leukaemia stem cells (LSCs). Further, we highlight recent advances in the development of drug candidates targeting CSCs/LSCs.
2. General Concept of Cancer Stem Cells
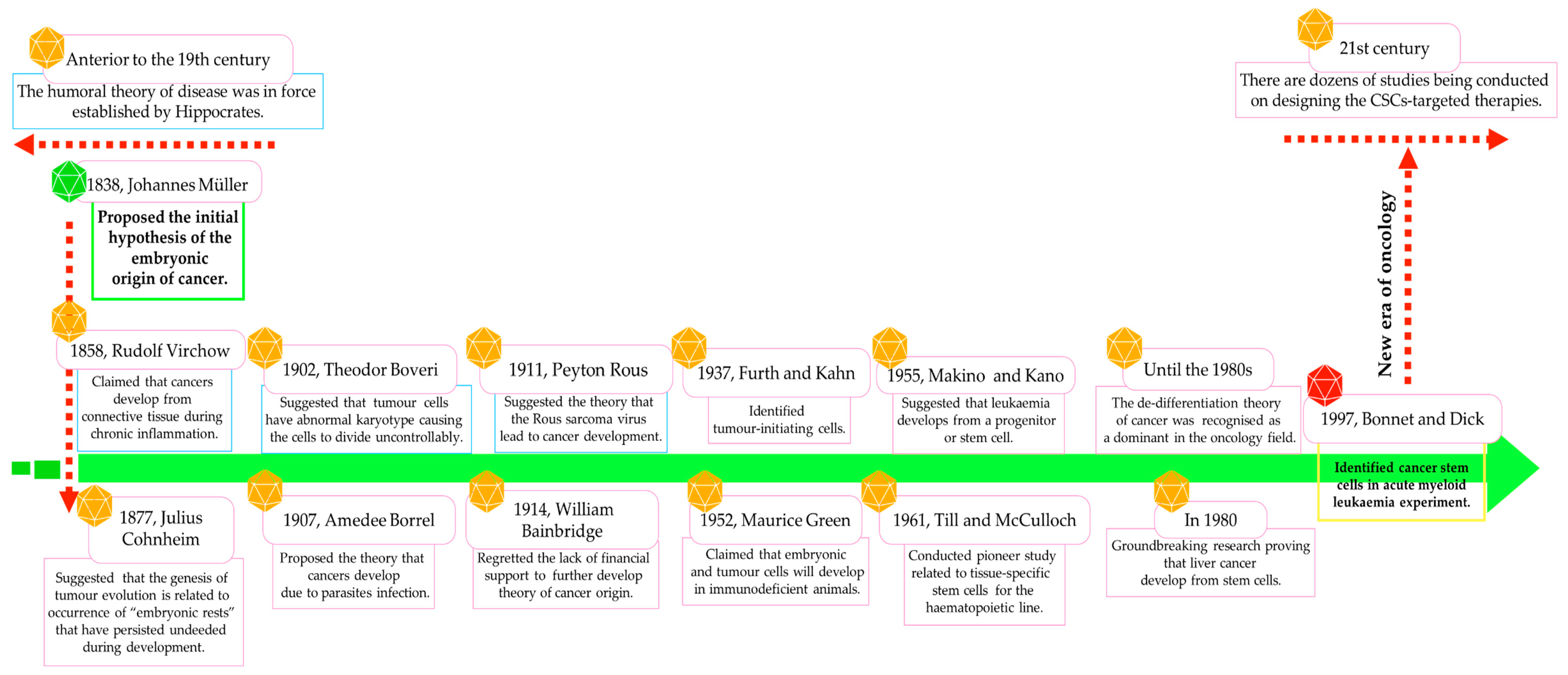
Already in 1838, Johannes Müller, and subsequently in 1858, Rudolf Virchow, suggested the hypothesis of the embryonic origin of tumour cells, which was confirmed by Julius Cohnheim in 1877 and further studies (Figure 1) [5,9,15,16]. CSCs were identified for the first time in an acute myeloid leukaemia (AML) model, and to this day, presenting in virtually all cancer types, by employing various cluster differentiation (CD) markers or through side population examination [6,9,10,17,18,19].
The discovery of CSCs/LSCs has modified the understanding of cancer’s nature and its response to anticancer drugs. Nowadays, it is believed that CSCs are responsible for the formation and expansion of cancerous tissue. CSCs, also called tumour-initiating cells [4,10] or stemness-high cancer cells [10], exhibit major stem-like properties, including self-renewal ability, pluripotent potential, and clonogenicity, which may promote the establishment of a metastatic foundation and resistance to standard chemotherapies and radiation [6,9,10]. Interestingly, Kreso and Dick demonstrated that only particular and more aggressive CSCs show the potential for tumour expansion and relapse, so various populations or even a hierarchy of CSCs may be present in the tumour bulk [20].
Figure 1.
Most representative milestones in the advancement of knowledge about the nature of cancer/leukaemia stem cells.
Figure 1.
Most representative milestones in the advancement of knowledge about the nature of cancer/leukaemia stem cells.
2.1. Cancer Stem Cells: Origin and Detailed Characteristics
Stem cells rarely divide under physiological conditions. In bone marrow, only 10% of stem cells are in the replication, pre-division, or mitosis phases at the same time, which proves that, despite unlimited proliferative activity, stem cells divide relatively rarely [3,21]. It is believed that CSCs may develop from normal stem cells or partially differentiated progenitor cells present in a given niche, or they may originate from fully differentiated somatic cells, so CSCs manifest a similar phenotype as normal stem cells (Table 1). However, the transformation of a normal stem cell into a CSC requires that a few conditions be met, including loss of cell cycle control and the accumulation of genetic and epigenetic alterations [2,17]. Toh et al. pointed out that epigenetic alterations (i.e., DNA methylation, chromatin remodelling, and histone modifications) are among the first events promoting the transition of stem cells into CSCs. Production of CSCs is also due to a decline in the expression of tumour suppressor genes, especially TP53, ATM, PTEN, and others [17].
CSCs are difficult to eradicate—they overexpress drug efflux pumps, secrete detoxifying enzymes, and demonstrate a potent ability to stimulate anti-apoptotic and pro-survival pathways, as well as DNA repair. The currently used chemotherapy, which acts primarily on rapidly dividing cells, is unable to adequately affect CSCs with a low replication index [4,6,18]. One of the proposed mechanisms of CSC resistance to anticancer agents is the fact that these cell populations can easily manoeuvre between different phases of the cell cycle in response to typical cell stimuli induced by anticancer drugs. Thus, CSCs in the G0 phase are insensitive to cell cycle blockade signals followed by failure of the apoptotic cascade, which gives them the potential to survive longer in a dormant state [22]. To understand this relationship between cancer progression and CSCs, we should first recognise the nature of CSCs. Thus, the most relevant differentiating and characterising features of normal stem cells and CSCs are included in Table 1 [2,3,4,6,9,10,13,23,24,25,26,27,28,29,30].

Table 1.
Features of normal stem cells versus cancer stem cells (CSCs).
Table 1.
Features of normal stem cells versus cancer stem cells (CSCs).
| Features | Normal Stem Cells | Cancer/Leukaemia Stem Cells |
|---|---|---|
| Localisation | In almost all physiological tissues | Periphery of the tumour |
| Composition | Hierarchical structure | Hierarchical structure |
| Characteristics | Primitive or undifferentiated precursors | Initiate and reconstitute tumour lesions |
| Function | To maintain tissue homeostasis | To maintain the unlimited growth of tumours and their morphological diversity |
| Self-renewal | Potent | Potent (tumour re-creation by metastasis) |
| Differentiation pattern | Pluripotent (differentiate into different kinds of normal cells) | Pluripotent (differentiate into different kinds of cancer cells) |
| Cell differentiation | Balanced | Dysregulated |
| Cell division | Mostly asymmetric | Subpopulations of CSCs: * Early-stage CSCs—mostly asymmetric * Late-stage CSCs—mostly symmetric |
| Cell cycle phase | G0/G1 phase | Ability to switch into any phase (mostly slow-cycling behaviour) |
| Proliferation index | Low, unlimited and well-controlled | Varied and uncontrolled |
| Morphology characteristics | High nuclear-to-cytoplasmic ratios | High nuclear-to-cytoplasmic ratios |
| Migration ability | High | High |
| Cell phenotypic potential (cell plasticity) | Stable | Heterogeneous |
| Partner of sld five 1 detection | Negative | Positive |
| Pro-angiogenic property | Limited | Unlimited |
| Drug sensitivity | Moderate | Strong resistance |
| Selected surface markers | CD24+, CD34+, CD44+, CD90+, CD133+ | CD24−/low, CD34+, CD44+, CD90+, CD133+, ALDH1high, ESA, EpCAM, side population cells |
| Immunosuppressive effect | Negative | Positive (via paracrine manner) |
| Survival rate | Prolong | Enhances their survival in an autocrine manner |
| Apoptosis | Antiapoptotic phenotype | Antiapoptotic phenotype (mediated by IL-4) |
| Chromosomal abnormality | Normal karyotype | Subpopulations of CSCs: * Early-stage CSCs—normal karyotype * Late-stage CSCs—an abnormal chromosome number |
| Telomerase activity | Potent | Potent |
| Histone H3 demethylation | Positive | Positive |
| Expression of Oct4, Notch, Sox1 genes | Positive | Positive |
| DNA repair ability | Potent | Potent |
| Genetic stability | Normal | Lost |
| Presence in peripheral blood | Trace amounts | Trace amounts |
| % of cells in specific tissue | 0.01 | 0.02–25 |
Notch, Oct4, Sox1: specific genes for all stem cells; ALDH1: aldehyde dehydrogenase 1; CD24: a small surface protein responsible for cell–extracellular matrix (ECM) and cell–cell interactions; CD34: a transmembrane glycoprotein expressed on early lymphohematopoietic stem cells, progenitor cells, and endothelial cells; CD44: a multifunctional glycoprotein responsible for cell adhesion, signalling, proliferation, migration, haematopoiesis, and lymphocyte activation; CD90: a glycophosphatidylinositol (GPI) anchored conserved cell surface protein; CD133: also known as prominin-1, a transmembrane cell surface glycoprotein commonly utilised as a hematopoietic stem cell marker; CSCs: cancer stem cells; DNA: deoxyribonucleic acid; EpCAM: epithelial cell adhesion molecule; ESA: epithelial-specific antigen; IL-4: interleukin 4; * It means that Early-stage CSCs and Late-stage CSCs are subpopulations of CSCs.
2.2. The Importance of the Tissue-Specific Microenvironment for the Maintenance of CSCs/LSCs
A niche as a specific microenvironment ensures suitable conditions for stem cell development and maintenance. The stem cell niche refers to the space in which stem cells are kept ready for the self-renewal, cell division, and differentiation necessary to maintain tissue homeostasis [2,27,31]. The specific features of niches for CSCs are disruption of the immune system and accumulation of malignant cells [2,24,29,32]. In this context, it is important to take into account that chronic inflammation is a natural driver in cancer-triggering niches. Specific characteristics of CSC niches are maintained by accumulation of cancer-associated fibroblasts, tumour-associated macrophages, tumour-associated neutrophils, and cell-mediated adhesion, which regulate cell–cell interactions and stromal, endothelial, and T cells [2,18,29,33,34]. Additional elements including extracellular vesicles, soluble factors, and the extracellular matrix support cancer-related surroundings [18]. Such a microenvironment favours specific features of CSCs, including infiltration, metastasis, and stimulation of tumour-associated neovasculature [18,35]. It is well known that neoangiogenesis is triggered in low-oxygen regions, but the neovasculature network is abnormal due higher permeability and a twisted, immature structure [36]. Additionally, hypoxic niches maintain undifferentiated CSCs via limiting cell cycling, followed by cell division rate reduction (stimulates switch into G0 phase) [24,28]. Interestingly, cancer-dependent hypoxia triggers a protective environment against DNA damage. According to Mohyeldin et al., 20% oxygen saturation was associated with significantly higher tissue damage compared to 3% O2 [37]. The above-mentioned mechanisms lead to the formation of pro-metastatic sites and also contribute to the insensitivity of hypoxic niches to chemotherapy [24]. Hypoxia-inducible factors (HIFs) affect the cell division, self-renewal, and cancerogenicity of CSCs. In accordance, higher CD44+ and CD133+ expression in hypoxic conditions was noted by Bai et al. and Won et al. [38,39].
Disruption of the bone morrow (BM) niche structure is a predictable state in blood malignancies. Accumulation and infiltration of leukaemia cells promotes elimination of normal haematopoietic progenitor cells from the BM niches and prepares an ideal microenvironment for them [40]. This modified BM microenvironment enables typical behaviours of LSCs including self-renewal, dormancy, and apoptosis evasion [41]. Moreover, the modified BM niche remains a space for LSCs, which is a reservoir for residual leukaemia cells and promotion of recurrence [42]. Interestingly, the BM niche demonstrates two separate microenvironmental regions (the osteoblastic niche and vascular niche) that likely modulate the cycling of LSCs [31,43]. Both niches effectively collaborate and promote the self-renewal, cell division, motility, and organisation of BM-related stem cells and LSCs [44].
2.3. Immunophenotypic Fingerprints of CSCs/LSCs
CSCs and non-CSCs can be distinguished via specific CD markers, but also based on their self-renewal ability [6]. Nevertheless, the disadvantage of the flow cytometry method is that selected surface markers are co-expressed in both populations. Additionally, in the analysis of the surface markers’ expression patterns, patients’ ethnicity or race must be taken into account in order to standardise the results [45]. Furthermore, considering both solid tumours and haematological malignancies, there is an intra-tumoral heterogeneity of surface markers among one type of cancer and stem cell plasticity, which often produces inconsistent results in this regard (Table 2). Thus, a specific phenotype of CSCs/LSCs is often not yet available in certain cancers. Therefore, this analysis should be extended by including enzymatic analysis (ALDHs) or CSC colony formation ability [45,46]. Table 2 presents composite profiles of CSC/LSC surface markers in solid tumours and haematological malignancies.

Table 2.
Summary of complex phenotypes of cancer stem cells (CSCs) and leukaemia stem cells (LSCs) in solid and non-solid tumours.
2.4. Detection of CSCs/LSCs
The current techniques for CSC identification include the estimation of surface markers or its functionality. The expression pattern of surface markers is commonly used for CSC/LSC determination and isolation using fluorescence-activated cell sorting (FACS) [46]. FACS based on detection of CSC/LSC-specific immunophenotype or surface antigens and further segregation of fluorescent vs. non-fluorescent cells can be implemented using a multicomponent assay [46,109,110]. However, determining one specific marker for CSCs or LSCs is very difficult (Table 2), and the method also requires aseptic conditions and vast number of cells. An alternative to FACS is magnetic-activated cell sorting (MACS), which is easy to perform and requires a smaller number of cells. MicroBeads with a typical diameter of 100 nm specifically bind to antigens enabling isolation of the targeted cells without further staining [46,110,111]. However, the selection of cells in mono-parameter mode is the greatest weakness of the method [112]. Undoubtedly, high intra- and inter-tumoral diversity limits the application of a well-defined immunophenotype for effortless detection of CSCs or LSCs, so there is still space for functional methods (Figure 2) [26]. It has been suggested that the assessment of surface markers is not sufficient for the detection of specific pro-metastatic CSCs; hence, a broader view of this issue is required and gene expression profiling of these subpopulations should be included [17]. Thus, the evaluation of CSCs/LSCs requires advanced analytical methods that demonstrate proper sensitivity and specificity as well as limit false positives and false negatives.
In the literature, several in vivo and in vitro functional methods have been proposed to recognise CSCs/LSCs in cancer tissues or cell lines. Figure 2 shows the application, benefits, and weakness of currently used methods for functional assessment of CSCs/LSCs [26,45,46,52,111,113,114,115,116,117]. It is accepted that new diagnostic techniques are indispensable for adequate recognition of these cells. Could the use of artificial intelligence (AI) or deep learning be the answer to the current need to identify CSCs (Figure 3)?
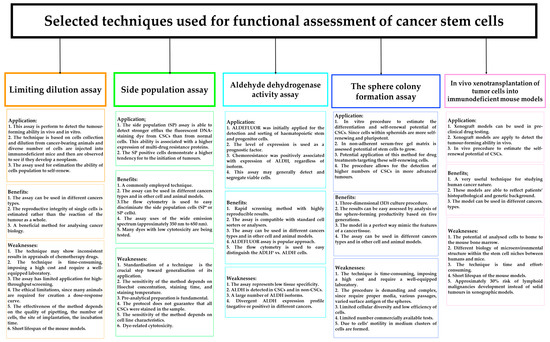
Figure 2.
In vivo and in vitro techniques assessing the functionality of cancer stem cells (CSCs) their application, benefits, and weaknesses.
Figure 2.
In vivo and in vitro techniques assessing the functionality of cancer stem cells (CSCs) their application, benefits, and weaknesses.
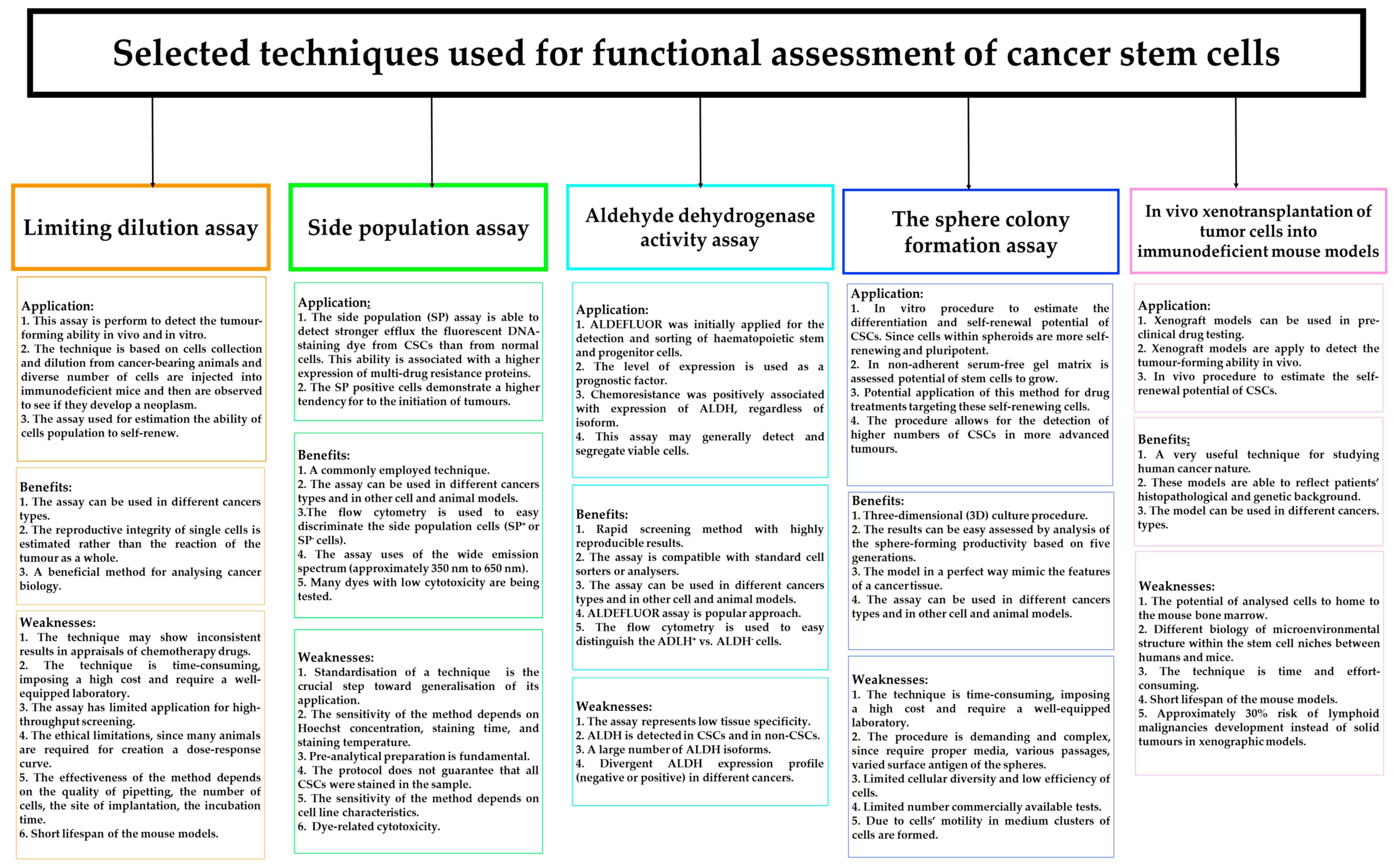
2.5. The Space for Artificial Intelligence in Cancer Stem Cell Detection
A new era of cancer diagnostic panels is opening or even forcing the space for AI technology in order to deliver fully automated identification of biological images of heterogeneous stem cell populations, including CSCs [9]. Considering the difficulties in laboratory practice in differentiating between normal and cancer stem cells, AI algorithms can find an important place in CSC detection. However, it is necessary to remember the appropriate and standardised method of selecting CSCs through qualitative and quantitative assessment of its morphological features. Deep learning algorithms are trained, tested, and validated to assess the proliferation, apoptosis, and dormant status of CSCs. The following factors may limit the use of standard AI algorithms: CSCs demonstrate different cell sizes with a different cytoplasmic-to-nuclear ratio in respect to non-CSCs, and the low number of CSCs in cancer tissue. Furthermore, insufficient image contrast and areas with blurry image features pose critical limitations in the training and testing stages of CSC recognition. New technologies open space for faster, automated diagnostics, but algorithms that are not fully developed still have limitations to overcome before being introduced into clinical practice. Figure 3 illustrates the applications of AI in the detection of cancer stem cells [9,23,114,118]. Ensuring proper identification of CSCs by advanced learning models able to include intra- and inter-tumoral heterogeneity will increase the application of AI in this field.
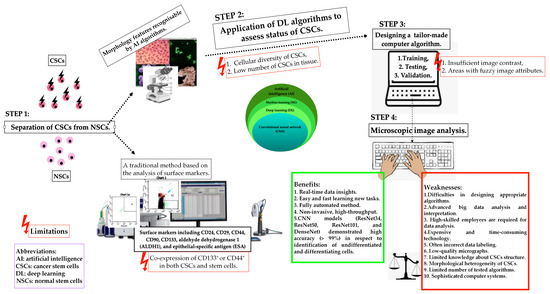
Figure 3.
Schematic representation of the applications of artificial intelligence in the detection of cancer stem cells (CSCs), their potential, and associated technical limitations.
Figure 3.
Schematic representation of the applications of artificial intelligence in the detection of cancer stem cells (CSCs), their potential, and associated technical limitations.
3. Cancer and Leukaemia Stem Cells in Disease Recurrence
Tumour cell dissemination, from primary origin to secondary sites, is strongly related to cancer-associated mortality in two out of every three solid tumours [119]. The CSC paradigm assumes that solid tumours and leukaemias are hierarchically defined, with CSCs at the top of this pyramid, leading to tumour development, spread, relapse, and drug resistance [6]. Interestingly, higher CSC counts have been detected in leukaemias and lymphomas, while solid tumours presented lower numbers [120]. However, it is considered that higher-grade tumours show higher percentages of CSCs [82,121]. Nevertheless, according to the CSC model, not all of those cells are able to trigger cancer progression (Table 3). Tumour spreading depends on a more anomalous and particular subpopulation of CSCs. Thus, there is a need to identify at least two CSC/LSC subpopulations: early-stage (pre-neoplastic) and late-stage (pro-metastatic) CSCs/LSCs. Table 3 summarises the key characteristics of CSC/LSC subfractions [2,44].

Table 3.
Characteristics of CSC/LSC subfractions.
In order to fully understand the relationship between CSCs and cancer progression, it is important to note that dysregulation of vascular homeostasis facilitates tumour progression [35,36]. Transcription factors specific for mesenchymal cells (Twist1, Slug, and Snail) and antigens (Vimentin and N-cadherin) are expressed on the surface of CSCs, helping them to undergo epithelial–mesenchymal transition (EMT) and trigger the formation of secondary malignant phenotypes, cell migration, and apoptosis-resistant CSCs [29,122,123]. Furthermore, the upregulation of stemness-related components, including Oct4, Notch, ALDH1, and SOX1, confirms the ability to effectively switch between CSC and non-CSC states [29].
The EMT is stimulated by mediators released from the niche, i.e., transforming growth factor β (TGF-β), hepatocyte growth factor (HGF), HIF, Hedgehog, Wnt, and Notch [30]. The Wnt/β-catenin, Hedgehog, Notch, and PI3K/Akt/mTOR signalling pathways are upregulated in all solid and non-solid tumours, leading to the enhancement of CSC/LSC-specific properties. The Wnt pathway enhances cancer cell division, motility, and drug resistance, while the self-renewal of CSCs/LSCs is mediated by the Hedgehog and Notch pathways [29,30,52,122,123]. However, the research so far has not allowed for us to fully understand and control the mechanism by which CSCs/LSCs contribute to cancer invasion. Nevertheless, the above-indicated signalling pathways provide a mechanism for explaining the differences in behaviour between early-stage (pre-tumorigenic stem cells) and late-stage CSCs/LSCs. The Wnt/ß-catenin pathway is fundamental to preserving the self-renewal ability of early-stage stem cells in leukaemias; breast, lung, and liver cancers; and melanomas, whereas the Notch signalling pathway has been implicated in stemness of late-stage cancer stem cells in AML, breast cancer, colon cancer, and glioblastoma [44,124,125]. Stemness of late-stage CSCs in glioblastoma, colon cancer, and pancreatic cancer involves the Hedgehog signalling pathway [44,126,127].
Haematological malignancies are highly heterogeneous in respect to diversity of clinical presentation, cytogenetics, and molecular profiles, as well as a future outcome that is associated with patient- and leukaemia-related factors [24]. Haematological malignancies arise not only from the genetic alterations in malignant cells, but also due to their communication/symbiotic relationship with the microenvironment. The evolution of the disease is strongly associated with reciprocal communication between stroma and malignant cells, which promotes anti-apoptotic signals in LSCs during their migration to the secondary space [128]. Many studies demonstrated that CSCs are quiescent or slowly dividing, whereas leukaemia progenitors are able to divide rapidly via escaping the dormant state [93]. Indisputably, LSCs hold great importance in the pathogenesis and relapse of leukaemia; thus, haematological malignancies should be treated based on stemness pattern [129]. Furthermore, the heterogeneous LSC population shows diversity at the level of functionality, since there exist sub-colonies that display the unfavourable phenotypes of dormancy, long-term neoplasm propagation, and drug insensitivity. This has modified the understanding of therapeutic needs in haematological malignancies, due to the fact that unfavourable phenotypes of dormancy are reversible and give space to use LSC-targeted treatments that prolong remission periods [130].
Table 4 shows the role of CSCs/LSCs in the recurrence of selected solid and non-solid tumours.

Table 4.
The role of cancer stem cells (CSCs) and leukaemia stem cells (LSCs) in the recurrence of solid and haematological cancers.
4. Perspectives and Modern Therapeutic Strategies Targeting CSCs in Solid Tumours
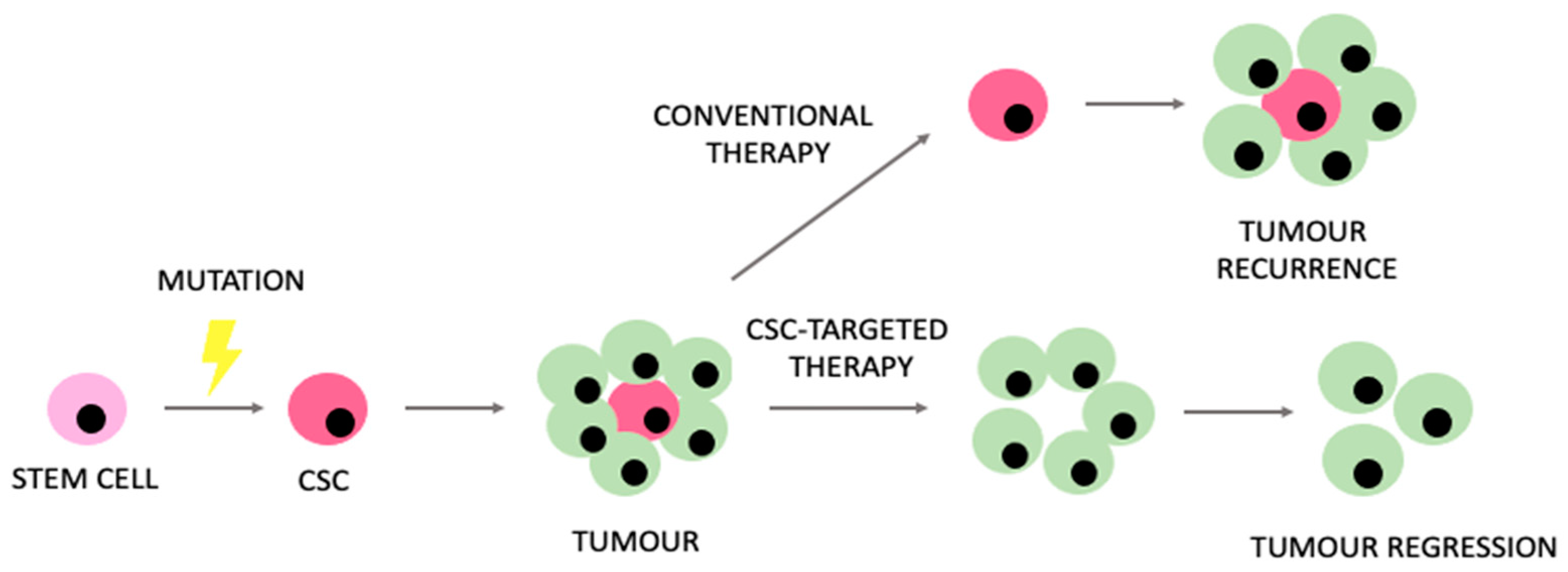
Despite prominent advances in modern oncology, relevant limitations and challenges still remain. Understanding the unique metabolic properties of CSCs might potentially enhance our ability to manage the therapeutic limitations that CSCs generate. The expected model of CSC-targeted therapies in comparison to conventional therapeutic approaches is presented in Figure 4.
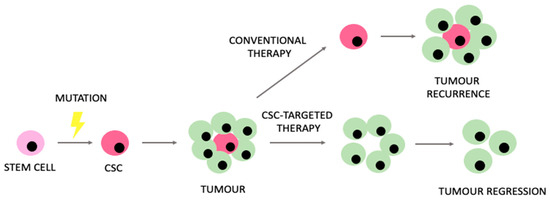
Figure 4.
Rationale encouraging the use of therapies targeting cancer stem cells (CSCs) in comparison to conventional therapeutic approaches.
Sekar et al. used liver cancer cell lines—Huh7—and found a reduced expression of CD133 and of the ABCG2 gene in cells treated with XAV939 and silenced with the EpCAM gene. Furthermore, cells treated with cisplatin alone formed spheroids, whereas the EpCAM gene-silenced cells and those treated with XAV939 in combination with cisplatin did not appear as spheroids. In a cytotoxicity assay, cisplatin alone and in combination with EpCAM silencing and XAV939-treated cells showed greater lactate dehydrogenase release than counterparts treated with the XAV939 silenced EpCAM cell group [150]. In their study, Miao et al. used oral squamous cell carcinoma (OSCC) cell lines and multicellular tumour spheroid models to generate CSC-like cells. They performed RNA sequencing to analyse the transcription levels of metabolic genes and analysed the single-cell transcriptome of six OSCC tumours to investigate the metabolic phenotypes of oral CSCs in their native microenvironment in humans. They concluded that CSCs were metabolically inactive compared to differentiated cancer cells and may be resistant to current metabolic therapeutic strategies [151].
In a different work, Huang et al. studied the antiproliferative effect of shikonin in a subpopulation of chemoresistant non-small cell lung cancer. They used A549 sublines to show shikonin’s antiproliferative properties. Shikonin also downregulated the PI3K/Akt/mTOR signalling pathway, inducing apoptosis. They discovered a synergistic action of modest dosages of shikonin and the dual inhibitor BEZ235, which suppressed the growth of lung CSCs and decreased the likelihood of lung cancer recurrence [152]. Furthermore, Santos et al. focused on the mechanism by which the ruthenium–xanthoxylin complex (RXC) targets the Hsp90 chaperone and eradicates colorectal cancer (CRC) stem cells. They demonstrated that RXC is very cytotoxic, inducing apoptosis in primary cancer cells as well as cancer cell lines [153]. In HCT116 CRC cells, Silva et al. investigated the mechanism of action of the ruthenium–5-fluorouracil (Ru/5-FU) complex. Ru/5-FU decreased colonosphere development, the percentage of CD133+ cells, and clonogenic survival, suggesting that Ru/5-FU can suppress stem cells in HCT116 cells. Additionally, in vivo HCT116 cell proliferation and experimental lung metastasis in mouse xenograft models were suppressed by Ru/5-FU. The complex inhibits Akt/mTOR signalling, making it a promising anti-CRC chemotherapeutic candidate [154]. Shang et al.’s research in CRC focused on tumour-associated macrophages (TAMs), specifically how they create niches for CSCs. The authors noted that poor treatment outcomes in CRC patients are associated with high expression of inhibitor of differentiation 1 (ID1) in TAMs. They showed that reducing ID1 expression increases the sensitivity of CRC to chemotherapy and immunotherapy [155]. Li et al. claimed that standard anticancer treatment is less effective against CSCs and can even enhance stemness gene expression. They discovered BBI608, a naphthofurandione, which is able to reduce metastasis and disease recurrence, via limitation of spherogenesis and Stat3-driven transcription. There was strong evidence that BBI608 reduced liver metastasis in a xenografted human CRC model and it robustly prevented recurrence in pancreatic cancer. Thus, an unconventional approach increases the range of treatment options for oncology patients, and the procedures based on cancer stemness inhibition open new possibilities for more effective treatment [10].
Zavareh et al. analysed the potential of the endemic plant Satureja bachtiarica in inhibiting and attacking CSCs in glioblastoma and breast cancer. They showed, especially in breast cancer, that S. bachtiarica can be an effective drug that reduces the viability and growth rate of cells, by inducing apoptosis, and it inhibits their migration [156]. Focusing on breast CSCs, Gil-Gas et al. investigated the role of the pigment epithelial-derived factor (PEDF) signalling. They designed a protein that blocks endogenous PEDF in cell culture tests and the modified PEDF interfered with CSC self-renewal and reduced the percentage of CSCs [157].
The aggressiveness of pancreatic cancer is believed to be closely related to a subpopulation of CSCs that have a greater evolutionary ability to escape the cytotoxic effects of chemotherapy compared to other cancer cells. In their work, Mouti et al. demonstrated that using the KMT2A-WDR5 inhibitor to target the protein subcomplex in pancreatic CSCs reduced the cells’ ability to self-renew, their survival, and their ability to cause tumours in vivo [158]. Interestingly, a recent study by Boudreault et al. analysed the role of the TGF-β signalling pathway in melanoma. They showed that TGF-β acts as a potent suppressor of tumour development, migration, and metastasis. Additionally, it has been shown that there is potential in the use of agents that stimulate or mimic TGF-β as new methods to fight melanoma [159].
The concept of CSC-targeting drugs must be taken into account for the reduction in adverse effects and dose-limiting toxicities. Also, for an efficacious therapy, all CSCs should be precisely eradicated to minimise risk of recurrence. In the past few years, a global effort has been made to design innovative therapeutic strategies against CSCs [10,150,151,152,153,154,155,156,157,158,159]. The results seem to be promising to improve long-term health outcomes; however, the biology of CSCs and their susceptibility to various types of therapy depend on the model on which the research was carried out. Additionally, the ability of CSCs to enter a dormant state, as well as the intra-tumoral diversity in surface markers expressed, makes it difficult to attain fully effective solutions. Last but not least, it is difficult to reproduce the real conditions of cancer development in experimental models, which would reflect the complexity of all components relevant for effective anticancer therapy [160]. Therefore, considering all these issues, further research will be necessary in this regard. In addition to discussing the essence of targeted therapies against CSCs in solid tumours, it is also necessary to emphasise the complex issue of anticancer agents that target LSCs in haematological malignancies.
5. Agents That Target Leukaemia Stem Cells in Haematological Malignancies
Conventional chemotherapy and stem cell transplantation have augmented the survival of patients with AML, multiple myeloma, and other haematological malignancies, but additional therapeutic strategies are needed [161,162,163]. Cancer stem cells are a logical target for novel drugs and the modulation of oncogenic cell signalling, and metabolic alterations in stem cells have attracted special attention [161,164,165,166]. The Wnt, Hedgehog, NF-κB, and Notch signalling routes play critical roles in the differentiation, proliferation, and survival of cancer stem cells [167,168], so various compounds have been developed targeting these pathways specifically. Cellular therapies have also provided good results in treating haematological malignancies, including targeting stem cells, but will not be covered in this article and are reviewed in the specialised literature [161].
5.1. Agents Targeting Wnt and Hedgehog Pathways
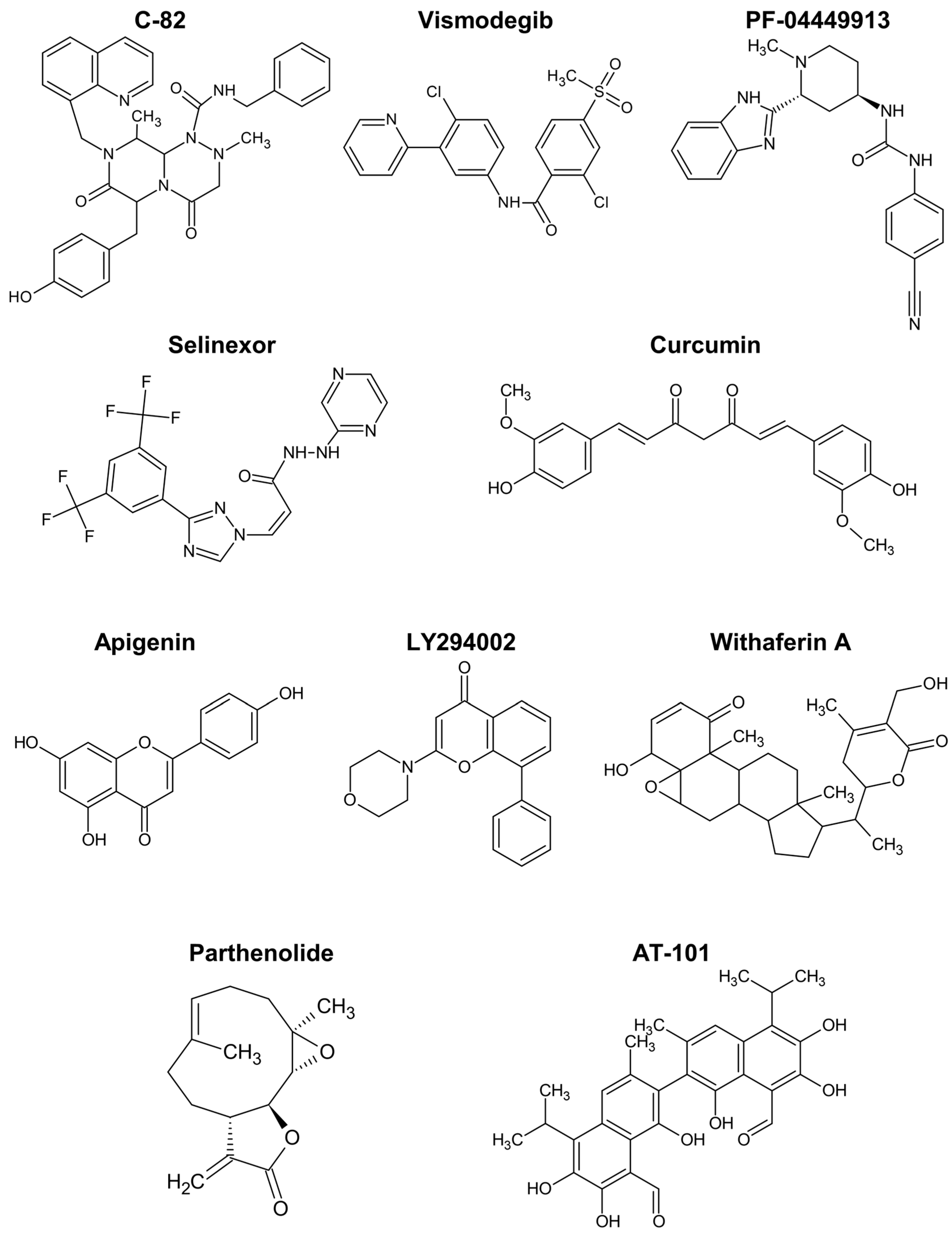
The Wnt/ß-catenin pathway promotes the expansion of haematopoietic stem cells and is activated in drug-resistant leukaemia-initiating cells, as demonstrated by different authors [161,169,170,171,172]. C-82 (Figure 5) and ICG-001 are β-catenin/CREB-binding protein (CBP) antagonists that block the interaction between the two proteins, downregulating Wnt-activated genes. Similar to β-catenin silencing with siRNA, those compounds restored the sensitivity of chronic myeloid leukaemia (CML) stem/progenitor cells to tyrosine kinase inhibitors [171,172].
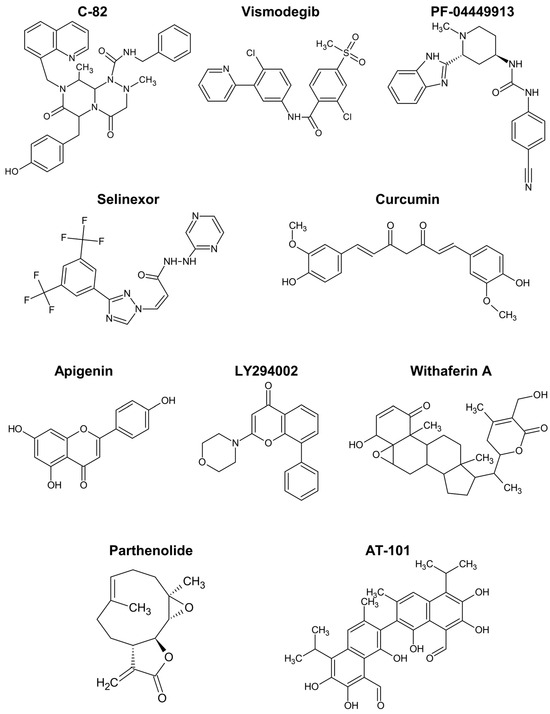
Figure 5.
Chemical structure of compounds described to target cancer stem cells in haematological malignancies.
The Hedgehog pathway has also been implicated in resistant phenotypes of CML cells [161,169,173,174]. Vismodegib is a drug targeting the Hedgehog pathway approved for cancer therapy. The incubation of CML cells with vismodegib decreased protein levels of relevant markers like MYC and induced autophagy [173]. Furthermore, the simultaneous inhibition of autophagy strongly enhanced the cell viability decrease induced by vismodegib and triggered apoptosis by way of caspase-3 and -9.
Glasdegib, or PF-04449913, is another clinical inhibitor of the Hedgehog pathway, approved for AML. Sadarangani et al. tested PF-04449913, an antagonist of the GLI2 transcriptional activator, smoothened (SMO), in dormant leukaemia stem cells. The treatment reduced the burden of GLI2-expressing leukaemia stem cells, their dormancy (enhancing cycling), and sensitised the cells to tyrosine kinase inhibition [174].
5.2. Agents Targeting NF-κB and Notch Pathways
NF-κB signalling is closely connected to cytokine/chemokine production and immune responses, being recognised a key role in cancer initiation, promotion, and progression [175,176]. The NF-κB pathway is stimulated in cancer stem cells [168,177], and one of the earliest pieces of evidence was the higher NF-κB DNA binding in AML samples compared to normal haematopoietic stem cells [175]. Alone or in cooperation with other signalling pathways, NF-κB promotes the expression of a wide variety of downstream targets, including stem factors (NANOG, SOX2, CD44, and others) and microRNAs, like let-7 and microRNA-21, contributing to self-renewal and expansion features of cancer stem cells [168,175,176,177].
Inhibition of NF-κB signalling with BMS-345541 reduced the stemness, self-renewal, and migration capacity of lung cancer stem cells [168]. BMS-345541 is an inhibitor of IκB kinase and reduced the expression of epithelial-to-mesenchymal transition genes and of the antiapoptotic BAX, along with decreasing the sphere-forming capacity of the cells.
The drug selinexor (Figure 5), described as interfering with NF-κB signalling, has been approved for the treatment of relapsed/refractory multiple myeloma [163]. It inhibits the protein exportin 1, the nuclear exporter of tumour suppressor proteins, the glucocorticoid receptor, and oncoprotein mRNAs, suppressing NF-κB activity, among other effects [178]. In spite of some safety concerns, selinexor in combination with dexamethasone resulted in treatment responses in patients with myeloma refractory to standard therapies [163]. Meanwhile, selinexor combinations with other chemotherapeutics showed the ability to inhibit cancer stem cell spheroids in pancreatic ductal adenocarcinoma [179], and interest in inhibitors of exportin 1 for haematological malignancies is growing [180].
Inflammation and NF-κB activity can crosstalk with the Notch pathway in different ways [177,181]. For example, IL-6-induces Notch1 activation and cancer stem cell proliferation by the assembly of γ-secretase at membrane caveolae [182]. Hence, controlling inflammatory and NF-κB signals can beneficially modulate the Notch-mediated stimulation of cancer stem cells. The addition of γ-secretase inhibitors, namely, MK-0752 [183] and RO4929097 [184], to chemo/radiotherapy gave indications of reducing cancer stem cell populations (CD44+, CD24−/low, ALDHhigh, and CD133+ cells), encouraging their assessment in haematological malignancies. Considering the key role of acute and chronic inflammation, the ability of the polyphenols discussed in the next section to regulate NF-κB signalling harbours great potential for controlling cancer stem cells.
5.3. Polyphenols
The chemopreventive action of polyphenols is supported by plenty of in vitro data, as well as by animal studies and epidemiological evidence [164,176]. These compounds usually affect multiple targets, modulating different interconnected biochemical processes, so they can put forward robust mechanisms of action against carcinogenesis and stemness-associated pathways [185,186]. The analysis of a group of 21 phenolic compounds and their interaction with cancer stem cell-related genes pointed to a selection of five high therapeutic potential compounds: resveratrol, curcumin, quercetin, epigallocatechin gallate (EGCG), and genistein [187]. Resveratrol is the chief stilbene present in grapes and wine, being one of the most established anticancer polyphenols [176]. Data from different works show that resveratrol and its methylated derivative pterostilbene can target cancer stem cells, regulating central mediators in signalling pathways [164,188]. Among the several mechanisms of action involved, resveratrol was reported to trigger autophagy via inactivation of Wnt/β-catenin pathway and suppresses the growth of cancer stem-like cells by inhibiting the fatty acid synthase [164,166,188].
Curcumin is another top anticancer polyphenol that displayed relevant effects on models of haematological malignancies [164,176]. It prevented the growth of CD34+CD38−/low cells isolated from AML patients by promoting the expression of osteopontin [189]. Burkitt lymphoma and AML cells incubated with low microM concentrations of curcumin exhibit a dose-dependent decrease in markers of cancer stem cells, namely, the ratio of ALDH-positive cells, inhibition of colony formation, and downregulation of Notch1, Gli1, and Cyclin D1 [190]. Curcumin showed strong cytotoxicity towards a human leukemic stem cell line (IC50 of 14 microM), and another curcuminoid, bisdemethoxycurcumin, greatly repressed the expression of Wilms’ tumour 1 and CD34 protein, warranting further studies to control leukaemia stem cells [191]. In this line, Nirachonkul et al. presented an alternative formulation of curcumin in nanoparticles targeting CD123 and, tested in the same leukaemia stem cells, it promoted the polyphenol interaction with the cells and induction of apoptosis, without apparent toxicity to peripheral blood mononuclear cells [192].
Quercetin is a prototypical flavonoid with antioxidant actions at low concentrations and is able to modulate diverse cellular processes underlying cancer initiation and progression. Regulation of microRNAs plays an important role in the anticancer activity of quercetin and, in particular, the upregulation of microRNA-200b-3p was implicated in the inhibition of cancer stem cells [164,193]. At high concentration (50 microM), quercetin interfered with the DNA damage response and inhibited the PI3K/AKT pathway in haematopoietic stem and progenitor cells [194]. Indeed, the inhibition of the PI3K/Akt/mTOR pathway was underlined as a key mechanism of quercetin for the elimination of cancer stem cells [164].
Green tea consumption shows beneficial effects, and EGCG is the component responsible for its stronger molecular anti-carcinogenic actions and results in human trials [176]. There is abundant evidence that EGCG can eliminate cancer stem cells of different types, decreasing stemness markers and inhibiting the Wnt/β-catenin pathway and proliferation indices, among other actions [164,195]. EGCG in combination with quercetin induced apoptosis of prostate cancer stem cells, and inhibited cancer stem cell proliferation phenotypes, in association to caspase activation and downregulation of cell survival mediators [195].
Genistein is a soy isoflavone able to protect haematopoietic stem cells from DNA damage [196]. Mechanistic studies with different cell models pointed to suppression of the Hedgehog/Gli1 pathway and/or upregulation of PTEN as key factors accounting for the anticancer activity of genistein [164].
Apigenin is another flavonoid with interesting anticancer activity, including targeting leukaemia stem cells responsible for failure in AML treatments [165,197]. The combination of apigenin with LY294002 (Figure 5) for treatment of CD34+CD38−/low leukaemia cells, including leukaemia stem cells, induced apoptosis in these cells associated to caspase activation, mitochondrial dysfunction, and downregulation of Bcl-xL and NF-κB [197]. Remarkably, these effects were not observed in healthy haematopoietic stem cells, suggesting an option for the safe eradication of leukaemia stem cells. Low sub-toxic concentrations of the two drugs were used and the potent synergistic action was rationalised on the basis of the simultaneous inhibition of the PI3K/Akt pathway by LY294002 and of protein kinase casein kinase 2 (CK2) by the flavone. In accordance, similar effects on caspase-3, antiapoptotic Bcl proteins, and NF-κB were reported with lung tumour and osteosarcoma models treated with apigenin or isovitexin (apigenin glucoside) in vivo [165]. These compounds were also shown to reduce stemness markers, namely, CD133, NANOG, MgSOD, and SOX2, in various in vitro and in vivo cancer models, with implication of c-Met signalling inhibition in the mechanism of action [165,186,198]. The increased expression of microRNA-34a was also associated to the stemness inhibition and apoptosis induction by isovitexin in hepatocellular carcinoma spheroids [199].
The inhibition of CK2, important for the maintenance of cancer stem cells, was also directly implicated in the reduction in self-renewal capability of HeLa sphere-forming cells by apigenin, while downregulation of survival and proliferation factors was accounted for by the sensitisation of CD44+ prostate cancer stem cells to cisplatin [165].
In overall, curcumin and apigenin (Figure 5) are the polyphenols showing stronger capacity for regulating cancer stem cells in haematological malignancies.
5.4. Other Natural Compounds and Derivatives
In addition to polyphenols, other natural compounds have been shown to inhibit the survival or growth of diverse cancer stem cells. An overview of the effects and mechanisms of action of compounds like sulforaphane, indole-3-carbinol, and phenethyl isothiocyanate can be found in the recent Chu et al. review [164].
Nevertheless, studies with models of haematological malignancies or those comparable uncovered some actions of specific relevance. Withaferin A is a steroidal lactone (Figure 5) and was found to induce cell cycle arrest and apoptotic death of multiple myeloma cancer stem cells [200]. Moreover, it was able to repress the growth and spheroid formation of diverse cancer cells [164].
The alkaloid berberine showed anticancer activity in various conditions, possibly by inhibiting HDACs and modulating the expression of stem cell-associated genes [162,201]. More potent than the flavonoids apigenin and wogonin, the naphthoquinone shikonin increased the apoptosis rate and inhibited invasiveness of renal carcinoma stem cells [186]. Shikonin reduced the expression of diverse cancer stem cell markers like ALDH3A1, CD133, EZH2, NANOG, and SOX2. Moreover, the combination of the phytochemical with an immune checkpoint inhibitor revealed a promising treatment strategy by regulating the T cell population [186]. Parthenolide is a sesquiterpene lactone (Figure 5) that prompted robust apoptosis in leukaemia stem cells, but not in normal haematopoietic cells, by a mechanism associated with inhibition of NF-κB and proapoptotic activation of p53 [202].
The modification of natural compounds is often employed to overcome limitations in the bioavailability or to improve the pharmacological potency. Li et al. synthesised derivatives of apigenin and found one—compound 15e—with strong activity (IC50 of 0.49 versus 44 microM of apigenin) against the growth of human renal carcinoma cells [203]. More recently, Fernando et al. reported a fatty acid ester of phloridzin that inhibited spheroid formation by breast cancer cells in vitro [204]. The conjugate could also inhibit the metabolic activity and induce cell death of paclitaxel-resistant variants, while investigation of the effects on stemness markers at low concentrations is warranted [204]. A synthetic analogue of genistein was also demonstrated to attenuate the expression of FoxM1 and other stemness features of gastric, ovarian, and lung cancer cells [164].
A more specific mechanism of action has been attributed to the gossypol enantiomer AT-101 (Figure 5). This compound is a BH3-mimetic pan-Bcl-2 inhibitor, binding to the BH3 motif of Bcl-2, Bcl-xL, and Mcl-1, in a way that inhibits their anti-apoptotic action, activates Bax and can trigger mitochondrial Smac release. In leukaemia stem cells, it inhibited proliferation and activated the intrinsic apoptotic pathway, with apparently low effects on normal CD34+ haematopoietic cells [205]. In accordance with the expected mechanism of action, AT-101 caused a decrease in mitochondrial membrane potential, and DNA damage partially dependent on caspase activity. The compound at microM concentration was also effective in ex vivo AML samples, offering a potential alternative therapy of relapsed and refractory conditions associated to cancer stem cells.
The multifunctional action and safety profile are usually regarded as important advantages of natural compounds like polyphenols for preventive and combination protocols [176,186,197,206,207]. Human trials showed that resveratrol and curcumin doses of up to a few g/day are tolerable, and some cases of hepatoxicity of EGCG have been reported only with oral doses equal or above 800 mg/day [176,206,208].
The above-mentioned astudies shown how important CSCs/LSCs are in cancer progression and as it stands a promising therapeutic target. Based on the works published in 2023–2024, we believe that this topic remains an open field for research and a consensus regarding the nature of CSCs/LSCs has still not been reached.
6. Top 10 Reasons Why This Manuscript Is Important in the Oncology Field
- Provides a historical graphical overview of research on the nature of CSCs/LSCs.
- Provides a clear, extensive, tabular presentation of the differences between normal stem cells and CSCs.
- Underlines the role of the tumour microenvironment in maintaining the pro-tumorigenic ability of CSCs/LSCs.
- Provides an organised summary of the knowledge regarding the functional methods of CSC/LSC detection including the application, benefits, and weaknesses of selected methods.
- Specifies the usefulness of the new technologies, including artificial intelligence and deep learning, in CSC examination.
- Provides an overview of the immunophenotypes of CSCs and LSCs in solid tumours and haematological malignancies.
- Discusses key characteristics of early-stage (pre-neoplastic) and late-stage (pro-metastatic) cancer and leukaemia stem cells.
- Indicates the importance of CSCs and LSCs in the recurrence of selected solid and non-solid cancers.
- Provides a concise analysis of the perspectives and modern therapeutic strategies targeting CSCs in solid tumours.
- Provides a broad analysis of candidate drugs for regulating LSCs in haematological malignancies, taking into account particular Wnt, Hedgehog, NF-κB, and Notch signalling pathways.
7. Methodology
To review the role of cancer stem cells in cancer biology, we carried out large-scale electronic searches within the following public databases: PubMed (U.S. National Library of Medicine) and Google Scholar. The following keywords were used alone or in combination: “cancer stem cells nature/biology”, “history of cancer stem cells”, ”epigenetics in cancer stem cells”, ”cancer stem cells immunophenotype in solid tumours and haematological neoplasms”, ”methods of cancer stem cells detection”, ”tumour microenvironment“, “cancer stem cell signalling pathways“, “epithelial-mesenchymal transition“, “artificial intelligence in cancer stem cells detection“, “cancer stem cells in solid and non-solid tumours recurrence”, “agents targeting leukaemia stem cells”, and “therapy targeting CSCs in solid tumours”.
Screening of the articles was made by four independent authors (BRC, RL, KK, and DM-d-S) and all inaccuracies were detected by final check by (BRC). The resulting literature was analysed and included in our review. The papers’ assessment was based on a critical reading. Only the full-text of articles in English was taken into consideration. Data from the current literature, up to January 2024, including clinical trials, prospective and retrospective observational studies, and review articles were reviewed. Most of the incorporated papers were published in the years 2015–2024 (75%). Since this review was based on previously published research, no ethical approval or patient consent was required.
8. Conclusions
Indisputably, CSCs/LSCs differ from normal/haematopoietic stem cells morphologically and functionally. CSCs/LSCs are heterogeneous populations and are influenced by the complex tumour microenvironment. Cancer progression and metastasis are strongly connected with CSCs/LSCs nature and biology. CSCs/LSCs constitute robust populations that can reversibly manoeuvre between different phases of the cell cycle, which gives them the ability to arrest pro-apoptotic signals and prolong the survival in quiescent state. Therefore, there is an urgent need for more sophisticated but easy-to-apply techniques to detect CSCs/LSCs, in order to identify patients who are at high risk for recurrent disease. Advances are also awaited in the clinical translation of the synthetic and natural drugs targeting CSCs discussed in this work.
Author Contributions
Conceptualisation, B.R.-C. and R.L., writing—original draft preparation, B.R.-C., R.L., D.M.-d.-S., and K.K., writing—review and editing, B.R.-C. and R.L., visualisation, B.R.-C., Supervision and Funding, B.R.-C. All authors have read and agreed to the published version of the manuscript.
Funding
D.M.-d.-S. and R.L. acknowledge the support to their research from LSRE-LCM and ALiCE, supported by national funds through Fundação para a Ciência e Tecnologia (FCT—Portugal): UIDB/50020/2020 (DOI: 10.54499/UIDB/50020/2020), UIDP/50020/2020 (DOI: 10.54499/UIDP/50020/2020), and LA/P/0045/2020 (DOI: 10.54499/LA/P/0045/2020).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- International Agency for Research on Cancer (IARC) Website. Available online: https://gco.iarc.fr/ (accessed on 22 January 2024).
- Afify, S.M.; Seno, M. Conversion of Stem Cells to Cancer Stem Cells: Undercurrent of Cancer Initiation. Cancers 2019, 11, 345. [Google Scholar] [CrossRef] [PubMed]
- Trosko, J.E. On the Potential Origin and Characteristics of Cancer Stem Cells. Carcinogenesis 2021, 42, 905–912. [Google Scholar] [CrossRef] [PubMed]
- Marzagalli, M.; Fontana, F.; Raimondi, M.; Limonta, P. Cancer Stem Cells-Key Players in Tumor Relapse. Cancers 2021, 13, 376. [Google Scholar] [CrossRef] [PubMed]
- Capp, J.P. Cancer Stem Cells: From Historical Roots to a New Perspective. J. Oncol. 2019, 2019, 5189232. [Google Scholar] [CrossRef] [PubMed]
- Toledo-Guzmán, M.E.; Bigoni-Ordóñez, G.D.; Ibáñez Hernández, M.; Ortiz-Sánchez, E. Cancer stem cell impact on clinical oncology. World J. Stem Cells 2018, 10, 183–195. [Google Scholar] [CrossRef] [PubMed]
- Hassan, G.; Afify, S.M.; Nair, N.; Kumon, K.; Osman, A.; Du, J.; Mansour, H.; Abu Quora, H.A.; Nawara, H.M.; Satoh, A.; et al. Hematopoietic Cells Derived from Cancer Stem Cells Generated from Mouse Induced Pluripotent Stem Cells. Cancers 2019, 12, 82. [Google Scholar] [CrossRef] [PubMed]
- Mei, X.; Chen, Y.S.; Chen, F.R.; Xi, S.Y.; Chen, Z.P. Glioblastoma stem cell differentiation into endothelial cells evidenced through live-cell imaging. Neuro Oncol. 2017, 19, 1109–1118. [Google Scholar] [CrossRef]
- Chen, J.; Xu, L.; Li, X.; Park, S. Deep learning models for cancer stem cell detection: A brief review. Front. Immunol. 2023, 14, 1214425. [Google Scholar] [CrossRef]
- Li, Y.; Rogoff, H.A.; Keates, S.; Gao, Y.; Murikipudi, S.; Mikule, K.; Leggett, D.; Li, W.; Pardee, A.B.; Li, C.J. Suppression of cancer relapse and metastasis by inhibiting cancer stemness. Proc. Natl. Acad. Sci. USA 2015, 112, 1839–1844. [Google Scholar] [CrossRef] [PubMed]
- Masciale, V.; Grisendi, G.; Banchelli, F.; D’Amico, R.; Maiorana, A.; Sighinolfi, P.; Stefani, A.; Morandi, U.; Dominici, M.; Aramini, B. Isolation and Identification of Cancer Stem-Like Cells in Adenocarcinoma and Squamous Cell Carcinoma of the Lung: A Pilot Study. Front. Oncol. 2019, 9, 1394. [Google Scholar] [CrossRef] [PubMed]
- Chan, B.A.; Hughes, B.G.M. Targeted therapy for non-small cell lung cancer: Current standards and the promise of the future. Transl. Lung Cancer Res. 2015, 4, 36–54. [Google Scholar] [CrossRef] [PubMed]
- Masciale, V.; Banchelli, F.; Grisendi, G.; D’Amico, R.; Maiorana, A.; Stefani, A.; Morandi, U.; Stella, F.; Dominici, M.; Aramini, B. The Influence of Cancer Stem Cells on the Risk of Relapse in Adenocarcinoma and Squamous Cell Carcinoma of the Lung: A Prospective Cohort Study. Stem Cells Transl. Med. 2022, 11, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Riedl, J.M.; Moik, F.; Esterl, T.; Kostmann, S.M.; Gerger, A.; Jost, P.J. Molecular diagnostics tailoring personalized cancer therapy-an oncologist’s view. Virchows Arch. 2023, 484, 169–179. [Google Scholar] [CrossRef]
- Sell, S. History of cancer stem cells. In Regulatory Networks in Stem Cells; Rajasekhar, V.K., Vemuri, M.C., Eds.; Humana Press: Totowa, NJ, USA, 2009; pp. 495–503. [Google Scholar]
- Bonnet, D.; Dick, J.E. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med. 1997, 3, 730–737. [Google Scholar] [CrossRef]
- Toh, T.B.; Lim, J.J.; Chow, E.K. Epigenetics in cancer stem cells. Mol. Cancer 2017, 16, 29. [Google Scholar] [CrossRef]
- Osman, A.; Afify, S.M.; Hassan, G.; Fu, X.; Seno, A.; Seno, M. Revisiting Cancer Stem Cells as the Origin of Cancer-Associated Cells in the Tumor Microenvironment: A Hypothetical View from the Potential of iPSCs. Cancers 2020, 12, 879. [Google Scholar] [CrossRef]
- Lapidot, T.; Sirard, C.; Vormoor, J.; Murdoch, B.; Hoang, T.; Caceres-Cortes, J.; Minden, M.; Paterson, B.; Caligiuri, M.A.; Dick, J.E. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature 1994, 367, 645–648. [Google Scholar] [CrossRef] [PubMed]
- Kreso, A.; Dick, J.E. Evolution of the Cancer Stem Cell Model. Cell Stem Cell 2014, 14, 275–291. [Google Scholar] [CrossRef] [PubMed]
- Gadre, P.; Nitsure, N.; Mazumdar, D.; Gupta, S.; Ray, K. The rates of stem cell division determine the cell cycle lengths of its lineage. iScience 2021, 24, 103232. [Google Scholar] [CrossRef] [PubMed]
- Moore, N.; Lyle, S. Quiescent, slow-cycling stem cell populations in cancer: A review of the evidence and discussion of significance. J. Oncol. 2011, 2011, 396076. [Google Scholar] [CrossRef] [PubMed]
- Aida, S.; Okugawa, J.; Fujisaka, S.; Kasai, T.; Kameda, H.; Sugiyama, T. Deep Learning of Cancer Stem Cell Morphology Using Conditional Generative Adversarial Networks. Biomolecules 2020, 10, 931. [Google Scholar] [CrossRef] [PubMed]
- Ju, F.; Atyah, M.M.; Horstmann, N.; Gul, S.; Vago, R.; Bruns, C.J.; Zhao, Y.; Dong, Q.Z.; Ren, N. Characteristics of the cancer stem cell niche and therapeutic strategies. Stem Cell Res. Ther. 2022, 13, 233. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.N.; Al-Karim, S.; Bora, R.S.; Chaudhary, A.G.; Saini, K.S. Cancer stem cells: A challenging paradigm for designing targeted drug therapies. Drug Discov. Today 2015, 20, 1205–1216. [Google Scholar] [CrossRef] [PubMed]
- Barreto, I.V.; Pessoa, F.M.C.P.; Machado, C.B.; Pantoja, L.D.C.; Ribeiro, R.M.; Lopes, G.S.; Amaral de Moraes, M.E.; de Moraes Filho, M.O.; de Souza, L.E.B.; Burbano, R.M.R.; et al. Leukemic Stem Cell: A Mini-Review on Clinical Perspectives. Front. Oncol. 2022, 12, 931050. [Google Scholar] [CrossRef] [PubMed]
- Bisht, S.; Nigam, M.; Kunjwal, S.S.; Sergey, P.; Mishra, A.P.; Sharifi-Rad, J. Cancer Stem Cells: From an Insight into the Basics to Recent Advances and Therapeutic Targeting. Stem Cells Int. 2022, 2022, 9653244. [Google Scholar] [CrossRef] [PubMed]
- Marcu, L.G.; Moghaddasi, L.; Bezak, E. Cannot Target What Cannot Be Seen: Molecular Imaging of Cancer Stem Cells. Int. J. Mol. Sci. 2023, 24, 1524. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.R.; Fang, Y.; Lyu, Z.; Zhu, Y.; Yang, L. Exploring the dynamic interplay between cancer stem cells and the tumor microenvironment: Implications for novel therapeutic strategies. J. Transl. Med. 2023, 21, 686. [Google Scholar] [CrossRef]
- Yang, L.; Shi, P.; Zhao, G.; Xu, J.; Peng, W.; Zhang, J.; Zhang, G.; Wang, X.; Dong, Z.; Chen, F.; et al. Targeting cancer stem cell pathways for cancer therapy. Signal Transduct. Target. Ther. 2020, 5, 8. [Google Scholar] [CrossRef] [PubMed]
- Marchand, T.; Pinho, S. Leukemic Stem Cells: From Leukemic Niche Biology to Treatment Opportunities. Front. Immunol. 2021, 12, 775128. [Google Scholar] [CrossRef]
- Schepers, K.; Campbell, T.B.; Passegue, E. Normal and leukemic stem cell niches: Insights and therapeutic opportunities. Cell Stem Cell 2015, 16, 254–267. [Google Scholar] [CrossRef]
- Nair, N.; Anna Sanchez Calle, A.S.; Zahra, M.H.; Prieto-Vila, M.; Oo, A.K.K.; Hurley, L.; Vaidyanath, A.; Seno, A.; Masuda, J.; Iwasaki, Y.; et al. A cancer stem cell model as the point of origin of cancer-associated fibroblasts in tumor microenvironment. Sci. Rep. 2017, 7, 6838. [Google Scholar] [CrossRef]
- Beier, D.; Hau, P.; Proescholdt, M.; Lohmeier, A.; Wischhusen, J.; Oefner, P.J.; Aigner, L.; Brawanski, A.; Bogdahn, U.; Beier, C.P. CD133+ and CD133− glioblastomaderived cancer stem cells show differential growth characteristics and molecular profiles. Cancer Res. 2007, 67, 4010–4015. [Google Scholar] [CrossRef]
- Matsuda, S.; Yan, T.; Mizutani, A.; Sota, T.; Hiramoto, Y.; Prieto-Vila, M.; Chen, L.; Satoh, A.; Kudoh, T.; Kasai, T.; et al. Cancer stem cells maintain a hierarchy of differentiation by creating their niche. Int. J. Cancer 2014, 135, 27–36. [Google Scholar] [CrossRef]
- Zarychta, E.; Ruszkowska-Ciastek, B. Cooperation between Angiogenesis, Vasculogenesis, Chemotaxis, and Coagulation in Breast CancerMetastases Development: Pathophysiological Point of View. Biomedicines 2022, 10, 300. [Google Scholar] [CrossRef]
- Mohyeldin, A.; Garzón-Muvdi, T.; Quiñones-Hinojosa, A. Oxygen in stem cell biology: A critical component of the stem cell niche. Cell Stem Cell 2010, 7, 150–161. [Google Scholar] [CrossRef]
- Bai, J.; Chen, W.B.; Zhang, X.Y.; Kang, X.N.; Jin, L.J.; Zhang, H.; Wang, Z.Y. HIF-2α regulates CD44 to promote cancer stem cell activation in triple-negative breast cancer via PI3K/AKT/mTOR signaling. World J. Stem Cells 2020, 12, 87–99. [Google Scholar] [CrossRef] [PubMed]
- Won, C.; Kim, B.H.; Yi, E.H.; Choi, K.J.; Kim, E.K.; Jeong, J.M.; Lee, J.H.; Jang, J.J.; Yoon, J.H.; Jeong, W.I.; et al. Signal transducer and activator of transcription 3-mediated CD133 up-regulation contributes to promotion of hepatocellular carcinoma. Hepatology 2015, 62, 1160–1173. [Google Scholar] [CrossRef]
- Colmone, A.; Amorim, M.; Pontier, A.L.; Wang, S.; Jablonski, E.; Sipkins, D.A. Leukemic cells create bone marrow niches that disrupt the behavior of normal hematopoietic progenitor cells. Science 2008, 322, 1861–1865. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Huang, S.; Chen, J.L. Understanding of leukemic stem cells and their clinical implications. Mol. Cancer. 2017, 16, 2. [Google Scholar] [CrossRef] [PubMed]
- Khaldoyanidi, S.K.; Hindoyan, A.; Stein, A.; Subklewe, M. Leukemic stem cells as a target for eliminating acute myeloid leukemia: Gaps in translational research. Crit. Rev. Oncol. Hematol. 2022, 175, 103710. [Google Scholar] [CrossRef]
- Konopleva, M.Y.; Jordan, C.T. Leukemia stem cells and microenvironment: Biology and therapeutic targeting. J. Clin. Oncol. 2011, 29, 591–599. [Google Scholar] [CrossRef] [PubMed]
- Schulenburg, A.; Blatt, K.; Cerny-Reiterer, S.; Sadovnik, I.; Herrmann, H.; Marian, B.; Grunt, T.W.; Zielinski, C.C.; Valent, P. Cancer stem cells in basic science and in translational oncology: Can we translate into clinical application? J. Hematol. Oncol. 2015, 8, 16. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Powell, K.; Li, L. Breast Cancer Stem Cells: Biomarkers, Identification and Isolation Methods, Regulating Mechanisms, Cellular Origin, and Beyond. Cancers 2020, 12, 3765. [Google Scholar] [CrossRef] [PubMed]
- Cianciosi, D.; Ansary, J.; Forbes-Hernandez, T.Y.; Regolo, L.; Quinzi, D.; Gracia Villar, S.; Garcia Villena, E.; Tutusaus Pifarre, K.; Alvarez-Suarez, J.M.; Battino, M.; et al. The Molecular Basis of Different Approaches for the Study of Cancer Stem Cells and the Advantages and Disadvantages of a Three-Dimensional Culture. Molecules 2021, 26, 2615. [Google Scholar] [CrossRef] [PubMed]
- Al-Hajj, M.; Wicha, M.S.; Benito-Hernandez, A.; Morrison, S.J.; Clarke, M.F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA 2003, 100, 3983–3988. [Google Scholar] [CrossRef] [PubMed]
- Enciso-Benavides, J.; Alfaro, L.; Castañeda-Altamirano, C.; Rojas, N.; González-Cabeza, J.; Enciso, N.; Riesco, F.; Castillo, M.; Enciso, J. Biological characteristics of a sub-population of cancer stem cells from two triple-negative breast tumour cell lines. Heliyon 2021, 7, e07273. [Google Scholar] [CrossRef] [PubMed]
- Ricardo, S.; Vieira, A.F.; Gerhard, R.; Leitão, D.; Pinto, R.; Cameselle-Teijeiro, J.F.; Milanezi, F.; Schmitt, F.; Paredes, J. Breast cancer stem cell markers CD44, CD24 and ALDH1: Expression distribution within intrinsic molecular subtype. J. Clin. Pathol. 2011, 64, 937–946. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Kim, M.-J.; Lee, J.W.; Son, B.H.; Kim, S.-B.; Ahn, J.H.; Noh, W.C.; Gong, G. Different prognostic significance of CD24 and CD44 expression in breast cancer according to hormone receptor status. Breast 2011, 20, 78–85. [Google Scholar] [CrossRef]
- Marcato, P.; Dean, C.A.; Pan, D.; Araslanova, R.; Gillis, M.; Joshi, M.; Helyer, L.; Pan, L.; Leidal, A.; Gujar, S.; et al. Aldehyde dehydrogenase activity of breast cancer stem cells is primarily due to isoform ALDH1A3 and its expression is predictive of metastasis. Stem Cells 2011, 29, 32–45. [Google Scholar] [CrossRef]
- Li, X.; Strietz, J.; Bleilevens, A.; Stickeler, E.; Maurer, J. Chemotherapeutic Stress Influences Epithelial–Mesenchymal Transition and Stemness in Cancer Stem Cells of Triple-Negative Breast Cancer. Int. J. Mol. Sci. 2020, 21, 404. [Google Scholar] [CrossRef]
- Xu, N.; Li, X.; Watanabe, M.; Ueki, H.; Hu, H.; Li, N.; Araki, M.; Wada, K.; Xu, A.; Liu, C.; et al. Induction of cells with prostate cancer stem-like properties from mouse induced pluripotent stem cells via conditioned medium. Am. J. Cancer Res. 2018, 8, 1624–1632. [Google Scholar] [PubMed]
- Zhang, Y.; Huang, Y.; Jin, Z.; Li, X.; Li, B.; Xu, P.; Huang, P.; Liu, C. A Convenient and Effective Strategy for the Enrichment of Tumor-Initiating Cell Properties in Prostate Cancer Cells. Tumour Biol. 2016, 37, 11973–11981. [Google Scholar] [CrossRef] [PubMed]
- Hurt, E.M.; Kawasaki, B.T.; Klarmann, G.J.; Thomas, S.B.; Farrar, W.L. CD44+ CD24− Prostate Cells Are Early Cancer Progenitor/Stem Cells That Provide a Model for Patients with Poor Prognosis. Br. J. Cancer 2008, 98, 756–765. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Su, Y.; Mei, Y.; Leng, Q.; Leng, B.; Liu, Z.; Stass, S.A.; Jiang, F. ALDH1A1 is a marker for malignant prostate stem cells and predictor of prostate cancer patients’ outcome. Lab. Investig. 2010, 90, 234–244. [Google Scholar] [CrossRef] [PubMed]
- Acikgoz, E.; Soner, B.C.; Ozdil, B.; Guven, M. CD133+/CD44+ prostate cancer stem cells exhibit embryo-like behavior patterns. Acta Histochem. 2021, 123, 151743. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.Y.; Zheng, P.S. High aldehyde dehydrogenase activity identifies cancer stem cells in human cervical cancer. Oncotarget 2013, 4, 2462–2475. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Sánchez, E.; Santiago-López, L.; Cruz-Domínguez, V.B.; Toledo-Guzmán, M.E.; Hernández-Cueto, D.; Muñiz-Hernández, S.; Garrido, E.; Cantú De León, D.; García-Carrancá, A. Characterization of cervical cancer stem cell-like cells: Phenotyping, stemness, and human papilloma virus co-receptor expression. Oncotarget 2016, 7, 31943–31954. [Google Scholar] [CrossRef]
- Zamulaeva, I.A.; Selivanova, E.I.; Matchuk, O.N.; Krikunova, L.I.; Mkrtchyan, L.S.; Kulieva, G.Z.; Kaprin, A.D. Quantitative Changes in the Population of Cancer Stem Cells after Radiation Exposure in a Dose of 10 Gy as a Prognostic Marker of Immediate Results of the Treatment of Squamous Cell Cervical Cancer. Bull. Exp. Biol. Med. 2019, 168, 156–159. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, A.; Vishnoi, K.; Mahata, S.; Verma, G.; Srivastava, Y.; Masaldan, S.; Roy, B.G.; Bharti, A.C.; Das, B.C. Cervical Cancer Stem Cells Selectively Overexpress HPV Oncoprotein E6 that Controls Stemness and Self-Renewal through Upregulation of HES1. Clin. Cancer Res. 2016, 22, 4170–4184. [Google Scholar] [CrossRef]
- Li, X.; Ding, J.; Li, N.; Liu, W.; Ding, F.; Zheng, H.; Ning, Y.; Wang, H.; Liu, R.; Ren, S. Synthesis and biological evaluation of celastrol derivatives as anti-ovarian cancer stem cell agents. Eur. J. Med. Chem. 2019, 179, 667–679. [Google Scholar] [CrossRef]
- Curley, M.D.; Therrien, V.A.; Cummings, C.L.; Sergent, P.A.; Koulouris, C.R.; Friel, A.M.; Roberts, D.J.; Seiden, M.V.; Scadden, D.T.; Rueda, B.R.; et al. CD133 expression defines a tumor initiating cell population in primary human ovarian cancer. Stem Cells 2009, 27, 2875–2883. [Google Scholar] [CrossRef] [PubMed]
- Meng, E.; Long, B.; Sullivan, P.; McClellan, S.; Finan, M.A.; Reed, E.; Shevde, L.; Rocconi, R.P. CD44+/CD24− ovarian cancer cells demonstrate cancer stem cell properties and correlate to survival. Clin. Exp. Metastasis 2012, 29, 939–948. [Google Scholar] [CrossRef] [PubMed]
- Kryczek, I.; Liu, S.; Roh, M.; Vatan, L.; Szeliga, W.; Wei, S.; Banerjee, M.; Mao, Y.; Kotarski, J.; Wicha, M.S.; et al. Expression of aldehyde dehydrogenase and CD133 defines ovarian cancer stem cells. Int. J. Cancer 2012, 130, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Balch, C.; Chan, M.W.; Lai, H.C.; Matei, D.; Schilder, J.M.; Yan, P.S.; Huang, T.H.; Nephew, K.P. Identification and characterization of ovarian cancer-initiating cells from primary human tumors. Cancer Res. 2008, 68, 4311–4320. [Google Scholar] [CrossRef] [PubMed]
- Folkins, C.; Shaked, Y.; Man, S.; Tang, T.; Lee, C.R.; Zhu, Z.; Hoffman, R.M.; Kerbel, R.S. Glioma tumor stem-like cells promote tumor angiogenesis and vasculogenesis via vascular endothelial growth factor and stromal-derived factor 1. Cancer Res. 2009, 69, 7243–7251. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.K.; Hawkins, C.; Clarke, I.D.; Squire, J.A.; Bayani, J.; Hide, T.; Henkelman, R.M.; Cusimano, M.D.; Dirks, P.B. Identification of human brain tumour initiating cells. Nature 2004, 432, 396–401. [Google Scholar] [CrossRef] [PubMed]
- Galdieri, L.; Jash, A.; Malkova, O.; Mao, D.D.; DeSouza, P.; Chu, Y.E.; Salter, A.; Campian, J.L.; Naegle, K.M.; Brennan, C.W.; et al. Defining phenotypic and functional heterogeneity of glioblastoma stem cells by mass cytometry. JCI Insight 2021, 6, e128456. [Google Scholar] [CrossRef] [PubMed]
- Acikgoz, E.; Tatar, C.; Oktem, G. Triptolide inhibits CD133+/CD44+ colon cancer stem cell growth and migration through triggering apoptosis and represses epithelial-mesenchymal transition via downregulating expressions of snail, slug, and twist. J. Cell Biochem. 2020, 121, 3313–3324. [Google Scholar] [CrossRef]
- Yang, M.H.; Imrali, A.; Heeschen, C. Circulating cancer stem cells: The importance to select. Chin. J. Cancer Res. 2015, 27, 437–449. [Google Scholar] [CrossRef]
- Jing, F.; Kim, H.J.; Kim, C.H.; Kim, Y.J.; Lee, J.H.; Kim, H.R. Colon cancer stem cell markers CD44 and CD133 in patients with colorectal cancer and synchronous hepatic metastases. Int. J. Oncol. 2015, 46, 1582–1588. [Google Scholar] [CrossRef]
- Huang, G.C.; Su, C.Y.; Chang, Y.C.; Chen, Y.J.; Fang, H.W. Establishment of surface marker expression profiles for colorectal cancer stem cells under different conditions. Transl. Cancer Res. 2020, 9, 2503–2510. [Google Scholar] [CrossRef] [PubMed]
- Tsunekuni, K.; Konno, M.; Haraguchi, N.; Koseki, J.; Asai, A.; Matsuoka, K.; Kobunai, T.; Takechi, T.; Doki, Y.; Mori, M.; et al. CD44/CD133-Positive Colorectal Cancer Stem Cells are Sensitive to Trifluridine Exposure. Sci. Rep. 2019, 9, 14861. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.; Zeng, Z.; Bai, B.; Zhu, J.; Song, Z. The prognostic value of CSCs biomarker CD133 in NSCLC: A meta-analysis. Oncotarget 2016, 7, 56526–56539. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, J.P.; Spinola, M.; Dodge, M.; Raso, M.G.; Behrens, C.; Gao, B.; Schuster, K.; Shao, C.; Larsen, J.E.; Sullivan, L.A.; et al. Aldehyde dehydrogenase activity selects for lung adenocarcinoma stem cells dependent on notch signaling. Cancer Res. 2010, 70, 9937–9948. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.-C.; Chao, Y.-J.; Hsieh, M.-H.; Tung, H.-L.; Wang, H.-C.; Shan, Y.-S. Low CD8+ T Cell Infiltration and High PD-L1 Expression Are Associated with Level of CD44+/CD133+ Cancer Stem Cells and Predict an Unfavorable Prognosis in Pancreatic Cancer. Cancers 2019, 11, 541. [Google Scholar] [CrossRef] [PubMed]
- Kawai, T.; Yasuchika, K.; Ishii, T.; Katayama, H.; Yoshitoshi, E.Y.; Ogiso, S.; Kita, S.; Yasuda, K.; Fukumitsu, K.; Mizumoto, M.; et al. Keratin 19, a Cancer Stem Cell Marker in Human Hepatocellular Carcinoma. Clin. Cancer Res. 2015, 21, 3081–3091. [Google Scholar] [CrossRef] [PubMed]
- Xia, Q.; Han, T.; Yang, P.; Wang, R.; Li, H.; Zhang, J.; Zhou, X. MicroRNA-28-5p Regulates Liver Cancer Stem Cell Expansion via IGF-1 Pathway. Stem Cells Int. 2019, 2019, 8734362. [Google Scholar] [CrossRef]
- Kahraman, D.C.; Kahraman, T.; Cetin-Atalay, R. Targeting PI3K/Akt/mTOR Pathway Identifies Differential Expression and Functional Role of IL8 in Liver Cancer Stem Cell Enrichment. Mol. Cancer Ther. 2019, 18, 2146–2157. [Google Scholar] [CrossRef] [PubMed]
- Škerlová, J.; Král, V.; Kachala, M.; Fábry, M.; Bumba, L.; Svergun, D.I.; Tošner, Z.; Veverka, V.; Řezáčová, P. Molecular mechanism for the action of the anti-CD44 monoclonal antibody MEM-85. J. Struct. Biol. 2015, 191, 214–223. [Google Scholar] [CrossRef]
- Chinn, S.B.; Darr, O.A.; Owen, J.H.; Bellile, E.; McHugh, J.B.; Spector, M.E.; Papagerakis, S.M.; Chepeha, D.B.; Bradford, C.R.; Carey, T.E.; et al. Cancer stem cells: Mediators of tumorigenesis and metastasis in head and neck squamous cell carcinoma. Head Neck 2015, 37, 317–326. [Google Scholar] [CrossRef]
- Ma, Z.; Zhang, C.; Liu, X.; Fang, F.; Liu, S.; Liao, X.; Tao, S.; Mai, H. Characterisation of a subpopulation of CD133+ cancer stem cells from Chinese patients with oral squamous cell carcinoma. Sci. Rep. 2020, 10, 8875. [Google Scholar] [CrossRef]
- Wang, J.; Wu, Y.; Gao, W.; Li, F.; Bo, Y.; Zhu, M.; Fu, R.; Liu, Q.; Wen, S.; Wang, B. Identification and characterization of CD133+ CD44+ cancer stem cells from human laryngeal squamous cell carcinoma cell lines. J. Cancer 2017, 8, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Terwijn, M.; Zeijlemaker, W.; Kelder, A.; Rutten, A.P.; Snel, A.N.; Scholten, W.J.; Pabst, T.; Verhoef, G.; Löwenberg, B.; Zweegman, S.; et al. Leukemic stem cell frequency: A strong biomarker for clinical outcome in acute myeloid leukemia. PLoS ONE 2014, 9, e107587. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, M.; Liedtke, M.; Gentles, A.J.; Cleary, M.L. CD93 Marks a Non-Quiescent Human Leukemia Stem Cell Population and Is Required for Development of MLL-Rearranged Acute Myeloid Leukemia. Cell Stem Cell 2015, 17, 412–421. [Google Scholar] [CrossRef] [PubMed]
- Boyd, A.L.; Lu, J.; Hollands, C.G.; Alsostovar, L.; Murali, S.; Reid, J.C.; Ye, W.; Vandersluis, S.; Johnson, P.; ElRafie, A.; et al. Leukemic progenitor compartment serves as a prognostic measure of cancer stemness in patients with acute myeloid leukemia. Cell Rep. Med. 2023, 4, 101108. [Google Scholar] [CrossRef] [PubMed]
- Zeijlemaker, W.; Kelder, A.; Wouters, R.; Valk, P.J.M.; Witte, B.I.; Cloos, J.; Ossenkoppele, G.J.; Schuurhuis, G.J. Absence of Leukaemic CD34+ Cells in Acute Myeloid Leukaemia is of High Prognostic Value: A Longstanding Controversy Deciphered. Br. J. Haematol. 2015, 171, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Vergez, F.; Green, A.S.; Tamburini, J.; Sarry, J.E.; Gaillard, B.; Cornillet-Lefebvre, P.; Pannetier, M.; Neyret, A.; Chapuis, N.; Ifrah, N.; et al. High Levels of CD34+CD38low/−CD123+ Blasts are Predictive of an Adverse Outcome in Acute Myeloid Leukemia: A Groupe Ouest-Est Des Leucemies Aigues Et Maladies Du Sang (GOELAMS) Study. Haematologica 2011, 96, 1792–1798. [Google Scholar] [CrossRef] [PubMed]
- Heo, S.K.; Noh, E.K.; Ju, L.J.; Sung, J.Y.; Jeong, Y.K.; Cheon, J.; Koh, S.J.; Min, Y.J.; Choi, Y.; Jo, J.C. CD45dimCD34+CD38−CD133+ cells have the potential as leukemic stem cells in acute myeloid leukemia. BMC Cancer 2020, 20, 285. [Google Scholar] [CrossRef]
- Ågerstam, H.; Hansen, N.; von Palffy, S.; Sanden, C.; Reckzeh, K.; Karlsson, C.; Lilljebjörn, H.; Landberg, N.; Askmyr, M.; Högberg, C.; et al. IL1RAP Antibodies Block IL-1-Induced Expansion of Candidate CML Stem Cells and Mediate Cell Killing in Xenograft Models. Blood 2016, 128, 2683–2693. [Google Scholar] [CrossRef] [PubMed]
- Warfvinge, R.; Geironson, L.; Sommarin, M.N.E.; Lang, S.; Karlsson, C.; Roschupkina, T.; Stenke, L.; Stentoft, J.; Olsson-Strömberg, U.; Hjorth-Hansen, H.; et al. Single-cell molecular analysis defines therapy response and immunophenotype of stem cell subpopulations in CML. Blood 2017, 129, 2384–2394. [Google Scholar] [CrossRef]
- Kinstrie, R.; Horne, G.A.; Morrison, H.; Irvine, D.; Munje, C.; Castañeda, E.G.; Moka, H.A.; Dunn, K.; Cassels, J.E.; Parry, N.; et al. CD93 is expressed on chronic myeloid leukemia stem cells and identifies a quiescent population which persists after tyrosine kinase inhibitor therapy. Leukemia 2020, 34, 1613–1625. [Google Scholar] [CrossRef] [PubMed]
- Houshmand, M.; Simonetti, G.; Circosta, P.; Gaidano, V.; Cignetti, A.; Martinelli, G.; Saglio, G.; Gale, R.P. Chronic myeloid leukemia stem cells. Leukemia 2019, 33, 1543–1556. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, H.; Sadovnik, I.; Cerny-Reiterer, S.; Rülicke, T.; Stefanzl, G.; Willmann, M.; Hoermann, G.; Bilban, M.; Blatt, K.; Herndlhofer, S.; et al. Dipeptidylpeptidase IV (CD26) defines leukemic stem cells (LSC) in chronic myeloid leukemia. Blood 2014, 123, 3951–3962. [Google Scholar] [CrossRef] [PubMed]
- Cox, C.V.; Diamanti, P.; Evely, R.S.; Kearns, P.R.; Blair, A. Expression of CD133 on leukemia-initiating cells in childhood ALL. Blood 2009, 113, 3287–3296. [Google Scholar] [CrossRef]
- Ji, H.; Chen, L.; Dai, Y.; Sun, X.; Li, X.; Wang, Q.; Ma, D.; Du, D.; Zhao, P.; Wang, Y. Aberrant expression of CD133 and CD82 in patients with pediatric acute lymphoblastic leukemia and the clinical significance. Oncol. Lett. 2017, 14, 5811–5818. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kong, Y.; Yoshida, S.; Saito, Y.; Doi, T.; Nagatoshi, Y.; Fukata, M.; Saito, N.; Yang, S.M.; Iwamoto, C.; Okamura, J.; et al. CD34+CD38+CD19+ as well as CD34+CD38−CD19+ cells are leukemia-initiating cells with self-renewal capacity in human B-precursor ALL. Leukemia 2008, 22, 1207–1213. [Google Scholar] [CrossRef] [PubMed]
- Aoki, Y.; Watanabe, T.; Saito, Y.; Kuroki, Y.; Hijikata, A.; Takagi, M.; Tomizawa, D.; Eguchi, M.; Eguchi-Ishimae, M.; Kaneko, A.; et al. Identification of CD34+ and CD34− leukemia-initiating cells in MLL-rearranged human acute lymphoblastic leukemia. Blood 2015, 125, 967–980. [Google Scholar] [CrossRef] [PubMed]
- Li, L.J.; Tao, J.L.; Fu, R.; Wang, H.Q.; Jiang, H.J.; Yue, L.Z.; Zhang, W.; Liu, H.; Shao, Z.H. Increased CD34+CD38−CD123+ cells in myelodysplastic syndrome displaying malignant features similar to those in AML. Int. J. Hematol. 2014, 100, 60–69. [Google Scholar] [CrossRef] [PubMed]
- Tehranchi, R.; Woll, P.S.; Anderson, K.; Buza-Vidas, N.; Mizukami, T.; Mead, A.J.; Astrand-Grundström, I.; Strömbeck, B.; Horvat, A.; Ferry, H.; et al. Persistent malignant stem cells in del(5q) myelodysplasia in remission. N. Engl. J. Med. 2010, 363, 1025–1037. [Google Scholar] [CrossRef]
- Will, B.; Zhou, L.; Vogler, T.O.; Ben-Neriah, S.; Schinke, C.; Tamari, R.; Yu, Y.; Bhagat, T.D.; Bhattacharyya, S.; Barreyro, L.; et al. Stem and progenitor cells in myelodysplastic syndromes show aberrant stage-specific expansion and harbor genetic and epigenetic alterations. Blood 2012, 120, 2076–2086. [Google Scholar] [CrossRef]
- Gao, M.; Bai, H.; Jethava, Y.; Wu, Y.; Zhu, Y.; Yang, Y.; Xia, J.; Cao, H.; Franqui-Machin, R.; Nadiminti, K.; et al. Identification and characterization of tumor-initiating cells in multiple myeloma. J. Natl. Cancer Inst. 2020, 112, 507–515. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Wang, H.; Chen, P.; Shen, X.; Zhang, B.; Liu, J.; Peng, H.; Xiao, X. Identification and Characterization of Multiple Myeloma Stem Cell-Like Cells. Cancers 2021, 13, 3523. [Google Scholar] [CrossRef] [PubMed]
- Matsui, W.; Wang, Q.; Barber, J.P.; Brennan, S.; Smith, B.D.; Borrello, I.; Mcniece, I.; Lin, L.; Ambinder, R.F.; Peacock, C.; et al. Clonogenic multiple myeloma progenitors, stem cell properties, and drug resistance. Cancer Res. 2008, 68, 190–197. [Google Scholar] [CrossRef] [PubMed]
- Samart, P.; Rojanasakul, Y.; Issaragrisil, S.; Luanpitpong, S. A novel E-cadherin/SOX9 axis regulates cancer stem cells in multiple myeloma by activating Akt and MAPK pathways. Exp. Hematol. Oncol. 2022, 11, 41. [Google Scholar] [CrossRef] [PubMed]
- McCarron, M.J.; Park, P.W.; Fooksman, D.R. CD138 mediates selection of mature plasma cells by regulating their survival. Blood 2017, 129, 2749–2759. [Google Scholar] [CrossRef] [PubMed]
- Reghunathan, R.; Bi, C.; Liu, S.C.; Loong, K.T.; Chung, T.H.; Huang, G.; Chng, W.J. Clonogenic multiple myeloma cells have shared stemness signature associated with patient survival. Oncotarget 2013, 4, 1230–1240. [Google Scholar] [CrossRef] [PubMed]
- Greve, B.; Kelsch, R.; Spaniol, K.; Eich, H.T.; Götte, M. Flow cytometry in cancer stem cell analysis and separation. Cytom. Part A 2012, 81, 284–293. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Borlak, J. Advances in Liver Cancer Stem Cell Isolation and their Characterization. Stem Cell Rev. Rep. 2021, 17, 1215–1238. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.I.; Czarnecka, A.M.; Helbrecht, I.; Bartnik, E.; Lian, F.; Szczylik, C. Current approaches in identification and isolation of human renal cell carcinoma cancer stem cells. Stem Cell Res. Ther. 2015, 6, 178. [Google Scholar] [CrossRef]
- Miltenyi, S.; Müller, W.; Weichel, W.; Radbruch, A. High gradient magnetic cell separation with MACS. Cytometry 1990, 11, 231–238. [Google Scholar] [CrossRef]
- Bahmad, H.F.; Cheaito, K.; Chalhoub, R.M.; Hadadeh, O.; Monzer, A.; Ballout, F.; El-Hajj, A.; Mukherji, D.; Liu, Y.N.; Daoud, G.; et al. Sphere-Formation Assay: Three-Dimensional in vitro Culturing of Prostate Cancer Stem/Progenitor Sphere-Forming Cells. Front. Oncol. 2018, 8, 347. [Google Scholar] [CrossRef] [PubMed]
- Chambost, A.J.; Berabez, N.; Cochet-Escartin, O.; Ducray, F.; Gabut, M.; Isaac, C.; Martel, S.; Idbaih, A.; Rousseau, D.; Meyronet, D.; et al. Machine learning-based detection of label-free cancer stem-like cell fate. Sci. Rep. 2022, 12, 19066. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liu, Y.; Wang, X.; Tang, Z.; Li, S.; Hu, Y.; Zong, X.; Wu, X.; Bu, Z.; Wu, A.; et al. The extent of inflammatory infiltration in primary cancer tissues is associated with lymphomagenesis in immunodeficient mice. Sci. Rep. 2015, 5, 9447. [Google Scholar] [CrossRef] [PubMed]
- Eid, R.A.; Alaa Edeen, M.; Shedid, E.M.; Kamal, A.S.S.; Warda, M.M.; Mamdouh, F.; Khedr, S.A.; Soltan, M.A.; Jeon, H.W.; Zaki, M.S.A.; et al. Targeting Cancer Stem Cells as the Key Driver of Carcinogenesis and Therapeutic Resistance. Int. J. Mol. Sci. 2023, 24, 1786. [Google Scholar] [CrossRef] [PubMed]
- Dobbin, Z.C.; Landen, C.N. Isolation and characterization of potential cancer stem cells from solid human tumors--potential applications. Curr. Protoc. Pharmacol. 2013, 63, 14–28. [Google Scholar] [CrossRef] [PubMed]
- Waisman, A.; La Greca, A.; Möbbs, A.M.; Scarafía, M.A.; Santín Velazque, N.L.; Neiman, G.; Moro, L.N.; Luzzani, C.; Sevlever, G.E.; Guberman, A.S.; et al. Deep Learning Neural Networks Highly Predict Very Early Onset of Pluripotent Stem Cell Differentiation. Stem Cell Rep. 2019, 12, 845–859. [Google Scholar] [CrossRef] [PubMed]
- Dillekås, H.; Rogers, M.S.; Straume, O. Are 90% of deaths from cancer caused by metastases? Cancer Med. 2019, 8, 5574–5576. [Google Scholar] [CrossRef] [PubMed]
- Visvader, J.E.; Lindeman, G.J. Cancer stem cells in solid tumours: Accumulating evidence and unresolved questions. Nat. Rev. Cancer 2008, 8, 755–768. [Google Scholar] [CrossRef] [PubMed]
- Quintana, E.; Shackleton, M.; Foster, H.R.; Fullen, D.R.; Sabel, M.S.; Johnson, T.M.; Morrison, S.J. Phenotypic heterogeneity among tumorigenic melanoma cells from patients that is reversible and not hierarchically organized. Cancer Cell 2010, 18, 510–523. [Google Scholar] [CrossRef]
- Xu, S.; Zhou, Y.; Biekemitoufu, H.; Wang, H.; Li, C.; Zhang, W.; Ma, Y. Expression of Twist, Slug and Snail in esophageal squamous cell carcinoma and their prognostic significance. Oncol. Lett. 2021, 21, 184. [Google Scholar] [CrossRef]
- Khales, S.A.; Mozaffari-Jovin, S.; Geerts, D.; Abbaszadegan, M.R. TWIST1 activates cancer stem cell marker genes to promote epithelial-mesenchymal transition and tumorigenesis in esophageal squamous cell carcinoma. BMC Cancer 2022, 22, 1272. [Google Scholar] [CrossRef] [PubMed]
- Wilson, B.J.; Saab, K.R.; Ma, J.; Schatton, T.; Putz, P.; Zhan, Q.; Murphy, G.F.; Gasser, M.; Waaga-Gasser, A.M.; Frank, N.Y.; et al. ABCB5 maintains melanoma-initiating cells through a proinflammatory cytokine signaling circuit. Cancer Res. 2014, 74, 4196–4207. [Google Scholar] [CrossRef] [PubMed]
- Láinez-González, D.; Serrano-López, J.; Alonso-Dominguez, J.M. Understanding the Notch Signaling Pathway in Acute Myeloid Leukemia Stem Cells: From Hematopoiesis to Neoplasia. Cancers 2022, 14, 1459. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.T.; Zhuan-Sun, Y.X.; Zhuang, Y.Y.; Wei, S.L.; Tang, J.; Chen, W.B.; Zhang, S.N. Inhibition of hedgehog signaling depresses self-renewal of pancreatic cancer stem cells and reverses chemoresistance. Int. J. Oncol. 2012, 41, 1707–1714. [Google Scholar] [CrossRef] [PubMed]
- Ferruzzi, P.; Mennillo, F.; De Rosa, A.; Giordano, C.; Rossi, M.; Benedetti, G.; Magrini, R.; Pericot Mohr, G.L.; Miragliotta, V.; Magnoni, L.; et al. In vitro and in vivo characterization of a novel Hedgehog signaling antagonist in human glioblastoma cell lines. Int. J. Cancer. 2012, 131, E33–E44. [Google Scholar] [CrossRef] [PubMed]
- Oskarsson, T.; Batlle, E.; Massagué, J. Metastatic stem cells: Sources, niches, and vital pathways. Cell Stem Cell. 2014, 14, 306–321. [Google Scholar] [CrossRef]
- Ng, S.W.K.; Mitchell, A.; Kennedy, J.A.; Chen, W.C.; McLeod, J.; Ibrahimova, N.; Arruda, A.; Popescu, A.; Gupta, V.; Schimmer, A.D.; et al. A 17-gene stemness score for rapid determination of risk in acute leukaemia. Nature 2016, 540, 433–437. [Google Scholar] [CrossRef] [PubMed]
- Senft, D.; Jeremias, I. A rare subgroup of leukemia stem cells harbors relapse-inducing potential in acute lymphoblastic leukemia. Exp. Hematol. 2019, 69, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Schwarz-Cruz, Y.; Celis, A.; Ceballos-Cancino, G.; Vazquez-Santillan, K.; Espinosa, M.; Zampedri, C.; Bahena, I.; Ruiz, V.; Maldonado, V.; Melendez-Zajgla, J. Basal-Type Breast Cancer Stem Cells Over-Express Chromosomal Passenger Complex Proteins. Cells 2020, 9, 709. [Google Scholar] [CrossRef]
- Jiang, Y.; Guo, Y.; Hao, J.; Guenter, R.; Lathia, J.; Beck, A.W.; Hattaway, R.; Hurst, D.; Wang, Q.J.; Liu, Y.; et al. Development of an arteriolar niche and self-renewal of breast cancer stem cells by lysophosphatidic acid/protein kinase D signaling. Commun. Biol. 2021, 4, 780. [Google Scholar] [CrossRef]
- Tran, T.T.; Lee, K. JAG1 Intracellular Domain Enhances AR Expression and Signaling and Promotes Stem-like Properties in Prostate Cancer Cells. Cancers 2022, 14, 5714. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.-C.; Chuang, I.-C.; Yang, Y.-C.; Chuang, P.-C.; Lin, H.; Ou, Y.-C.; Chang Chien, C.-C.; Huang, H.-S.; Kang, H.-Y. Low P16INK4A Expression Associated with High Expression of Cancer Stem Cell Markers Predicts Poor Prognosis in Cervical Cancer after Radiotherapy. Int. J. Mol. Sci. 2018, 19, 2541. [Google Scholar] [CrossRef] [PubMed]
- Iżycka, N.; Zaborowski, M.P.; Ciecierski, Ł.; Jaz, K.; Szubert, S.; Miedziarek, C.; Rezler, M.; Piątek-Bajan, K.; Synakiewicz, A.; Jankowska, A.; et al. Cancer Stem Cell Markers—Clinical Relevance and Prognostic Value in High-Grade Serous Ovarian Cancer (HGSOC) Based on The Cancer Genome Atlas Analysis. Int. J. Mol. Sci. 2023, 24, 12746. [Google Scholar] [CrossRef] [PubMed]
- Ardizzoia, A.; Jemma, A.; Redaelli, S.; Silva, M.; Bentivegna, A.; Lavitrano, M.; Conconi, D. AhRR and PPP1R3C: Potential Prognostic Biomarkers for Serous Ovarian Cancer. Int. J. Mol. Sci. 2023, 24, 11455. [Google Scholar] [CrossRef] [PubMed]
- Howard, C.M.; Zgheib, N.B.; Bush, S., 2nd; de Eulis, T.; Cortese, A.; Mollo, A.; Lirette, S.T.; Denning, K.; Valluri, J.; Claudio, P.P. Clinical relevance of cancer stem cell chemotherapeutic assay for recurrent ovarian cancer. Transl. Oncol. 2020, 13, 100860. [Google Scholar] [CrossRef] [PubMed]
- Vora, P.; Venugopal, C.; Salim, S.K.; Tatari, N.; Bakhshinyan, D.; Singh, M.; Seyfrid, M.; Upreti, D.; Rentas, S.; Wong, N.; et al. The Rational Development of CD133-Targeting Immunotherapies for Glioblastoma. Cell Stem Cell 2020, 26, 832–844.e6. [Google Scholar] [CrossRef] [PubMed]
- Hattermann, K.; Flüh, C.; Engel, D.; Mehdorn, H.M.; Synowitz, M.; Mentlein, R.; Held-Feindt, J. Stem cell markers in glioma progression and recurrence. Int. J. Once. 2016, 49, 1899–1910. [Google Scholar] [CrossRef] [PubMed]
- Miller, T.J.; McCoy, M.J.; Lee-Pullen, T.F.; Anyaegbu, C.C.; Hemmings, C.; Bulsara, M.K.; Platell, C.F. The Prognostic and Predictive Value of SOX2+ Cell Densities in Patients Treated for Colorectal Cancer. Cancers 2020, 12, 1110. [Google Scholar] [CrossRef]
- Obermayr, E.; Koppensteiner, N.; Heinzl, N.; Schuster, E.; Holzer, B.; Fabikan, H.; Weinlinger, C.; Illini, O.; Hochmair, M.; Zeillinger, R. Cancer Stem Cell-Like Circulating Tumor Cells Are Prognostic in Non-Small Cell Lung Cancer. J. Pers. Med. 2021, 11, 1225. [Google Scholar] [CrossRef] [PubMed]
- Kuo, Y.-C.; Kou, H.-W.; Hsu, C.-P.; Lo, C.-H.; Hwang, T.-L. Identification and Clinical Significance of Pancreatic Cancer Stem Cells and Their Chemotherapeutic Drug Resistance. Int. J. Mol. Sci. 2023, 24, 7331. [Google Scholar] [CrossRef]
- Zhang, X.; Li, J.; Ghoshal, K.; Fernandez, S.; Li, L. Identification of a Subtype of Hepatocellular Carcinoma with Poor Prognosis Based on Expression of Genes within the Glucose Metabolic Pathway. Cancers 2019, 11, 2023. [Google Scholar] [CrossRef]
- Motegi, A.; Fujii, S.; Zenda, S.; Arahira, S.; Tahara, M.; Hayashi, R.; Akimoto, T. Impact of Expression of CD44, a Cancer Stem Cell Marker, on the Treatment Outcomes of Intensity Modulated Radiation Therapy in Patients With Oropharyngeal Squamous Cell Carcinoma. Int. J. Radiat. Oncol. Biol. Phys. 2016, 94, 461–468. [Google Scholar] [CrossRef] [PubMed]
- Jakob, M.; Sharaf, K.; Schirmer, M.; Leu, M.; Küffer, S.; Bertlich, M.; Ihler, F.; Haubner, F.; Canis, M.; Kitz, J. Role of cancer stem cell markers ALDH1, BCL11B, BMI-1, and CD44 in the prognosis of advanced HNSCC. Strahlenther. Onkol. 2021, 197, 231–245. [Google Scholar] [CrossRef] [PubMed]
- Rao, R.S.; Raju, K.L.; Augustine, D.; Patil, S. Prognostic Significance of ALDH1, Bmi1, and OCT4 Expression in Oral Epithelial Dysplasia and Oral Squamous Cell Carcinoma. Cancer Control. 2020, 27, 1073274820904959. [Google Scholar] [CrossRef] [PubMed]
- Ho, T.C.; LaMere, M.; Stevens, B.M.; Ashton, J.M.; Myers, J.R.; O’Dwyer, K.M.; Liesveld, J.L.; Mendler, J.H.; Guzman, M.; Morrissette, J.D.; et al. Evolution of acute myelogenous leukemia stem cell properties after treatment and progression. Blood 2016, 128, 1671–1678. [Google Scholar] [CrossRef] [PubMed]
- Chu, S.; McDonald, T.; Lin, A.; Chakraborty, S.; Huang, Q.; Snyder, D.S.; Bhatia, R. Persistence of leukemia stem cells in chronic myelogenous leukemia patients in prolonged remission with imatinib treatment. Blood 2011, 118, 5565–5572. [Google Scholar] [CrossRef] [PubMed]
- Ebinger, S.; Özdemir, E.Z.; Ziegenhain, C.; Tiedt, S.; Castro Alves, C.; Grunert, M.; Dworzak, M.; Lutz, C.; Turati, V.A.; Enver, T.; et al. Characterization of Rare, Dormant, and Therapy-Resistant Cells in Acute Lymphoblastic Leukemia. Cancer Cell 2016, 30, 849–862. [Google Scholar] [CrossRef] [PubMed]
- Sekar, V.; Veerabathiran, R.; Pandian, A.; Sivamani, G. Targeting liver cancer stem cell through EpCAM therapy targeted with chemotherapy endorse enhanced progression in hepatocellular carcinoma. Egypt. Liver J. 2023, 13, 29. [Google Scholar] [CrossRef]
- Miao, Y.; Wang, P.; Huang, J.; Qi, X.; Liang, Y.; Zhao, W.; Wang, H.; Lyu, J.; Zhu, H. Metabolomics, Transcriptome and Single-Cell RNA Sequencing Analysis of the Metabolic Heterogeneity between Oral Cancer Stem Cells and Differentiated Cancer Cells. Cancers 2024, 16, 237. [Google Scholar] [CrossRef]
- Huang, Y.-H.; Chiu, L.-Y.; Tseng, J.-S.; Hsu, K.-H.; Chen, C.-H.; Sheu, G.-T.; Yang, T.-Y. Attenuation of PI3K-Akt-mTOR Pathway to Reduce Cancer Stemness on Chemoresistant Lung Cancer Cells by Shikonin and Synergy with BEZ235 Inhibitor. Int. J. Mol. Sci. 2024, 25, 616. [Google Scholar] [CrossRef]
- Santos, L.S.; Silva, V.R.; de Castro, M.V.L.; Dias, R.B.; Valverde, L.F.; Rocha, C.A.G.; Soares, M.B.P.; Quadros, C.A.; Dos Santos, E.R.; Oliveira, R.M.M.; et al. New ruthenium-xanthoxylin complex eliminates colorectal cancer stem cells by targeting the heat shock protein 90 chaperone. Cell Death Dis. 2023, 14, 832. [Google Scholar] [CrossRef] [PubMed]
- Silva, V.R.; Santos, L.S.; de Castro, M.V.L.; Dias, R.B.; Valverde, L.F.; Rocha, C.A.G.; Soares, M.B.P.; Quadros, C.A.; Correa, R.S.; Batista, A.A.; et al. A novel ruthenium complex with 5-fluorouracil suppresses colorectal cancer stem cells by inhibiting Akt/mTOR signaling. Cell Death Discov. 2023, 9, 460. [Google Scholar] [CrossRef] [PubMed]
- Shang, S.; Yang, C.; Chen, F.; Xiang, R.S.; Zhang, H.; Dai, S.Y.; Liu, J.; Lv, X.X.; Zhang, C.; Liu, X.T.; et al. ID1 expressing macrophages support cancer cell stemness and limit CD8+ T cell infiltration in colorectal cancer. Nat. Commun. 2023, 14, 7661. [Google Scholar] [CrossRef] [PubMed]
- Zavareh, V.A.; Gharibi, S.; Hosseini Rizi, M.; Nekookar, A.; Mirhendi, H.; Rahimmalek, M.; Szumny, A. Satureja bachtiarica Induces Cancer Cell Death in Breast and Glioblastoma Cancer in 2D/3D Models and Suppresses Breast Cancer Stem Cells. Cells 2023, 12, 2713. [Google Scholar] [CrossRef] [PubMed]
- Gil-Gas, C.; Sánchez-Díez, M.; Honrubia-Gómez, P.; Sánchez-Sánchez, J.L.; Alvarez-Simón, C.B.; Sabater, S.; Sánchez-Sánchez, F.; Ramírez-Castillejo, C. Self-Renewal Inhibition in Breast Cancer Stem Cells: Moonlight Role of PEDF in Breast Cancer. Cancers 2023, 15, 5422. [Google Scholar] [CrossRef] [PubMed]
- Mouti, M.A.; Deng, S.; Pook, M.; Malzahn, J.; Rendek, A.; Militi, S.; Nibhani, R.; Soonawalla, Z.; Oppermann, U.; Hwang, C.I.; et al. KMT2A associates with PHF5A-PHF14-HMG20A-RAI1 subcomplex in pancreatic cancer stem cells and epigenetically regulates their characteristics. Nat. Commun. 2023, 14, 5685. [Google Scholar] [CrossRef] [PubMed]
- Boudreault, J.; Wang, N.; Ghozlan, M.; Lebrun, J.-J. Transforming Growth Factor-β/Smad Signaling Inhibits Melanoma Cancer Stem Cell Self-Renewal, Tumor Formation and Metastasis. Cancers 2024, 16, 224. [Google Scholar] [CrossRef]
- Zeng, Z.; Fu, M.; Hu, Y.; Wei, Y.; Wei, X.; Luo, M. Regulation and signaling pathways in cancer stem cells: Implications for targeted therapy for cancer. Mol. Cancer. 2023, 22, 172. [Google Scholar] [CrossRef] [PubMed]
- Walcher, L.; Kistenmacher, A.K.; Suo, H.; Kitte, R.; Dluczek, S.; Strauß, A.; Blaudszun, A.R.; Yevsa, T.; Fricke, S.; Kossatz-Boehlert, U. Cancer Stem Cells-Origins and Biomarkers: Perspectives for Targeted Personalized Therapies. Front. Immunol. 2020, 11, 1280. [Google Scholar] [CrossRef]
- Pal, D.; Raj, K.; Nandi, S.S.; Sinha, S.; Mishra, A.; Mondal, A.; Lagoa, R.; Burcher, J.T.; Bishayee, A. Potential of Synthetic and Natural Compounds as Novel Histone Deacetylase Inhibitors for the Treatment of Hematological Malignancies. Cancers 2023, 15, 2808. [Google Scholar] [CrossRef] [PubMed]
- XPO1 Inhibitor Approved for Multiple Myeloma. Cancer Discov. 2019, 9, 1150–1151. [CrossRef] [PubMed]
- Chu, M.; Zheng, C.; Chen, C.; Song, G.; Hu, X.; Wang, Z.W. Targeting cancer stem cells by nutraceuticals for cancer therapy. Semin. Cancer Biol. 2022, 85, 234–245. [Google Scholar] [CrossRef] [PubMed]
- Ghanbari-Movahed, M.; Shafiee, S.; Burcher, J.T.; Lagoa, R.; Farzaei, M.H.; Bishayee, A. Anticancer Potential of Apigenin and Isovitexin with Focus on Oncogenic Metabolism in Cancer Stem Cells. Metabolites 2023, 13, 404. [Google Scholar] [CrossRef] [PubMed]
- Chakravarti, B.; Akhtar Siddiqui, J.; Anthony Sinha, R.; Raza, S. Targeting autophagy and lipid metabolism in cancer stem cells. Biochem. Pharmacol. 2023, 212, 115550. [Google Scholar] [CrossRef] [PubMed]
- Takebe, N.; Miele, L.; Harris, P.J.; Jeong, W.; Bando, H.; Kahn, M.; Yang, S.X.; Ivy, S.P. Targeting Notch, Hedgehog, and Wnt pathways in cancer stem cells: Clinical update. Nat. Rev. Clin. Oncol. 2015, 12, 445–464. [Google Scholar] [CrossRef] [PubMed]
- Zakaria, N.; Mohd Yusoff, N.; Zakaria, Z.; Widera, D.; Yahaya, B.H. Inhibition of NF-κB Signaling Reduces the Stemness Characteristics of Lung Cancer Stem Cells. Front. Oncol. 2018, 8, 166. [Google Scholar] [CrossRef]
- Stuart, S.A.; Minami, Y.; Wang, J.Y. The CML stem cell: Evolution of the progenitor. Cell Cycle 2009, 8, 1338–1343. [Google Scholar] [CrossRef] [PubMed]
- Grainger, S.; Traver, D.; Willert, K. Wnt Signaling in Hematological Malignancies. Prog. Mol. Biol. Transl. Sci. 2018, 153, 321–341. [Google Scholar] [CrossRef]
- Zhou, H.; Mak, P.Y.; Mu, H.; Mak, D.H.; Zeng, Z.; Cortes, J.; Liu, Q.; Andreeff, M.; Carter, B.Z. Combined inhibition of β-catenin and Bcr-Abl synergistically targets tyrosine kinase inhibitor-resistant blast crisis chronic myeloid leukemia blasts and progenitors in vitro and in vivo. Leukemia 2017, 31, 2065–2074. [Google Scholar] [CrossRef]
- Zhao, Y.; Wu, K.; Wu, Y.; Melendez, E.; Smbatyan, G.; Massiello, D.; Kahn, M. Characterization of Imatinib Resistant CML Leukemic Stem/Initiating Cells and Their Sensitivity to CBP/Catenin Antagonists. Curr. Mol. Pharmacol. 2018, 11, 113–121. [Google Scholar] [CrossRef]
- Zeng, X.; Zhao, H.; Li, Y.; Fan, J.; Sun, Y.; Wang, S.; Wang, Z.; Song, P.; Ju, D. Targeting Hedgehog signaling pathway and autophagy overcomes drug resistance of BCR-ABL-positive chronic myeloid leukemia. Autophagy 2015, 11, 355–372. [Google Scholar] [CrossRef]
- Sadarangani, A.; Pineda, G.; Lennon, K.M.; Chun, H.J.; Shih, A.; Schairer, A.E.; Court, A.C.; Goff, D.J.; Prashad, S.L.; Geron, I.; et al. GLI2 inhibition abrogates human leukemia stem cell dormancy. J. Transl. Med. 2015, 13, 98. [Google Scholar] [CrossRef] [PubMed]
- Rinkenbaugh, A.L.; Baldwin, A.S. The NF-κB Pathway and Cancer Stem Cells. Cells 2016, 5, 16. [Google Scholar] [CrossRef] [PubMed]
- Lagoa, R.; Marques-da-Silva, D.; Diniz, M.; Daglia, M.; Bishayee, A. Molecular mechanisms linking environmental toxicants to cancer development: Significance for protective interventions with polyphenols. Semin. Cancer Biol. 2022, 80, 118–144. [Google Scholar] [CrossRef] [PubMed]
- Garner, J.M.; Fan, M.; Yang, C.H.; Du, Z.; Sims, M.; Davidoff, A.M.; Pfeffer, L.M. Constitutive activation of signal transducer and activator of transcription 3 (STAT3) and nuclear factor κB signaling in glioblastoma cancer stem cells regulates the Notch pathway. J. Biol. Chem. 2013, 288, 26167–26176. [Google Scholar] [CrossRef] [PubMed]
- Chari, A.; Vogl, D.T.; Gavriatopoulou, M.; Nooka, A.K.; Yee, A.J.; Huff, C.A.; Moreau, P.; Dingli, D.; Cole, C.; Lonial, S.; et al. Oral Selinexor-Dexamethasone for Triple-Class Refractory Multiple Myeloma. N. Engl. J. Med. 2019, 381, 727–738. [Google Scholar] [CrossRef] [PubMed]
- Azmi, A.S.; Khan, H.Y.; Muqbil, I.; Aboukameel, A.; Neggers, J.E.; Daelemans, D.; Mahipal, A.; Dyson, G.; Kamgar, M.; Al-Hallak, M.N.; et al. Preclinical Assessment with Clinical Validation of Selinexor with Gemcitabine and Nab-Paclitaxel for the Treatment of Pancreatic Ductal Adenocarcinoma. Clin. Cancer Res. 2020, 26, 1338–1348. [Google Scholar] [CrossRef] [PubMed]
- Wanga, H.; Yanga, J.; Wanga, L. The potential of XPO1 inhibitors as a game changer in relapsed/refractory hematologic malignancies. Aging Pathobiol. Ther. 2020, 2, 109–113. [Google Scholar] [CrossRef]
- Hossain, F.; Sorrentino, C.; Ucar, D.A.; Peng, Y.; Matossian, M.; Wyczechowska, D.; Crabtree, J.; Zabaleta, J.; Morello, S.; Del Valle, L.; et al. Notch Signaling Regulates Mitochondrial Metabolism and NF-κB Activity in Triple-Negative Breast Cancer Cells via IKKα-Dependent Non-canonical Pathways. Front. Oncol. 2018, 8, 575. [Google Scholar] [CrossRef]
- Hsu, E.C.; Kulp, S.K.; Huang, H.L.; Tu, H.J.; Salunke, S.B.; Sullivan, N.J.; Sun, D.; Wicha, M.S.; Shapiro, C.L.; Chen, C.S. Function of Integrin-Linked Kinase in Modulating the Stemness of IL-6-Abundant Breast Cancer Cells by Regulating γ-Secretase-Mediated Notch1 Activation in Caveolae. Neoplasia 2015, 17, 497–508. [Google Scholar] [CrossRef]
- Schott, A.F.; Landis, M.D.; Dontu, G.; Griffith, K.A.; Layman, R.M.; Krop, I.; Paskett, L.A.; Wong, H.; Dobrolecki, L.E.; Lewis, M.T.; et al. Preclinical and clinical studies of gamma secretase inhibitors with docetaxel on human breast tumors. Clin. Cancer Res. 2013, 19, 1512–1524. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Shimizu, F.; Hovinga, K.; Beal, K.; Karimi, S.; Droms, L.; Peck, K.K.; Gutin, P.; Iorgulescu, J.B.; Kaley, T.; et al. Molecular and Clinical Effects of Notch Inhibition in Glioma Patients: A Phase 0/I Trial. Clin. Cancer Res. 2016, 22, 4786–4796. [Google Scholar] [CrossRef] [PubMed]
- Silva, J.S.; Marques-Da-Silva, D.; Lagoa, R. Towards the Development of Delivery Systems of Bioactive Compounds with Eyes Set on Pharmacokinetics. In Modeling and Control of Drug Delivery Systems; Elsevier: Amsterdam, The Netherlands, 2021; pp. 125–144. [Google Scholar]
- Lyu, C.; Stadlbauer, B.; Wang, L.; Buchner, A.; Pohla, H. Identification of a novel combination treatment strategy in clear cell renal cell carcinoma stem cells with shikonin and ipilimumab. Front. Immunol. 2023, 14, 1186388. [Google Scholar] [CrossRef] [PubMed]
- Mandal, M.; Sahoo, S.K.; Patra, P.; Mallik, S.; Zhao, Z. In silico ranking of phenolics for therapeutic effectiveness on cancer stem cells. BMC Bioinform. 2020, 21, 499. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wen, X.; Li, M.; Li, S.; Zhao, H. Targeting cancer stem cells and signaling pathways by resveratrol and pterostilbene. Biofactors 2018, 44, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, S.; Ghaffari, S.H.; Shaiegan, M.; Nikogoftar Zarif, M.; Nikbakht, M.; Alimoghaddam, K.; Ghavamzadeh, A. Curcumin Veto the Effects of Osteopontin (OPN) Specific Inhibitor on Leukemic Stem Cell Colony Forming Potential via Promotion of OPN Overexpression. Int. J. Hematol. Oncol. Stem Cell Res. 2016, 10, 120–129. [Google Scholar] [PubMed]
- Li, Y.; Domina, A.; Lim, G.; Chang, T.; Zhang, T. Evaluation of curcumin, a natural product in turmeric, on Burkitt lymphoma and acute myeloid leukemia cancer stem cell markers. Future Oncol. 2018, 14, 2353–2360. [Google Scholar] [CrossRef]
- Panyajai, P.; Tima, S.; Chiampanichayakul, S.; Anuchapreeda, S. Dietary Turmeric Bisdemethoxycurcumin Suppresses Wilms’ Tumor 1 and CD34 Protein Expressions in KG-1a Leukemic Stem Cells. Nutr. Cancer. 2019, 71, 1189–1200. [Google Scholar] [CrossRef] [PubMed]
- Nirachonkul, W.; Ogonoki, S.; Thumvijit, T.; Chiampanichayakul, S.; Panyajai, P.; Anuchapreeda, S.; Tima, S.; Chiampanichayakul, S. CD123-Targeted Nano-Curcumin Molecule Enhances Cytotoxic Efficacy in Leukemic Stem Cells. Nanomaterials 2021, 11, 2974. [Google Scholar] [CrossRef]
- Kim, D.H.; Khan, H.; Ullah, H.; Hassan, S.T.S.; Šmejkal, K.; Efferth, T.; Mahomoodally, M.F.; Xu, S.; Habtemariam, S.; Filosa, R.; et al. MicroRNA targeting by quercetin in cancer treatment and chemoprotection. Pharmacol. Res. 2019, 147, 104346. [Google Scholar] [CrossRef]
- Biechonski, S.; Gourevich, D.; Rall, M.; Aqaqe, N.; Yassin, M.; Zipin-Roitman, A.; Trakhtenbrot, L.; Olender, L.; Raz, Y.; Jaffa, A.J.; et al. Quercetin alters the DNA damage response in human hematopoietic stem and progenitor cells via TopoII- and PI3K-dependent mechanisms synergizing in leukemogenic rearrangements. Int. J. Cancer 2017, 140, 864–876. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.N.; Singh, C.; Nall, D.; Meeker, D.; Shankar, S.; Srivastava, R.K. The dietary bioflavonoid quercetin synergizes with epigallocathechin gallate (EGCG) to inhibit prostate cancer stem cell characteristics, invasion, migration and epithelial-mesenchymal transition. J. Mol. Signal. 2010, 5, 14. [Google Scholar] [CrossRef] [PubMed]
- Souza, L.R.; Silva, E.; Calloway, E.; Kucuk, O.; Rossi, M.; McLemore, M.L. Genistein protects hematopoietic stem cells against G-CSF-induced DNA damage. Cancer Prev. Res. 2014, 7, 534–544. [Google Scholar] [CrossRef] [PubMed]
- Cheong, J.W.; Min, Y.H.; Eom, J.I.; Kim, S.J.; Jeung, H.K.; Kim, J.S. Inhibition of CK2α and PI3K/Akt synergistically induces apoptosis of CD34+CD38− leukaemia cells while sparing haematopoietic stem cells. Anticancer Res. 2010, 30, 4625–4634. [Google Scholar] [PubMed]
- Kim, B.; Jung, N.; Lee, S.; Sohng, J.K.; Jung, H.J. Apigenin Inhibits Cancer Stem Cell-Like Phenotypes in Human Glioblastoma Cells via Suppression of c-Met Signaling. Phytother. Res. 2016, 30, 1833–1840. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Cao, X.; Cao, X.; Liu, L.; Qiu, Y.; Li, X.; Zhou, L.; Ning, Y.; Ren, K.; Cao, J. Isovitexin Inhibits Stemness and Induces Apoptosis in Hepatocellular Carcinoma SK-Hep-1 Spheroids by Upregulating miR-34a Expression. Anticancer. Agents Med. Chem. 2020, 20, 1654–1663. [Google Scholar] [CrossRef] [PubMed]
- Issa, M.E.; Cuendet, M. Withaferin A induces cell death and differentiation in multiple myeloma cancer stem cells. Medchemcomm. 2016, 8, 112–121. [Google Scholar] [CrossRef] [PubMed]
- Park, S.H.; Sung, J.H.; Chung, N. Berberine diminishes side population and down-regulates stem cell-associated genes in the pancreatic cancer cell lines PANC-1 and MIA PaCa-2. Mol. Cell Biochem. 2014, 394, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Guzman, M.L.; Rossi, R.M.; Karnischky, L.; Li, X.; Peterson, D.R.; Howard, D.S.; Jordan, C.T. The sesquiterpene lactone parthenolide induces apoptosis of human acute myelogenous leukemia stem and progenitor cells. Blood 2005, 105, 4163–4169. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Tan, G.; Cai, Y.; Liu, R.; Xiong, X.; Gu, B.; He, W.; Liu, B.; Ren, Q.; Wu, J.; et al. A novel Apigenin derivative suppresses renal cell carcinoma via directly inhibiting wild-type and mutant MET. Biochem. Pharmacol. 2021, 190, 114620. [Google Scholar] [CrossRef] [PubMed]
- Fernando, W.; Clark, R.F.; Rupasinghe, H.P.V.; Hoskin, D.W.; Coombs, M.R.P. Phloridzin Docosahexaenoate Inhibits Spheroid Formation by Breast Cancer Stem Cells and Exhibits Cytotoxic Effects against Paclitaxel-Resistant Triple Negative Breast Cancer Cells. Int. J. Mol. Sci. 2023, 24, 14577. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhou, Y.; Chen, K.; Shi, P.; Li, Y.; Deng, M.; Jiang, Z.; Wang, X.; Li, P.; Xu, B. The pan-Bcl2 Inhibitor AT101 Activates the Intrinsic Apoptotic Pathway and Causes DNA Damage in Acute Myeloid Leukemia Stem-Like Cells. Target. Oncol. 2017, 12, 677–687. [Google Scholar] [CrossRef] [PubMed]
- James, M.I.; Iwuji, C.; Irving, G.; Karmokar, A.; Higgins, J.A.; Griffin-Teal, N.; Thomas, A.; Greaves, P.; Cai, H.; Patel, S.R.; et al. Curcumin inhibits cancer stem cell phenotypes in ex vivo models of colorectal liver metastases, and is clinically safe and tolerable in combination with FOLFOX chemotherapy. Cancer Lett. 2015, 364, 135–141. [Google Scholar] [CrossRef]
- Marques-Da-Silva, D.; Rodrigues, J.R.; Lagoa, R. Anthocyanins, Effects in Mitochondria and Metabolism. In Mitochondrial Physiology and Vegetal Molecules Therapeutic Potential of Natural Compounds on Mitochondrial Health; de Oliveira, M.R., Ed.; Academic Press: Cambridge, MA, USA, 2021; pp. 267–300. [Google Scholar]
- EFSA Panel on Food Additives and Nutrient Sources added to Food (ANS); Younes, M.; Aggett, P.; Aguilar, F.; Crebelli, R.; Dusemund, B.; Filipič, M.; Frutos, M.J.; Galtier, P.; Gott, D.; et al. Scientific opinion on the safety of green tea catechins. EFSA J. 2018, 16, e05239. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).