Wide-Targeted Semi-Quantitative Analysis of Acidic Glycosphingolipids in Cell Lines and Urine to Develop Potential Screening Biomarkers for Renal Cell Carcinoma
Abstract
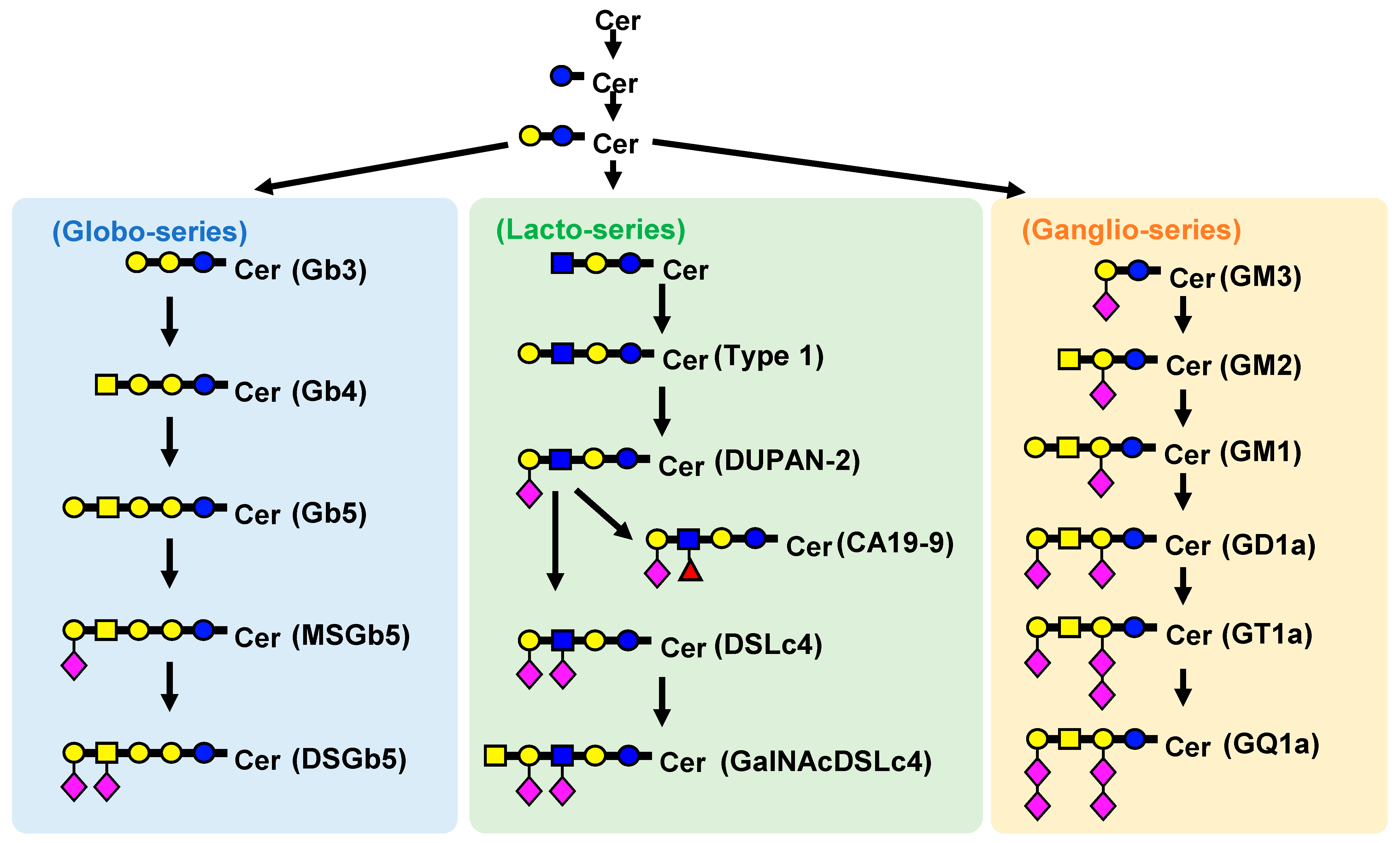
1. Introduction
2. Results and Discussion
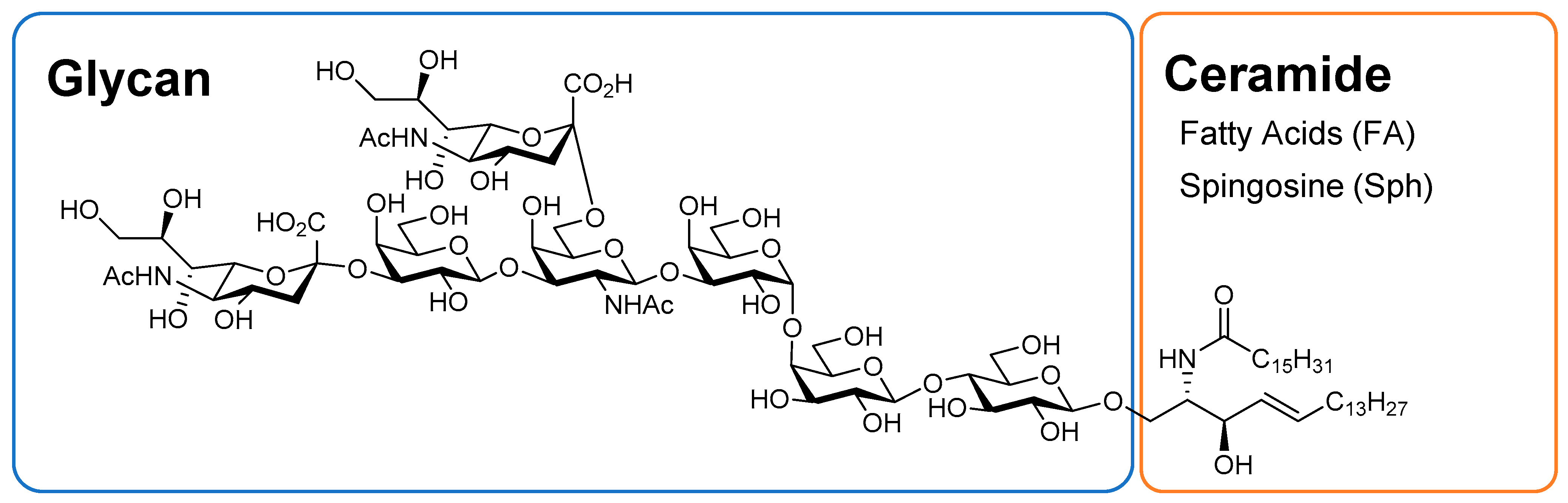
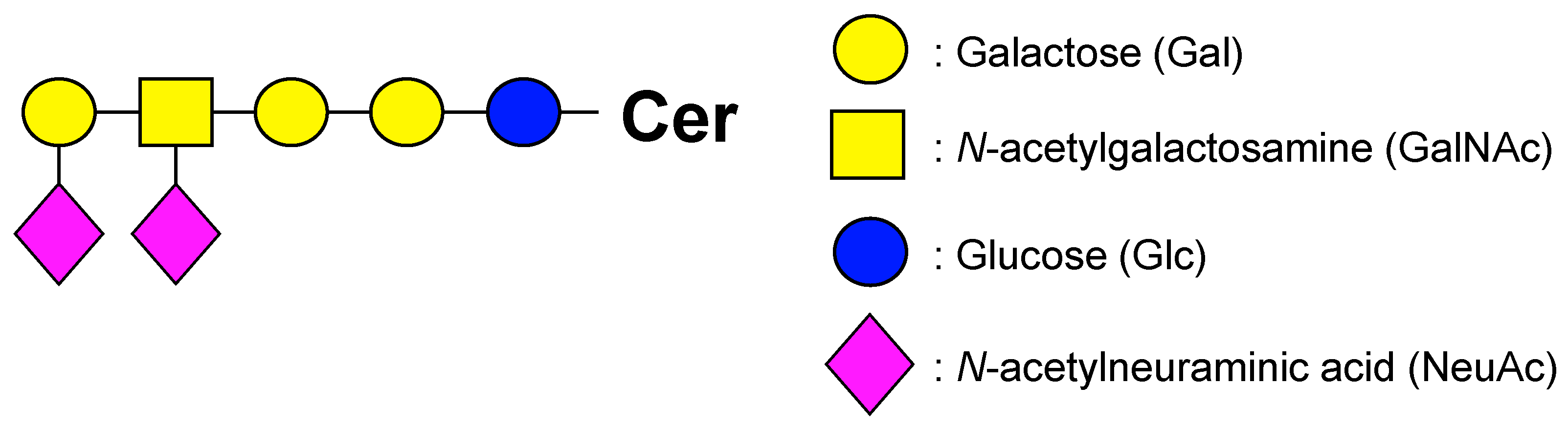
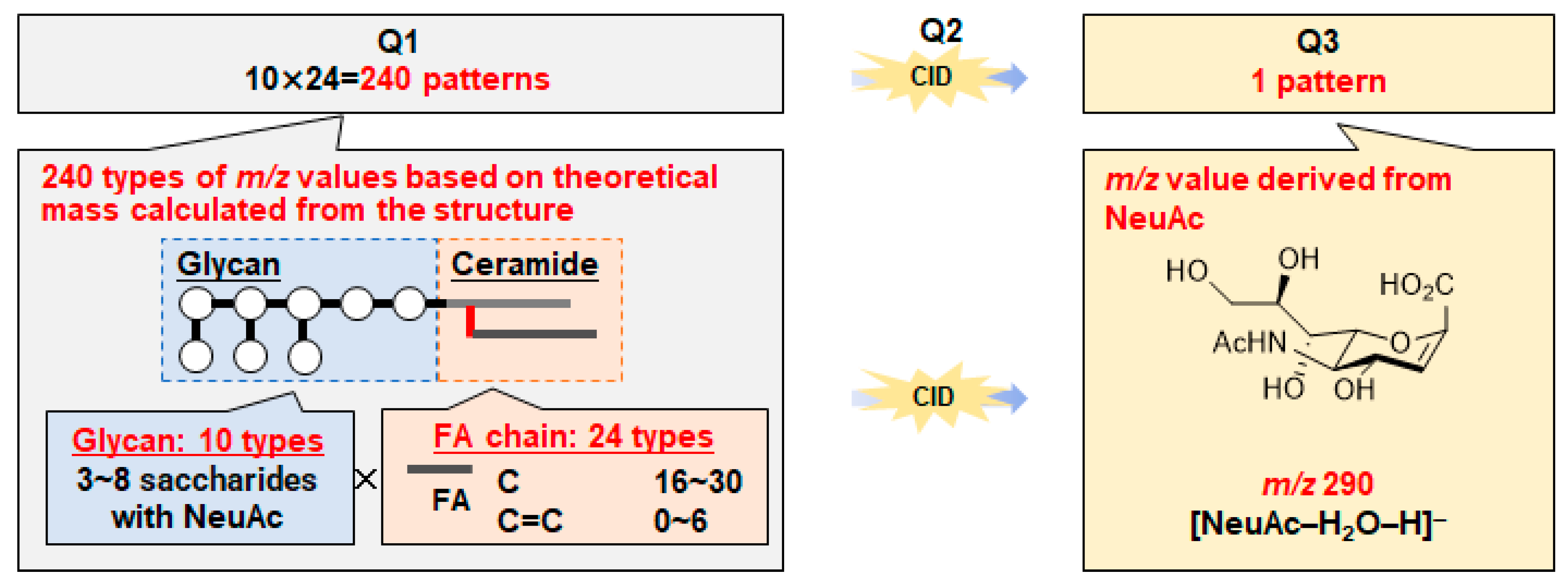
2.1. Optimization of LC–MS/MS for Wide-Targeted GSL Analysis
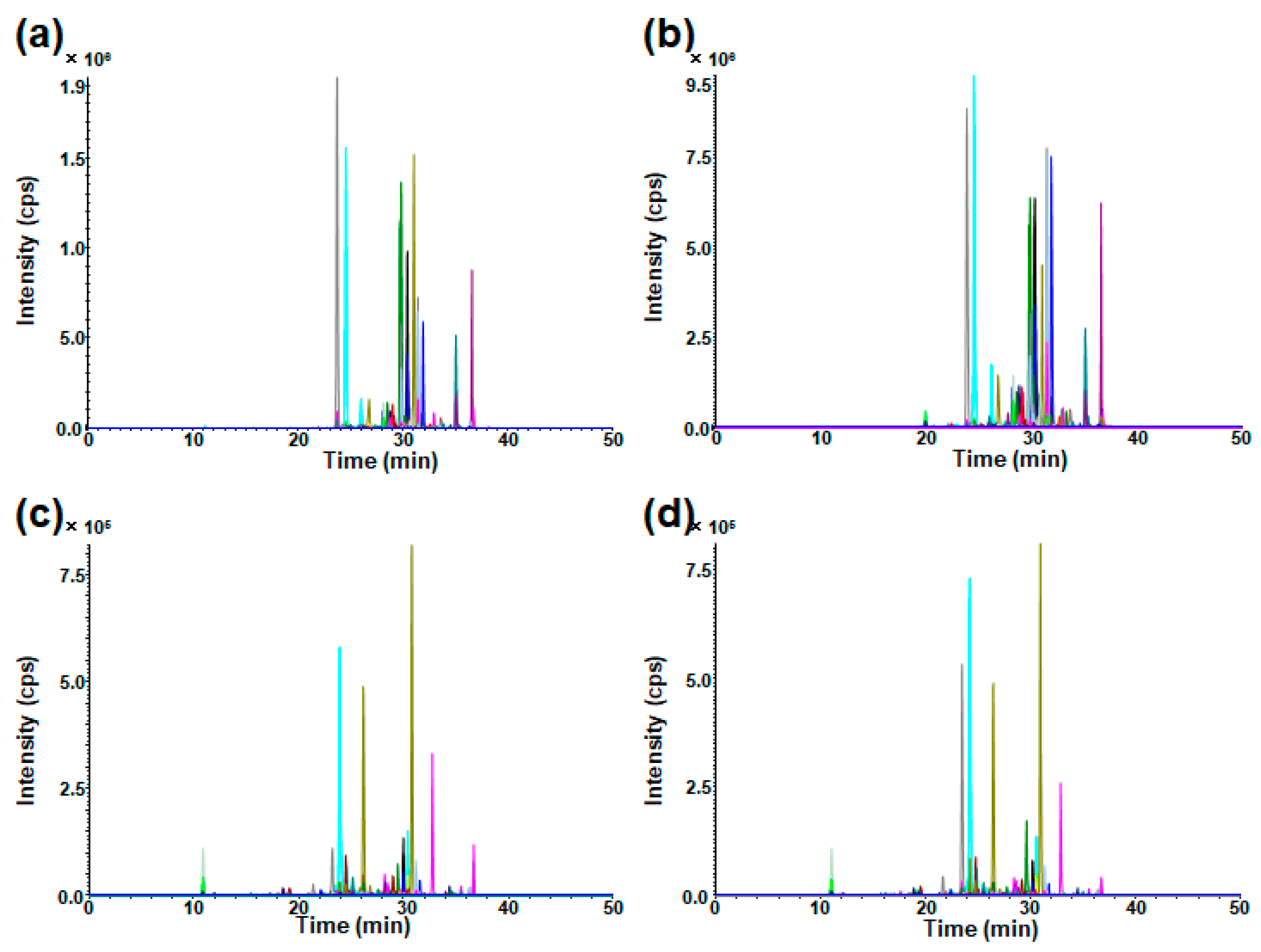
2.2. Measurement of GSLs in Cultured Cells
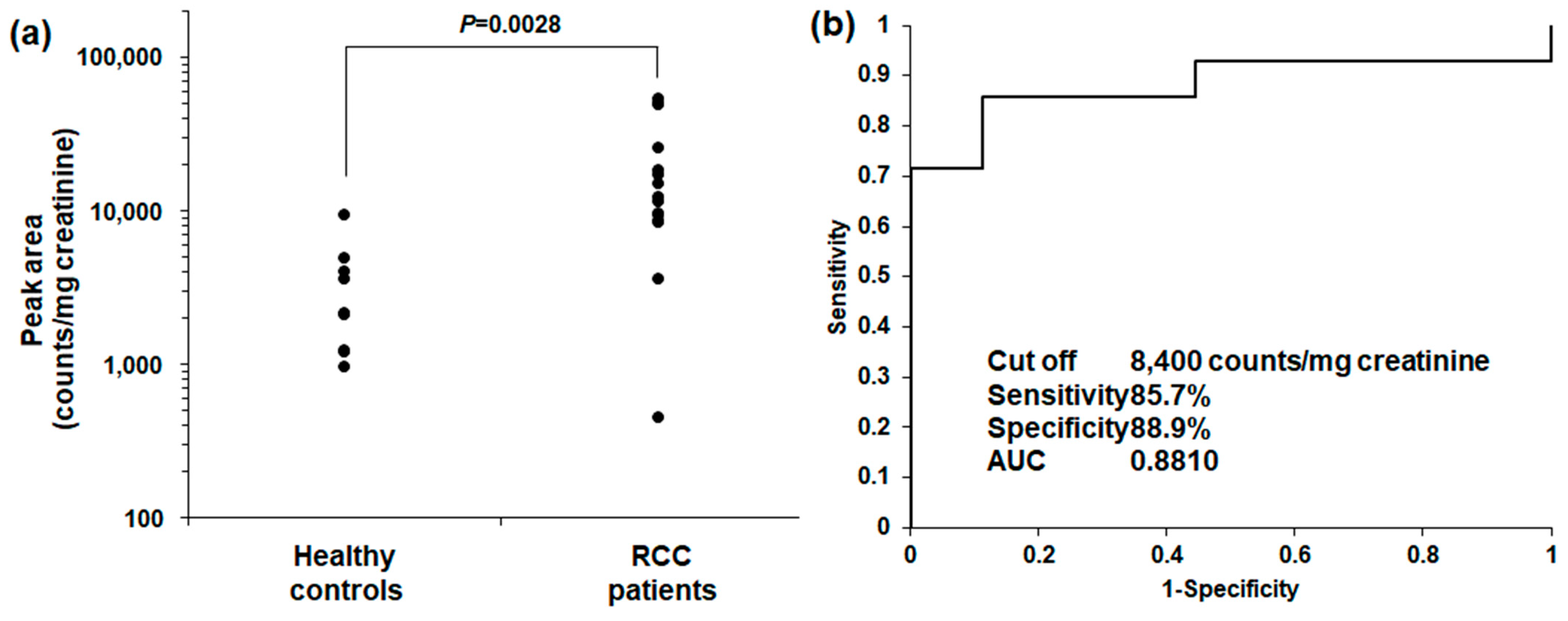
2.3. GSL Urinary DSGb5 (d18:1/16:0) Analysis in RCC Patients and Healthy Controls
3. Materials and Methods
3.1. Chemicals and Reagents
3.2. LC–MS/MS Equipment
3.3. Optimization of MS/MS Conditions for Targeted GSL Analysis
3.4. LC Conditions for Targeted GSL Analysis
3.5. Setting Virtual SRM Conditions
3.6. Preparation of Standard Solution of GD1a (d18:1/18:0)
3.7. Linearity of SRM Analysis in GD1a Analysis
3.8. Investigation of Solvent for Redissolution of Targeted GSL Analysis Using GD1a (d18:1/18:0) Standard Solution
3.9. Investigation of Pretreatment Using GD1a (d18:1/18:0) Standard Solution
3.10. Investigation of Matrix Effect Using GD1a (d18:1/18:0) Standard Solution and Cell Medium
3.11. Cell Culturing and Sample Preparation
3.12. Urine Samples
3.13. Urine Pretreatment
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cohen, H.T.; McGovern, F.J. Renal-Cell Carcinoma. N. Engl. J. Med. 2005, 353, 2477–2490. [Google Scholar] [CrossRef] [PubMed]
- Motzer, R.J.; Bander, N.H.; Nanus, D.M. Renal-Cell Carcinoma. N. Engl. J. Med. 1996, 335, 865–875. [Google Scholar] [CrossRef] [PubMed]
- Barata, P.C.; Rini, B.I. Treatment of Renal Cell Carcinoma: Current Status and Future Directions. CA Cancer J. Clin. 2017, 67, 507–524. [Google Scholar] [CrossRef] [PubMed]
- International, W.C.R.F. Kidney Cancer Statistics. Available online: https://www.wcrf.org/cancer-trends/kidney-cancer-statistics/ (accessed on 28 January 2024).
- Tran, J.; Ornstein, M.C. Clinical Review on the Management of Metastatic Renal Cell Carcinoma. JCO Oncol. Pract. 2022, 18, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Rathmell, W.K.; Rumble, R.B.; Van Veldhuizen, P.J.; Al-Ahmadie, H.; Emamekhoo, H.; Hauke, R.J.; Louie, A.V.; Milowsky, M.I.; Molina, A.M.; Rose, T.L.; et al. Management of Metastatic Clear Cell Renal Cell Carcinoma: ASCO Guideline. J. Clin. Oncol. 2022, 40, 2957–2995. [Google Scholar] [CrossRef] [PubMed]
- Ljungberg, B.; Bensalah, K.; Canfield, S.; Dabestani, S.; Hofmann, F.; Hora, M.; Kuczyk, M.A.; Lam, T.; Marconi, L.; Merseburger, A.S.; et al. EAU Guidelines on Renal Cell Carcinoma: 2014 Update. Eur. Urol. 2015, 67, 913–924. [Google Scholar] [CrossRef] [PubMed]
- Ljungberg, B.; Albiges, L.; Abu-Ghanem, Y.; Bedke, J.; Capitanio, U.; Dabestani, S.; Fernández-Pello, S.; Giles, R.H.; Hofmann, F.; Hora, M.; et al. European Association of Urology Guidelines on Renal Cell Carcinoma: The 2022 Update. Eur. Urol. 2022, 82, 399–410. [Google Scholar] [CrossRef] [PubMed]
- Kuchar, L.; Sikora, J.; Gulinello, M.E.; Poupetova, H.; Lugowska, A.; Malinova, V.; Jahnova, H.; Asfaw, B.; Ledvinova, J. Quantitation of Plasmatic Lysosphingomyelin and Lysosphingomyelin-509 for Differential Screening of Niemann-Pick A/B and C Diseases. Anal. Biochem. 2017, 525, 73–77. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Iwamoto, T.; Hossain, M.A.; Akiyama, K.; Igarashi, J.; Miyajima, T.; Eto, Y. A Combination of 7-Ketocholesterol, Lysosphingomyelin and Bile Acid-408 to Diagnose Niemann-Pick Disease Type C Using LC-MS/MS. PLoS ONE 2020, 15, e0238624. [Google Scholar] [CrossRef]
- Ishikawa, I.; Honda, R.; Yamada, Y.; Kakuma, T. Renal Cell Carcinoma Detected by Screening Shows Better Patient Survival than That Detected Following Symptoms in Dialysis Patients. Ther. Apher. Dial. 2004, 8, 468–473. [Google Scholar] [CrossRef]
- Gong, J.; Maia, M.C.; Dizman, N.; Govindarajan, A.; Pal, S.K. Metastasis in Renal Cell Carcinoma: Biology and Implications for Therapy. Asian J. Urol. 2016, 3, 286–292. [Google Scholar] [CrossRef]
- Zheng, H.; Ji, J.; Zhao, L.; Chen, M.; Shi, A.; Pan, L.; Huang, Y.; Zhang, H.; Dong, B.; Gao, H. Prediction and Diagnosis of Renal Cell Carcinoma Using Nuclear Magnetic Resonance-Based Serum Metabolomics and Self-Organizing Maps. Oncotarget 2016, 7, 59189–59198. [Google Scholar] [CrossRef]
- Kroeger, N.; Xie, W.; Lee, J.L.; Bjarnason, G.A.; Knox, J.J.; MacKenzie, M.J.; Wood, L.; Srinivas, S.; Vaishamayan, U.N.; Rha, S.Y.; et al. Metastatic Non-Clear Cell Renal Cell Carcinoma Treated with Targeted Therapy Agents: Characterization of Survival Outcome and Application of the International MRCC Database Consortium Criteria. Cancer 2013, 119, 2999–3006. [Google Scholar] [CrossRef]
- Hakomori, S. Glycosphingolipids in Cellular Interaction, Differentiation, and Oncogenesis. Annu. Rev. Biochem. 1981, 50, 733–764. [Google Scholar] [CrossRef] [PubMed]
- Bremer, E.G.; Schlessinger, J.; Hakomori, S. Ganglioside-Mediated Modulation of Cell Growth. Specific Effects of GM3 on Tyrosine Phosphorylation of the Epidermal Growth Factor Receptor. J. Biol. Chem. 1986, 261, 2434–2440. [Google Scholar] [CrossRef]
- Varki, A.; Cummings, R.D.; Aebi, M.; Packer, N.H.; Seeberger, P.H.; Esko, J.D.; Stanley, P.; Hart, G.; Darvill, A.; Kinoshita, T.; et al. Symbol Nomenclature for Graphical Representations of Glycans. Glycobiology 2015, 25, 1323–1324. [Google Scholar] [CrossRef] [PubMed]
- Neelamegham, S.; Aoki-Kinoshita, K.; Bolton, E.; Frank, M.; Lisacek, F.; Lütteke, T.; O’Boyle, N.; Packer, N.H.; Stanley, P.; Toukach, P.; et al. Updates to the Symbol Nomenclature for Glycans Guidelines. Glycobiology 2019, 29, 620–624. [Google Scholar] [CrossRef] [PubMed]
- Hannun, Y.A.; Obeid, L.M. Principles of Bioactive Lipid Signalling: Lessons from Sphingolipids. Nat. Rev. Mol. Cell Biol. 2008, 9, 139–150. [Google Scholar] [CrossRef] [PubMed]
- Hannun, Y.A. Functions of Ceramide in Coordinating Cellular Responses to Stress. Science 1996, 274, 1855–1859. [Google Scholar] [CrossRef]
- Hanada, K.; Kumagai, K.; Yasuda, S.; Miura, Y.; Kawano, M.; Fukasawa, M.; Nishijima, M. Molecular Machinery for Non-Vesicular Trafficking of Ceramide. Nature 2003, 426, 803–809. [Google Scholar] [CrossRef]
- Ogretmen, B. Sphingolipid Metabolism in Cancer Signalling and Therapy. Nat. Rev. Cancer 2017, 18, 33–50. [Google Scholar] [CrossRef] [PubMed]
- Sokolowska, E.; Blachnio-Zabielska, A. The Role of Ceramides in Insulin Resistance. Front. Endocrinol. 2019, 10, 577. [Google Scholar] [CrossRef] [PubMed]
- Meikopoulos, T.; Begou, O.; Theodoridis, G.; Gika, H. Ceramides Biomarkers Determination in Quantitative Dried Blood Spots by UHPLC-MS/MS. Anal. Chim. Acta 2023, 1255, 341131. [Google Scholar] [CrossRef] [PubMed]
- Kagan, T.; Stoyanova, G.; Lockshin, R.A.; Zakeri, Z. Ceramide from Sphingomyelin Hydrolysis Induces Neuronal Differentiation, Whereas de Novo Ceramide Synthesis and Sphingomyelin Hydrolysis Initiate Apoptosis after NGF Withdrawal in PC12 Cells. Cell Commun. Signal. 2022, 20, 15. [Google Scholar] [CrossRef] [PubMed]
- Saito, S.; Aoki, H.; Ito, A.; Ueno, S.; Wada, T.; Mitsuzuka, K.; Satoh, M.; Arai, Y.; Miyagi, T. Human A2,3-Sialyltransferase (ST3Gal II) Is a Stage-Specific Embryonic Antigen-4 Synthase. J. Biol. Chem. 2003, 278, 26474–26479. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, Y.; Ito, A.; Withers, D.A.; Taima, T.; Kakoi, N.; Saito, S.; Arai, Y. Ganglioside DSGb5, Preferred Ligand for Siglec-7, Inhibits NK Cell Cytotoxicity against Renal Cell Carcinoma Cells. Glycobiology 2010, 20, 1373–1379. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, Y.; Ito, A.; Kakoi, N.; Shimada, S.; Itoh, J.; Mitsuzuka, K.; Arai, Y. Ganglioside, Disialosyl Globopentaosylceramide (DSGb5), Enhances the Migration of Renal Cell Carcinoma Cells. Tohoku J. Exp. Med. 2015, 236, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Satoh, M.; Handa, K.; Saito, S.; Tokuyama, S.; Ito, A.; Miyao, N.; Orikasa, S.; Hakomori, S. Disialosyl Galactosylgloboside as an Adhesion Molecule Expressed on Renal Cell Carcinoma and Its Relationship to Metastatic Potential. Cancer Res. 1996, 56, 1932–1938. [Google Scholar] [PubMed]
- Tsuchida, A.; Senda, M.; Ito, A.; Saito, S.; Kiso, M.; Ando, T.; Harduin-Lepers, A.; Matsuda, A.; Furukawa, K.; Furukawa, K. Roles of GalNAc-Disialyl Lactotetraosyl Antigens in Renal Cancer Cells. Sci. Rep. 2018, 8, 7017. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, T.; Tsubakihara, Y.; Fushimi, H.; Yamaguchi, S.; Takabatake, Y.; Rakugi, H.; Kawakami, H.; Isaka, Y. Histochemical and Immunoelectron Microscopic Analysis of Ganglioside GM3 in Human Kidney. Clin. Exp. Nephrol. 2015, 19, 403–410. [Google Scholar] [CrossRef]
- Saito, S.; Nojiri, H.; Satoh, M.; Ito, A.; Ohyama, C.; Orikasa, S. Inverse Relationship of Expression between GM3 and Globo-Series Ganglioside in Human Renal Cell Carcinoma. Tohoku J. Exp. Med. 2000, 190, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Ueno, S.; Saito, S.; Wada, T.; Yamaguchi, K.; Satoh, M.; Arai, Y.; Miyagi, T. Plasma Membrane-Associated Sialidase Is up-Regulated in Renal Cell Carcinoma and Promotes Interleukin-6-Induced Apoptosis Suppression and Cell Motility. J. Biol. Chem. 2006, 281, 7756–7764. [Google Scholar] [CrossRef]
- Banerjee, A.; Mahata, B.; Dhir, A.; Mandal, T.K.; Biswas, K. Elevated Histone H3 Acetylation and Loss of the Sp1–HDAC1 Complex de-Repress the GM2-Synthase Gene in Renal Cell Carcinoma. J. Biol. Chem. 2019, 294, 1005–1018. [Google Scholar] [CrossRef] [PubMed]
- Aoki, H.; Satoh, M.; Mitsuzuka, K.; Ito, A.; Saito, S.; Funato, T.; Endoh, M.; Takahashi, T.; Arai, Y. Inhibition of Motility and Invasiveness of Renal Cell Carcinoma Induced by Short Interfering RNA Transfection of Β1,4GalNAc Transferase. FEBS Lett. 2004, 567, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Biswas, K.; Richmond, A.; Rayman, P.; Biswas, S.; Thornton, M.; Sa, G.; Das, T.; Zhang, R.; Chahlavi, A.; Tannenbaum, C.S.; et al. GM2 Expression in Renal Cell Carcinoma: Potential Role in Tumor-Induced T-Cell Dysfunction. Cancer Res. 2006, 66, 6816–6825. [Google Scholar] [CrossRef] [PubMed]
- Higashi, T.; Shimada, K. Application of Cookson-Type Reagents for Biomedical HPLC and LC/MS Analyses: A Brief Overview. Biomed. Chromatogr. 2017, 31, e3808. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, S.; Takafuji, K.; Tsubuku, S.; Horie, Y.; Ikegawa, S.; Higashi, T. Isotope-Coded Derivatization Based LC/ESI-MS/MS Methods Using a Pair of Novel Reagents for Quantification of Hydroxycinnamic Acids and Hydroxybenzoic Acids in Fermented Brown Rice Product. J. Pharm. Biomed. Anal. 2017, 142, 162–170. [Google Scholar] [CrossRef] [PubMed]
- Higashi, T.; Ogawa, S. Chemical Derivatization for Enhancing Sensitivity during LC/ESI–MS/MS Quantification of Steroids in Biological Samples: A Review. J. Steroid Biochem. Mol. Biol. 2016, 162, 57–69. [Google Scholar] [CrossRef] [PubMed]
- Taguchi, R.; Houjou, T.; Nakanishi, H.; Yamazaki, T.; Ishida, M.; Imagawa, M.; Shimizu, T. Focused Lipidomics by Tandem Mass Spectrometry. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2005, 823, 26–36. [Google Scholar] [CrossRef]
- Iino, T.; Watanabe, S.; Yamashita, K.; Tamada, E.; Hasegawa, T.; Irino, Y.; Iwanaga, S.; Harada, A.; Noda, K.; Suto, K.; et al. Quantification of Amyloid-β in Plasma by Simple and Highly Sensitive Immunoaffinity Enrichment and LC-MS/MS Assay. J. Appl. Lab. Med. 2021, 6, 834–845. [Google Scholar] [CrossRef]
- Honda, A.; Yamashita, K.; Miyazaki, H.; Shirai, M.; Ikegami, T.; Xu, G.; Numazawa, M.; Hara, T.; Matsuzaki, Y. Highly Sensitive Analysis of Sterol Profiles in Human Serum by LC-ESI-MS/MS. J. Lipid Res. 2008, 49, 2063–2073. [Google Scholar] [CrossRef] [PubMed]
- Honda, A.; Yamashita, K.; Numazawa, M.; Ikegami, T.; Doy, M.; Matsuzaki, Y.; Miyazaki, H. Highly Sensitive Quantification of 7α-Hydroxy-4-Cholesten-3-One in Human Serum by LC-ESI-MS/MS. J. Lipid Res. 2007, 48, 458–464. [Google Scholar] [CrossRef]
- Maekawa, M.; Miyoshi, K.; Narita, A.; Sato, T.; Sato, Y.; Kumondai, M.; Kikuchi, M.; Higaki, K.; Okuyama, T.; Eto, Y.; et al. Development of a Highly Sensitive and Rapid Liquid Chromatography–Tandem Mass Spectrometric Method Using a Basic Mobile Phase Additive to Determine the Characteristics of the Urinary Metabolites for Niemann–Pick Disease Type C. Biol. Pharm. Bull. 2022, 45, 1259–1268. [Google Scholar] [CrossRef]
- Yamashita, K.; Takahashi, M.; Tsukamoto, S.; Numazawa, M.; Okuyama, M.; Honma, S. Use of Novel Picolinoyl Derivatization for Simultaneous Quantification of Six Corticosteroids by Liquid Chromatography-Electrospray Ionization Tandem Mass Spectrometry. J. Chromatogr. A 2007, 1173, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, K.; Shimizu, T.; Taguchi, R. Targeted Analysis of Ganglioside and Sulfatide Molecular Species by LC/ESI-MS/MS with Theoretically Expanded Multiple Reaction Monitoring. J. Lipid Res. 2008, 49, 2678–2689. [Google Scholar] [CrossRef]
- Shoji, S.; Maekawa, M.; Ogura, J.; Sato, T.; Mano, N. Identification Cholesterol Metabolites Altered before the Onset of Nonalcoholic Steatohepatitis by Targeted Metabolomics. Biochim. Et Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2022, 1867, 159135. [Google Scholar] [CrossRef] [PubMed]
- Hishinuma, E.; Shimada, M.; Matsukawa, N.; Saigusa, D.; Li, B.; Kudo, K.; Tsuji, K.; Shigeta, S.; Tokunaga, H.; Kumada, K.; et al. Wide-Targeted Metabolome Analysis Identifies Potential Biomarkers for Prognosis Prediction of Epithelial Ovarian Cancer. Toxins 2021, 13, 461. [Google Scholar] [CrossRef]
- Nakatani, K.; Izumi, Y.; Hata, K.; Bamba, T. An Analytical System for Single-Cell Metabolomics of Typical Mammalian Cells Based on Highly Sensitive Nano-Liquid Chromatography Tandem Mass Spectrometry. Mass Spectrom. 2020, 9, A0080. [Google Scholar] [CrossRef]
- Soma, Y.; Takahashi, M.; Fujiwara, Y.; Tomiyasu, N.; Goto, M.; Hanai, T.; Izumi, Y.; Bamba, T. Quantitative Metabolomics for Dynamic Metabolic Engineering Using Stable Isotope Labeled Internal Standards Mixture (SILIS). J. Biosci. Bioeng. 2022, 133, 46–55. [Google Scholar] [CrossRef]
- Nakatani, K.; Izumi, Y.; Takahashi, M.; Bamba, T. Unified-Hydrophilic-Interaction/Anion-Exchange Liquid Chromatography Mass Spectrometry (Unified-HILIC/AEX/MS): A Single-Run Method for Comprehensive and Simultaneous Analysis of Polar Metabolome. Anal. Chem. 2022, 94, 16877–16886. [Google Scholar] [CrossRef]
- Bligh, E.G.; Dyer, W.J. A Rapid Method of Total Lipid Extraction and Purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef] [PubMed]
- Iverson, S.J.; Lang, S.L.C.; Cooper, M.H. Comparison of the Bligh and Dyer and Folch Methods for Total Lipid Determination in a Broad Range of Marine Tissue. Lipids 2001, 36, 1283–1287. [Google Scholar] [CrossRef] [PubMed]
- Welford, R.W.D.; Garzotti, M.; Marques Lourenço, C.; Mengel, E.; Marquardt, T.; Reunert, J.; Amraoui, Y.; Kolb, S.A.; Morand, O.; Groenen, P. Plasma Lysosphingomyelin Demonstrates Great Potential as a Diagnostic Biomarker for Niemann-Pick Disease Type C in a Retrospective Study. PLoS ONE 2014, 9, e114669. [Google Scholar] [CrossRef] [PubMed]
- Ohtsu, Y.; Tanaka, S.; Igarashi, H.; Kakehi, M.; Mori, T.; Nakamura, T.; Ohashi, R.; Shimizu, H.; Yasuda, Y.; Okayama, T.; et al. Analytical Method Validation for Biomarkers as a Drug Development Tool: Points to Consider. Bioanalysis 2021, 13, 1379–1389. [Google Scholar] [CrossRef]
- Patel, N.P.; Sanyal, M.; Sharma, N.; Patel, D.S.; Shrivastav, P.S.; Patel, B.N. Determination of Asenapine in Presence of Its Inactive Metabolites in Human Plasma by LC-MS/MS. J. Pharm. Anal. 2018, 8, 341–347. [Google Scholar] [CrossRef] [PubMed]
- Minnikin, D.E.; O’Donnell, A.G.; Goodfellow, M.; Alderson, G.; Athalye, M.; Schaal, A.; Parlett, J.H. An Integrated Procedure for the Extraction of Bacterial Isoprenoid Quinones and Polar Lipids. J. Microbiol. Methods 1984, 2, 233–241. [Google Scholar] [CrossRef]
- Rakete, S.; Schuster, C.; Paal, M.; Vogeser, M. An Isotope-Dilution LC-MS/MS Method for the Simultaneous Quantification of Meropenem and Its Open-Ring Metabolite in Serum. J. Pharm. Biomed. Anal. 2021, 197, 113944. [Google Scholar] [CrossRef]
- Sato, T.; Kawasaki, Y.; Maekawa, M.; Takasaki, S.; Morozumi, K.; Sato, M.; Shimada, S.; Kawamorita, N.; Yamashita, S.; Mitsuzuka, K.; et al. Metabolomic Analysis to Elucidate Mechanisms of Sunitinib Resistance in Renal Cell Carcinoma. Metabolites 2020, 11, 1. [Google Scholar] [CrossRef]

| No | Component | Q1 (m/z) | HK-2 (Counts) | ACHN (Counts) | Ratio (%) |
|---|---|---|---|---|---|
| 1 | DSGb5 (d18:1/16:0) | 984.5 | 997,593 | 4,577,916 | 459 |
| 2 | GD1a (d18:1/16:0) | 903.5 | 6,356,833 | 7,462,803 | 117 |
| 3 | GD1a (d18:1/18:0) | 917.5 | 4,503,695 | 4,343,382 | 96.4 |
| 4 | GM3 (d18:1/16:0) | 1151.7 | 6,482,229 | 5,807,321 | 89.6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maekawa, M.; Sato, T.; Kanno, C.; Sakamoto, I.; Kawasaki, Y.; Ito, A.; Mano, N. Wide-Targeted Semi-Quantitative Analysis of Acidic Glycosphingolipids in Cell Lines and Urine to Develop Potential Screening Biomarkers for Renal Cell Carcinoma. Int. J. Mol. Sci. 2024, 25, 4098. https://doi.org/10.3390/ijms25074098
Maekawa M, Sato T, Kanno C, Sakamoto I, Kawasaki Y, Ito A, Mano N. Wide-Targeted Semi-Quantitative Analysis of Acidic Glycosphingolipids in Cell Lines and Urine to Develop Potential Screening Biomarkers for Renal Cell Carcinoma. International Journal of Molecular Sciences. 2024; 25(7):4098. https://doi.org/10.3390/ijms25074098
Chicago/Turabian StyleMaekawa, Masamitsu, Tomonori Sato, Chika Kanno, Izumi Sakamoto, Yoshihide Kawasaki, Akihiro Ito, and Nariyasu Mano. 2024. "Wide-Targeted Semi-Quantitative Analysis of Acidic Glycosphingolipids in Cell Lines and Urine to Develop Potential Screening Biomarkers for Renal Cell Carcinoma" International Journal of Molecular Sciences 25, no. 7: 4098. https://doi.org/10.3390/ijms25074098
APA StyleMaekawa, M., Sato, T., Kanno, C., Sakamoto, I., Kawasaki, Y., Ito, A., & Mano, N. (2024). Wide-Targeted Semi-Quantitative Analysis of Acidic Glycosphingolipids in Cell Lines and Urine to Develop Potential Screening Biomarkers for Renal Cell Carcinoma. International Journal of Molecular Sciences, 25(7), 4098. https://doi.org/10.3390/ijms25074098






