Sulfate Availability and Hormonal Signaling in the Coordination of Plant Growth and Development
Abstract
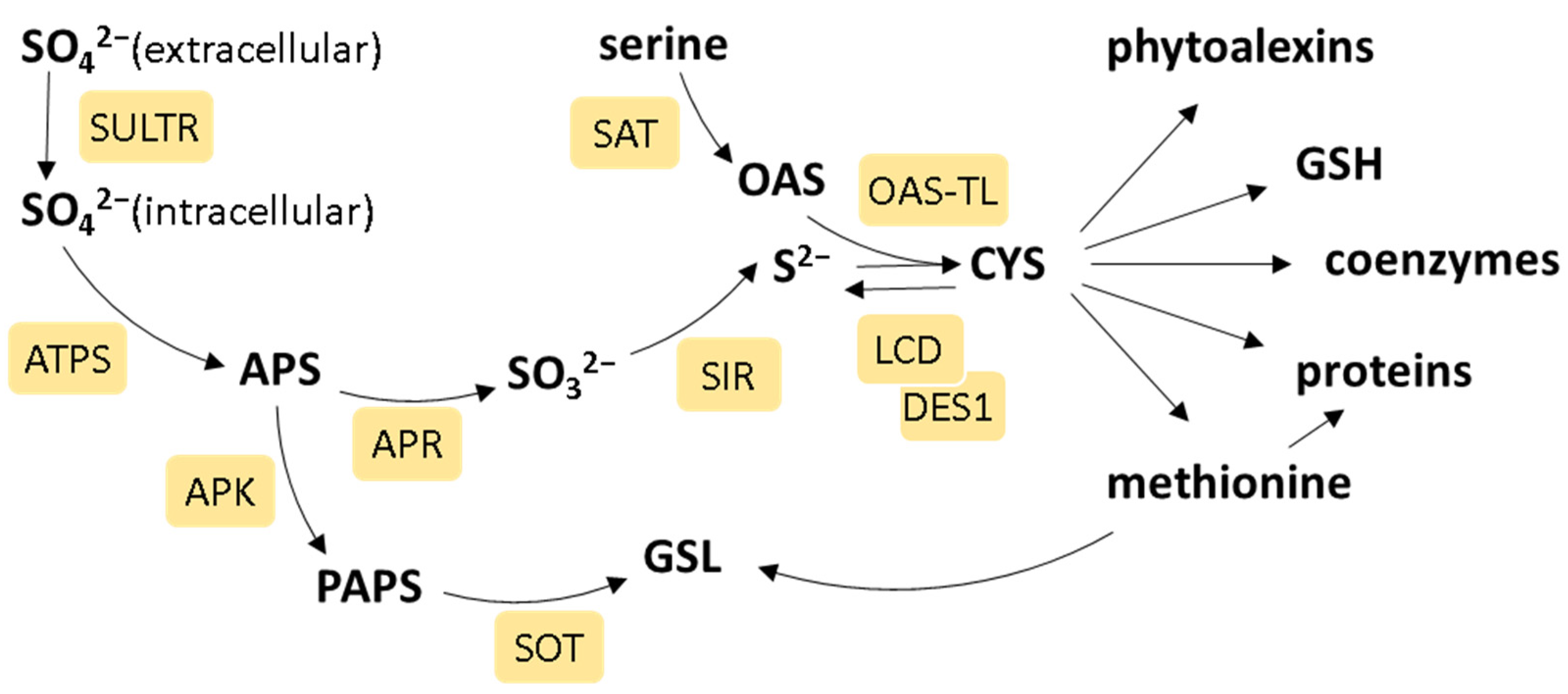
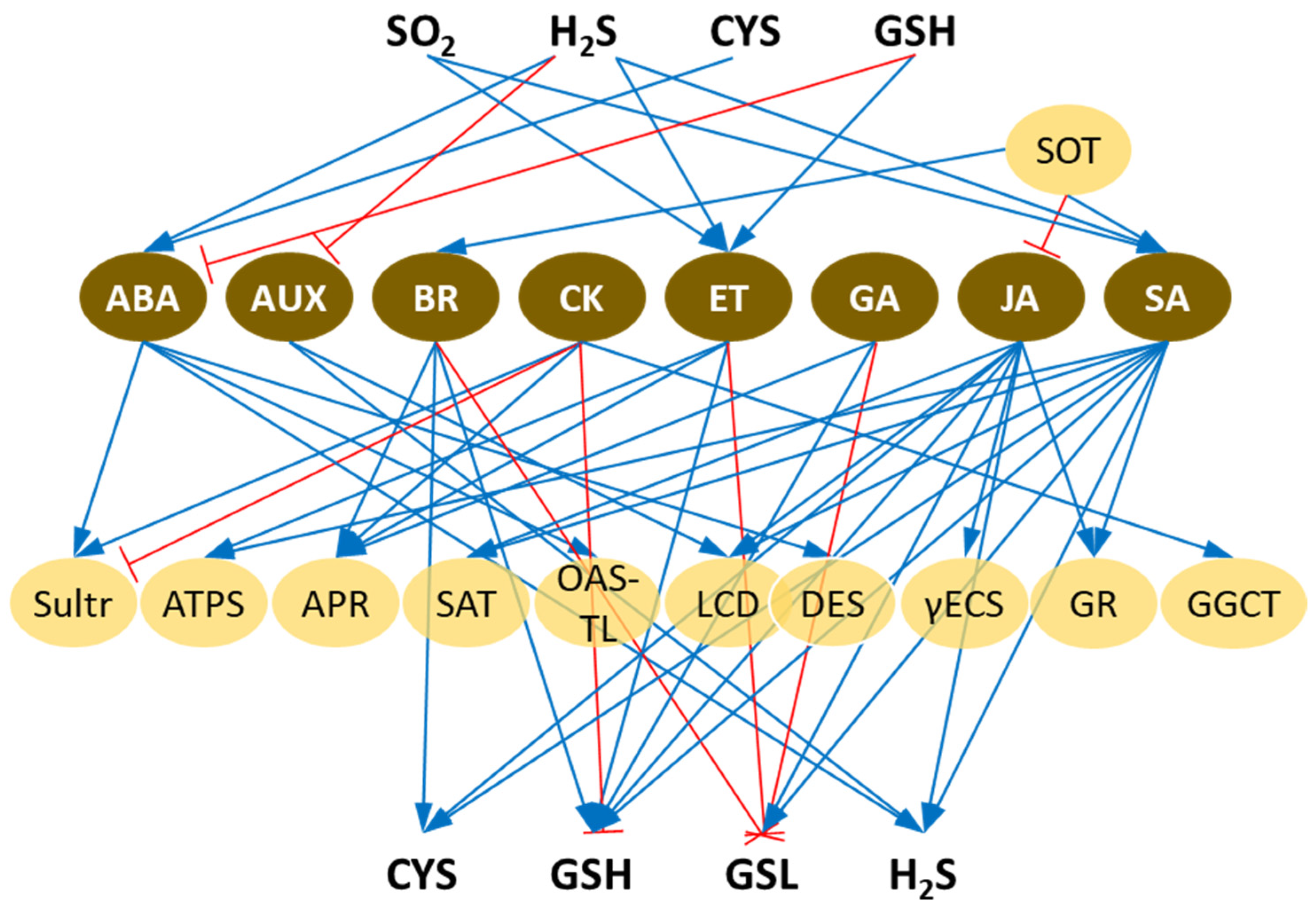
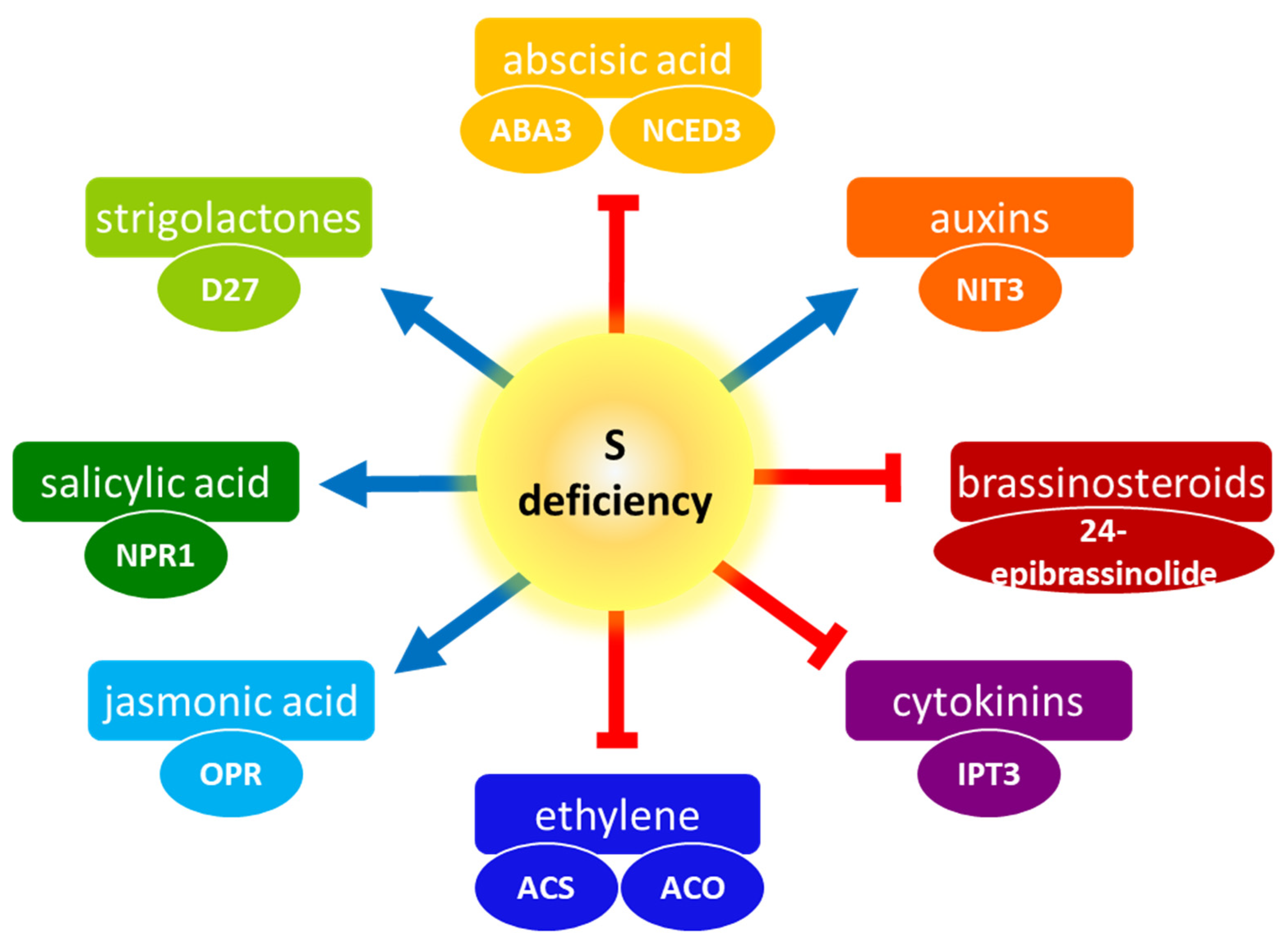
1. Introduction
2. Abscisic Acid Crosstalk with S Metabolism
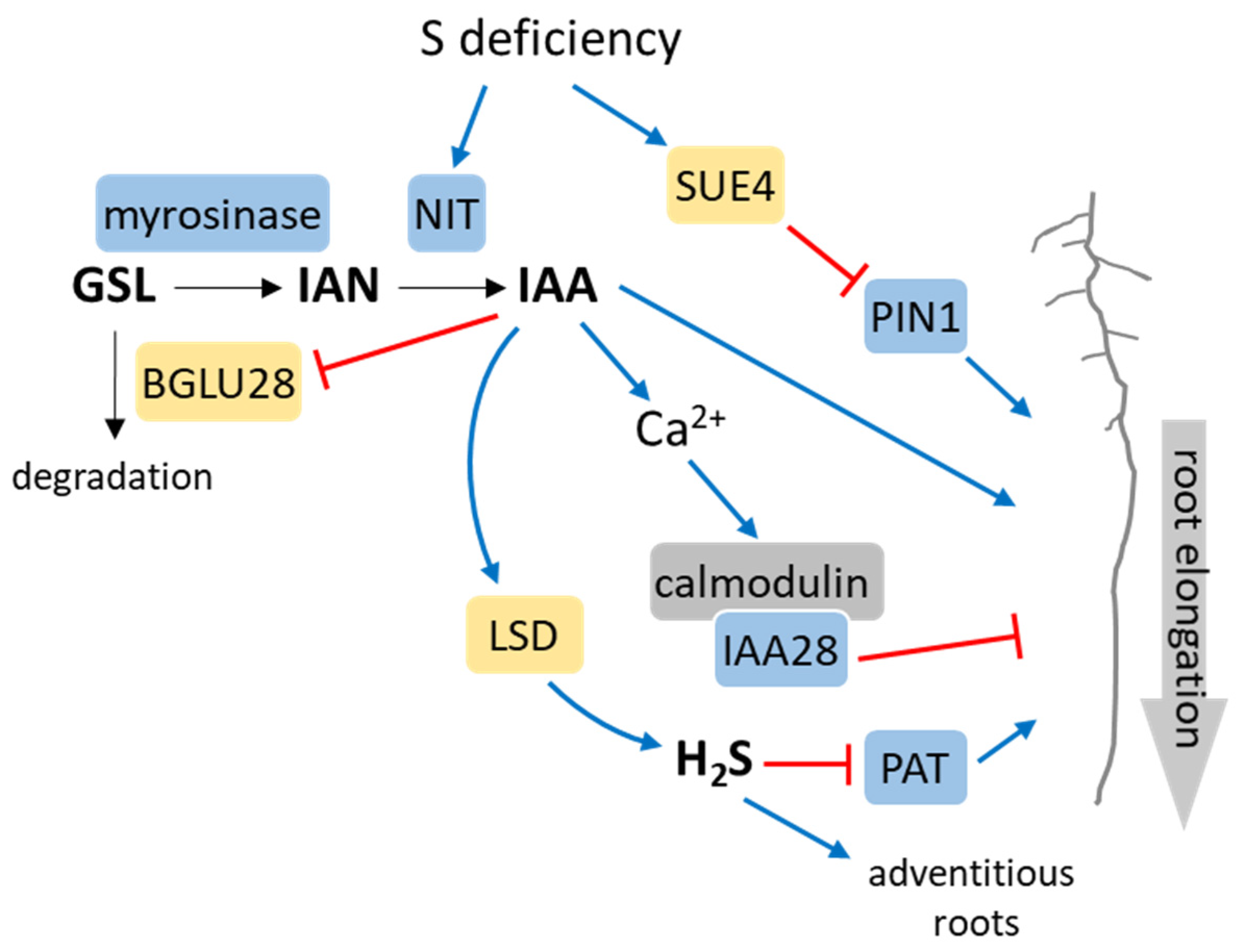
3. Auxin Crosstalk with S Metabolism
4. Brassinosteroid Crosstalk with S Metabolism
5. Cytokinin Crosstalk with S Metabolism
6. Ethylene Crosstalk with S Metabolism
7. Gibberellic Acid Crosstalk with S Metabolism
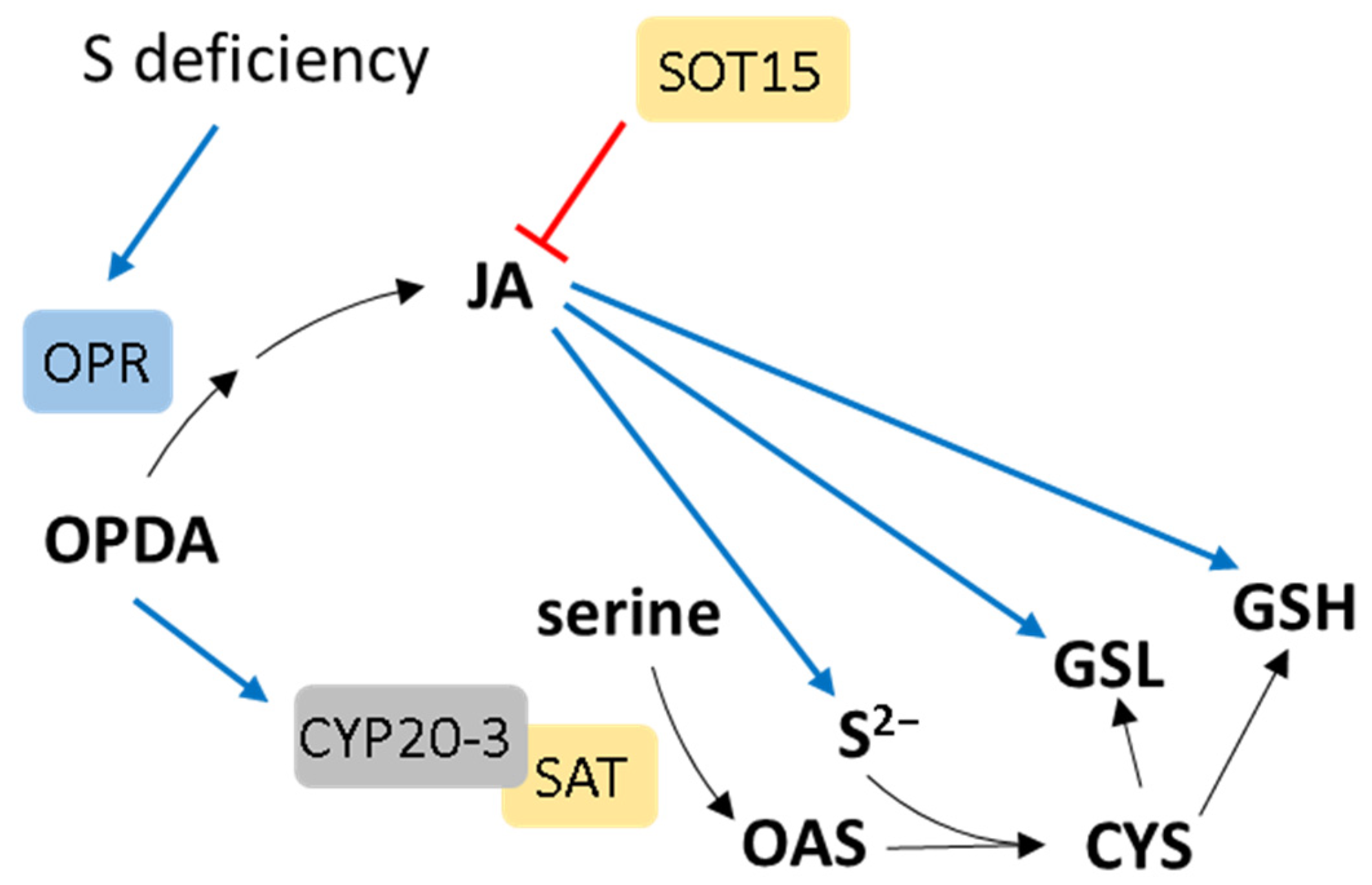
8. Jasmonic Acid Crosstalk with S Metabolism
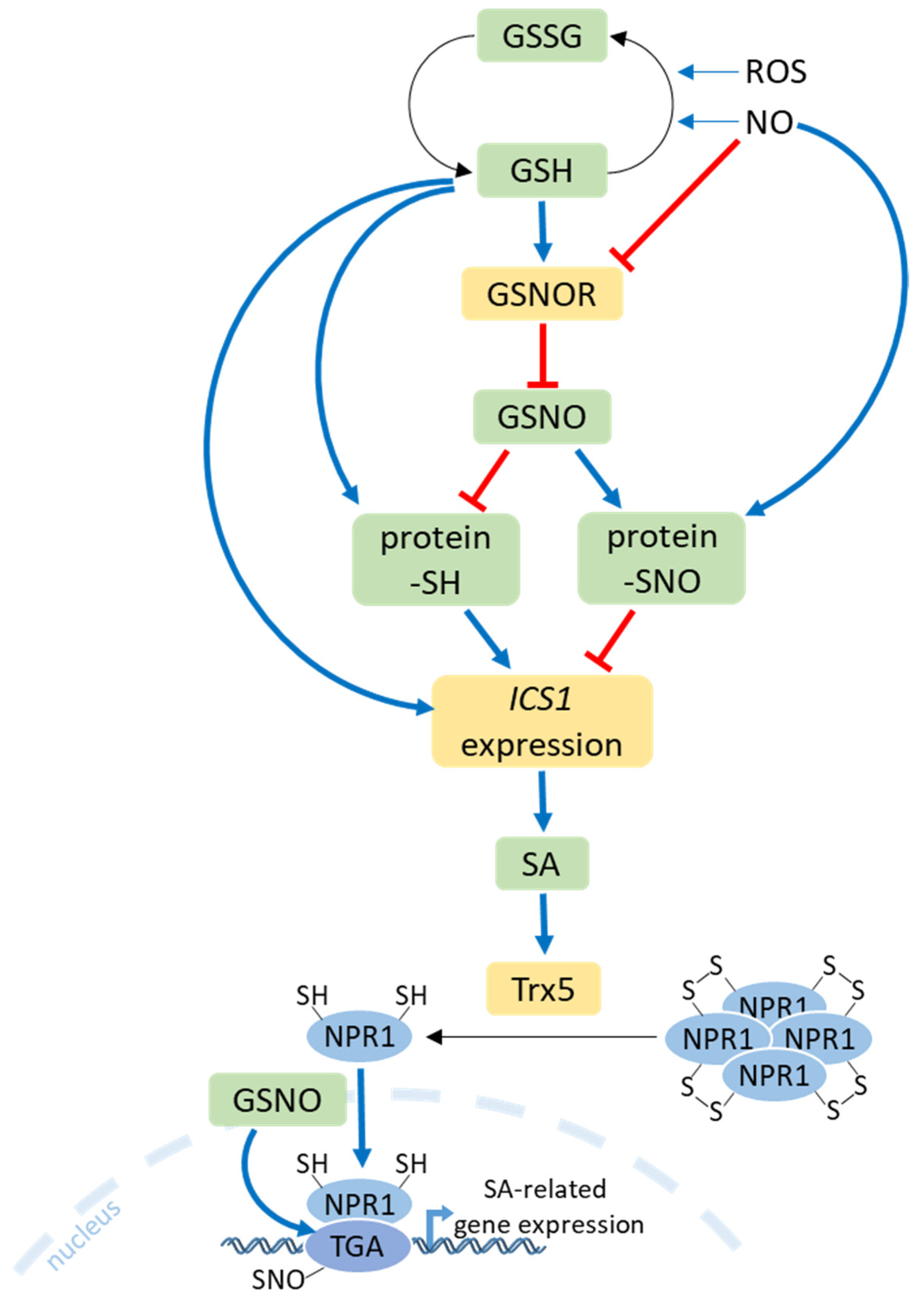
9. Salicylic Acid Crosstalk with S Metabolism
10. Strigolactone Crosstalk with S Metabolism
11. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Takahashi, H.; Kopriva, S.; Giordano, M.; Saito, K.; Hell, R. Sulfur assimilation in photosynthetic organisms: Molecular functions and regulations of transporters and assimilatory enzymes. Annu. Rev. Plant Biol. 2011, 62, 157–184. [Google Scholar] [CrossRef]
- Prioretti, L.; Gontero, B.; Hell, R.; Giordano, M. Diversity and regulation of ATP sulfurylase in photosynthetic organisms. Front. Plant Sci. 2014, 5, 597. [Google Scholar] [CrossRef]
- Mugford, S.G.; Yoshimoto, N.; Reichelt, M.; Wirtz, M.; Hill, L.; Mugford, S.T.; Nakazato, Y.; Noji, M.; Takahashi, H.; Kramell, R.; et al. Disruption of Adenosine-5′-Phosphosulfate Kinase in Arabidopsis Reduces Levels of Sulfated Secondary Metabolites. Plant Cell 2009, 21, 910–927. [Google Scholar] [CrossRef]
- Kopriva, S. Regulation of sulfate assimilation in arabidopsis and beyond. Ann. Bot. 2006, 97, 479–495. [Google Scholar] [CrossRef]
- Nakai, Y.; Maruyama-Nakashita, A. Biosynthesis of Sulfur-Containing Small Biomolecules in Plants. Int. J. Mol. Sci. 2020, 21, 3470. [Google Scholar] [CrossRef]
- Li, Q.; Gao, Y.; Yang, A. Sulfur Homeostasis in Plants. Int. J. Mol. Sci. 2020, 21, 8926. [Google Scholar] [CrossRef]
- Aroca, A.; Zhang, J.; Xie, Y.; Romero, L.C.; Gotor, C. Hydrogen sulfide signaling in plant adaptations to adverse conditions: Molecular mechanisms. J. Exp. Bot. 2021, 72, 5893–5904. [Google Scholar] [CrossRef]
- Aroca, A.; Gotor, C.; Romero, L.C. Hydrogen Sulfide Signaling in Plants: Emerging Roles of Protein Persulfidation. Front. Plant Sci. 2018, 9, 1369. [Google Scholar] [CrossRef]
- Capaldi, F.R.; Gratão, P.L.; Reis, A.R.; Lima, L.W.; Azevedo, R.A. Sulfur Metabolism and Stress Defense Responses in Plants. Trop. Plant Biol. 2015, 8, 60–73. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Bhuyan, M.; Mahmud, J.A.; Nahar, K.; Mohsin, S.M.; Parvin, K.; Fujita, M. Interaction of sulfur with phytohormones and signaling molecules in conferring abiotic stress tolerance to plants. Plant Signal. Behav. 2018, 13, e1477905. [Google Scholar] [CrossRef]
- Kopriva, S.; Malagoli, M.; Takahashi, H. Sulfur nutrition: Impacts on plant development, metabolism, and stress responses. J. Exp. Bot. 2019, 70, 4069–4073. [Google Scholar] [CrossRef]
- Chen, K.; Li, G.J.; Bressan, R.A.; Song, C.P.; Zhu, J.K.; Zhao, Y. Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 2020, 62, 25–54. [Google Scholar] [CrossRef]
- Batool, S.; Uslu, V.V.; Rajab, H.; Ahmad, N.; Waadt, R.; Geiger, D.; Malagoli, M.; Xiang, C.-B.; Hedrich, R.; Rennenberg, H.; et al. Sulfate is Incorporated into Cysteine to Trigger ABA Production and Stomatal Closure. Plant Cell 2018, 30, 2973–2987. [Google Scholar] [CrossRef]
- Malcheska, F.; Ahmad, A.; Batool, S.; Müller, H.M.; Ludwig-Müller, J.; Kreuzwieser, J.; Randewig, D.; Hänsch, R.; Mendel, R.R.; Hell, R.; et al. Drought-Enhanced Xylem Sap Sulfate Closes Stomata by Affecting ALMT12 and Guard Cell ABA Synthesis. Plant Physiol. 2017, 174, 798–814. [Google Scholar] [CrossRef]
- Cao, M.J.; Wang, Z.; Zhao, Q.; Mao, J.L.; Speiser, A.; Wirtz, M.; Hell, R.; Zhu, J.K.; Xiang, C.B. Sulfate availability affects ABA levels and germination response to ABA and salt stress in Arabidopsis thaliana. Plant J. 2014, 77, 604–615. [Google Scholar] [CrossRef]
- Ren, Z.; Wang, R.-Y.; Huang, X.-Y.; Wang, Y. Sulfur Compounds in Regulation of Stomatal Movement. Front. Plant Sci. 2022, 13, 846518. [Google Scholar] [CrossRef]
- Chao, Q.; Rothenberg, M.; Solano, R.; Roman, G.; Terzaghi, W.; Ecker, J.R. Activation of the ethylene gas response pathway in arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 1997, 89, 1133–1144. [Google Scholar] [CrossRef]
- Chen, Z.; Zhao, P.-X.; Miao, Z.-Q.; Qi, G.-F.; Wang, Z.; Yuan, Y.; Ahmad, N.; Cao, M.-J.; Hell, R.; Wirtz, M.; et al. SULTR3s Function in Chloroplast Sulfate Uptake and Affect ABA Biosynthesis and the Stress Response. Plant Physiol. 2019, 180, 593–604. [Google Scholar] [CrossRef]
- Ruiz, J.; Blumwald, E. Salinity-induced glutathione synthesis in Brassica napus. Planta 2002, 214, 965. [Google Scholar] [CrossRef]
- Koprivova, A.; North, K.A.; Kopriva, S. Complex signaling network in regulation of adenosine 5′-phosphosulfate reductase by salt stress in arabidopsis roots. Plant Physiol. 2008, 146, 1408–1420. [Google Scholar] [CrossRef]
- Zhu, J.-K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef]
- Barroso, C.; Romero, L.C.; Cejudo, F.J.; Vega, J.M.; Gotor, C. Salt-specific regulation of the cytosolic O-acetylserine(thiol)lyase gene from Arabidopsis thaliana is dependent on abscisic acid. Plant Mol. Biol. 1999, 40, 729–736. [Google Scholar] [CrossRef]
- Liu, H.; Xue, S. Interplay between hydrogen sulfide and other signaling molecules in the regulation of guard cell signaling and abiotic/biotic stress response. Plant Commun. 2021, 2, 100179. [Google Scholar] [CrossRef]
- Scuffi, D.; Alvarez, C.; Laspina, N.; Gotor, C.; Lamattina, L.; Garcia-Mata, C. Hydrogen sulfide generated by L-cysteine desulfhydrase acts upstream of nitric oxide to modulate abscisic acid-dependent stomatal closure. Plant Physiol. 2014, 166, 2065–2076. [Google Scholar] [CrossRef]
- Aroca, A.; Benito, J.M.; Gotor, C.; Romero, L.C. Persulfidation proteome reveals the regulation of protein function by hydrogen sulfide in diverse biological processes in Arabidopsis. J. Exp. Bot. 2017, 68, 4915–4927. [Google Scholar] [CrossRef]
- Chen, S.; Jia, H.; Wang, X.; Shi, C.; Wang, X.; Ma, P.; Wang, J.; Ren, M.; Li, J. Hydrogen Sulfide Positively Regulates Abscisic Acid Signaling through Persulfidation of SnRK2.6 in Guard Cells. Mol. Plant 2020, 13, 732–744. [Google Scholar] [CrossRef]
- Zhou, H.; Zhou, Y.; Zhang, F.; Guan, W.; Su, Y.; Yuan, X.; Xie, Y. Persulfidation of Nitrate Reductase 2 Is Involved in l-Cysteine Desulfhydrase-Regulated Rice Drought Tolerance. Int. J. Mol. Sci. 2021, 22, 12119. [Google Scholar] [CrossRef]
- Zhang, J.; Zhou, H.; Zhou, M.; Ge, Z.; Zhang, F.; Foyer, C.H.; Yuan, X.; Xie, Y. The coordination of guard-cell autonomous ABA synthesis and DES1 function regulates plant water deficit responses. J. Adv. Res. 2021, 27, 191–197. [Google Scholar] [CrossRef]
- Zhou, M.; Zhang, J.; Shen, J.; Zhou, H.; Zhao, D.; Gotor, C.; Romero, L.C.; Fu, L.; Li, Z.; Yang, J.; et al. Hydrogen sulfide-linked persulfidation of ABI4 controls ABA responses through the transactivation of MAPKKK18 in Arabidopsis. Mol. Plant 2021, 14, 921–936. [Google Scholar] [CrossRef]
- Li, K.; Yang, F.; Miao, Y.; Song, C.-P. Abscisic acid signaling is involved in regulating the mitogen-activated protein kinase cascade module, AIK1-MKK5-MPK6. Plant Signal. Behav. 2017, 12, e1321188. [Google Scholar] [CrossRef] [PubMed]
- Okuma, E.; Jahan, S.; Munemasa, S.; Hossain, M.A.; Muroyama, D.; Islam, M.M.; Ogawa, K.; Watanabe-Sugimoto, M.; Nakamura, Y.; Shimoishi, Y.; et al. Negative regulation of abscisic acid-induced stomatal closure by glutathione in Arabidopsis. J. Plant Physiol. 2011, 168, 2048–2055. [Google Scholar] [CrossRef] [PubMed]
- Noctor, G.; Mhamdi, A.; Chaouch, S.; Han, Y.; Neukermans, J.; Marquez-Garcia, B.; Queval, G.; Foyer, C.H. Glutathione in plants: An integrated overview. Plant Cell Environ. 2011, 35, 454–484. [Google Scholar] [CrossRef] [PubMed]
- Lu, B.; Luo, X.; Gong, C.; Bai, J. Overexpression of gamma-glutamylcysteine synthetase gene from Caragana korshinskii decreases stomatal density and enhances drought tolerance. BMC Plant Biol. 2021, 21, 444. [Google Scholar] [CrossRef] [PubMed]
- Bekturova, A.; Oshanova, D.; Tiwari, P.; Nurbekova, Z.; Kurmanbayeva, A.; Soltabayeva, A.; Yarmolinsky, D.; Srivastava, S.; Turecková, V.; Strnad, M.; et al. Adenosine 5′ phosphosulfate reductase and sulfite oxidase regulate sulfite-induced water loss in Arabidopsis. J. Exp. Bot. 2021, 72, 6447–6466. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.-H.; Jiang, H.-W.; Hsieh, E.-J.; Chen, H.-Y.; Chien, C.-T.; Hsieh, H.-L.; Lin, T.-P. Drought and salt stress tolerance of an Arabidopsis glutathione S-transferase U17 knockout mutant are attributed to the combined effect of glutathione and abscisic acid. Plant Physiol. 2012, 158, 340–351. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.-W.; Liu, M.-J.; Chen, I.-C.; Huang, C.-H.; Chao, L.-Y.; Hsieh, H.-L. A Glutathione S-transferase regulated by light and hormones participates in the modulation of arabidopsis seedling development. Plant Physiol. 2010, 154, 1646–1658. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.; Lv, D.; Wang, P.; Wang, X.-C.; Chen, J.; Miao, C.; Song, C.-P. An Arabidopsis Glutathione Peroxidase Functions as Both a Redox Transducer and a Scavenger in Abscisic Acid and Drought Stress Responses. Plant Cell 2006, 18, 2749–2766. [Google Scholar] [CrossRef]
- Ruan, M.-B.; Yang, Y.-L.; Li, K.-M.; Guo, X.; Wang, B.; Yu, X.-L.; Peng, M. Identification and characterization of drought-responsive CC-type glutaredoxins from cassava cultivars reveals their involvement in ABA signalling. BMC Plant Biol. 2018, 18, 329. [Google Scholar] [CrossRef]
- Caumon, H.; Vernoux, T. A matter of time: Auxin signaling dynamics and the regulation of auxin responses during plant development. J. Exp. Bot. 2023, 74, 3887–3902. [Google Scholar] [CrossRef]
- Nikiforova, V.; Freitag, J.; Kempa, S.; Adamik, M.; Hesse, H.; Hoefgen, R. Transcriptome analysis of sulfur depletion in Arabidopsis thaliana: Interlacing of biosynthetic pathways provides response specificity. Plant J. 2003, 33, 633–650. [Google Scholar] [CrossRef]
- Abdel-Massih, R.M.; Debs, E.; Othman, L.; Attieh, J.; Cabrerizo, F.M. Glucosinolates, a natural chemical arsenal: More to tell than the myrosinase story. Front. Microbiol. 2023, 14, 1130208. [Google Scholar] [CrossRef] [PubMed]
- Vorwerk, S.; Biernacki, S.; Hillebrand, H.; Janzik, I.; Muller, A.; Weiler, E.W.; Piotrowski, M. Enzymatic characterization of the recombinant Arabidopsis thaliana nitrilase subfamily encoded by the NIT2/NIT1/NIT3-gene cluster. Planta 2001, 212, 508–516. [Google Scholar] [CrossRef] [PubMed]
- Kutz, A.; Müller, A.; Hennig, P.; Kaiser, W.M.; Piotrowski, M.; Weiler, E.W. A role for nitrilase 3 in the regulation of root morphology in sulphur-starving Arabidopsis thaliana. Plant J. 2002, 30, 95–106. [Google Scholar] [CrossRef] [PubMed]
- Falkenberg, B.; Witt, I.; Zanor, M.I.; Steinhauser, D.; Mueller-Roeber, B.; Hesse, H.; Hoefgen, R. Transcription factors relevant to auxin signalling coordinate broad-spectrum metabolic shifts including sulphur metabolism. J. Exp. Bot. 2008, 59, 2831–2846. [Google Scholar] [CrossRef] [PubMed]
- Dan, H.; Yang, G.; Zheng, Z.-L. A negative regulatory role for auxin in sulphate deficiency response in Arabidopsis thaliana. Plant Mol. Biol. 2007, 63, 221–235. [Google Scholar] [CrossRef] [PubMed]
- Hoefgen, R.; Nikiforova, V.J. Metabolomics integrated with transcriptomics: Assessing systems response to sulfur-deficiency stress. Physiol. Plant. 2008, 132, 190–198. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Wu, Y.; Gao, L.; Ma, J.; Li, C.; Xiang, C. Sulfur nutrient availability regulates root elongation by affecting root indole-3-acetic acid levels and the stem cell niche. J. Integr. Plant Biol. 2014, 56, 1151–1163. [Google Scholar] [CrossRef]
- Zhao, Q.; Zhao, P.; Wu, Y.; Zhong, C.; Liao, H.; Li, C.; Fu, X.; Fang, P.; Xu, P.; Xiang, C. SUE4, a novel PIN1-interacting membrane protein, regulates acropetal auxin transport in response to sulfur deficiency. New Phytol. 2023, 237, 78–87. [Google Scholar] [CrossRef] [PubMed]
- Rogg, L.E.; Lasswell, J.; Bartel, B. A gain-of-function mutation in IAA28 suppresses lateral root development. Plant Cell 2001, 13, 465–480. [Google Scholar] [CrossRef]
- Shane, M.W.; Stigter, K.; Fedosejevs, E.T.; Plaxton, W.C. Senescence-inducible cell wall and intracellular purple acid phosphatases: Implications for phosphorus remobilization in Hakea prostrata (Proteaceae) and Arabidopsis thaliana (Brassicaceae). J. Exp. Bot. 2014, 65, 6097–6106. [Google Scholar] [CrossRef]
- Kasajima, I.; Ohkama-Ohtsu, N.; Ide, Y.; Hayashi, H.; Yoneyama, T.; Suzuki, Y.; Naito, S.; Fujiwara, T. The BIG gene is involved in regulation of sulfur deficiency-responsive genes in Arabidopsis thaliana. Physiol. Plant. 2007, 129, 351–363. [Google Scholar] [CrossRef]
- Normanly, J. Approaching cellular and molecular resolution of auxin biosynthesis and metabolism. Cold Spring Harb. Perspect. Biol. 2010, 2, a001594. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Luo, Q.; Wang, R.; Xu, J. Hydrogen sulfide toxicity inhibits primary root growth through the ROS-NO pathway. Sci. Rep. 2017, 7, 868. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Hu, Y.; Fan, T.; Li, J. Hydrogen sulfide modulates actin-dependent auxin transport via regulating ABPs results in changing of root development in Arabidopsis. Sci. Rep. 2015, 5, 8251. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.-J.; Shi, Z.-Q.; Gan, L.-J.; Chen, J. Hydrogen sulfide is a novel gasotransmitter with pivotal role in regulating lateral root formation in plants. Plant Signal. Behav. 2014, 9, e29127. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Maruyama-Nakashita, A.; Nakamura, Y.; Watanabe-Takahashi, A.; Inoue, E.; Yamaya, T.; Takahashi, H. Identification of a novel cis-acting element conferring sulfur deficiency response in Arabidopsis roots. Plant J. 2005, 42, 305–314. [Google Scholar] [CrossRef]
- Wei, Z.; Li, J. Brassinosteroids Regulate Root Growth, Development, and Symbiosis. Mol. Plant 2016, 9, 86–100. [Google Scholar] [CrossRef]
- Sharma, I.; Kaur, N.; Pati, P.K. Brassinosteroids: A Promising Option in Deciphering Remedial Strategies for Abiotic Stress Tolerance in Rice. Front. Plant Sci. 2017, 8, 2151. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Lee, J.; Kim, H.; Chae, W.B.; Kim, S.-J.; Lim, Y.P.; Oh, M.-H. Brassinosteroids regulate glucosinolate biosynthesis in Arabidopsis thaliana. Physiol. Plant. 2018, 163, 450–458. [Google Scholar] [CrossRef]
- Lv, Q.; Li, X.; Fan, B.; Zhu, C.; Chen, Z. The Cellular and Subcellular Organization of the Glucosinolate–Myrosinase System against Herbivores and Pathogens. Int. J. Mol. Sci. 2022, 23, 1577. [Google Scholar] [CrossRef]
- Wang, M.; Cai, C.; Li, Y.; Tao, H.; Meng, F.; Sun, B.; Miao, H.; Wang, Q. Brassinosteroids fine-tune secondary and primary sulfur metabolism through BZR1-mediated transcriptional regulation. J. Integr. Plant Biol. 2023, 65, 1153–1169. [Google Scholar] [CrossRef] [PubMed]
- Nazir, F.; Jahan, B.; Kumari, S.; Iqbal, N.; Albaqami, M.; Sofo, A.; Khan, M.I.R. Brassinosteroid modulates ethylene synthesis and antioxidant metabolism to protect rice (Oryza sativa) against heat stress-induced inhibition of source—sink capacity and photosynthetic and growth attributes. J. Plant Physiol. 2023, 289, 154096. [Google Scholar] [CrossRef] [PubMed]
- Marsolais, F.; Boyd, J.; Paredes, Y.; Schinas, A.-M.; Garcia, M.; Elzein, S.; Varin, L. Molecular and biochemical characterization of two brassinosteroid sulfotransferases from Arabidopsis, AtST4a (At2g14920) and AtST1 (At2g03760). Planta 2007, 225, 1233–1244. [Google Scholar] [CrossRef] [PubMed]
- Rouleau, M.; Marsolais, F.; Richard, M.; Nicolle, L.; Voigt, B.; Adam, G.; Varin, L. Inactivation of Brassinosteroid Biological Activity by a Salicylate-inducible Steroid Sulfotransferase from Brassica napus. J. Biol. Chem. 1999, 274, 20925. [Google Scholar] [CrossRef] [PubMed]
- Sandhu, K.S.; Neff, M.M. The Arabidopsis geneATST4ais not a typical brassinosteroids catabolic gene. Plant Signal. Behav. 2013, 8, e26847. [Google Scholar] [CrossRef]
- Symons, G.M.; Ross, J.J.; Jager, C.E.; Reid, J.B. Brassinosteroid transport. J. Exp. Bot. 2008, 59, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Argueso, C.T.; Kieber, J.J. Cytokinin: From autoclaved DNA to two-component signaling. Plant Cell 2024. [Google Scholar] [CrossRef] [PubMed]
- Hirose, N.; Takei, K.; Kuroha, T.; Kamada-Nobusada, T.; Hayashi, H.; Sakakibara, H. Regulation of cytokinin biosynthesis, compartmentalization and translocation. J. Exp. Bot. 2008, 59, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Honsel, A.; Kojima, M.; Haas, R.; Frank, W.; Sakakibara, H.; Herschbach, C.; Rennenberg, H. Sulphur limitation and early sulphur deficiency responses in poplar: Significance of gene expression, metabolites, and plant hormones. J. Exp. Bot. 2012, 63, 1873–1893. [Google Scholar] [CrossRef]
- Bhargava, A.; Clabaugh, I.; To, J.P.; Maxwell, B.B.; Chiang, Y.-H.; Schaller, G.E.; Loraine, A.; Kieber, J.J. Identification of cytokinin-responsive genes using microarray meta-analysis and RNA-Seq in Arabidopsis. Plant Physiol. 2013, 162, 272–294. [Google Scholar] [CrossRef]
- Werner, T.; Nehnevajova, E.; Köllmer, I.; Novák, O.; Strnad, M.; Krämer, U.; Schmülling, T. Root-specific reduction of cytokinin causes enhanced root growth, drought tolerance, and leaf mineral enrichment in Arabidopsis and tobacco. Plant Cell 2010, 22, 3905–3920. [Google Scholar] [CrossRef] [PubMed]
- Ohkama, N.; Takei, K.; Sakakibara, H.; Hayashi, H.; Yoneyama, T.; Fujiwara, T. Regulation of sulfur-responsive gene expression by exogenously applied cytokinins in Arabidopsis thaliana. Plant Cell Physiol. 2002, 43, 1493–1501. [Google Scholar] [CrossRef]
- Ahanger, M.A.; Alyemeni, M.N.; Wijaya, L.; Alamri, S.A.; Alam, P.; Ashraf, M.; Ahmad, P. Potential of exogenously sourced kinetin in protecting Solanum lycopersicum from NaCl-induced oxidative stress through up-regulation of the antioxidant system, ascorbate-glutathione cycle and glyoxalase system. PLoS ONE 2018, 13, e0202175. [Google Scholar] [CrossRef] [PubMed]
- Pavlů, J.; Kerchev, P.; Černý, M.; Novák, J.; Berka, M.; Jobe, T.O.; Ramos, J.M.L.; Saiz-Fernández, I.; Rashotte, A.M.; Kopriva, S.; et al. Cytokinin modulates the metabolic network of sulfur and glutathione. J. Exp. Bot. 2022, 73, 7417–7433. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, N.; Khan, N.A.; Ferrante, A.; Trivellini, A.; Francini, A.; Khan, M.I.R. Ethylene Role in Plant Growth, Development and Senescence: Interaction with Other Phytohormones. Front. Plant Sci. 2017, 8, 475. [Google Scholar] [CrossRef]
- Sauter, M.; Moffatt, B.; Saechao, M.C.; Hell, R.; Wirtz, M. Methionine salvage and S-adenosylmethionine: Essential links between sulfur, ethylene and polyamine biosynthesis. Biochem. J. 2013, 451, 145–154. [Google Scholar] [CrossRef]
- Hatzfeld, Y.; Maruyama, A.; Schmidt, A.; Noji, M.; Ishizawa, K.; Saito, K. beta-Cyanoalanine synthase is a mitochondrial cysteine synthase-like protein in spinach and Arabidopsis. Plant Physiol. 2000, 123, 1163–1171. [Google Scholar] [CrossRef] [PubMed]
- Ghanta, S.; Bhattacharyya, D.; Sinha, R.; Banerjee, A.; Chattopadhyay, S. Nicotiana tabacum overexpressing gamma-ECS exhibits biotic stress tolerance likely through NPR1-dependent salicylic acid-mediated pathway. Planta 2011, 233, 895–910. [Google Scholar] [CrossRef] [PubMed]
- Datta, R.; Kumar, D.; Sultana, A.; Hazra, S.; Bhattacharyya, D.; Chattopadhyay, S. Glutathione Regulates 1-Aminocyclopropane-1-Carboxylate Synthase Transcription via WRKY33 and 1-Aminocyclopropane-1-Carboxylate Oxidase by Modulating Messenger RNA Stability to Induce Ethylene Synthesis during Stress. Plant Physiol. 2015, 169, 2963–2981. [Google Scholar]
- Moniuszko, G.; Skoneczny, M.; Zientara-Rytter, K.; Wawrzyńska, A.; Głów, D.; Cristescu, S.M.; Harren, F.J.M.; Sirko, A. Tobacco LSU-like protein couples sulphur-deficiency response with ethylene signalling pathway. J. Exp. Bot. 2013, 64, 5173–5182. [Google Scholar] [CrossRef]
- Sirko, A.; Wawrzynska, A.; Rodriguez, M.C.; Sektas, P. The family of LSU-like proteins. Front. Plant Sci. 2014, 5, 774. [Google Scholar] [CrossRef]
- Canales, J.; Arenas, M.A.; Medina, J.; Vidal, E.A. A Revised View of the LSU Gene Family: New Functions in Plant Stress Responses and Phytohormone Signaling. Int. J. Mol. Sci. 2023, 24, 2819. [Google Scholar] [CrossRef]
- Lewandowska, M.; Wawrzyńska, A.; Moniuszko, G.; Łukomska, J.; Zientara, K.; Piecho, M.; Hodurek, P.; Zhukov, I.; Liszewska, F.; Nikiforova, V.; et al. A Contribution to Identification of novel regulators of plant response to sulfur deficiency: Characteristics of a tobacco gene UP9C, its protein product and the effects of UP9C silencing. Mol. Plant 2010, 3, 347–360. [Google Scholar] [CrossRef]
- Iven, V.; Vanbuel, I.; Hendrix, S.; Cuypers, A. The glutathione-dependent alarm triggers signalling responses involved in plant acclimation to cadmium. J. Exp. Bot. 2023, 74, 3300–3312. [Google Scholar] [CrossRef]
- Zhang, L.; Kawaguchi, R.; Enomoto, T.; Nishida, S.; Burow, M.; Maruyama-Nakashita, A. Glucosinolate Catabolism Maintains Glucosinolate Profiles and Transport in Sulfur-Starved Arabidopsis. Plant Cell Physiol. 2023, 64, 1534–1550. [Google Scholar] [CrossRef] [PubMed]
- Hildreth, S.B.; Foley, E.E.; Muday, G.K.; Helm, R.F.; Winkel, B.S.J. The dynamic response of the Arabidopsis root metabolome to auxin and ethylene is not predicted by changes in the transcriptome. Sci. Rep. 2020, 10, 679. [Google Scholar] [CrossRef] [PubMed]
- Uhrig, R.G.; O’Leary, B.; Spang, H.E.; MacDonald, J.A.; She, Y.-M.; Plaxton, W.C. Coimmunopurification of phosphorylated bacterial- and plant-type phosphoenolpyruvate carboxylases with the plastidial pyruvate dehydrogenase complex from developing castor oil seeds. Plant Physiol. 2008, 146, 1346–1357. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jahan, B.; Iqbal, N.; Fatma, M.; Sehar, Z.; Masood, A.; Sofo, A.; D’Ippolito, I.; Khan, N.A. Ethylene Supplementation Combined with Split Application of Nitrogen and Sulfur Protects Salt-Inhibited Photosynthesis through Optimization of Proline Metabolism and Antioxidant System in Mustard (Brassica juncea L.). Plants 2021, 10, 1303. [Google Scholar] [CrossRef]
- Iqbal, N.; Masood, A.; Khan, M.I.R.; Asgher, M.; Fatma, M.; Khan, N.A. Cross-talk between sulfur assimilation and ethylene signaling in plants. Plant Signal. Behav. 2013, 8, e22478. [Google Scholar] [CrossRef]
- Al Murad, M.; Razi, K.; Benjamin, L.K.; Lee, J.H.; Kim, T.H.; Muneer, S. Ethylene regulates sulfur acquisition by regulating the expression of sulfate transporter genes in oilseed rape. Physiol. Plant. 2021, 171, 533–545. [Google Scholar] [CrossRef]
- Meldau, D.G.; Meldau, S.; Hoang, L.H.; Underberg, S.; Wunsche, H.; Baldwin, I.T. Dimethyl disulfide produced by the naturally associated bacterium bacillus sp B55 promotes nicotiana attenuata growth by enhancing sulfur nutrition. Plant Cell 2013, 25, 2731–2747. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Li, J.; Li, Z.; Pei, Y. Hydrogen sulfide inhibits ethylene-induced petiole abscission in tomato (Solanum lycopersicum L.). Hortic. Res. 2020, 7, 14. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.; Hu, K.D.; Wang, S.S.; Hu, L.Y.; Chen, X.Y.; Li, Y.H.; Yang, Y.; Yang, F.; Zhang, H. Hydrogen sulfide alleviates postharvest ripening and senescence of banana by antagonizing the effect of ethylene. PLoS ONE 2017, 12, e0180113. [Google Scholar] [CrossRef] [PubMed]
- Giraud, E.; Ivanova, A.; Gordon, C.S.; Whelan, J.; Considine, M.J. Sulphur dioxide evokes a large scale reprogramming of the grape berry transcriptome associated with oxidative signalling and biotic defence responses. Plant Cell Environ. 2012, 35, 405–417. [Google Scholar] [CrossRef] [PubMed]
- Dietzen, C.; Koprivova, A.; Whitcomb, S.J.; Langen, G.; Jobe, T.O.; Hoefgen, R.; Kopriva, S. The Transcription Factor EIL1 Participates in the Regulation of Sulfur-Deficiency Response. Plant Physiol. 2020, 184, 2120–2136. [Google Scholar] [CrossRef] [PubMed]
- Maruyama-Nakashita, A.; Nakamura, Y.; Tohge, T.; Saito, K.; Takahashi, H. Arabidopsis SLIM1 Is a central transcriptional regulator of plant sulfur response and metabolism. Plant Cell 2006, 18, 3235–3251. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.H.; Islam, S.; Mohammad, F.; Siddiqui, M.H. Gibberellic Acid: A Versatile Regulator of Plant Growth, Development and Stress Responses. J. Plant Growth Regul. 2023, 42, 7352–7373. [Google Scholar] [CrossRef]
- Masood, A.; Khan, M.I.R.; Fatma, M.; Asgher, M.; Per, T.S.; Khan, N.A. Involvement of ethylene in gibberellic acid-induced sulfur assimilation, photosynthetic responses, and alleviation of cadmium stress in mustard. Plant Physiol. Biochem. 2016, 104, 1–10. [Google Scholar] [CrossRef]
- Hasan, S.; Sehar, Z.; Khan, N.A. Gibberellic Acid and Sulfur-Mediated Reversal of Cadmium-Inhibited Photosynthetic Performance in Mungbean (Vigna radiata L.) Involves Nitric Oxide. J. Plant Growth Regul. 2020, 39, 1605–1615. [Google Scholar] [CrossRef]
- Wang, S.-S.; Zhang, Y.-X.; Yang, F.; Huang, Z.-Q.; Tang, J.; Hu, K.-D.; Zhang, H. Sulfur dioxide alleviates programmed cell death in barley aleurone by acting as an antioxidant. PLoS ONE 2017, 12, e0188289. [Google Scholar] [CrossRef]
- Yu, Q.; Hao, G.; Zhou, J.; Wang, J.; Evivie, E.R.; Li, J. Identification and expression pattern analysis of BoMYB51 involved in indolic glucosinolate biosynthesis from broccoli (Brassica oleracea var. italica). Biochem. Biophys. Res. Commun. 2018, 501, 598–604. [Google Scholar] [CrossRef] [PubMed]
- Miao, H.-Y.; Wang, M.-Y.; Chang, J.-Q.; Tao, H.; Sun, B.; Wang, Q.-M. Effects of glucose and gibberellic acid on glucosinolate content and antioxidant properties of Chinese kale sprouts. J. Zhejiang Univ. Sci. B 2017, 18, 1093–1100. [Google Scholar] [CrossRef] [PubMed]
- Bhat, B.A.T.L.; Nissar, S.; Hamdani, S.S.; Dar, M.A.; Mehraj, S.; Dar, T.H. Role of Methyl Jasmonate in Mitigating Plant Stress and Its Interaction with Salicylic Acid. In Plant Abiotic Stress Physiology; Apple Academic Press: Burlington, ON, Canada, 2022; p. 23. [Google Scholar]
- Frerigmann, H.; Gigolashvili, T. MYB34, MYB51, and MYB122 distinctly regulate Indolic Glucosinolate biosynthesis in Arabidopsis thaliana. Mol. Plant 2014, 7, 814–828. [Google Scholar] [CrossRef] [PubMed]
- Frerigmann, H.; Berger, B.; Gigolashvili, T. bHLH05 is an interaction partner of MYB51 and a novel regulator of glucosinolate biosynthesis in Arabidopsis. Plant Physiol. 2014, 166, 349–369. [Google Scholar] [CrossRef] [PubMed]
- Jost, R.; Altschmied, L.; Bloem, E.; Bogs, J.; Gershenzon, J.; Hähnel, U.; Hänsch, R.; Hartmann, T.; Kopriva, S.; Kruse, C.; et al. Expression profiling of metabolic genes in response to methyl jasmonate reveals regulation of genes of primary and secondary sulfur-related pathways in Arabidopsis thaliana. Photosynth. Res. 2005, 86, 491–508. [Google Scholar] [CrossRef]
- Han, Y.; Mhamdi, A.; Chaouch, S.; Noctor, G. Regulation of basal and oxidative stress-triggered jasmonic acid-related gene expression by glutathione. Plant Cell Environ. 2013, 36, 1135–1146. [Google Scholar] [CrossRef]
- Maruyama-Nakashita, A.; Inoue, E.; Watanabe-Takahashi, A.; Yamaya, T.; Takahashi, H. Transcriptome profiling of sulfur-responsive genes in Arabidopsis reveals global effects of sulfur nutrition on multiple metabolic pathways. Plant Physiol. 2003, 132, 597–605. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-W.; Li, W.; Viehhauser, A.; He, B.; Kim, S.; Nilsson, A.K.; Andersson, M.X.; Kittle, J.D.; Ambavaram, M.M.R.; Luan, S.; et al. Cyclophilin 20-3 relays a 12-oxo-phytodienoic acid signal during stress responsive regulation of cellular redox homeostasis. Proc. Natl. Acad. Sci. USA 2013, 110, 9559–9564. [Google Scholar] [CrossRef] [PubMed]
- Miersch, O.; Neumerkel, J.; Dippe, M.; Stenzel, I.; Wasternack, C. Hydroxylated jasmonates are commonly occurring metabolites of jasmonic acid and contribute to a partial switch-off in jasmonate signaling. New Phytol. 2008, 177, 114–127. [Google Scholar] [CrossRef]
- Shan, C.; Wang, T.; Zhou, Y.; Wang, W. Hydrogen sulfide is involved in the regulation of ascorbate and glutathione metabolism by jasmonic acid in Arabidopsis thaliana. Biol. Plant. 2018, 62, 188–193. [Google Scholar] [CrossRef]
- Deng, G.; Zhou, L.; Wang, Y.; Zhang, G.; Chen, X. Hydrogen sulfide acts downstream of jasmonic acid to inhibit stomatal development in Arabidopsis. Planta 2020, 251, 42. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.; Kumar, D.; Tikoria, R.; Sharma, R.; Parkirti, P.; Vikram, V.; Kaushal, K.; Ohri, P. Exploring the potential role of hydrogen sulfide and jasmonic acid in plants during heavy metal stress. Nitric Oxide 2023, 140–141, 16–29. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.-G.; Xiang, R.-H.; Wang, J.-Q. Hydrogen Sulfide–Phytohormone Interaction in Plants Under Physiological and Stress Conditions. J. Plant Growth Regul. 2021, 40, 2476–2484. [Google Scholar] [CrossRef]
- Song, W.; Shao, H.; Zheng, A.; Zhao, L.; Xu, Y. Advances in Roles of Salicylic Acid in Plant Tolerance Responses to Biotic and Abiotic Stresses. Plants 2023, 12, 3475. [Google Scholar] [CrossRef] [PubMed]
- Pál, M.; Kovács, V.; Szalai, G.; Soós, V.; Ma, X.; Liu, H.; Mei, H.; Janda, T. Salicylic Acid and Abiotic Stress Responses in Rice. J. Agron. Crop Sci. 2014, 200, 1–11. [Google Scholar] [CrossRef]
- Nazar, R.; Umar, S.; Khan, N.A. Exogenous salicylic acid improves photosynthesis and growth through increase in ascorbate-glutathione metabolism and S assimilation in mustard under salt stress. Plant Signal. Behav. 2015, 10, e1003751. [Google Scholar] [CrossRef]
- Kiddle, G.A.; Doughty, K.J.; Wallsgrove, R.M. Salicylic acid-induced accumulation of glucosinolates in oilseed rape (Brassica napus L.) leaves. J. Exp. Bot. 1994, 45, 1343–1346. [Google Scholar] [CrossRef]
- Baek, D.; Pathange, P.; Chung, J.; Jiang, J.; Gao, L.; Oikawa, A.; Hirai, M.Y.; Saito, K.; Pare, P.W.; Shi, H. A stress-inducible sulphotransferase sulphonates salicylic acid and confers pathogen resistance in Arabidopsis. Plant Cell Environ. 2010, 33, 1383–1392. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Chaouch, S.; Mhamdi, A.; Queval, G.; Zechmann, B.; Noctor, G. Functional Analysis of arabidopsis mutants points to novel roles for glutathione in coupling H2O2 to activation of salicylic acid accumulation and signaling. Antioxid. Redox Signal. 2013, 18, 2106–2121. [Google Scholar] [CrossRef]
- Künstler, A.; Király, L.; Kátay, G.; Enyedi, A.J.; Gullner, G. Glutathione Can Compensate for Salicylic Acid Deficiency in Tobacco to Maintain Resistance to Tobacco Mosaic Virus. Front. Plant Sci. 2019, 10, 1115. [Google Scholar] [CrossRef]
- Lindermayr, C. Crosstalk between reactive oxygen species and nitric oxide in plants: Key role of S-nitrosoglutathione reductase. Free Radic. Biol. Med. 2018, 122, 110–115. [Google Scholar] [CrossRef] [PubMed]
- Feechan, A.; Kwon, E.; Yun, B.-W.; Wang, Y.; Pallas, J.A.; Loake, G.J. A central role for S-nitrosothiols in plant disease resistance. Proc. Natl. Acad. Sci. USA 2005, 102, 8054–8059. [Google Scholar] [CrossRef]
- Zhang, T.; Ma, M.; Chen, T.; Zhang, L.; Fan, L.; Zhang, W.; Wei, B.; Li, S.; Xuan, W.; Noctor, G.; et al. Glutathione-dependent denitrosation of GSNOR1 promotes oxidative signalling downstream of H2O2. Plant Cell Environ. 2020, 43, 1175–1191. [Google Scholar] [CrossRef] [PubMed]
- Mou, Z.; Fan, W.; Dong, X. Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 2003, 113, 935–944. [Google Scholar] [CrossRef]
- Wu, Y.; Zhang, D.; Chu, J.Y.; Boyle, P.; Wang, Y.; Brindle, I.D.; De Luca, V.; Després, C. The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Rep. 2012, 1, 639–647. [Google Scholar] [CrossRef] [PubMed]
- Lindermayr, C.; Sell, S.; Müller, B.; Leister, D.; Durner, J. Redox Regulation of the NPR1-TGA1 System of Arabidopsis thaliana by Nitric Oxide. Plant Cell 2010, 22, 2894–2907. [Google Scholar] [CrossRef]
- Criollo-Arteaga, S.; Moya-Jimenez, S.; Jimenez-Meza, M.; Gonzalez-Vera, V.; Gordon-Nunez, J.; Llerena-Llerena, S.; Ramirez-Villacis, D.X.; van’t Hof, P.; Leon-Reyes, A. Sulfur Deprivation Modulates Salicylic Acid Responses via Nonexpressor of Pathogenesis-Related Gene 1 in Arabidopsis thaliana. Plants 2021, 10, 1065. [Google Scholar] [CrossRef]
- Rasheed, F.; Anjum, N.A.; Masood, A.; Sofo, A.; Khan, N.A. The key roles of salicylic acid and sulfur in plant salinity stress tolerance. J. Plant Growth Regul. 2022, 41, 1891–1904. [Google Scholar] [CrossRef]
- Li, Z.-G.; Xie, L.-R.; Li, X.-J. Hydrogen sulfide acts as a downstream signal molecule in salicylic acid-induced heat tolerance in maize (Zea mays L.) seedlings. J. Plant Physiol. 2015, 177, 121–127. [Google Scholar] [CrossRef]
- Qiao, Z.; Jing, T.; Liu, Z.; Zhang, L.; Jin, Z.; Liu, D.; Pei, Y. H2S acting as a downstream signaling molecule of SA regulates Cd tolerance in Arabidopsis. Plant Soil 2015, 393, 137–146. [Google Scholar] [CrossRef]
- Shi, H.; Ye, T.; Han, N.; Bian, H.; Liu, X.; Chan, Z. Hydrogen sulfide regulates abiotic stress tolerance and biotic stress resistance in Arabidopsis. J. Integr. Plant Biol. 2015, 57, 628–640. [Google Scholar] [CrossRef] [PubMed]
- Rai, K.K.; Pandey, N.; Rai, S.P. Salicylic acid and nitric oxide signaling in plant heat stress. Physiol. Plant. 2020, 168, 241–255. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Wang, M.; Chen, J.; Wu, J.; Xia, Z. Sulfur dioxide improves the thermotolerance of maize seedlings by regulating salicylic acid biosynthesis. Ecotoxicol. Environ. Saf. 2023, 254, 114746. [Google Scholar] [CrossRef] [PubMed]
- Rasheed, F.; Mir, I.R.; Sehar, Z.; Fatma, M.; Gautam, H.; Khan, S.; Anjum, N.A.; Masood, A.; Sofo, A.; Khan, N.A. Nitric Oxide and Salicylic Acid Regulate Glutathione and Ethylene Production to Enhance Heat Stress Acclimation in Wheat Involving Sulfur Assimilation. Plants 2022, 11, 3131. [Google Scholar] [CrossRef] [PubMed]
- Bano, K.; Kumar, B.; Tenguria, R.K.; Alsahli, A.A.; Chen, Y. Salicylic acid and sulfur synergism ameliorates arsenic toxicity in Brassica napus through regulating carbohydrate accumulation and ethylene production. S. Afr. J. Bot. 2023, 160, 246–259. [Google Scholar] [CrossRef]
- Al-Babili, S.; Bouwmeester, H.J. Strigolactones, a Novel carotenoid-derived plant hormone. Annu. Rev. Plant Biol. 2015, 66, 161–186. [Google Scholar] [CrossRef] [PubMed]
- Miyata, K.; Umehara, M. Roles of Arbuscular Mycorrhizal Fungi for Essential Nutrient Acquisition Under Nutrient Deficiency in Plants. In Arbuscular Mycorrhizal Fungi and Higher Plants: Fundamentals and Applications; Ahammed, G.J., Hajiboland, R., Eds.; Springer Nature Singapore: Singapore, 2024; pp. 123–148. [Google Scholar]
- Shindo, M.; Shimomura, K.; Yamaguchi, S.; Umehara, M. Upregulation of DWARF27 is associated with increased strigolactone levels under sulfur deficiency in rice. Plant Direct 2018, 2, e00050. [Google Scholar] [CrossRef] [PubMed]
- Shindo, M.; Yamamoto, S.; Shimomura, K.; Umehara, M. Strigolactones Decrease Leaf Angle in Response to Nutrient Deficiencies in Rice. Front. Plant Sci. 2020, 11, 135. [Google Scholar] [CrossRef] [PubMed]
- Allen, J.W.; Shachar-Hill, Y. Sulfur Transfer through an Arbuscular Mycorrhiza. Plant Physiol. 2009, 149, 549–560. [Google Scholar] [CrossRef]
- Shindo, M.; Nagasaka, S.; Kashiwada, S.; Shimomura, K.; Umehara, M. Shoot has important roles in strigolactone production of rice roots under sulfur deficiency. Plant Signal. Behav. 2021, 16, 1880738. [Google Scholar] [CrossRef]
- Quaggiotti, S.; Buzzicotti, L.; Koch, K.E.; Guan, J.C.; Trevisan, S.; Varotto, S.; Ruperti, B.; Ravazzolo, L. Strigolactone roles in maize tolerance to low nitrogen involve shifts in acquisition and partitioning of protein, sulfur, and iron. Plant Soil 2024. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wawrzyńska, A.; Sirko, A. Sulfate Availability and Hormonal Signaling in the Coordination of Plant Growth and Development. Int. J. Mol. Sci. 2024, 25, 3978. https://doi.org/10.3390/ijms25073978
Wawrzyńska A, Sirko A. Sulfate Availability and Hormonal Signaling in the Coordination of Plant Growth and Development. International Journal of Molecular Sciences. 2024; 25(7):3978. https://doi.org/10.3390/ijms25073978
Chicago/Turabian StyleWawrzyńska, Anna, and Agnieszka Sirko. 2024. "Sulfate Availability and Hormonal Signaling in the Coordination of Plant Growth and Development" International Journal of Molecular Sciences 25, no. 7: 3978. https://doi.org/10.3390/ijms25073978
APA StyleWawrzyńska, A., & Sirko, A. (2024). Sulfate Availability and Hormonal Signaling in the Coordination of Plant Growth and Development. International Journal of Molecular Sciences, 25(7), 3978. https://doi.org/10.3390/ijms25073978






