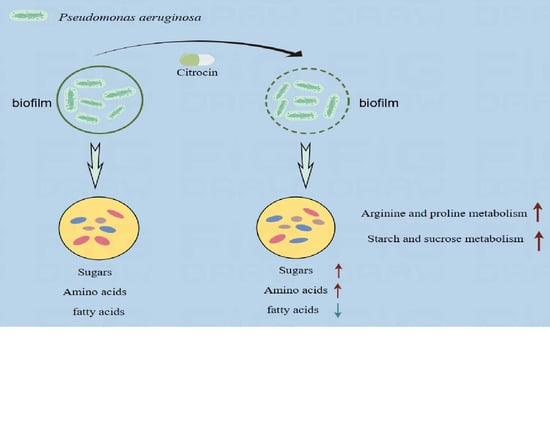
The Anti-Microbial Peptide Citrocin Controls Pseudomonas aeruginosa Biofilms by Breaking Down Extracellular Polysaccharide
Abstract
:1. Introduction
2. Results
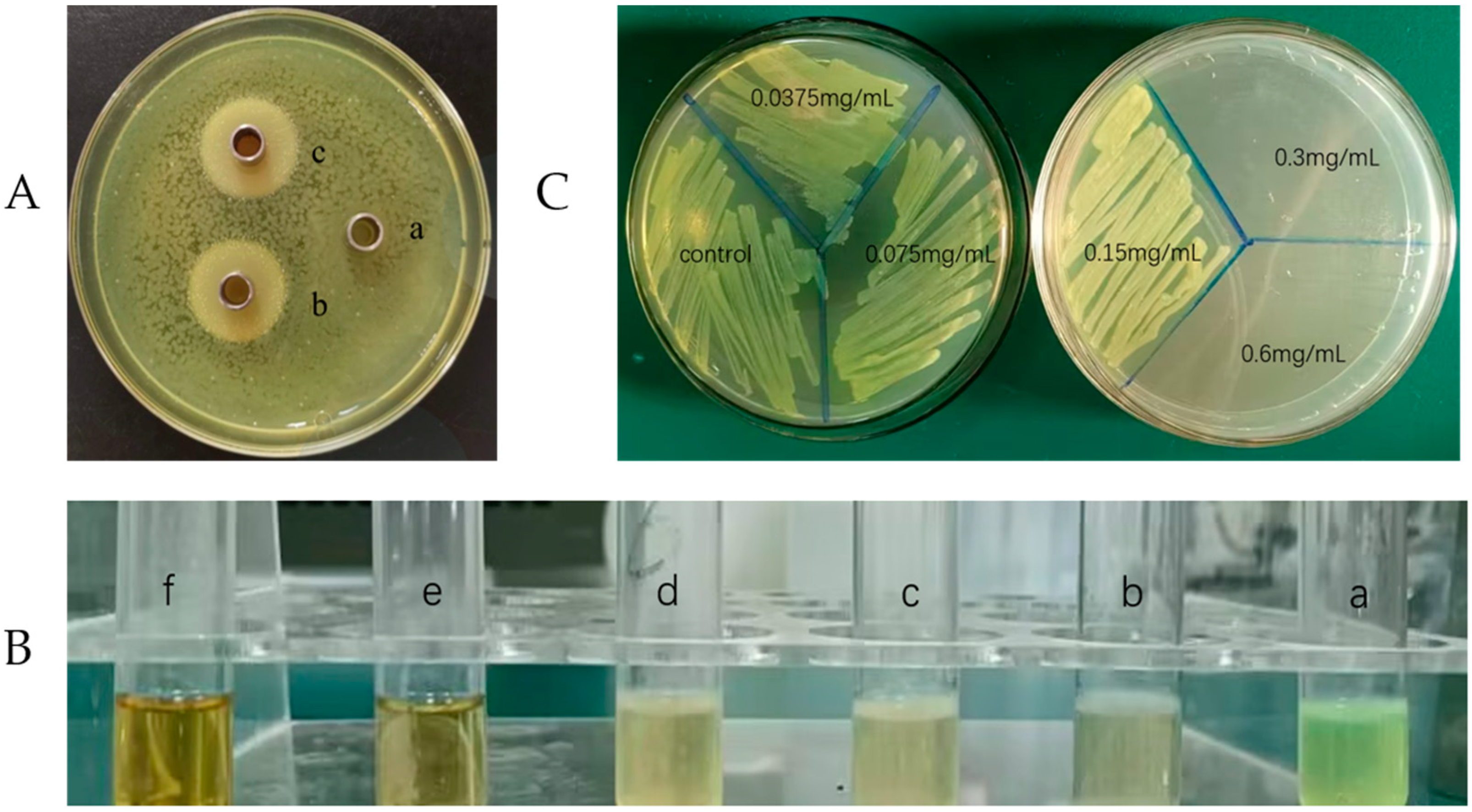
2.1. Evaluation of Inhibitory Effect
2.2. Minimum Inhibitory Concentration (MIC) and Minimum Bactericidal Concentration (MBC)
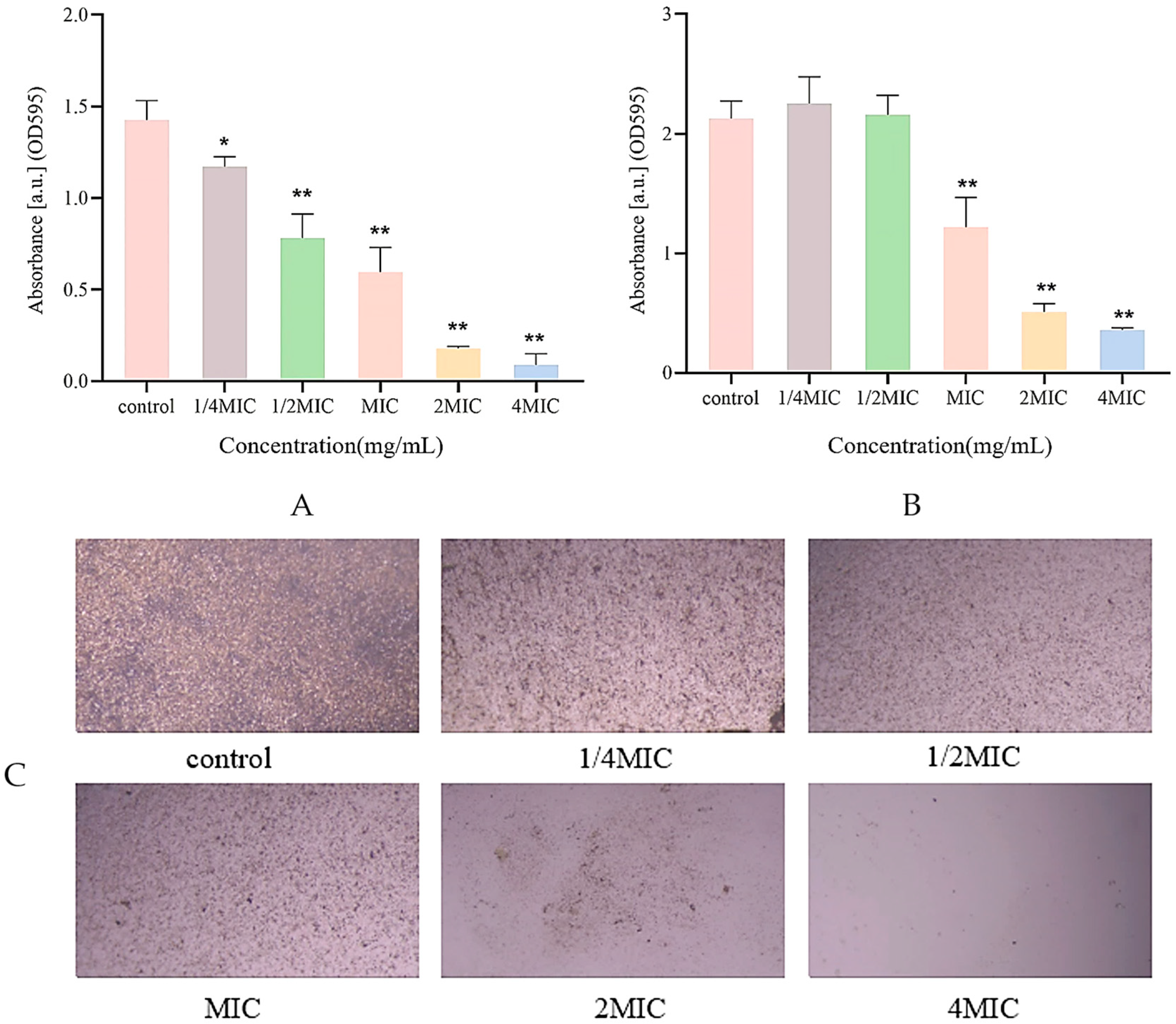
2.3. Biofilm Inhibition Assay
2.4. Biofilm Structure Detection
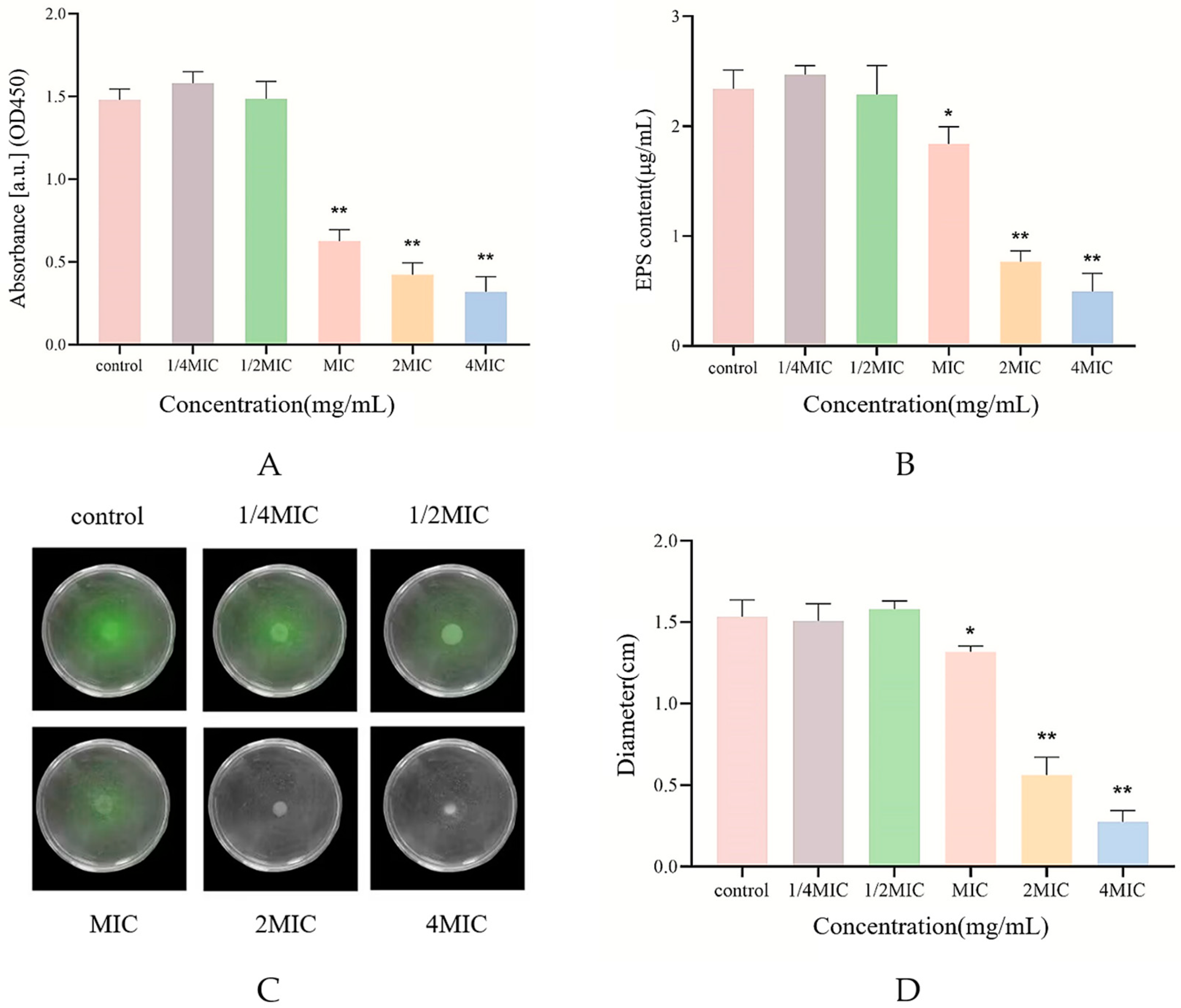
2.5. Measurement of Biofilm Biomass
2.6. Biofilm Metabolic Activity Assay
2.7. Swarming Motility Assay
2.8. Determination of Extracellular Polysaccharide Content
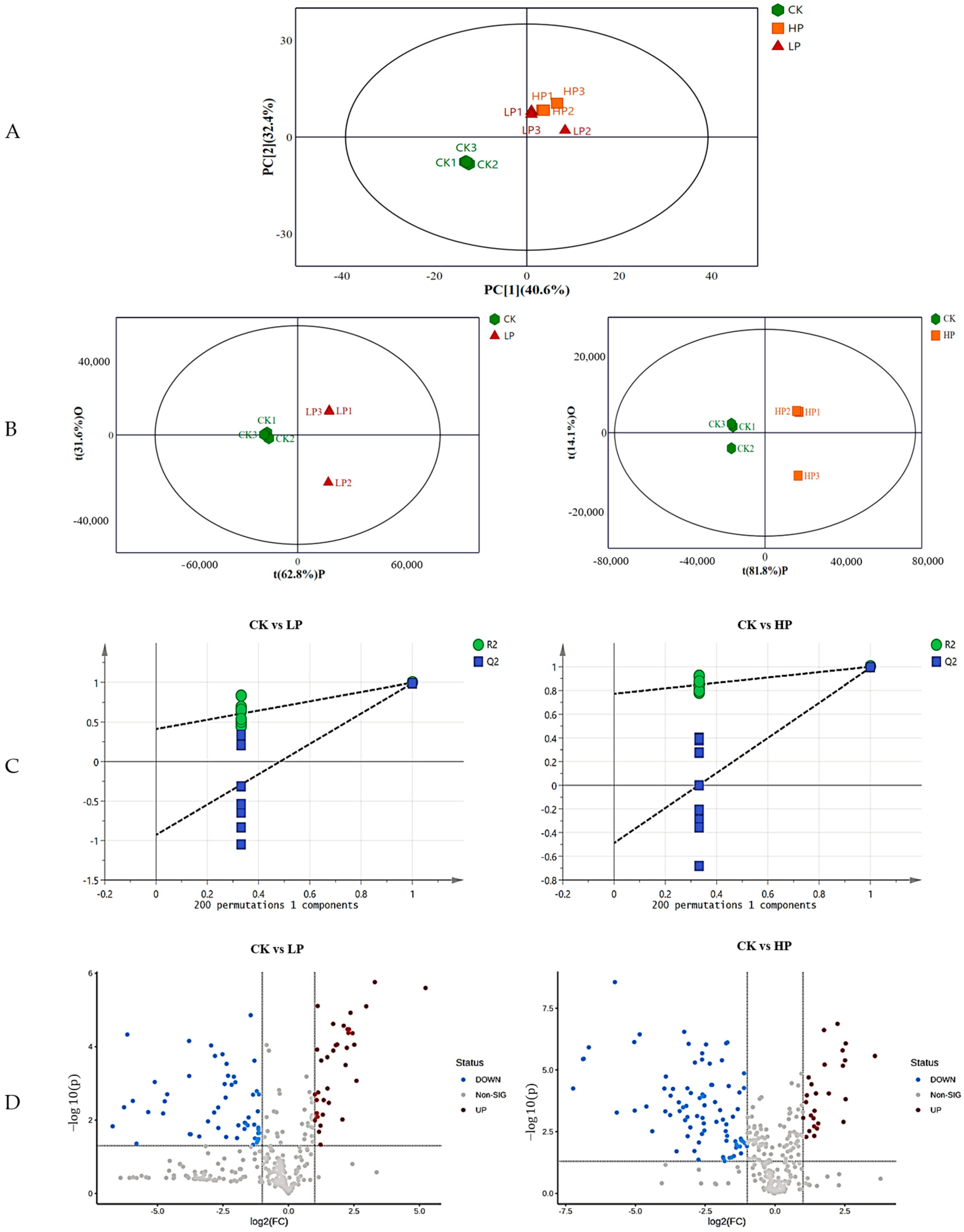
2.9. Metabolomics Analysis
2.9.1. Effect of Different Treatments on the Metabolic Profile of Biofilm
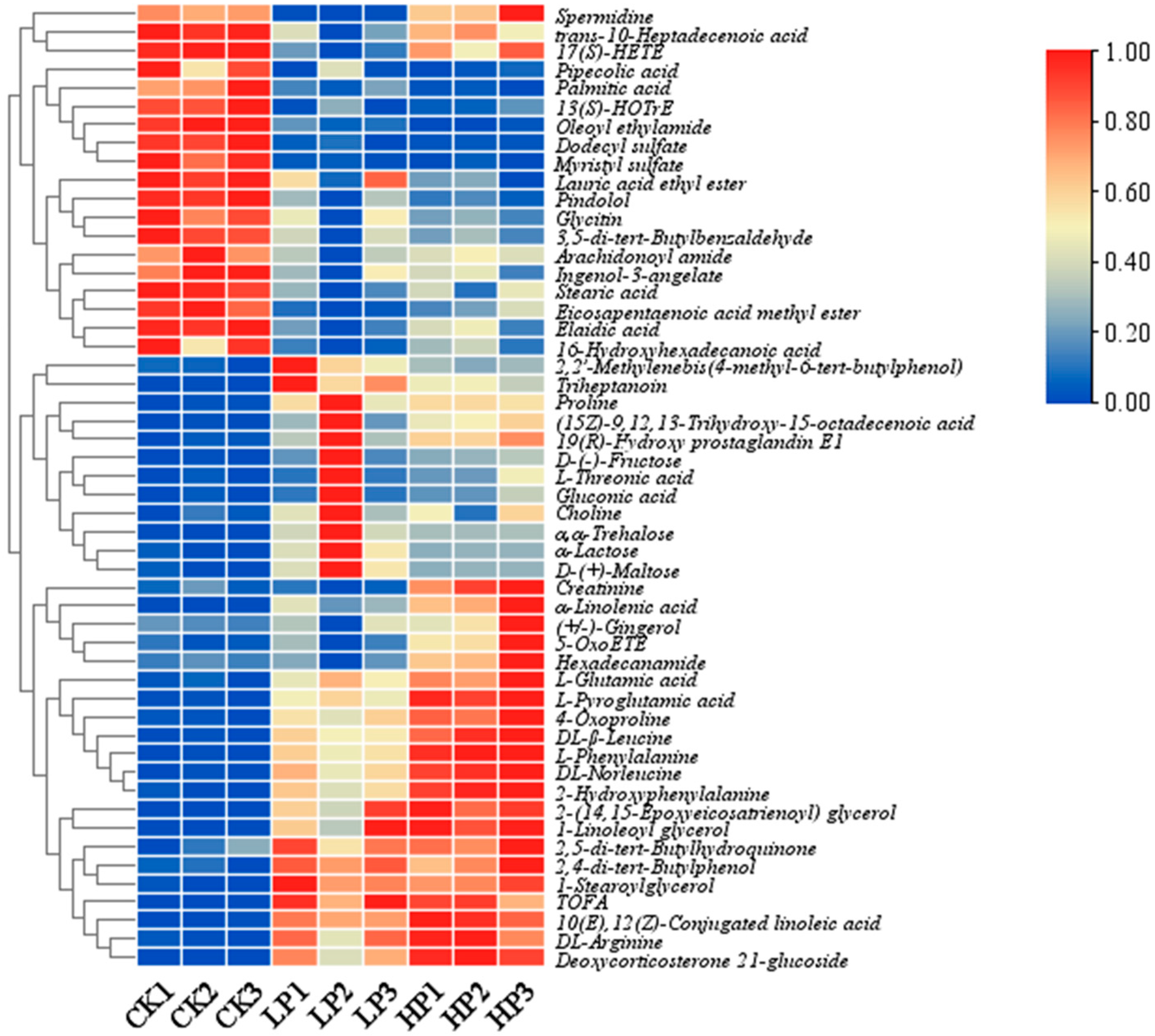
2.9.2. Identification of Differential Metabolites
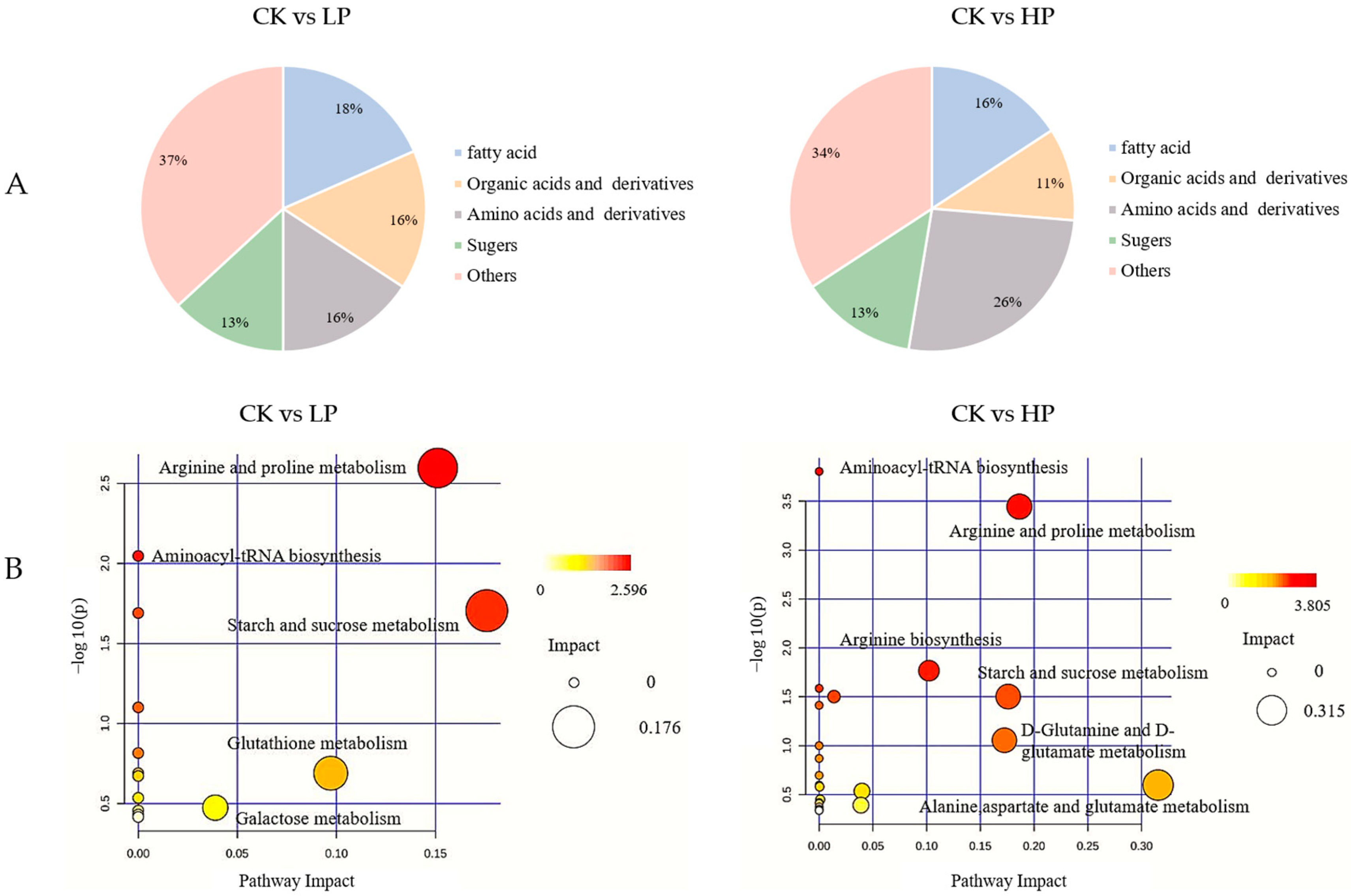
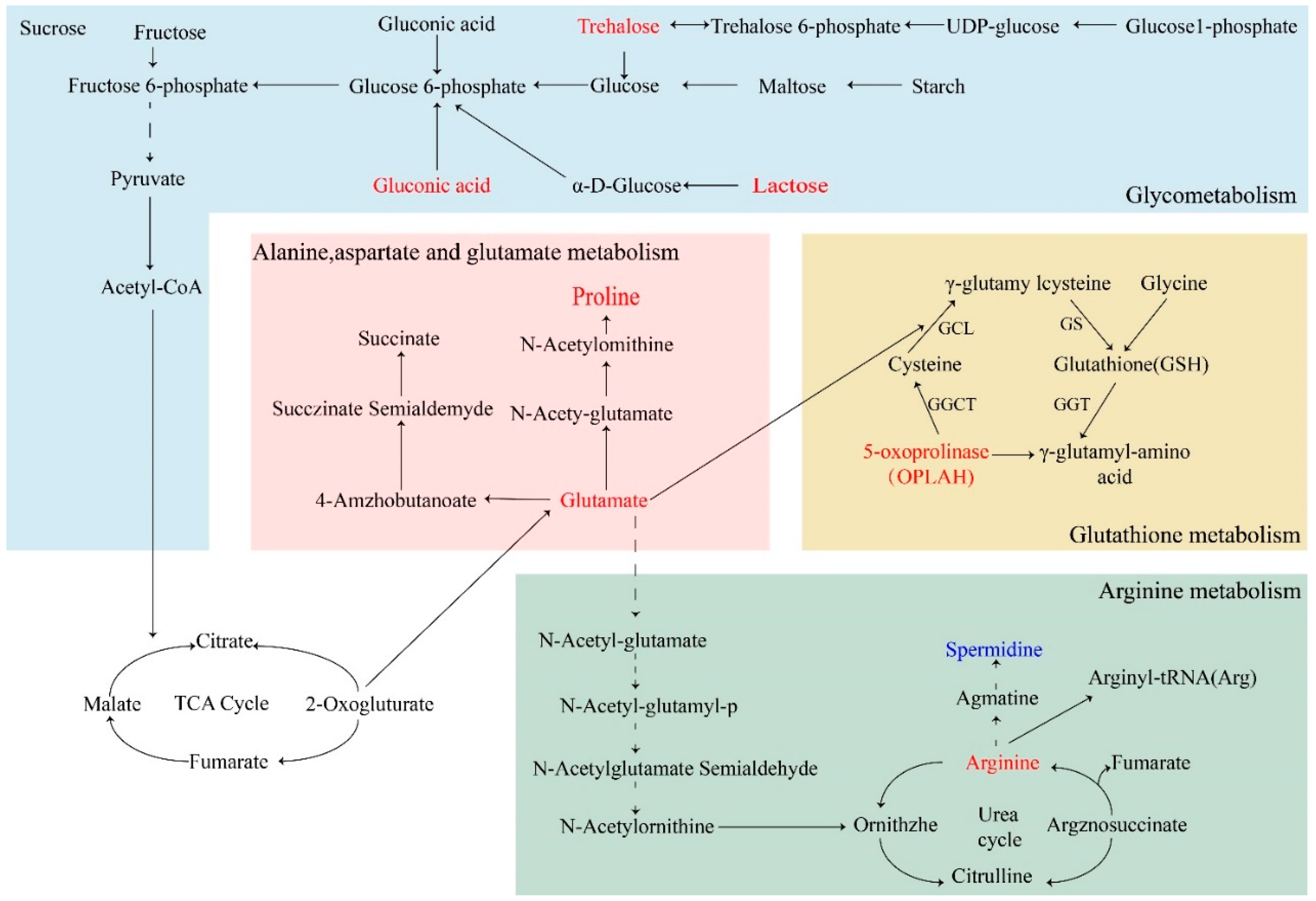
2.9.3. Metabolic Pathway Analysis
3. Discussion
4. Materials and Methods
4.1. Strain and Culture
4.2. Anti-Microbial Activity
4.2.1. Agar Diffusion Test
4.2.2. Minimum Inhibitory Concentration (MIC) and Minimum Bactericidal Concentration (MBC)
4.3. Biofilm Inhibition Test
4.3.1. Biofilm Biomass Assay
4.3.2. Biofilm Structure Detection
4.4. Biofilm Elimination Tests
4.4.1. Biofilm Biomass Determination
4.4.2. Biofilm Metabolic Activity Assay
4.5. Plate-Based Assay for Swarming Motility
4.6. Determination of Extracellular Polysaccharide Content
4.6.1. Preparation of Standard Curve
4.6.2. Preparation of Crude Polysaccharide
4.7. Metabolite Analysis
4.7.1. Collection of Biofilm
4.7.2. Extraction of Metabolites
4.7.3. LC-MS/MS Analysis
4.8. Data Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sharma, G.; Rao, S.; Bansal, A.; Dang, S.; Gupta, S.; Gabrani, R. Pseudomonas aeruginosa biofilm: Potential therapeutic targets. Biologicals 2014, 42, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Balasubramanian, D.; Schneper, L.; Kumari, H.; Mathee, K. A dynamic and intricate regulatory network determines Pseudomonas aeruginosa virulence. Nucleic Acids Res. 2013, 41, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Skariyachan, S.; Sridhar, V.S.; Packirisamy, S.; Kumargowda, S.T.; Challapilli, S.B. Recent perspectives on the molecular basis of biofilm formation by Pseudomonas aeruginosa and approaches for treatment and biofilm dispersal. Folia Microbiol. 2018, 63, 413–432. [Google Scholar] [CrossRef] [PubMed]
- Dosler, S.; Karaaslan, E. Inhibition and destruction of Pseudomonas aeruginosa biofilms by antibiotics and antimicrobial peptides. Peptides 2014, 62, 32–37. [Google Scholar] [CrossRef] [PubMed]
- Page, M.G.; Heim, J. Prospects for the next anti-Pseudomonas drug. Curr. Opin. Pharmacol. 2009, 9, 558–565. [Google Scholar] [CrossRef] [PubMed]
- Favre-Bonté, S.; Chamot, E.; Köhler, T.; Romand, J.-A.; van Delden, C. Autoinducer production and quorum-sensing dependent phenotypes of Pseudomonas aeruginosa vary according to isolation site during colonization of intubated patients. BMC Microbiol. 2007, 7, 33. [Google Scholar] [CrossRef] [PubMed]
- Marmont, L.S.; Whitfield, G.B.; Rich, J.D.; Yip, P.; Giesbrecht, L.B.; Stremick, C.A.; Whitney, J.C.; Parsek, M.R.; Harrison, J.J.; Howell, P.L. PelA and PelB proteins form a modification and secretion complex essential for Pel polysaccharide-dependent biofilm formation in Pseudomonas aeruginosa. J. Biol. Chem. 2017, 292, 19411–19422. [Google Scholar] [CrossRef]
- Annous, B.A.; Fratamico, P.M.; Smith, J.L. Scientific Status Summary. J. Food Sci. 2009, 74, R24–R37. [Google Scholar] [CrossRef]
- Byrd, M.S.; Pang, B.; Mishra, M.; Swords, W.E.; Wozniak, D.J. The Pseudomonas aeruginosa Exopolysaccharide Psl Facilitates Surface Adherence and NF-κB Activation in A549 Cells. mBio 2010, 1, 10–1128. [Google Scholar] [CrossRef]
- Colvin, K.M.; Alnabelseya, N.; Baker, P.; Whitney, J.C.; Howell, P.L.; Parsek, M.R. PelA Deacetylase Activity Is Required for Pel Polysaccharide Synthesis in Pseudomonas aeruginosa. J. Bacteriol. 2013, 195, 2329–2339. [Google Scholar] [CrossRef]
- Jennings, L.K.; Storek, K.M.; Ledvina, H.E.; Coulon, C.; Marmont, L.S.; Sadovskaya, I.; Secor, P.R.; Tseng, B.S.; Scian, M.; Filloux, A.; et al. Pel is a cationic exopolysaccharide that cross-links extracellular DNA in the Pseudomonas aeruginosa biofilm matrix. Proc. Natl. Acad. Sci. USA 2015, 112, 11353–11358. [Google Scholar] [CrossRef] [PubMed]
- Wei, Q.; Ma, L.Z. Biofilm Matrix and Its Regulation in Pseudomonas aeruginosa. Int. J. Mol. Sci. 2013, 14, 20983–21005. [Google Scholar] [CrossRef] [PubMed]
- Hay, I.D.; Remminghorst, U.; Rehm, B.H.A. MucR, a Novel Membrane-Associated Regulator of Alginate Biosynthesis in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2009, 75, 1110–1120. [Google Scholar] [CrossRef] [PubMed]
- Campodónico, V.L.; Llosa, N.J.; Grout, M.; Döring, G.; Maira-Litrán, T.; Pier, G.B. Evaluation of Flagella and Flagellin of Pseudomonas aeruginosa as Vaccines. Infect. Immun. 2010, 78, 746–755. [Google Scholar] [CrossRef] [PubMed]
- Hazes, B.; Sastry, P.A.; Hayakawa, K.; Read, R.J.; Irvin, R.T. Crystal structure of Pseudomonas aeruginosa PAK pilin suggests a main-chain-dominated mode of receptor binding. J. Mol. Biol. 2000, 299, 1005–1017. [Google Scholar] [CrossRef] [PubMed]
- Rybtke, M.; Jensen, P.Ø.; Nielsen, C.H.; Tolker-Nielsen, T. The Extracellular Polysaccharide Matrix of Pseudomonas aeruginosa Biofilms Is a Determinant of Polymorphonuclear Leukocyte Responses. Infect. Immun. 2020, 89, e00631-20. [Google Scholar] [CrossRef] [PubMed]
- Aboagye, C.I.; Ampomah, G.B.; Mensah, J.O.; Mensah, C.N.; Nartey, D.; Gasu, E.N.; Borquaye, L.S. N-benzylimidazoles as potential antimicrobial and antibiofilm agents—Syntheses, bioactivity and molecular docking studies. Sci. Afr. 2023, 19, e01529. [Google Scholar] [CrossRef]
- Hills, O.J.; Smith, J.; Scott, A.J.; Devine, D.A.; Chappell, H.F. Cation complexation by mucoid Pseudomonas aeruginosa extracellular polysaccharide. PLoS ONE 2021, 16, e0257026. [Google Scholar] [CrossRef]
- Daboor, S.M.; Rohde, J.R.; Cheng, Z. Disruption of the extracellular polymeric network of Pseudomonas aeruginosa biofilms by alginate lyase enhances pathogen eradication by antibiotics. J. Cyst. Fibros. 2021, 20, 264–270. [Google Scholar] [CrossRef]
- Fernández-Billón, M.; Llambías-Cabot, A.E.; Jordana-Lluch, E.; Oliver, A.; Macià, M.D. Mechanisms of antibiotic resistance in Pseudomonas aeruginosa biofilms. Biofilm 2023, 5, 100129. [Google Scholar] [CrossRef]
- Qin, S.; Xiao, W.; Zhou, C.; Pu, Q.; Deng, X.; Lan, L.; Liang, H.; Song, X.; Wu, M. Pseudomonas aeruginosa: Pathogenesis, virulence factors, antibiotic resistance, interaction with host, technology advances and emerging therapeutics. Signal Transduct. Target. Ther. 2022, 7, 199. [Google Scholar] [CrossRef] [PubMed]
- Cheung-Lee, W.L.; Parry, M.E.; Cartagena, A.J.; Darst, S.A.; Link, A.J. Discovery and structure of the antimicrobial lasso. J. Biol. Chem. 2019, 294, 6822–6830. [Google Scholar] [CrossRef] [PubMed]
- Deb, S.; Dutta, T.K. Characterization of an antimicrobial pentapeptide produced by a drug-sensitive Pseudomonas aeruginosa strain PAST18. Biochem. Biophys. Res. Commun. 2023, 663, 78–86. Available online: https://www.sciencedirect.com/science/article/pii/S0006291X23004862 (accessed on 25 April 2023). [CrossRef] [PubMed]
- Shi, N.; Gao, Y.; Yin, D.; Song, Y.; Kang, J.; Li, X.; Zhang, Z.; Feng, X.; Duan, J. The effect of the sub-minimal inhibitory concentration and the concentrations within resistant mutation window of ciprofloxacin on MIC, swimming motility and biofilm formation of Pseudomonas aeruginosa. Microb. Pathog. 2019, 137, 103765. [Google Scholar] [CrossRef] [PubMed]
- Batoni, G.; Maisetta, G.; Esin, S. Antimicrobial peptides and their interaction with biofilms of medically relevant bacteria. Biochim. Biophys. Acta (BBA) Biomembr. 2016, 1858, 1044–1060. [Google Scholar] [CrossRef] [PubMed]
- Verdon, J.; Coutos-Thevenot, P.; Rodier, M.-H.; Landon, C.; Depayras, S.; Noel, C.; La Camera, S.; Moumen, B.; Greve, P.; Bouchon, D.; et al. Armadillidin H, a Glycine-Rich Peptide from the Terrestrial Crustacean Armadillidium vulgare, Displays an Unexpected Wide Antimicrobial Spectrum with Membranolytic Activity. Front. Microbiol. 2016, 7, 1484. [Google Scholar] [CrossRef] [PubMed]
- Jamal, M.; Ahmad, W.; Andleeb, S.; Jalil, F.; Imran, M.; Nawaz, M.A.; Hussain, T.; Ali, M.; Rafiq, M.; Kamil, M.A. Bacterial biofilm and associated infections. J. Chin. Med. Assoc. 2018, 81, 7–11. [Google Scholar] [CrossRef] [PubMed]
- Olivares, E.; Badel-Berchoux, S.; Provot, C.; Prévost, G.; Bernardi, T.; Jehl, F. Clinical Impact of Antibiotics for the Treatment of Pseudomonas aeruginosa Biofilm Infections. Front. Microbiol. 2020, 10, 2894. [Google Scholar] [CrossRef] [PubMed]
- Srey, S.; Jahid, I.K.; Ha, S.-D. Biofilm formation in food industries: A food safety concern. Food Control 2013, 31, 572–585. [Google Scholar] [CrossRef]
- Noori, H.G.; Tadjrobehkar, O.; Moazamian, E. Biofilm formation capacity of Pseudomonas aeruginosa is significantly enhanced by sub-inhibitory concentrations of Tomatidine. Gene Rep. 2022, 27, 101570. [Google Scholar] [CrossRef]
- Koo, H.; Allan, R.N.; Howlin, R.P.; Hall-Stoodley, L.; Stoodley, P. Targeting microbial biofilms: Current and prospective therapeutic strategies. Nat. Rev. Microbiol. 2017, 15, 740–755. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Tsuji, G.; Opoku-Temeng, C.; Sintim, H.O. Inhibition of P. aeruginosa c-di-GMP phosphodiesterase RocR and swarming motility by a benzoisothiazolinone derivative. Chem. Sci. 2016, 7, 6238–6244. [Google Scholar] [CrossRef] [PubMed]
- Verderosa, A.D.; Totsika, M.; Fairfull-Smith, K.E. Bacterial Biofilm Eradication Agents: A Current Review. Front. Chem. 2019, 7, 824. [Google Scholar] [CrossRef] [PubMed]
- Palmer, S.R.; Ren, Z.; Hwang, G.; Liu, Y.; Combs, A.; Söderström, B.; Vasquez, P.L.; Khosravi, Y.; Brady, L.J.; Koo, H.; et al. Streptococcus mutans yidC1 and yidC2 Impact Cell Envelope Biogenesis, the Biofilm Matrix, and Biofilm Biophysical Properties. J. Bacteriol. 2018, 201, e00396-18. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Hu, K.; Hong, B.; Lu, X.; Liu, Y.; Xie, J.; Jin, S.; Zhou, S.; Zhao, Q.; Lu, H.; et al. The inhibitory effect of Bacillus amyloliquefaciens L1 on Aeromonas hydrophila and its mechanism. Aquaculture 2021, 539, 736590. [Google Scholar] [CrossRef]
- El-Telbany, M.; El-Sharaki, A. Antibacterial and anti-biofilm activity of silver nanoparticles on multi-drug resistance pseudomonas aeruginosa isolated from dental-implant. J. Oral Biol. Craniofacial Res. 2022, 12, 199–203. [Google Scholar] [CrossRef]
- El Khatib, K.; Hadeer, R.A.; Saad, A.; Kalaydjian, A.; Fayad, E.; Mahfouz, Y.; Dougnon, V.; Daoud, Z.; Abdel-Massih, R.M. Determination of MIC, MPC, and MSW of Ilex paraguariensis against non-typhoidal Salmonella with identification of the mechanisms of resistance and pathogenicity factors. Microb. Pathog. 2023, 174, 105905. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.J.; Kim, Y.J.; Yu, H.H.; Lee, N.-K.; Paik, H.-D. Cell-free supernatants of Bacillus subtilis and Bacillus polyfermenticus inhibit Listeria monocytogenes biofilm formation. Food Control 2023, 144, 109387. [Google Scholar] [CrossRef]
- Djordjevic, D.; Wiedmann, M.; McLandsborough, L.A. Microtiter Plate Assay for Assessment of Listeria monocytogenes Biofilm Formation. Appl. Environ. Microbiol. 2002, 68, 2950–2958. [Google Scholar] [CrossRef]
- Guo, L.; Zhang, C.; Chen, G.; Wu, M.; Liu, W.; Ding, C.; Dong, Q.; Fan, E.; Liu, Q. Reactive oxygen species inhibit biofilm formation of Listeria monocytogenes. Microb. Pathog. 2019, 127, 183–189. [Google Scholar] [CrossRef]
- Alonso, B.; Cruces, R.; Pérez, A.; Sánchez-Carrillo, C.; Guembe, M. Comparison of the XTT and resazurin assays for quantification of the metabolic activity of Staphylococcus aureus biofilm. J. Microbiol. Methods 2017, 139, 135–137. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Li, C.; Abdel-Samie, M.A.; Cui, H.; Lin, L. Unraveling the inhibitory mechanism of clove essential oil against Listeria monocytogenes biofilm and applying it to vegetable surfaces. LWT 2020, 134, 110210. [Google Scholar] [CrossRef]
- Khan, F.; Jeong, M.-C.; Park, S.-K.; Kim, S.-K.; Kim, Y.-M. Contribution of chitooligosaccharides to biofilm formation, antibiotics resistance and disinfectants tolerance of Listeria monocytogenes. Microb. Pathog. 2019, 136, 103673. [Google Scholar] [CrossRef] [PubMed]
- Cuesta, G.; Suarez, N.; Bessio, M.I.; Ferreira, F.; Massaldi, H. Quantitative determination of pneumococcal capsular polysaccharide serotype 14 using a modification of phenol-sulfuric acid method. J. Microbiol. Methods 2003, 52, 69–73. [Google Scholar] [CrossRef] [PubMed]
- Saha, A.K.; Brewer, C.F. Determination of the concentrations of oligosaccharides, complex type carbohydrates, and glycoproteins using the phenol-sulfuric acid method. Carbohydr. Res. 1994, 254, 157–167. [Google Scholar] [CrossRef]
- Song, J.; Lee, H.; Lee, H. Extensive analysis of monosaccharide diversity in fermented soybean paste using LC-MS/MS. J. Food Compos. Anal. 2024, 130, 106149. [Google Scholar] [CrossRef]
| Paradigm | Time | Ingredient |
|---|---|---|
| E+ | 0–10 min | 5% A and 95% A |
| 10–12 min | 5% A and 95% B | |
| 12–13 min | 5% A and 95% B | |
| 13–14 min | 95% A and 5% B | |
| E− | 0–2.5 min | 95% C and 5% D |
| 2.5–16.5 min | 95% D and 5% C | |
| 16.5–19 min | 95% D and 5% C | |
| 19–20 min | 95% C and 5% D |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Zheng, J.; Hou, W.; Zhang, C.; Zhang, J.; Fan, X.; Zhang, H.; Han, Y. The Anti-Microbial Peptide Citrocin Controls Pseudomonas aeruginosa Biofilms by Breaking Down Extracellular Polysaccharide. Int. J. Mol. Sci. 2024, 25, 4122. https://doi.org/10.3390/ijms25074122
Wang L, Zheng J, Hou W, Zhang C, Zhang J, Fan X, Zhang H, Han Y. The Anti-Microbial Peptide Citrocin Controls Pseudomonas aeruginosa Biofilms by Breaking Down Extracellular Polysaccharide. International Journal of Molecular Sciences. 2024; 25(7):4122. https://doi.org/10.3390/ijms25074122
Chicago/Turabian StyleWang, Liyao, Jiaqi Zheng, Wenchao Hou, Chaowen Zhang, Jie Zhang, Xuanbo Fan, Hongliang Zhang, and Yuzhu Han. 2024. "The Anti-Microbial Peptide Citrocin Controls Pseudomonas aeruginosa Biofilms by Breaking Down Extracellular Polysaccharide" International Journal of Molecular Sciences 25, no. 7: 4122. https://doi.org/10.3390/ijms25074122
APA StyleWang, L., Zheng, J., Hou, W., Zhang, C., Zhang, J., Fan, X., Zhang, H., & Han, Y. (2024). The Anti-Microbial Peptide Citrocin Controls Pseudomonas aeruginosa Biofilms by Breaking Down Extracellular Polysaccharide. International Journal of Molecular Sciences, 25(7), 4122. https://doi.org/10.3390/ijms25074122