Overexpression of miR-25 Downregulates the Expression of ROBO2 in Idiopathic Intellectual Disability
Abstract
1. Introduction
2. Results
2.1. Confirmation of the Neural Origin of Progenitors of the NPC Cell Cultures Derived from the NEO of IID Patients and Controls
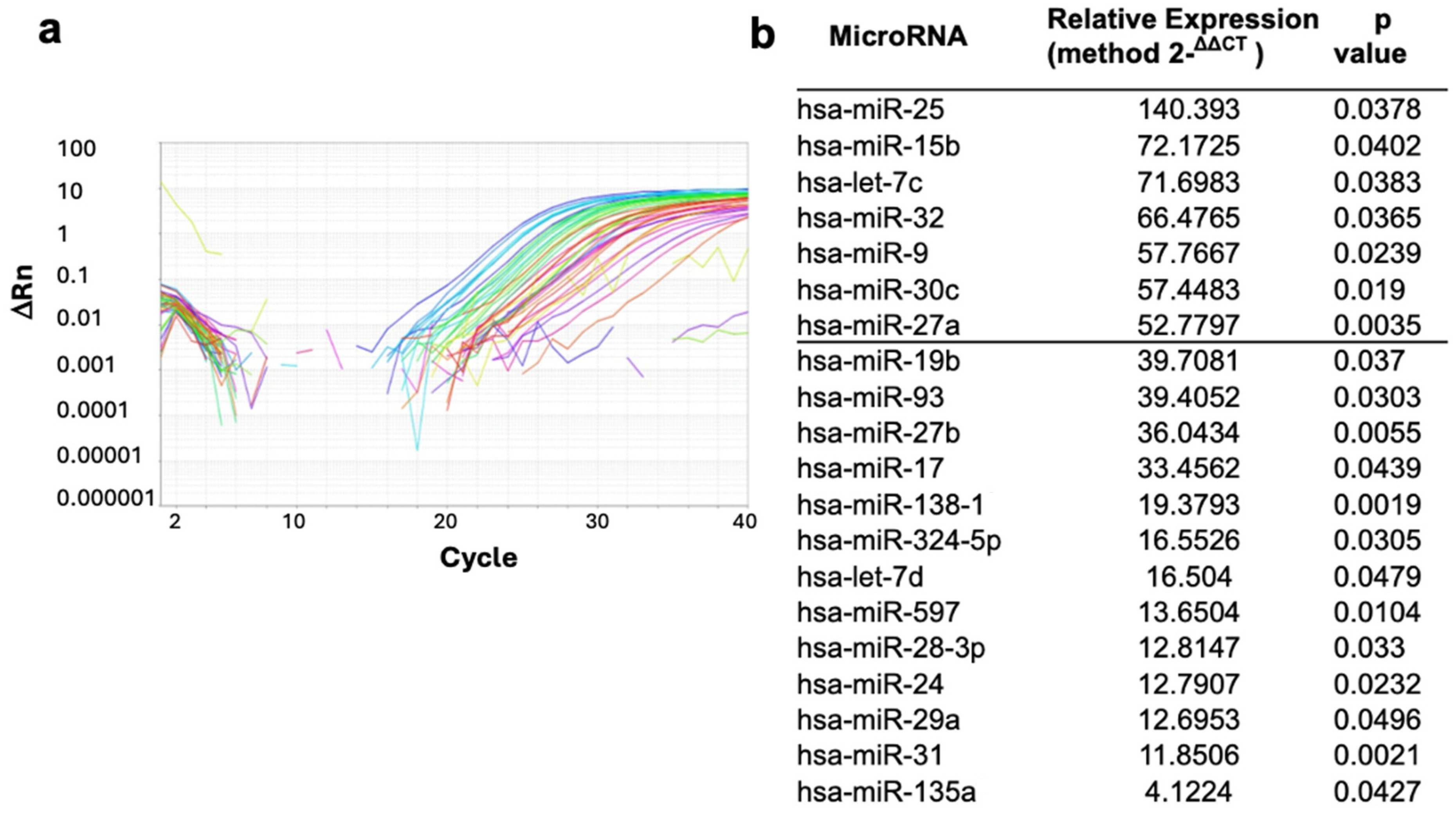
2.2. Overexpression of miRNAs in NPCs of IID Patients
2.3. mRNA Expression Profile and Selection of Targets for miR-25
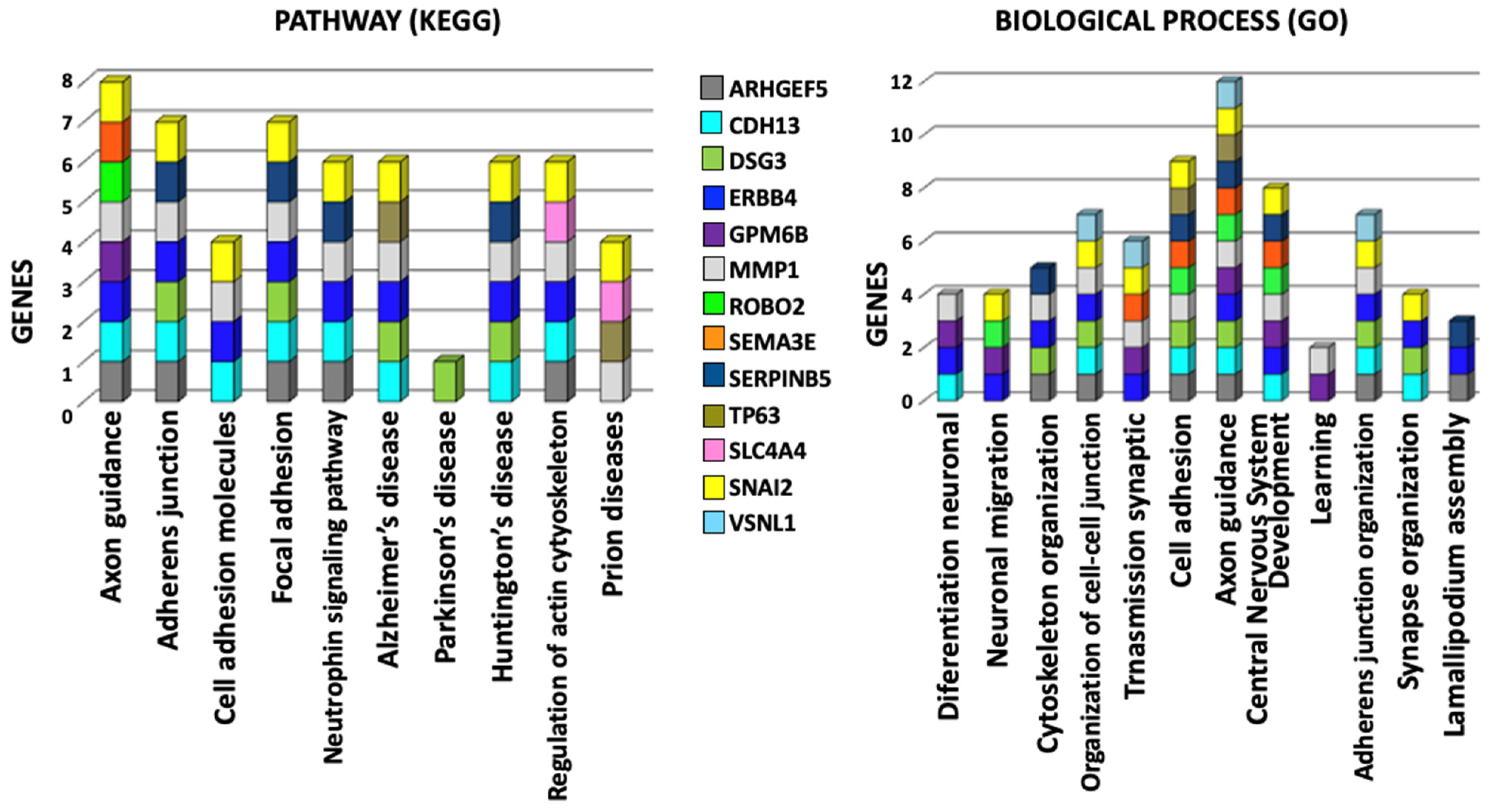
2.4. KEGG and GO Functional Annotation and Enrichment Analysis
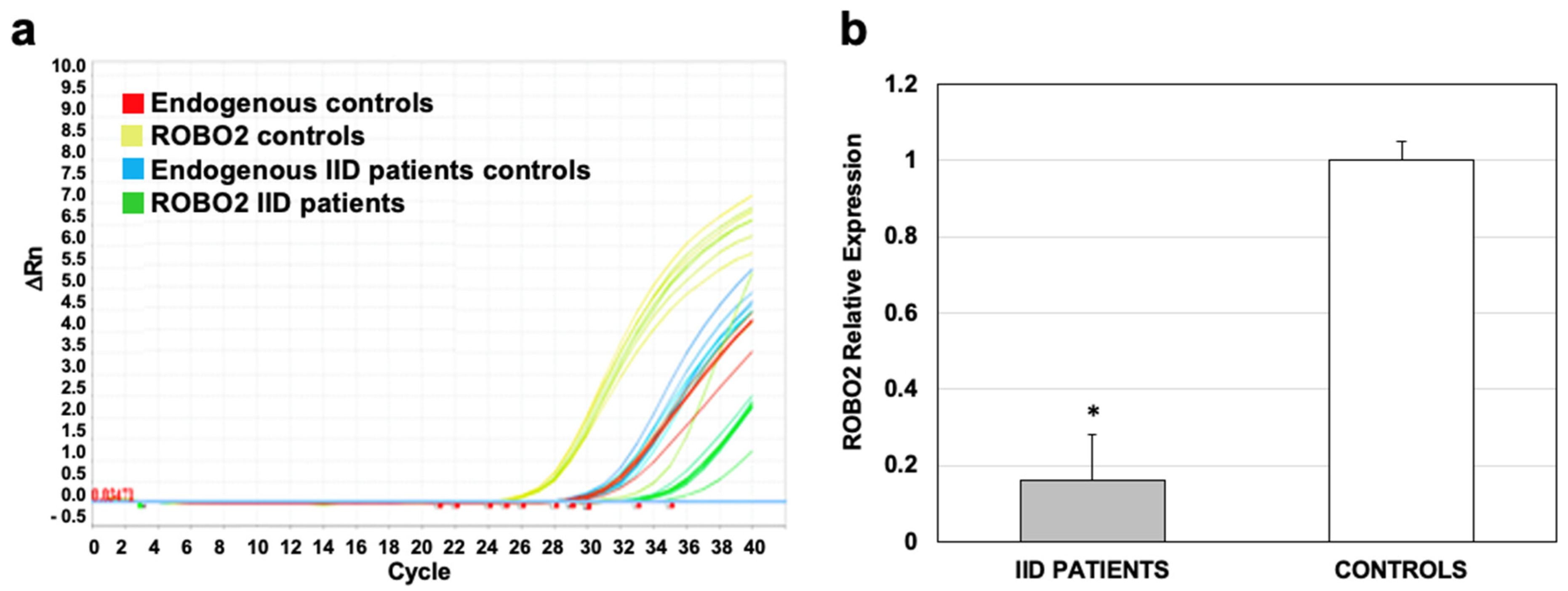
2.5. Validation of Expression Obtained in the Microarray of the ROBO2 mRNA by qRT-PCR
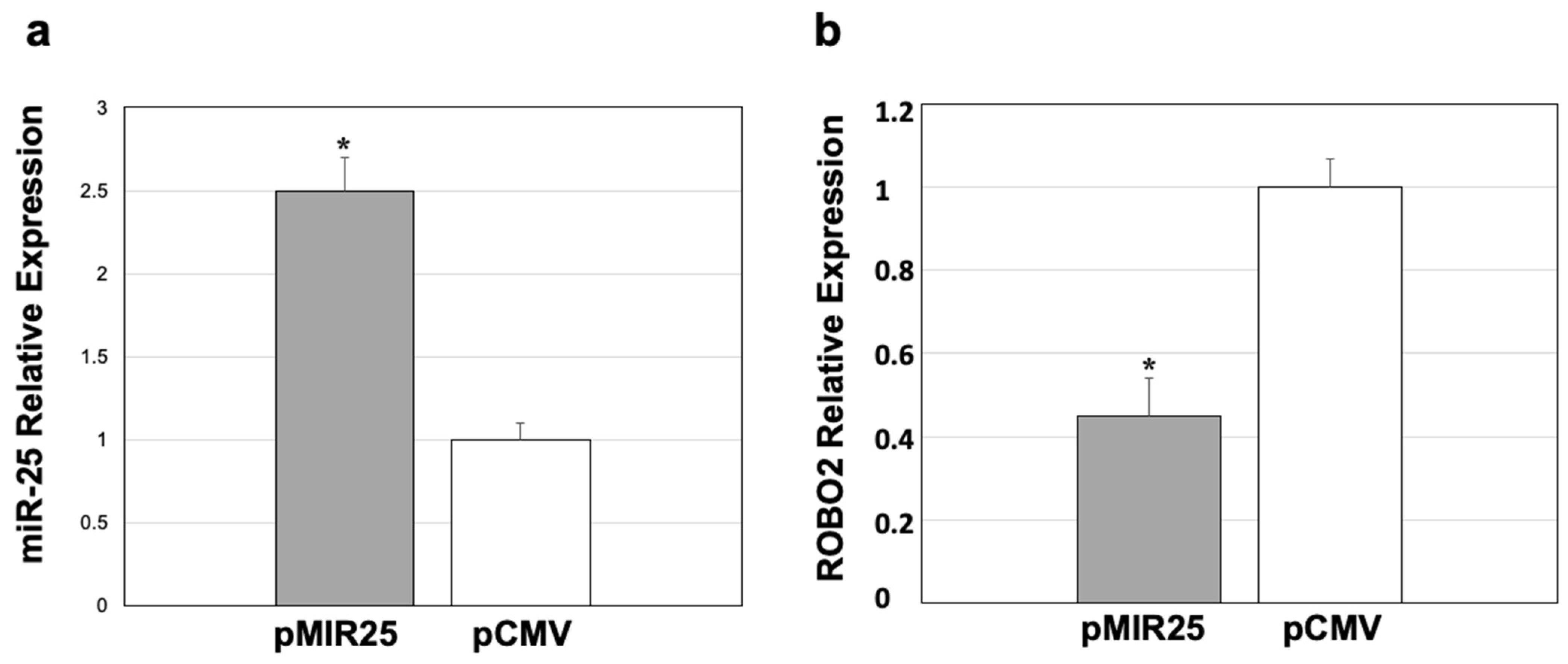
2.6. Down-Regulated ROBO2 Expression by miR-25
3. Discussion
4. Materials and Methods
4.1. Study Subjects
4.2. Sampling and Human Progenitor Neural Cell (NPC) Culture Characterization
4.3. MicroRNA Expression Profile
4.4. mRNA Expression Profile
4.5. Selection of Differentially-Expressed Genes with Function in the CNS and Neurodevelopment
4.6. In Silico Prediction of Target Genes for miR-25
4.7. Functional Annotation (KEGG) and GO Enrichment Analysis
4.8. Validation of ROBO2 Expression by qRT-PCR
4.9. In Vitro Assay of miR-25 Overexpression and Its Effect on ROBO2 Expression
- (a)
- Culture of the Human Lung Carcinoma Cell Line NCI-H2087
- (b)
- Transfection of pMIR25 and pCMV plasmids in NCI-H2087 Cells
- (c)
- Relative Expression of miR-25 Transfected with the Plasmids
- (d)
- Relative Expression of ROBO2 in the NCI-H2087 Cell Line Transfected with pMIR25 and pCMV
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Galasso, C.; Lo-Castro, A.; El-Malhany, N.; Curatolo, P. “Idiopathic” mental retardation and new chromosomal abnormalities. Ital. J. Pediatr. 2010, 14, 17. [Google Scholar] [CrossRef] [PubMed]
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders, 5th ed.; American Psychological Association: Washington, DC, USA, 2013; pp. 33–40. [Google Scholar] [CrossRef]
- Vissers, L.E.; de Vries, B.B.; Veltman, J.A. Genomic microarrays in mental retardation: From copy number variation to gene, from research to diagnosis. J. Med. Genet. 2010, 47, 289–297. [Google Scholar] [CrossRef]
- Colleaux, L.; Rio, M.; Heuertz, S.; Moindrault, S.; Turleau, C.; Ozilou, C.; Gosset, P.; Raoult, O.; Lyonnet, S.; Cormier-Daire, V.; et al. A novel automated strategy for screening cryptic telomeric rearrangements in children with idiopathic mental retardation. Eur. J. Hum. Genet. 2001, 9, 319–327. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chelly, J.; Mandel, J.L. Monogenic causes of X-linked mental retardation. Nat. Rev. Genet. 2001, 2, 669–680. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Ingolia, N.T.; Weissman, J.S.; Bartel, D.P. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 2010, 466, 835–840. [Google Scholar] [CrossRef]
- Kosik, K. The neuronal microRNA system. Nat. Rev. Neurosci. 2006, 7, 911–920. [Google Scholar] [CrossRef]
- Siew, W.H.; Tan, K.L.; Babaei, M.A.; Cheah, P.S.; Ling, K.H. MicroRNAs and intellectual disability (ID) in Down syndrome, X-linked ID, and Fragile X syndrome. Front. Cell Neurosci. 2013, 7, 41. [Google Scholar] [CrossRef] [PubMed]
- Palacios-Reyes, C.; Espinosa, A.; Contreras, A.; Ordonez, R.; Hidalgo-Miranda, A.; Rubio-Gayosso, I.; Garcia-Alonso, P.; Benitez-King, G.; Ramirez-Rodriguez, G.; Najera, N.; et al. Williams’ neural stem cells: New model for insight into microRNA dysregulation. Front. Biosci. (Elite Ed). 2013, 5, 1057–1073. [Google Scholar] [CrossRef] [PubMed]
- Edbauer, D.; Neilson, J.R.; Foster, K.A.; Wang, C.F.; Seeburg, D.P.; Batterton, M.N.; Tada, T.; Dolan, B.M.; Sharp, P.A.; Sheng, M. Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron 2010, 65, 373–384. [Google Scholar] [CrossRef]
- Gaughwin, P.; Ciesla, M.; Yang, H.; Lim, B.; Brundin, P. Stage-specific modulation of cortical neuronal development by Mmu-miR-134. Cereb. Cortex 2011, 21, 1857–1869. [Google Scholar] [CrossRef]
- Szulwach, K.E.; Li, X.; Smrt, R.D.; Li, Y.; Luo, Y.; Lin, L.; Santistevan, N.J.; Li, W.; Zhao, X.; Jin, P. Cross talk between microRNA and epigenetic regulation in adult neurogenesis. J. Cell Biol. 2010, 189, 127–141. [Google Scholar] [CrossRef]
- Brett, J.O.; Renault, V.M.; Rafalski, V.A.; Webb, A.E.; Brunet, A. The microRNA cluster miR-106b~25 regulates adult neural stem/progenitor cell proliferation and neuronal differentiation. Aging 2011, 3, 108–124. [Google Scholar] [CrossRef]
- Kim, Y.K.; Yu, J.; Han, T.S.; Park, S.Y.; Namkoong, B.; Kim, D.H.; Hur, K.; Yoo, M.W.; Lee, H.J.; Yang, H.K.; et al. Functional links between clustered microRNAs: Suppression of cell-cycle inhibitors by microRNA clusters in gastric cancer. Nucleic Acids Res. 2009, 37, 1672–1681. [Google Scholar] [CrossRef]
- Graziadei, P.P.; Graziadei, G.A. Neurogenesis and neuron regeneration in the olfactory system of mammals. I. Morphological aspects of differentiation and structural organization of the olfactory sensory neurons. J. Neurocytol. 1979, 8, 1–18. [Google Scholar] [CrossRef]
- Graziadei, G.A.; Graziadei, P.P. Neurogenesis and neuron regeneration in the olfactory system of mammals. II. Degeneration and reconstitution of the olfactory sensory neurons after axotomy. J. Neurocytol. 1979, 8, 197–213. [Google Scholar] [CrossRef]
- Graziadei, P.P.; Monti Graziadei, G.A. Neurogenesis and neuron regeneration in the olfactory system of mammals. III. Deafferentation and reinnervation of the olfactory bulb following section of the fila olfactoria in rat. J. Neurocytol. 1980, 9, 145–162. [Google Scholar] [CrossRef]
- Jiménez-Vaca, A.L.; Benitez-King, G.; Ruiz, V.; Ramírez-Rodríguez, G.B.; Hernández-de la Cruz, B.; Salamanca-Gómez, F.A.; González-Márquez, H.; Ramírez-Sánchez, I.; Ortíz-López, L.; Vélez-Del Valle, C.; et al. Exfoliated Human Olfactory Neuroepithelium: A Source of Neural Progenitor Cells. Mol. Neurobiol. 2018, 55, 2516–2523. [Google Scholar] [CrossRef]
- Benítez-King, G.; Riquelme, A.; Ortíz-López, L.; Berlanga, C.; Rodríguez-Verdugo, M.S.; Romo, F.; Calixto, E.; Solís-Chagoyán, H.; Jímenez, M.; Montaño, L.M.; et al. A non-invasive method to isolate the neuronal linage from the nasal epithelium from schizophrenic and bipolar diseases. J. Neurosci. Methods 2011, 201, 35–45. [Google Scholar] [CrossRef]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W.; DAVID Bioinformatics Database. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef]
- McGeary, S.E.; Lin, K.S.; Shi, C.Y.; Pham, T.M.; Bisaria, N.; Kelley, G.M.; Bartel, D.P. The biochemical basis of microRNA targeting efficacy. Science 2019, 366, eaav1741. [Google Scholar] [CrossRef]
- Enright, A.J.; John, B.; Gaul, U.; Tuschl, T.; Sander, C.; Marks, D.S. MicroRNA targets in Drosophila (2020 update). Genome Biol. 2003, 5, R1. [Google Scholar] [CrossRef]
- Tastsoglou, S.; Alexiou, A.; Karagkouni, D.; Skoufos, G.; Zacharopoulou, E.; Hatzigeorgiou, A.G. DIANA-microT 2023: Including predicted targets of virally encoded miRNAs. Nucleic Acids Res. 2023, 51, W148–W153. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Kawashima, M.; Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2023, 49, D545–D551. [Google Scholar] [CrossRef]
- The Gene Ontology Consortium. The Gene Ontology knowledgebase in 2023. Genetics 2023, 224, iyad031. [Google Scholar] [CrossRef] [PubMed]
- Thomas, P.D.; Ebert, D.; Muruganujan, A.; Mushayahama, T.; Albou, L.P.; Mi, H. PANTHER: Making genome-scale phylogenetics accessible to all. Protein Sci. 2022, 31, 8–22. [Google Scholar] [CrossRef]
- Pontén, F.; Jirström, K.; Uhlen, M. The Human Protein Atlas—A tool for pathology. J. Pathol. 2008, 216, 387–393. [Google Scholar] [CrossRef]
- Liu, B.; Sun, X. miR-25 promotes invasion of human non-small cell lung cancer via CDH1. Bioengineered 2019, 10, 271–281. [Google Scholar] [CrossRef] [PubMed]
- Gonda, Y.; Namba, T.; Hanashima, C. Beyond Axon Guidance: Roles of Slit-Robo Signaling in Neocortical Formation. Front. Cell Dev. Biol. 2020, 23, 607415. [Google Scholar] [CrossRef]
- Blockus, H.; Rolotti, S.V.; Szoboszlay, M.; Peze-Heidsieck, E.; Ming, T.; Schroeder, A.; Apostolo, N.; Vennekens, K.M.; Katsamba, P.S.; Bahna, F.; et al. Synaptogenic activity of the axon guidance molecule Robo2 underlies hippocampal circuit function. Cell Rep. 2021, 37, 109828. [Google Scholar] [CrossRef]
- Nguyen Ba-Charvet, K.T.; Brose, K.; Marillat, V.; Kidd, T.; Goodman, C.S.; Tessier-Lavigne, M.; Sotelo, C.; Chédotal, A. Slit2-Mediated chemorepulsion and collapse of developing forebrain axons. Neuron 1999, 22, 463–473. [Google Scholar] [CrossRef]
- Wu, W.; Wong, K.; Chen, J.; Jiang, Z.; Dupuis, S.; Wu, J.Y.; Rao, Y. Directional guidance of neuronal migration in the olfactory system by the protein Slit. Nature 1999, 400, 331–336. [Google Scholar] [CrossRef]
- Eastwood, S.L.; Law, A.J.; Everall, I.P.; Harrison, P.J. The axonal chemorepellant semaphorin 3A is increased in the cerebellum in schizophrenia and may contribute to its synaptic pathology. Mol. Psychiatry 2003, 8, 148–155. [Google Scholar] [CrossRef]
- Fisher, S.E.; Francks, C. Genes, cognition and dyslexia: Learning to read the genome. Trends Cogn. Sci. 2006, 10, 250–257. [Google Scholar] [CrossRef]
- Yaron, A.; Zheng, B. Navigating their way to the clinic: Emerging roles for axon guidance molecules in neurological disorders and injury. Dev. Neurobiol. 2007, 67, 1216–1231. [Google Scholar] [CrossRef]
- Diaz-Martinez, N.E.; Tamariz EDiaz, N.F.; Garcia-Peña, C.M.; Varela-Echavarria, A.; Velasco, I. Recovery from experimental parkinsonism by semaphorin-guided axonal growth of grafted dopamine neurons. Mol. Ther. 2013, 21, 1579–1591. [Google Scholar] [CrossRef]
- Long, H.; Sabatier, C.; Ma, L.; Plump, A.; Yuan, W.; Ornitz, D.M.; Tamada, A.; Murakami, F.; Goodman, C.S.; Tessier-Lavigne, M. Conserved roles for Slit and Robo proteins in midline commissural axon guidance. Neuron 2004, 42, 213–223. [Google Scholar] [CrossRef]
- Muller, C.; Anacker, A.; Veenstra-VanderWeele, J. The serotonin system in autism spectrum disorder: From biomarker to animal models. Neuroscience 2016, 321, 24–41. [Google Scholar] [CrossRef]
- Anitha, A.; Nakamura, K.; Yamada, K.; Suda, S.; Thanseem, I.; Tsujii, M.; Iwayama, Y.; Hattori, E.; Toyota, T.; Miyachi, T.; et al. Genetic analyses of Roundabout (ROBO) axon guidance receptors in autism. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2008, 147, 1019–1027. [Google Scholar] [CrossRef]
- Cárdenas, A.; Villalba, A.; de Juan Romero, C.; Picó, E.; Kyrousi, C.; Tzika, A.C.; Tessier-Lavigne, M.; Ma, L.; Drukker, M.; Cappello, S.; et al. Evolution of Cortical Neurogenesis in Amniotes Controlled by Robo Signaling Levels. Cell 2018, 174, 590–606. [Google Scholar] [CrossRef]
- Anselmo, M.A.; Dalvin, S.; Prodhan, P.; Komatsuzaki, K.; Aidlen, J.T.; Schnitzer, J.J.; Wu, J.Y.; Kinane, T.B. Slit and robo: Expression patterns in lung development. Gene Expr. Patterns. 2003, 3, 13–19. [Google Scholar] [CrossRef]
- Sundaresan, V.; Chung, G.; Heppell-Parton, A.; Xiong, J.; Grundy, C.; Roberts, I.; James, L.; Cahn, A.; Bench, A.; Douglas, J.; et al. Homozygous deletions at 3p12 in breast and lung cancer. Oncogene 1998, 17, 1723–1729. [Google Scholar] [CrossRef] [PubMed]
- Ypsilanti, A.R.; Zagar, Y.; Chédotal, A. Moving away from the midline: New developments for Slit and Robo. Development 2010, 137, 1939–1952. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Xiao, Y.; Ding, B.B.; Zhang, N.; Yuan Xb Gui, L.; Qian, K.X.; Duan, S.; Chen, Z.; Rao, Y.; Geng, J.G. Induction of tumor angiogenesis by Slit-Robo signaling and inhibition of cancer growth by blocking Robo activity. Cancer Cell 2003, 4, 19–29. [Google Scholar] [CrossRef] [PubMed]

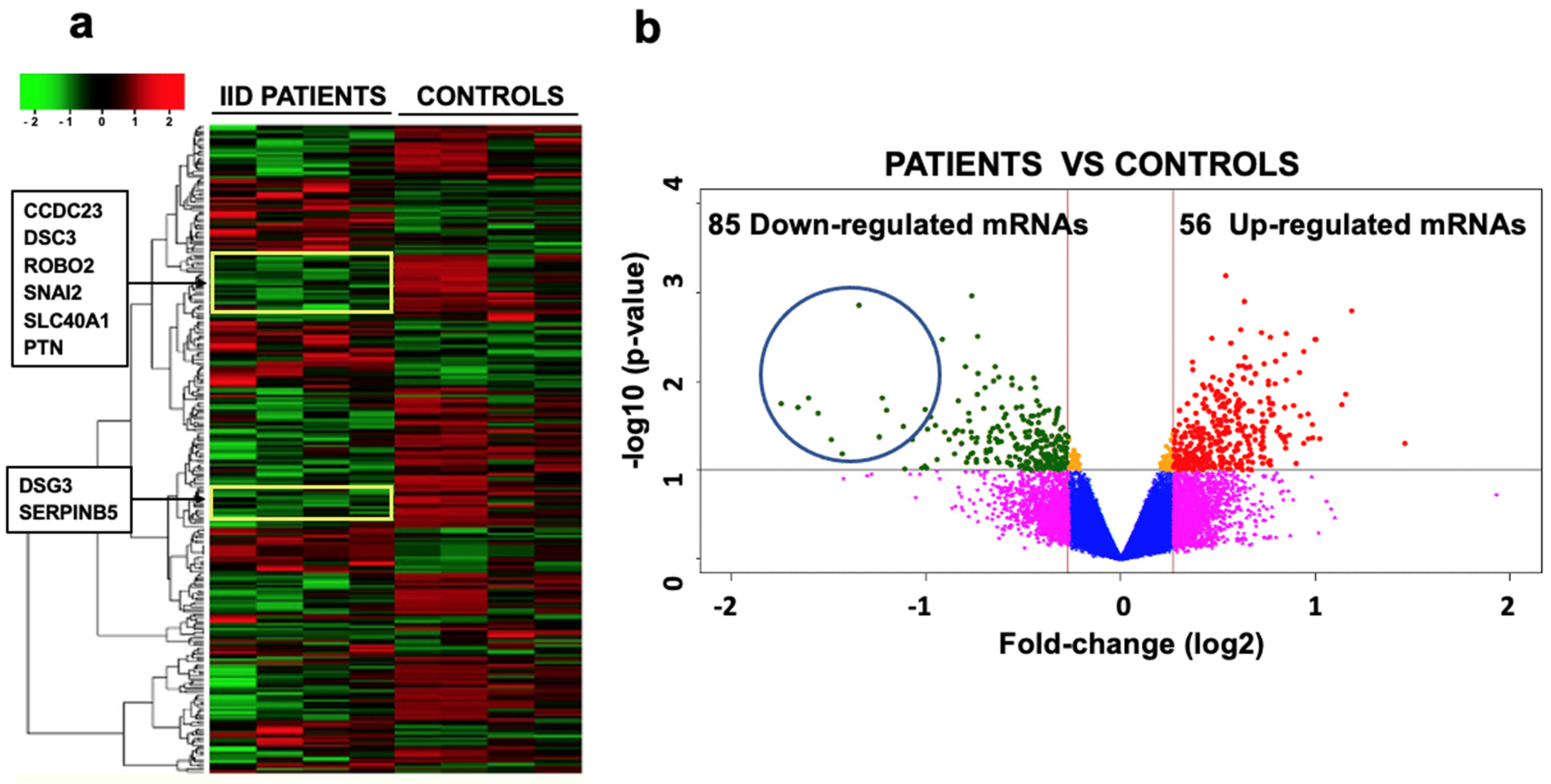
| mRNA | Expression | p Value | Function |
|---|---|---|---|
| ROBO2 | −1.50 ± 0.0294 | 0.006 | Axon guidance and neurogenesis. |
| PTN | −1.60 ± 0.0937 | 0.008 | Promotes neurites growth. |
| PAX7 | −1.57 ± 0.1212 | 0.010 | Neurogenesis. |
| TIMP1 | −1.42 ± 0.0901 | 0.019 | Formation of the cerebral cortex and memory. |
| ARHGEF5 | −1.50 ± 0.0654 | 0.019 | Associated with Rho protein (important in Alzheimer’s). |
| MMP1 | −1.50 ± 0.0450 | 0.023 | Projection of neurons and dendrites. |
| CDH13 | −1.40 ± 0.0972 | 0.024 | Neural projection/signaling receptor. |
| SERPINB5 | −1.97 ± 0.6061 | 0.025 | Adherent union in axon guide and dendrite formation. |
| GPM6B | −1.50 ± 0.0681 | 0.025 | Neurogenesis. |
| SNAI2 | −1.70 ± 0.2608 | 0.028 | Cell adhesion and migration of the neural tube. |
| CTTNBP2 | −1.39 ± 0.4588 | 0.028 | Regulation of synaptic signaling. |
| CCDC23 | −1.30 ± 0.5006 | 0.029 | Synapse formation. |
| ERBB4 | −1.37 ± 0.1877 | 0.029 | Neural crest migration and axon guidance. |
| TP63 | −1.93 ± 0.3308 | 0.031 | Apoptosis in neurodevelopment. |
| VSNL1 | −2.16 ± 0.1474 | 0.032 | Neuronal morphology (Schizophrenia). |
| DSG3 | −1.98 ± 0.2170 | 0.034 | Junction adherents in the axon guide. |
| SLC4A4 | −1.59 ± 0.1901 | 0.037 | Synaptic vesicle formation. |
| SEMA3E | −1.59 ± 0.1877 | 0.042 | Axon guide. |
| mRNA | Alignment | mirSVR | PhastCons |
|---|---|---|---|
| ROBO2 | 15nt, 12nt, 6nt (×2) | −0.9934 | 0.8038 |
| PTN | - | - | - |
| PAX7 | - | - | - |
| TIMP1 | 10nt, 6nt | −0.1734 | 0.6056 |
| ARHGEF5 | - | - | - |
| MMP1 | - | - | - |
| CDH13 | 6nt | −0.0564 | 0.7202 |
| SERPINB5 | - | - | - |
| GPM6B | - | - | - |
| SNAI2 | 16nt, 10nt | −0.1044 | 0.6056 |
| CTTNBP2 | 6nt | ||
| CCDC23 | - | - | - |
| ERBB4 | 16nt, 13nt | −0.0143 | 0.5669 |
| TP63 | 6nt | −0.0580 | 0.6197 |
| VSNL1 | - | - | - |
| DSG3 | 12nt, 6nt | ||
| SLC4A4 | 11nt, 6nt (×2) | −0.1022 | 0.5950 |
| SEMA3E | 6nt (×2) | −0.0233 | 0.5001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ordoñez-Razo, R.M.; Gutierrez-López, Y.; Araujo-Solis, M.A.; Benitez-King, G.; Ramírez-Sánchez, I.; Galicia, G. Overexpression of miR-25 Downregulates the Expression of ROBO2 in Idiopathic Intellectual Disability. Int. J. Mol. Sci. 2024, 25, 3953. https://doi.org/10.3390/ijms25073953
Ordoñez-Razo RM, Gutierrez-López Y, Araujo-Solis MA, Benitez-King G, Ramírez-Sánchez I, Galicia G. Overexpression of miR-25 Downregulates the Expression of ROBO2 in Idiopathic Intellectual Disability. International Journal of Molecular Sciences. 2024; 25(7):3953. https://doi.org/10.3390/ijms25073953
Chicago/Turabian StyleOrdoñez-Razo, Rosa María, Yessica Gutierrez-López, María Antonieta Araujo-Solis, Gloria Benitez-King, Israel Ramírez-Sánchez, and Gabriela Galicia. 2024. "Overexpression of miR-25 Downregulates the Expression of ROBO2 in Idiopathic Intellectual Disability" International Journal of Molecular Sciences 25, no. 7: 3953. https://doi.org/10.3390/ijms25073953
APA StyleOrdoñez-Razo, R. M., Gutierrez-López, Y., Araujo-Solis, M. A., Benitez-King, G., Ramírez-Sánchez, I., & Galicia, G. (2024). Overexpression of miR-25 Downregulates the Expression of ROBO2 in Idiopathic Intellectual Disability. International Journal of Molecular Sciences, 25(7), 3953. https://doi.org/10.3390/ijms25073953






