Deoxyxylulose 5-Phosphate Synthase Does Not Play a Major Role in Regulating the Methylerythritol 4-Phosphate Pathway in Poplar
Abstract
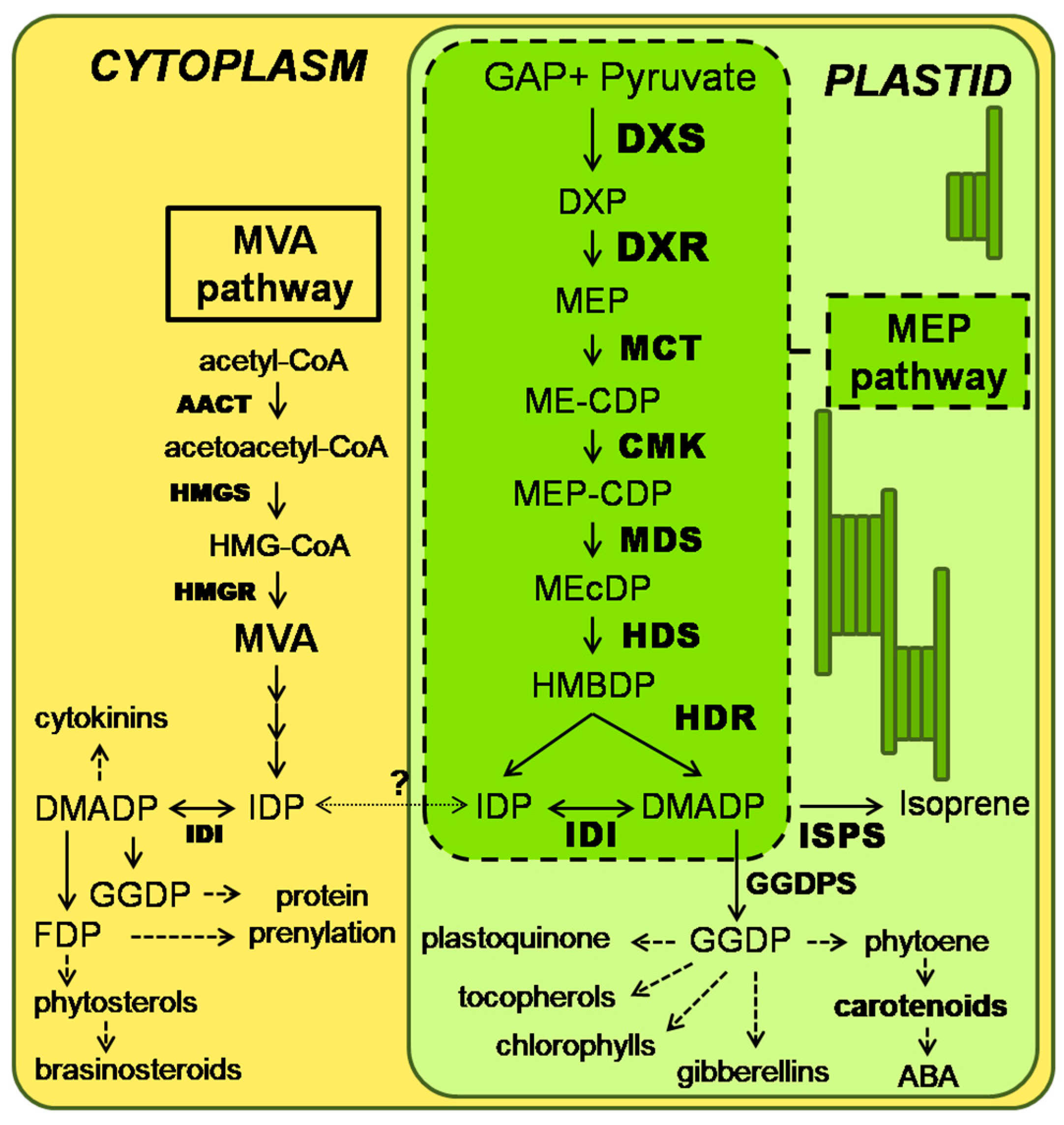
1. Introduction
2. Results
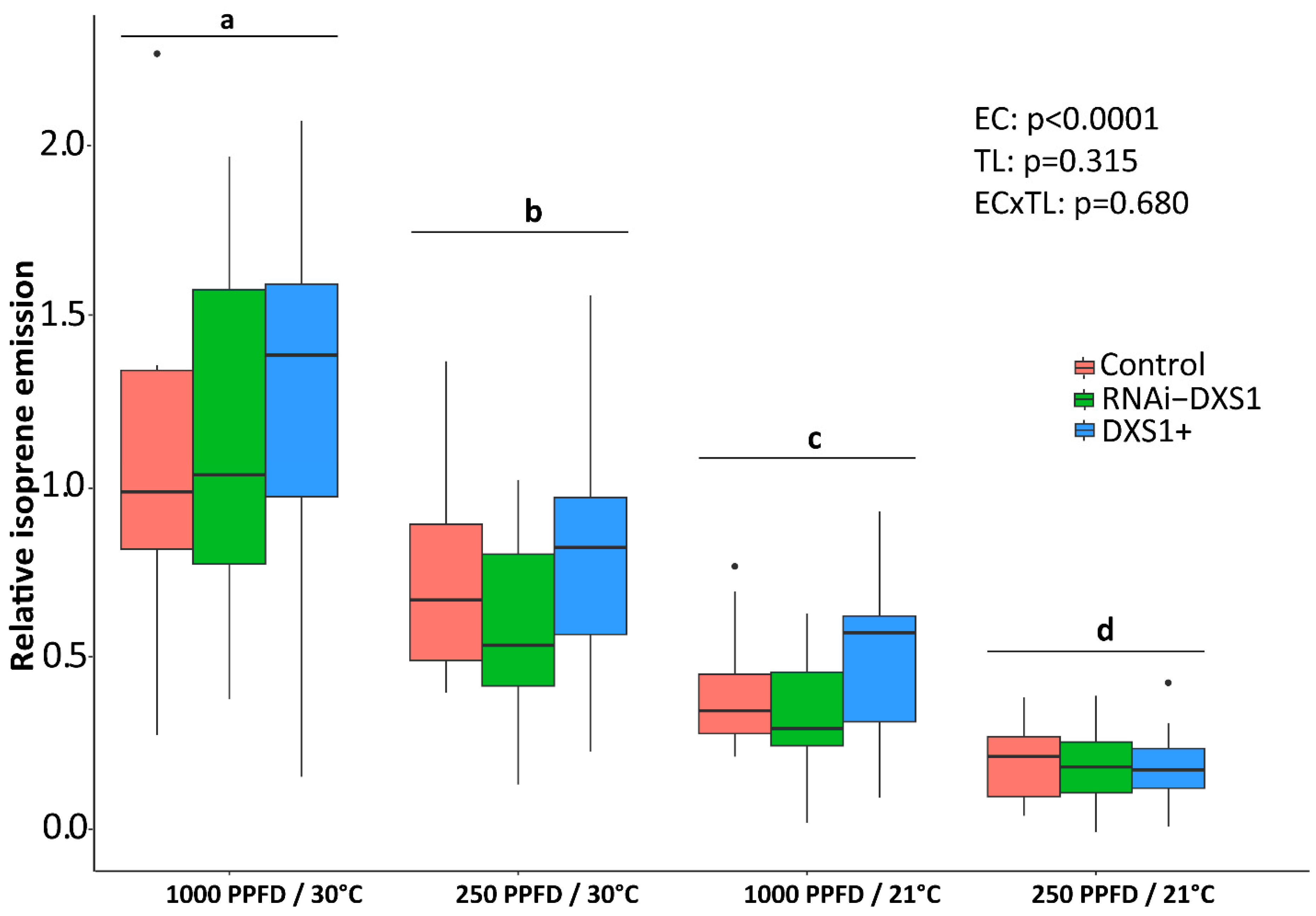
2.1. Varying Light and Temperature Conditions Have Significant Effects on Isoprene Emission
2.2. The MEP Pathway to Isoprene Maintains Metabolic Steady State during 13CO2 Labeling
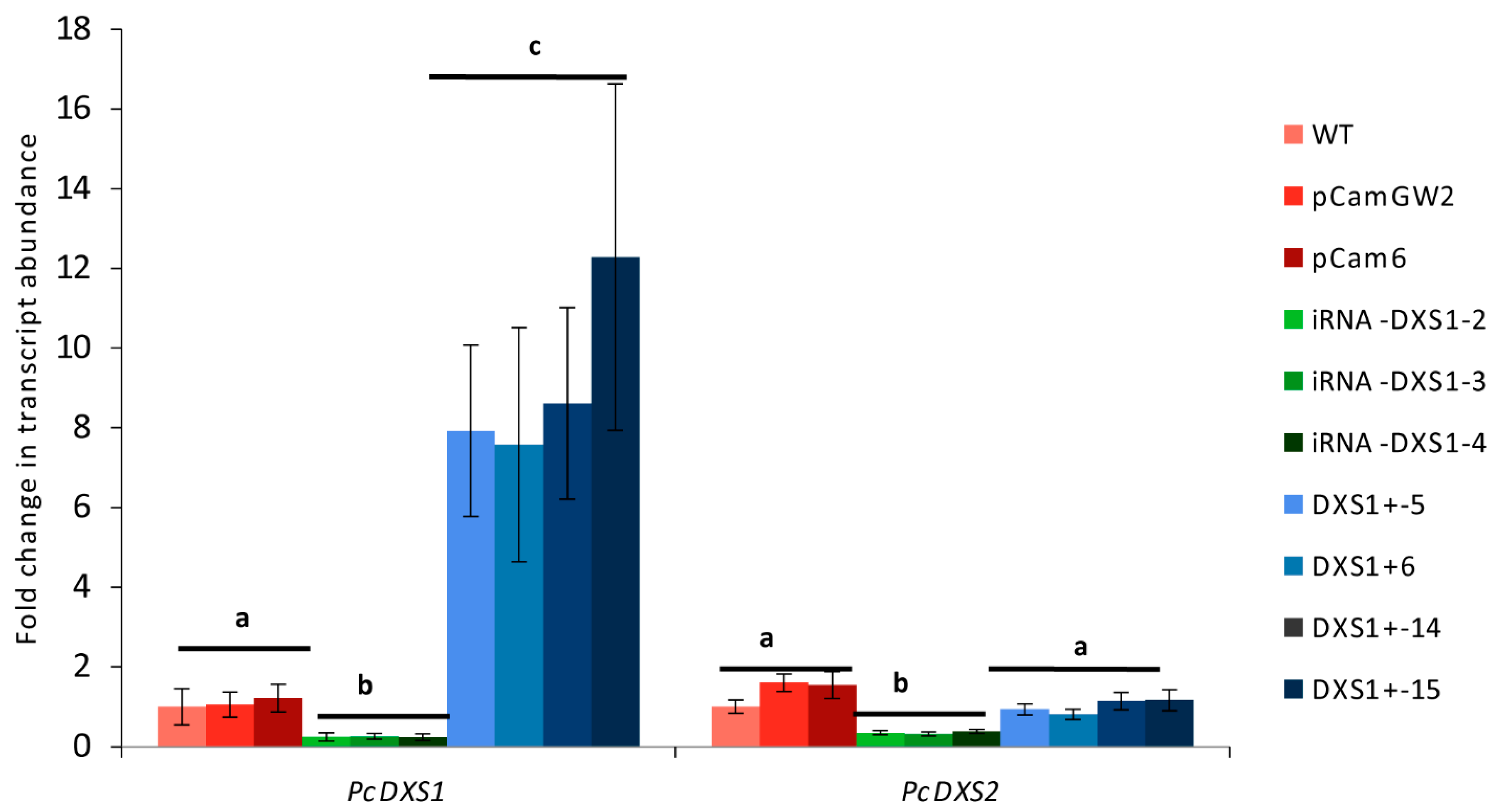
2.3. DXS Overexpression or RNA Interference Have No Effect on Transcript Levels of Other MEP or Mevalonate Pathway Genes
2.4. DXS-Silenced Lines Have Reduced DXS Enzyme Activity While Overexpression of DXS Increases Activity
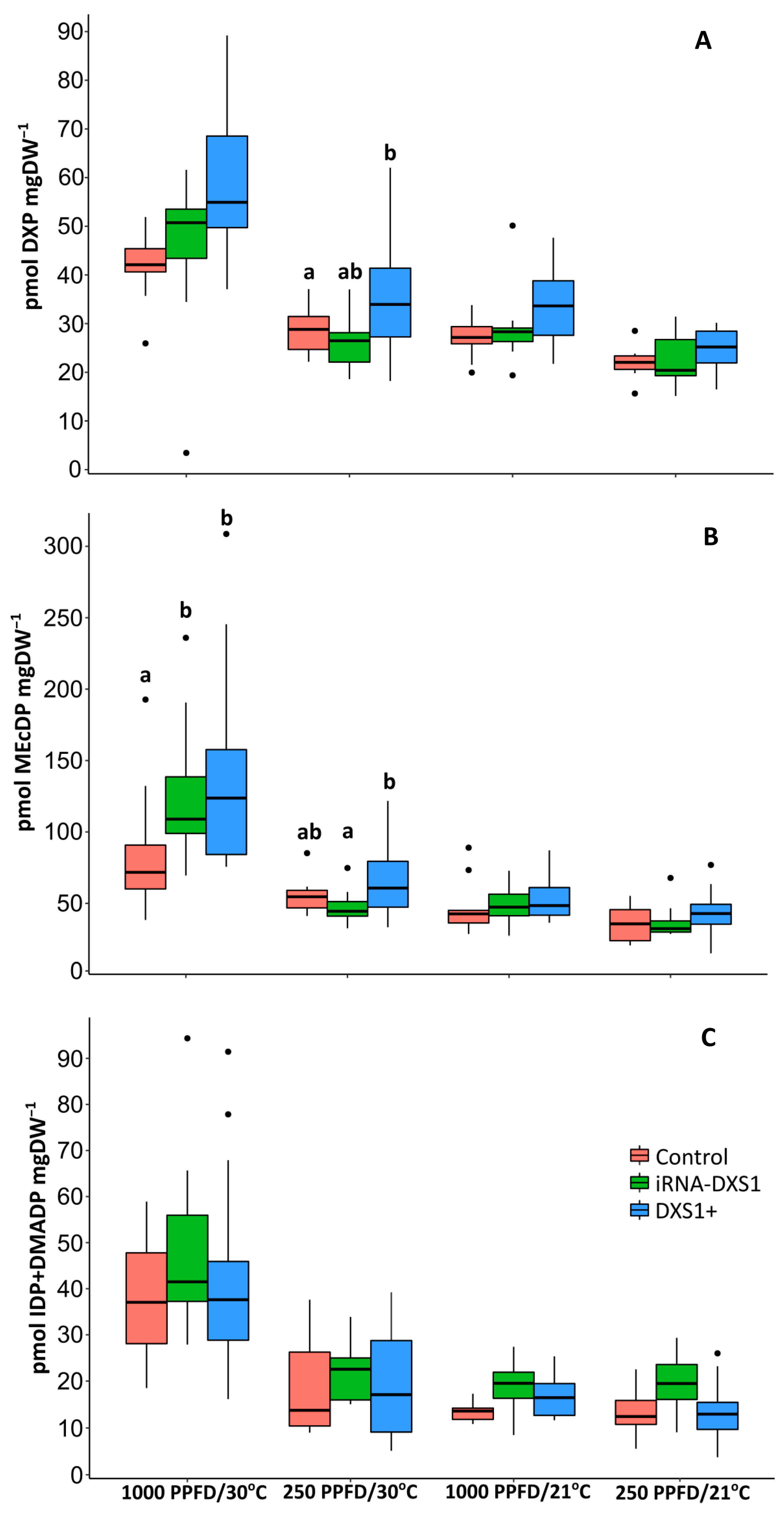
2.5. The Rate of 13C Incorporation into Isoprene and the Pool Sizes of the Three Major Metabolites of the MEP Pathway Were Used to Calculate the Carbon Flux
2.6. The Flux Control Coefficients for DXS Are Low under All Environmental Conditions Tested
2.7. Silencing DXS Results in Decreases in Some Isoprenoid End Products Compared with Overexpression Lines
3. Discussion
3.1. Photosynthetically Fixed Carbon Supplies the MEP Pathway in Grey Poplar Leaves
3.2. DXS Is Not a Major Rate-Controlling Enzyme of the MEP Pathway in Poplar
3.3. The MEP Pathway and the Role of Isoprene
4. Materials and Methods
4.1. Plant Material and Experimental Set-Up
4.2. Vector Construction and Transformation of Populus × canescens
4.3. Quantification and Label Incorporation Measurements of DXP, MEcDP and IDP/DMADP
4.4. Measurement of Isoprene Emission and Determination of Stable 13C Isotopes of Isoprene with PTR-MS
4.5. Determination of Flux by Label Incorporation through the MEP Pathway and Isoprene
4.6. Determination of In Vitro PcDXS Activity
4.7. Calculation of Flux Control Coefficients
4.8. RNA Extraction, cDNA Synthesis, and Quantitative Real-Time PCR
4.9. HPLC Analysis of Plant Pigments
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gershenzon, J.; Dudareva, N. The function of terpene natural products in the natural world. Nat. Chem. Biol. 2007, 3, 408–414. [Google Scholar] [CrossRef] [PubMed]
- Croteau, R.; Kutchan, T.M.; Lewis, N.G. Natural Products (Secondary Metabolites). Biochem. Mol. Biol. Plants 2000, 24, 1250–1319. [Google Scholar]
- Croteau, R. Biosynthesis and Catabolism of Monoterpenoids. Chem. Rev. 1987, 87, 929–954. [Google Scholar] [CrossRef]
- Eisenreich, W.; Schwarz, M.; Cartayrade, A.; Arigoni, D.; Zenk, M.H.; Bacher, A. The deoxyxylulose phosphate pathway of terpenoid biosynthesis in plants and microorganisms. Chem. Biol. 1998, 5, R22–R233. [Google Scholar] [CrossRef] [PubMed]
- Rohmer, M.; Knani, M.; Simonin, P.; Sutter, B.; Sahm, H. Isoprenoid biosynthesis in bacteria: A novel pathway for the early steps leading to isopentenyl diphosphate. Biochem. J. 1993, 295 Pt 2, 517–524. [Google Scholar] [CrossRef] [PubMed]
- Rohmer, M.; Seemann, M.; Horbach, S.; BringerMeyer, S.; Sahm, H. Glyceraldehyde 3-phosphate and pyruvate as precursors of isoprenic units in an alternative non-mevalonate pathway for terpenoid biosynthesis. J. Am. Chem. Soc. 1996, 118, 2564–2566. [Google Scholar] [CrossRef]
- Flesch, G.; Rohmer, M. Prokaryotic Hopanoids—The Biosynthesis of the Bacteriohopane Skeleton—Formation of Isoprenic Units from 2 Distinct Acetate Pools and a Novel Type of Carbon Carbon Linkage between a Triterpene and D-Ribose. Eur. J. Biochem. 1988, 175, 405–411. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, M.K. Terpen-Biosynthese in Ginkgo biloba: Eine überraschende Geschichte. Ph.D. Thesis, ETH Zürich, Zürich, Switzerland, 1994. [Google Scholar]
- Cordoba, E.; Salmi, M.; Leon, P. Unravelling the regulatory mechanisms that modulate the MEP pathway in higher plants. J. Exp. Bot. 2009, 60, 2933–2943. [Google Scholar] [CrossRef] [PubMed]
- Bouvier, F.; Rahier, A.; Camara, B. Biogenesis, molecular regulation and function of plant isoprenoids. Prog. Lipid Res. 2005, 44, 357–429. [Google Scholar] [CrossRef]
- Rodriguez-Concepcion, M.; Fores, O.; Martinez-Garcia, J.F.; Gonzalez, V.; Phillips, M.A.; Ferrer, A.; Boronat, A. Distinct light-mediated pathways regulate the biosynthesis and exchange of isoprenoid precursors during Arabidopsis seedling development. Plant Cell 2004, 16, 144–156. [Google Scholar] [CrossRef]
- Boronat, A.; Rodriguez-Concepcion, M. Terpenoid Biosynthesis in Prokaryotes. Biotechnol. Isoprenoids 2015, 148, 3–18. [Google Scholar] [CrossRef]
- Rodriguez-Concepcion, A.; Boronat, A. Isoprenoid Biosynthesis in Prokaryotic Organisms. In Isoprenoid Synthesis in Plants and Microorganisms: New Concepts and Experimental Approaches; Bach, T.J., Rohmer, M., Eds.; Springer: New York, NY, USA, 2012; pp. 1–16. [Google Scholar]
- Rodriguez-Concepcion, A. Early steps in isoprenoid biosynthesis: Multilevel regulation of the supply of common precursors in plant cells. In Phytochemistry Reviews; Springer: Berlin/Heidelberg, Germany, 2006; Volume 5, pp. 1–15. [Google Scholar]
- Laule, O.; Furholz, A.; Chang, H.S.; Zhu, T.; Wang, X.; Heifetz, P.B.; Gruissem, W.; Lange, B.M. Crosstalk between cytosolic and plastidial pathways of isoprenoid biosynthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2003, 100, 6866–6871. [Google Scholar] [CrossRef] [PubMed]
- Flugge, U.I.; Gao, W. Transport of isoprenoid intermediates across chloroplast envelope membranes. Plant Biol. 2005, 7, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Bick, J.A.; Lange, B.M. Metabolic cross talk between cytosolic and plastidial pathways of isoprenoid biosynthesis: Unidirectional transport of intermediates across the chloroplast envelope membrane. Arch. Biochem. Biophys. 2003, 415, 146–154. [Google Scholar] [CrossRef] [PubMed]
- Sprenger, G.A.; Schorken, U.; Wiegert, T.; Grolle, S.; deGraaf, A.A.; Taylor, S.V.; Begley, T.P.; BringerMeyer, S.; Sahm, H. Identification of a thiamin-dependent synthase in Escherichia coli required for the formation of the 1-deoxy-D-xylulose 5-phosphate precursor to isoprenoids, thiamin, and pyridoxol. Proc. Natl. Acad. Sci. USA 1997, 94, 12857–12862. [Google Scholar] [CrossRef]
- Lange, B.M.; Wildung, M.R.; McCaskill, D.; Croteau, R. A family of transketolases that directs isoprenoid biosynthesis via a mevalonate-independent pathway. Proc. Natl. Acad. Sci. USA 1998, 95, 2100–2104. [Google Scholar] [CrossRef] [PubMed]
- Hunter, W.N. The non-mevalonate pathway of isoprenoid precursor biosynthesis. J. Biol. Chem. 2007, 282, 21573–21577. [Google Scholar] [CrossRef]
- Phillips, M.A.; Leon, P.; Boronat, A.; Rodriguez-Concepcion, M. The plastidial MEP pathway: Unified nomenclature and resources. Trends Plant Sci. 2008, 13, 619–623. [Google Scholar] [CrossRef]
- Ward, J.L.; Baker, J.M.; Llewellyn, A.M.; Hawkins, N.D.; Beale, M.H. Metabolomic analysis of Arabidopsis reveals hemiterpenoid glycosides as products of a nitrate ion-regulated, carbon flux overflow. Proc. Natl. Acad. Sci. USA 2011, 108, 10762–10767. [Google Scholar] [CrossRef]
- Xiao, Y.M.; Savchenko, T.; Baidoo, E.E.K.; Chehab, W.E.; Hayden, D.M.; Tolstikov, V.; Corwin, J.A.; Kliebenstein, D.J.; Keasling, J.D.; Dehesh, K. Retrograde Signaling by the Plastidial Metabolite MEcPP Regulates Expression of Nuclear Stress-Response Genes. Cell 2012, 149, 1525–1535. [Google Scholar] [CrossRef]
- Gonzalez-Cabanelas, D.; Wright, L.P.; Paetz, C.; Onkokesung, N.; Gershenzon, J.; Rodriguez-Concepcion, M.; Phillips, M.A. The diversion of 2-C-methyl-D-erythritol-2,4-cyclodiphosphate from the 2-C-methyl-D-erythritol 4-phosphate pathway to hemiterpene glycosides mediates stress responses in Arabidopsis thaliana. Plant J. 2015, 82, 122–137. [Google Scholar] [CrossRef] [PubMed]
- Lemos, M.; Xiao, Y.M.; Bjornson, M.; Wang, J.Z.; Hicks, D.; de Souza, A.; Wang, C.Q.; Yang, P.Y.; Ma, S.S.; Dinesh-Kumar, S.; et al. The plastidial retrograde signal methyl erythritol cyclopyrophosphate is a regulator of salicylic acid and jasmonic acid crosstalk. J. Exp. Bot. 2016, 67, 1557–1566. [Google Scholar] [CrossRef] [PubMed]
- Onkokesung, N.; Reichelt, M.; Wright, L.P.; Phillips, M.A.; Gershenzon, J.; Dicke, M. The plastidial metabolite 2-C-methyl-D-erythritol-2,4-cyclodiphosphate modulates defence responses against aphids. Plant Cell Environ. 2019, 42, 2309–2323. [Google Scholar] [CrossRef] [PubMed]
- Zuo, Z.; Weraduwage, S.M.; Lantz, A.T.; Sanchez, L.M.; Weise, S.E.; Wang, J.; Childs, K.L.; Sharkey, T.D. Isoprene acts as a signaling molecule in gene networks important for stress responses and plant growth. Plant Physiol. 2019, 180, 124–152. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Sharkey, T. Methylerythritol 4-phosphate (MEP) pathway metabolic regulation. Nat. Prod. Rep. 2014, 31, 1043–1055. [Google Scholar] [CrossRef] [PubMed]
- Wright, L.P.; Rohwer, J.M.; Ghirardo, A.; Hammerbacher, A.; Ortiz-Alcaide, M.; Raguschke, B.; Schnitzler, J.P.; Gershenzon, J.; Phillips, M.A. Deoxyxylulose 5-Phosphate Synthase Controls Flux through the Methylerythritol 4-Phosphate Pathway in Arabidopsis. Plant Physiol. 2014, 165, 1488–1504. [Google Scholar] [CrossRef]
- Krause, T.; Wiesinger, P.; González-Cabanelas, D.; Lackus, N.; Köllner, T.G.; Klüpfel, T.; Williams, J.; Rohwer, J.; Gershenzon, J.; Schmidt, A. HDR, the last enzyme in the MEP pathway, differently regulates isoprenoid biosynthesis in two woody plants. Plant Physiol. 2023, 192, 767–788. [Google Scholar] [CrossRef]
- Ma, D.; Li, G.; Zhu, Y.; Xie, D.-Y. Overexpression and suppression of Artemisia annua 4-hydroxy-3-methylbut-2-enyl diphosphate reductase 1 gene (AaHDR1) differentially regulate artemisinin and terpenoid biosynthesis. Front. Plant Sci. 2017, 8, 77. [Google Scholar] [CrossRef]
- Lee, Y.J.; Kim, J.K.; Baek, S.-A.; Yu, J.-S.; You, M.K.; Ha, S.-H. Differential regulation of an OsIspH1, the functional 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, for photosynthetic pigment biosynthesis in rice leaves and seeds. Front. Plant Sci. 2022, 13, 861036. [Google Scholar] [CrossRef]
- Banerjee, A.; Wu, Y.; Banerjee, R.; Li, Y.; Yan, H.; Sharkey, T.D. Feedback inhibition of deoxy-D-xylulose-5-phosphate synthase regulates the methylerythritol 4-phosphate pathway. J. Biol. Chem. 2013, 288, 16926–16936. [Google Scholar] [CrossRef]
- Perreca, E.; Rohwer, J.; González-Cabanelas, D.; Loreto, F.; Schmidt, A.; Gershenzon, J.; Wright, L.P. Effect of drought on the methylerythritol 4-phosphate (MEP) pathway in the isoprene emitting conifer Picea glauca. Front. Plant Sci. 2020, 11, 546295. [Google Scholar] [CrossRef]
- Mitra, S.; Estrada-Tejedor, R.; Volke, D.C.; Phillips, M.A.; Gershenzon, J.; Wright, L.P. Negative regulation of plastidial isoprenoid pathway by herbivore-induced β-cyclocitral in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2021, 118, e2008747118. [Google Scholar] [CrossRef] [PubMed]
- Carretero-Paulet, L.; Cairo, A.; Talavera, D.; Saura, A.; Imperial, S.; Rodriguez-Concepcion, M.; Campos, N.; Boronat, A. Functional and evolutionary analysis of DXL1, a non-essential gene encoding a 1-deoxy-D-xylulose 5-phosphate synthase like protein in Arabidopsis thaliana. Gene 2013, 524, 40–53. [Google Scholar] [CrossRef] [PubMed]
- Estevez, J.M.; Cantero, A.; Reindl, A.; Reichler, S.; Leon, P. 1-deoxy-D-xylulose-5-phosphate synthase, a limiting enzyme for plastidic isoprenoid biosynthesis in plants. J. Biol. Chem. 2001, 276, 22901–22909. [Google Scholar] [CrossRef] [PubMed]
- Munoz-Bertomeu, J.; Arrillaga, I.; Ros, R.; Segura, J. Up-regulation of 1-deoxy-D-xylulose-5-phosphate synthase enhances production of essential oils in transgenic spike lavender. Plant Physiol. 2006, 142, 890–900. [Google Scholar] [CrossRef] [PubMed]
- Lois, L.M.; Rodriguez-Concepcion, M.; Gallego, F.; Campos, N.; Boronat, A. Carotenoid biosynthesis during tomato fruit development: Regulatory role of 1-deoxy-D-xylulose 5-phosphate synthase. Plant J. 2000, 22, 503–513. [Google Scholar] [CrossRef] [PubMed]
- Enfissi, E.M.A.; Fraser, P.D.; Lois, L.M.; Boronat, A.; Schuch, W.; Bramley, P.M. Metabolic engineering of the mevalonate and non-mevalonate isopentenyl diphosphate-forming pathways for the production of health-promoting isoprenoids in tomato. Plant Biotechnol. J. 2005, 3, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Morris, W.L.; Ducreux, L.J.M.; Hedden, P.; Millam, S.; Taylor, M.A. Overexpression of a bacterial 1-deoxy-D-xylulose 5-phosphate synthase gene in potato tubers perturbs the isoprenoid metabolic network: Implications for the control of the tuber life cycle. J. Exp. Bot. 2006, 57, 3007–3018. [Google Scholar] [CrossRef] [PubMed]
- Gong, Y.F.; Liao, Z.H.; Guo, B.H.; Sun, X.F.; Tang, K.X. Molecular cloning and expression profile analysis of Ginkgo biloba DXS gene encoding 1-deoxy-D-xylulose 5-phosphate synthase, the first committed enzyme of the 2-C-methyl-D-erythritol 4-phosphate pathway. Planta Medica 2006, 72, 329–335. [Google Scholar] [CrossRef]
- Kiendler-Scharr, A.; Andres, S.; Bachner, M.; Behnke, K.; Broch, S.; Hofzumahaus, A.; Holland, F.; Kleist, E.; Mentel, T.F.; Rubach, F.; et al. Isoprene in poplar emissions: Effects on new particle formation and OH concentrations. Atmos. Chem. Phys. 2012, 12, 1021–1030. [Google Scholar] [CrossRef]
- Fuentes, J.D.; Lerdau, M.; Atkinson, R.; Baldocchi, D.; Bottenheim, J.W.; Ciccioli, P.; Lamb, B.; Geron, C.; Gu, L.; Guenther, A.; et al. Biogenic hydrocarbons in the atmospheric boundary layer: A review. Bull. Am. Meteorol. Soc. 2000, 81, 1537–1575. [Google Scholar] [CrossRef]
- Guenther, A. Upscaling biogenic volatile compound emissions from leaves to landscapes. In Biology, Controls and Models of Tree Volatile Organic Compound Emissions; Niinemets, U., Monson, R.K., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; Volume 5, pp. 391–414. [Google Scholar]
- Loreto, F.; Mannozzi, M.; Maris, C.; Nascetti, P.; Ferranti, F.; Pasqualini, S. Ozone quenching properties of isoprene and its antioxidant role in leaves. Plant Physiol. 2001, 126, 993–1000. [Google Scholar] [CrossRef] [PubMed]
- Vickers, C.E.; Gershenzon, J.; Lerdau, M.T.; Loreto, F. A unified mechanism of action for volatile isoprenoids in plant abiotic stress. Nat. Chem. Biol. 2009, 5, 283–291. [Google Scholar] [CrossRef]
- Velikova, V.; Sharkey, T.D.; Loreto, F. Stabilization of thylakoid membranes in isoprene-emitting plants reduces formation of reactive oxygen species. Plant Signal Behav. 2012, 7, 139–141. [Google Scholar] [CrossRef]
- Perreca, E.; Gershenzon, J.; Eberl, F. Tree volatiles: Effects of biotic and abiotic factors on emission and biological roles. In Biology of Plant Volatiles; CRC Press: Boca Raton, FL, USA, 2020; pp. 361–375. [Google Scholar]
- Velikova, V.; Loreto, F.; Tsonev, T.; Brilli, F.; Edreva, A. Isoprene prevents the negative consequences of high temperature stress in Platanus orientalis leaves. Funct. Plant Biol. 2006, 33, 931–940. [Google Scholar] [CrossRef]
- Ghirardo, A.; Wright, L.P.; Bi, Z.; Rosenkranz, M.; Pulido, P.; Rodriguez-Concepcion, M.; Niinemets, U.; Bruggemann, N.; Gershenzon, J.; Schnitzler, J.P. Metabolic Flux Analysis of Plastidic Isoprenoid Biosynthesis in Poplar Leaves Emitting and Nonemitting Isoprene. Plant Physiol. 2014, 165, 37–51. [Google Scholar] [CrossRef]
- Behnke, K.; Loivamäki, M.; Zimmer, I.; Rennenberg, H.; Schnitzler, J.-P.; Louis, S. Isoprene emission protects photosynthesis in sunfleck exposed Grey poplar. Photosynth. Res. 2010, 104, 5–17. [Google Scholar] [CrossRef] [PubMed]
- Rasulov, B.; Huve, K.; Bichele, I.; Laisk, A.; Niinemets, U. Temperature Response of Isoprene Emission In Vivo Reflects a Combined Effect of Substrate Limitations and Isoprene Synthase Activity: A Kinetic Analysis. Plant Physiol. 2010, 154, 1558–1570. [Google Scholar] [CrossRef] [PubMed]
- Schnitzler, J.P.; Zimmer, I.; Bachl, A.; Arend, M.; Fromm, J.; Fischbach, R.J. Biochemical properties of isoprene synthase in poplar (Populus × canescens). Planta 2005, 222, 777–786. [Google Scholar] [CrossRef]
- Weraduwage, S.M.; Whitten, D.; Kulke, M.; Sahu, A.; Vermaas, J.V.; Sharkey, T.D. The isoprene-responsive phosphoproteome provides new insights into the putative signalling pathways and novel roles of isoprene. Plant Cell Environ. 2024, 47, 1099–1117. [Google Scholar] [CrossRef]
- Dani, K.G.S.; Pollastri, S.; Pinosio, S.; Reichelt, M.; Sharkey, T.D.; Schnitzler, J.P.; Loreto, F. Isoprene enhances leaf cytokinin metabolism and induces early senescence. New Phytol. 2022, 234, 961–974. [Google Scholar] [CrossRef] [PubMed]
- Loreto, F.; Velikova, V. Isoprene produced by leaves protects the photosynthetic apparatus against ozone damage, quenches ozone products, and reduces lipid peroxidation of cellular membranes. Plant Physiol. 2001, 127, 1781–1787. [Google Scholar] [CrossRef]
- Li, Z.R.; Sharkey, T.D. Metabolic profiling of the methylerythritol phosphate pathway reveals the source of post-illumination isoprene burst from leaves. Plant Cell Environ. 2013, 36, 429–437. [Google Scholar] [CrossRef]
- Fares, S.; Brilli, F.; Nogues, I.; Velikova, V.; Tsonev, T.; Dagli, S.; Loreto, F. Isoprene emission and primary metabolism in Phragmites australis grown under different phosphorus levels. Plant Biol. 2007, 9, e99–e104. [Google Scholar] [CrossRef] [PubMed]
- Fortunati, A.; Barta, C.; Brilli, F.; Centritto, M.; Zimmer, I.; Schnitzler, J.P.; Loreto, F. Isoprene emission is not temperature-dependent during and after severe drought-stress: A physiological and biochemical analysis. Plant J. 2008, 55, 687–697. [Google Scholar] [CrossRef]
- Centritto, M.; Brilli, F.; Fodale, R.; Loreto, F. Different sensitivity of isoprene emission, respiration and photosynthesis to high growth temperature coupled with drought stress in black poplar (Populus nigra) saplings. Tree Physiol. 2011, 31, 275–286. [Google Scholar] [CrossRef]
- Sharkey, T.D.; Loreto, F.; Delwiche, C.F. High-Carbon Dioxide and Sun Shade Effects on Isoprene Emission from Oak and Aspen Tree Leaves. Plant Cell Environ. 1991, 14, 333–338. [Google Scholar] [CrossRef]
- Hasunuma, T.; Harada, K.; Miyazawa, S.I.; Kondo, A.; Fukusaki, E.; Miyake, C. Metabolic turnover analysis by a combination of in vivo C-13-labelling from (CO2)-C-13 and metabolic profiling with CE-MS/MS reveals rate-limiting steps of the C-3 photosynthetic pathway in Nicotiana tabacum leaves. J. Exp. Bot. 2010, 61, 1041–1051. [Google Scholar] [CrossRef]
- Szecowka, M.; Heise, R.; Tohge, T.; Nunes-Nesi, A.; Vosloh, D.; Huege, J.; Feil, R.; Lunn, J.; Nikoloski, Z.; Stitt, M.; et al. Metabolic Fluxes in an Illuminated Arabidopsis Rosette. Plant Cell 2013, 25, 694–714. [Google Scholar] [CrossRef]
- Delgado, J.; Liao, J.C. Metabolic Control Analysis Using Transient Metabolite Concentrations—Determination of Metabolite Concentration Control Coefficients. Biochem. J. 1992, 285, 965–972. [Google Scholar] [CrossRef]
- Kacser, H.; Burns, J.A.; Fell, D.A. The Control of Flux. Biochem. Soc. Trans. 1995, 23, 341–366. [Google Scholar] [CrossRef]
- Fell, D.A. Metabolic Control Analysis—A Survey of Its Theoretical and Experimental Development. Biochem. J. 1992, 286, 313–330. [Google Scholar] [CrossRef]
- Rohwer, J.M. Kinetic modelling of plant metabolic pathways. J. Exp. Bot. 2012, 63, 2275–2292. [Google Scholar] [CrossRef] [PubMed]
- Monson, R.K.; Grote, R.; Niinemets, U.; Schnitzler, J.P. Modeling the isoprene emission rate from leaves. New Phytol. 2012, 195, 541–559. [Google Scholar] [CrossRef] [PubMed]
- Bruggemann, N.; Schnitzler, J.P. Relationship of isopentenyl diphosphate (IDP) isomerase activity to isoprene emission of oak leaves. Tree Physiol. 2002, 22, 1011–1018. [Google Scholar] [CrossRef] [PubMed]
- Magel, E.; Mayrhofer, S.; Muller, A.; Zimmer, I.; Hampp, R.; Schnitzler, J.P. Photosynthesis and substrate supply for isoprene biosynthesis in poplar leaves. Atmos. Environ. 2006, 40, S138–S151. [Google Scholar] [CrossRef]
- Loreto, F.; Pinelli, P.; Brancaleoni, E.; Ciccioli, P. C-13 labeling reveals chloroplastic and extrachloroplastic pools of dimethylallyl pyrophosphate and their contribution to isoprene formation. Plant Physiol. 2004, 135, 1903–1907. [Google Scholar] [CrossRef] [PubMed]
- Wolfertz, M.; Sharkey, T.D.; Boland, W.; Kuhnemann, F. Rapid regulation of the methylerythritol 4-phosphate pathway during isoprene synthesis. Plant Physiol. 2004, 135, 1939–1945. [Google Scholar] [CrossRef]
- Wu, S.Q.; Schalk, M.; Clark, A.; Miles, R.B.; Coates, R.; Chappell, J. Redirection of cytosolic or plastidic isoprenoid precursors elevates terpene production in plants. Nat. Biotechnol. 2006, 24, 1441–1447. [Google Scholar] [CrossRef]
- Loivamaki, M.; Louis, S.; Cinege, G.; Zimmer, I.; Fischbach, R.J.; Schnitzler, J.P. Circadian rhythms of isoprene biosynthesis in grey poplar leaves. Plant Physiol. 2007, 143, 540–551. [Google Scholar] [CrossRef]
- Covington, M.F.; Maloof, J.N.; Straume, M.; Kay, S.A.; Harmer, S.L. Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol. 2008, 9, R130. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Holm, S. A Simple Sequentially Rejective Multiple Test Procedure. Scand. J. Stat. 1979, 6, 65–70. [Google Scholar]
- Fell, D. Understanding the Control of Metabolism: Frontiers in Metabolism; Portland Press: London, UK, 1997; Volume 2. [Google Scholar]
- Funk, J.L.; Mak, J.E.; Lerdau, M.T. Stress-induced changes in carbon sources for isoprene production in Populus deltoides. Plant Cell Environ. 2004, 27, 747–755. [Google Scholar] [CrossRef]
- Affek, H.P.; Yakir, D. Natural abundance carbon isotope composition of isoprene reflects incomplete coupling between isoprene synthesis and photosynthetic carbon flow. Plant Physiol. 2003, 131, 1727–1736. [Google Scholar] [CrossRef] [PubMed]
- Brilli, F.; Barta, C.; Fortunati, A.; Lerdau, M.; Loreto, F.; Centritto, M. Response of isoprene emission and carbon metabolism to drought in white poplar (Populus alba) saplings. New Phytol. 2007, 175, 244–254. [Google Scholar] [CrossRef]
- Trowbridge, A.M.; Asensio, D.; Eller, A.S.; Way, D.A.; Wilkinson, M.J.; Schnitzler, J.-P.; Jackson, R.B.; Monson, R.K. Contribution of various carbon sources toward isoprene biosynthesis in poplar leaves mediated by altered atmospheric CO2 concentrations. PLoS ONE 2012, 7, e32387. [Google Scholar] [CrossRef]
- Karl, T.; Fall, R.; Rosenstiel, T.N.; Prazeller, P.; Larsen, B.; Seufert, G.; Lindinger, W. On-line analysis of the (CO2)-C-13 labeling of leaf isoprene suggests multiple subcellular origins of isoprene precursors. Planta 2002, 215, 894–905. [Google Scholar] [CrossRef]
- Hemmerlin, A.; Tritsch, D.; Hartmann, M.; Pacaud, K.; Hoeffler, J.F.; van Dorsselaer, A.; Rohmer, M.; Bach, T.J. A cytosolic arabidopsis D-xylulose kinase catalyzes the phosphorylation of 1-deoxy-D-xylulose into a precursor of the plastidial isoprenoid pathway. Plant Physiol. 2006, 142, 441–457. [Google Scholar] [CrossRef]
- Schnitzler, J.P.; Graus, M.; Kreuzwieser, J.; Heizmann, U.; Rennenberg, H.; Wisthaler, A.; Hansel, A. Contribution of different carbon sources to isoprene biosynthesis in poplar leaves. Plant Physiol. 2004, 135, 152–160. [Google Scholar] [CrossRef]
- Kreuzwieser, J.; Graus, M.; Wisthaler, A.; Hanse, A.; Rennenberg, H.; Schnitzler, J.P. Xylem-transported glucose as an additional carbon source for leaf isoprene formation in Quercus robur. New Phytol. 2002, 156, 171–178. [Google Scholar] [CrossRef]
- Sharkey, T.D.; Preiser, A.L.; Weraduwage, S.M.; Gog, L. Source of 12C in Calvin–Benson cycle intermediates and isoprene emitted from plant leaves fed with 13CO2. Biochem. J. 2020, 477, 3237–3252. [Google Scholar] [CrossRef] [PubMed]
- Sharkey, T.D.; Yeh, S.S. Isoprene emission from plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001, 52, 407–436. [Google Scholar] [CrossRef] [PubMed]
- Rasulov, B.; Bichele, I.; Laisk, A.; Niinemets, U. Competition between isoprene emission and pigment synthesis during leaf development in aspen. Plant Cell Environ. 2014, 37, 724–741. [Google Scholar] [CrossRef]
- Velikova, V.; Várkonyi, Z.; Szabó, M.; Maslenkova, L.; Nogues, I.; Kovács, L.; Peeva, V.; Busheva, M.; Garab, G.; Sharkey, T.D.; et al. Increased Thermostability of Thylakoid Membranes in Isoprene-Emitting Leaves Probed with Three Biophysical Techniques. Plant Physiol. 2011, 157, 905–916. [Google Scholar] [CrossRef]
- Harvey, C.M.; Li, Z.; Tjellström, H.; Blanchard, G.J.; Sharkey, T.D. Concentration of isoprene in artificial and thylakoid membranes. J. Bioenerg. Biomembr. 2015, 47, 419–429. [Google Scholar] [CrossRef]
- Vanzo, E.; Merl-Pham, J.; Velikova, V.; Ghirardo, A.; Lindermayr, C.; Hauck, S.M.; Bernhardt, J.; Riedel, K.; Durner, J.; Schnitzler, J.-P. Modulation of Protein S-Nitrosylation by Isoprene Emission in Poplar. Plant Physiol. 2016, 170, 1945–1961. [Google Scholar] [CrossRef] [PubMed]
- Tattini, M.; Velikova, V.; Vickers, C.; Brunetti, C.; Di Ferdinando, M.; Trivellini, A.; Fineschi, S.; Agati, G.; Ferrini, F.; Loreto, F. Isoprene production in transgenic tobacco alters isoprenoid, non-structural carbohydrate and phenylpropanoid metabolism, and protects photosynthesis from drought stress. Plant Cell Environ. 2014, 37, 1950–1964. [Google Scholar] [CrossRef]
- Harvey, C.M.; Sharkey, T.D. Exogenous isoprene modulates gene expression in unstressed Arabidopsis thaliana plants. Plant Cell Environ. 2016, 39, 1251–1263. [Google Scholar] [CrossRef]
- Rosenstiel, T.; Ebbets, A.; Khatri, W.; Fall, R.; Monson, R. Induction of poplar leaf nitrate reductase: A test of extrachloroplastic control of isoprene emission rate. Plant Biol. 2004, 7, 12–21. [Google Scholar] [CrossRef]
- Behnke, K.; Ehlting, B.; Teuber, M.; Bauerfeind, M.; Louis, S.; Hasch, R.; Polle, A.; Bohlmann, J.; Schnitzler, J.P. Transgenic, non-isoprene emitting poplars don’t like it hot. Plant J. 2007, 51, 485–499. [Google Scholar] [CrossRef]
- Kuzma, J.; Fall, R. Leaf Isoprene Emission Rate Is Dependent on Leaf Development and the Level of Isoprene Synthase. Plant Physiol. 1993, 101, 435–440. [Google Scholar] [CrossRef] [PubMed]
- Sharkey, T.D.; Wiberley, A.E.; Donohue, A.R. Isoprene emission from plants: Why and how. Ann. Bot-Lond. 2008, 101, 5–18. [Google Scholar] [CrossRef]
- Wiberley, A.E.; Donohue, A.R.; Westphal, M.M.; Sharkey, T.D. Regulation of isoprene emission from poplar leaves throughout a day. Plant Cell Environ. 2009, 32, 939–947. [Google Scholar] [CrossRef] [PubMed]
- Levee, V.; Major, I.; Levasseur, C.; Tremblay, L.; MacKay, J.; Seguin, A. Expression profiling and functional analysis of Populus WRKY23 reveals a regulatory role in defense. New Phytol. 2009, 184, 48–70. [Google Scholar] [CrossRef] [PubMed]
- Meilan, R.; Ma, C. Populus. In Methods in Molecular Biology, Agrobacterium Protocols, 2nd ed.; Humana Press: Totowa, NJ, USA, 2006; Volume 344, pp. 143–152. [Google Scholar]
- Gonzalez-Cabanelas, D.; Hammerbacher, A.; Raguschke, B.; Gershenzon, J.; Wright, L.P. Quantifying the Metabolites of the Methylerythritol 4-Phosphate (MEP) Pathway in Plants and Bacteria by Liquid Chromatography-Triple Quadrupole Mass Spectrometry. Methods Enzym. 2016, 576, 225–249. [Google Scholar] [CrossRef]
- Yuan, J.; Bennett, B.D.; Rabinowitz, J.D. Kinetic flux profiling for quantitation of cellular metabolic fluxes. Nat. Protoc. 2008, 3, 1328–1340. [Google Scholar] [CrossRef] [PubMed]
- Newville, M.; Stensitzki, T.; Allen, D.B.; Ingargiola, A.; Nelson, A. LMFIT: Non-linear least-square minimization and curve-fitting for Python. Zenodo 2014. [Google Scholar] [CrossRef]
- Wright, L.P.; Phillips, M.A. Measuring the Activity of 1-Deoxy-D-Xylulose 5-Phosphate Synthase, the First Enzyme in the MEP Pathway, in Plant Extracts. Methods Mol. Biol. 2014, 1153, 9–20. [Google Scholar] [CrossRef]
- Small, J.R.; Kacser, H. Responses of Metabolic Systems to Large Changes in Enzyme-Activities and Effectors. 1. The Linear Treatment of Unbranched Chains. Eur. J. Biochem. 1993, 213, 613–624. [Google Scholar] [CrossRef]
- Pinheiro, J.; Bates, D.; DebRoy, S.; Sarkar, D.; R Core Team. nlme: Linear and Nonlinear Mixed Effects Models; R. Package version 3.1-137. 2018. Available online: https://CRAN.R-project.org/package=nlme (accessed on 21 February 2024).
- Zuur, A.; Fleno, E.N.; Walker, N.; Saveliev, A.; Smith, G.M. Mixed Effects Models and Extensions in Ecology with R; Springer: New York, NY, USA, 2009. [Google Scholar]
- Crawley, M. The R Book; John Wiley and Sons, Ltd.: Hoboken, NJ, USA, 2013. [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing, 3.5.1; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]




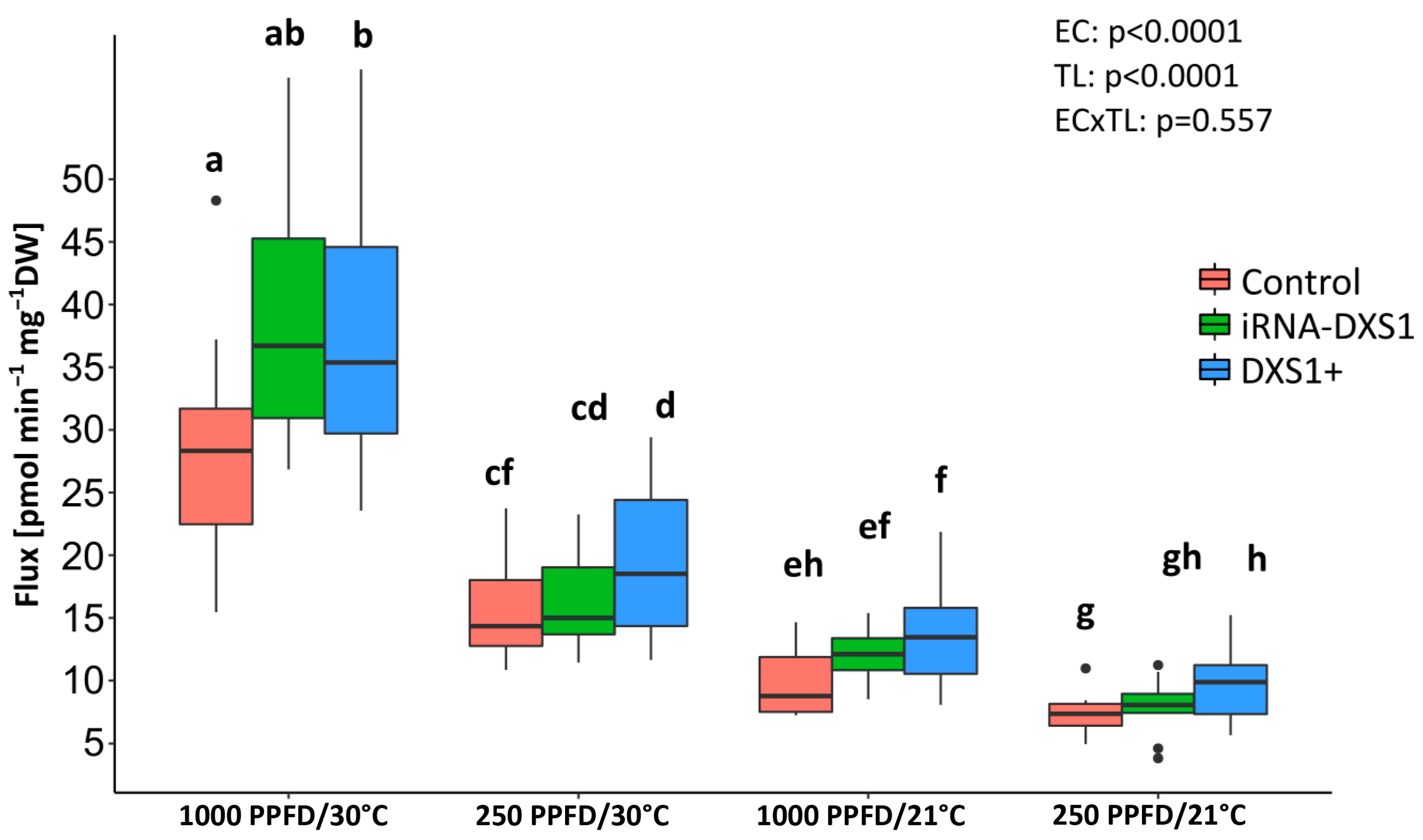
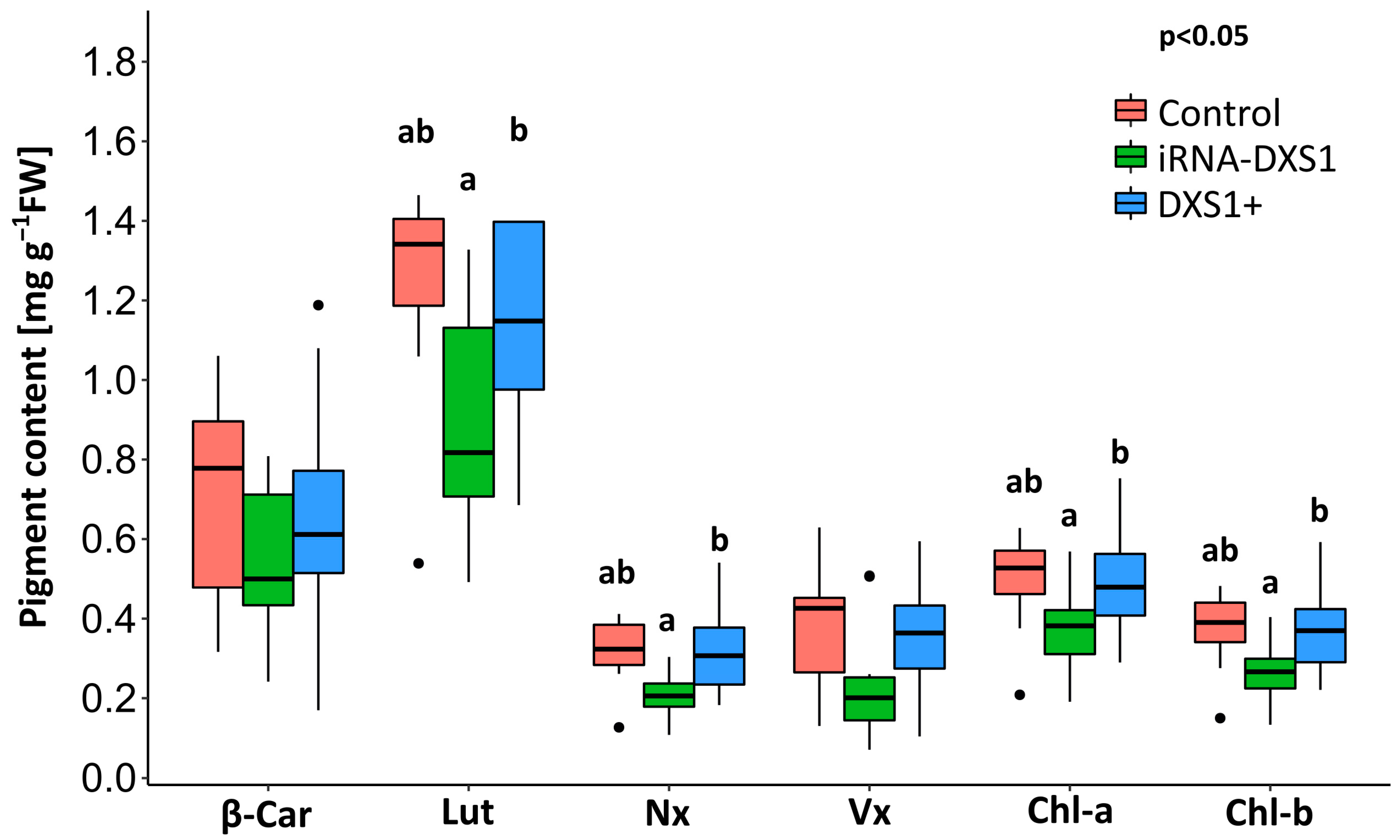
| Environmental Condition | Plant Line | n | Maximum Isoprene Labeling | DXP Pool Size | MEcDP Pool Size | IDP/DMADP Pool Size | Flux |
|---|---|---|---|---|---|---|---|
| pmol mg−1 DW | pmol mg−1 DW | pmol mg−1 DW | pmol min−1 mg−1 DW | ||||
| 1000 PPFD 30 °C | EV | 10 | 0.912 ± 0.007 | 41.65 ± 2.22 | 85.59 ± 14.43 | 38.09 ± 4.01 | 28.65 ± 2.95 |
| RNAi-DXS1 | 14 | 0.912 ± 0.005 | 46.01 ± 3.86 | 124.9 ± 12.14 | 47.74 ± 4.67 | 38.94 ± 2.62 | |
| DXS1+ | 18 | 0.902 ± 0.006 | 58.61 ± 3.21 | 139.4 ± 15.89 | 41.20 ± 4.76 | 38.52 ± 2.56 | |
| 250 PPFD 30 °C | EV | 10 | 0.823 ± 0.036 | 28.53 ± 1.49 | 54.98 ± 4.06 | 18.47 ± 3.23 | 15.66 ± 1.42 |
| RNAi-DXS1 | 15 | 0.870 ± 0.008 | 25.61 ± 1.29 | 46.99 ± 2.66 | 22.03 ± 1.68 | 16.49 ± 0.96 | |
| DXS1+ | 20 | 0.865 ± 0.018 | 35.97 ± 2.55 | 65.43 ± 5.23 | 19.41 ± 2.44 | 19.69 ± 1.24 | |
| 1000 PPFD 21 °C | EV | 9 | 0.932 ± 0.008 | 27.18 ± 1.45 | 48.57 ± 6.53 | 13.64 ± 0.78 | 9.79 ± 0.87 |
| RNAi-DXS1 | 13 | 0.929 ± 0.009 | 28.64 ± 1.96 | 48.19 ± 3.39 | 19.44 ± 1.35 | 12.15 ± 0.55 | |
| DXS1+ | 16 | 0.943 ±0.005 | 33.51 ± 1.93 | 53.36 ± 3.79 | 16.90 ± 1.14 | 13.50 ± 0.93 | |
| 250 PPFD 21 °C | EV | 8 | 0.946 ± 0.009 | 21.99 ± 1.29 | 35.74 ± 4.54 | 13.31 ± 1.89 | 7.43 ± 0.65 |
| RNAi-DXS1 | 10 | 0.940 ± 0.005 | 22.29 ± 1.70 | 36.92 ± 3.85 | 19.57 ± 2.04 | 7.91 ± 0.73 | |
| DXS1+ | 19 | 0.957 ± 0.006 | 24.59 ± 0.97 | 43.21 ± 3.37 | 13.18 ± 1.32 | 9.55 ± 0.64 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
González-Cabanelas, D.; Perreca, E.; Rohwer, J.M.; Schmidt, A.; Engl, T.; Raguschke, B.; Gershenzon, J.; Wright, L.P. Deoxyxylulose 5-Phosphate Synthase Does Not Play a Major Role in Regulating the Methylerythritol 4-Phosphate Pathway in Poplar. Int. J. Mol. Sci. 2024, 25, 4181. https://doi.org/10.3390/ijms25084181
González-Cabanelas D, Perreca E, Rohwer JM, Schmidt A, Engl T, Raguschke B, Gershenzon J, Wright LP. Deoxyxylulose 5-Phosphate Synthase Does Not Play a Major Role in Regulating the Methylerythritol 4-Phosphate Pathway in Poplar. International Journal of Molecular Sciences. 2024; 25(8):4181. https://doi.org/10.3390/ijms25084181
Chicago/Turabian StyleGonzález-Cabanelas, Diego, Erica Perreca, Johann M. Rohwer, Axel Schmidt, Tobias Engl, Bettina Raguschke, Jonathan Gershenzon, and Louwrance P. Wright. 2024. "Deoxyxylulose 5-Phosphate Synthase Does Not Play a Major Role in Regulating the Methylerythritol 4-Phosphate Pathway in Poplar" International Journal of Molecular Sciences 25, no. 8: 4181. https://doi.org/10.3390/ijms25084181
APA StyleGonzález-Cabanelas, D., Perreca, E., Rohwer, J. M., Schmidt, A., Engl, T., Raguschke, B., Gershenzon, J., & Wright, L. P. (2024). Deoxyxylulose 5-Phosphate Synthase Does Not Play a Major Role in Regulating the Methylerythritol 4-Phosphate Pathway in Poplar. International Journal of Molecular Sciences, 25(8), 4181. https://doi.org/10.3390/ijms25084181








