Mitochondrial Fraction of Circulating Cell-Free DNA as an Indicator of Human Pathology
Abstract
1. Introduction
2. Results
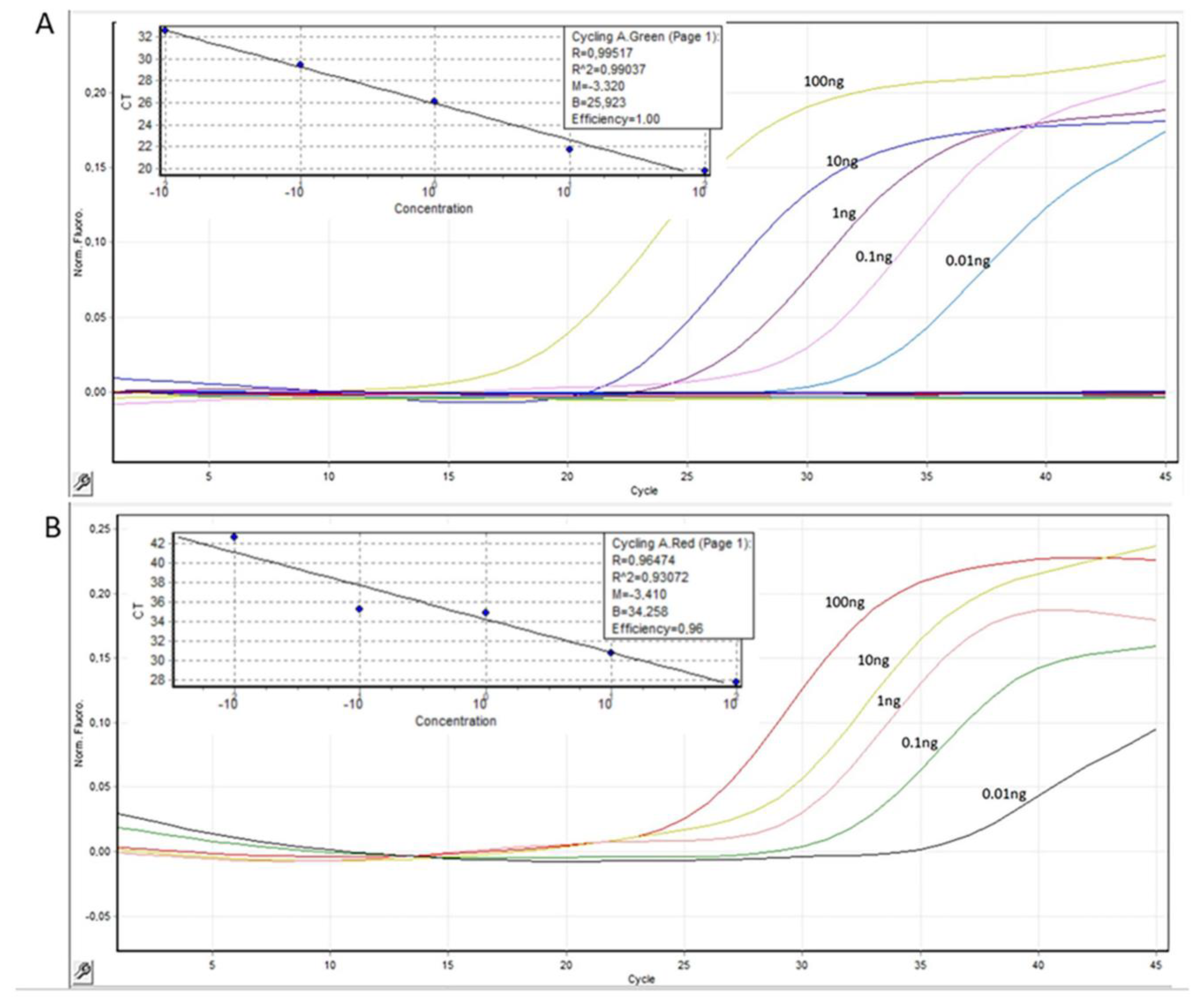
2.1. Development of Dual PCR Assay for Detecting ccf-mtDNA and Reference Measurements
2.2. ccf-mtDNA Fraction in Diseases
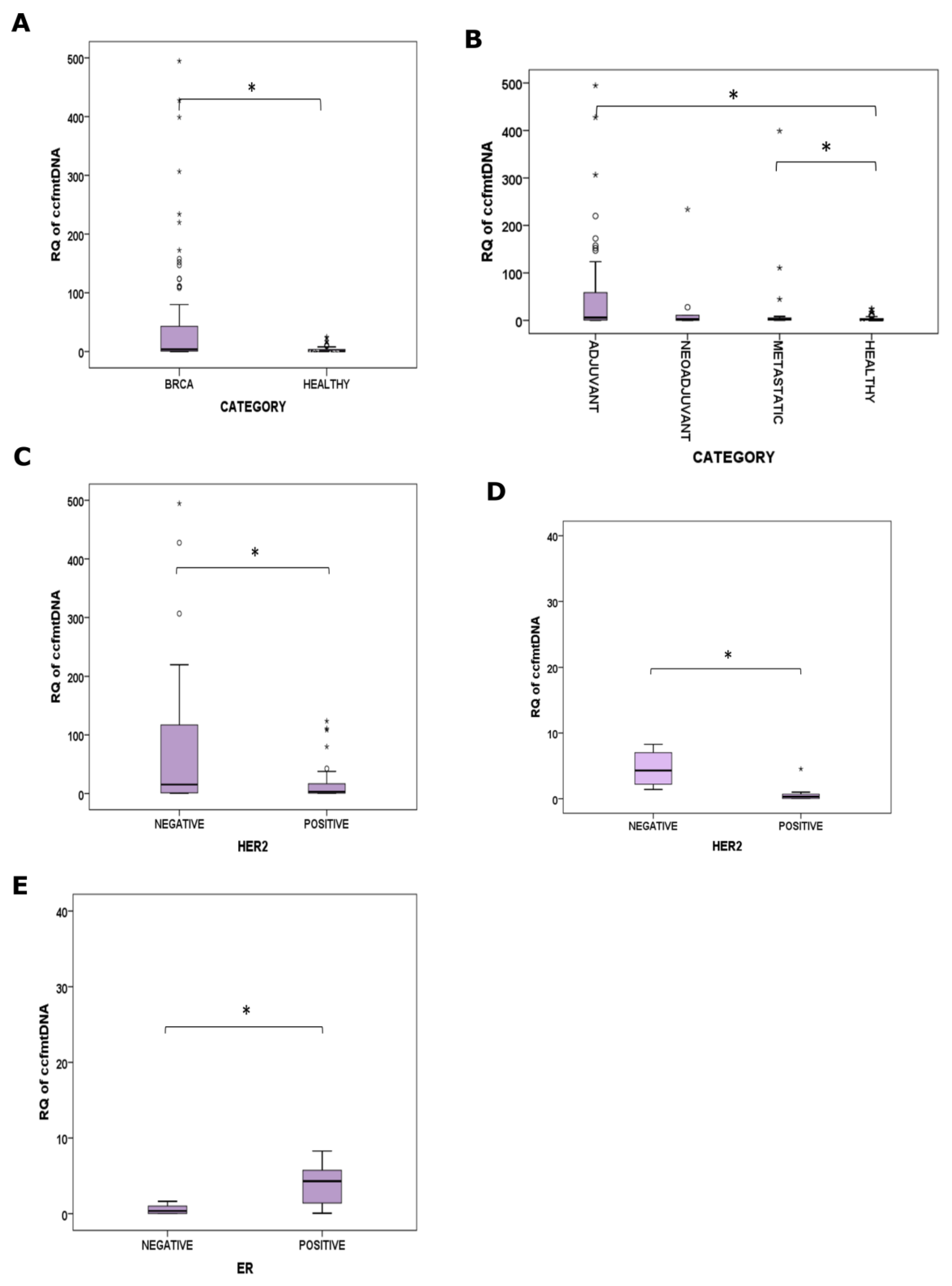
2.2.1. BrCa
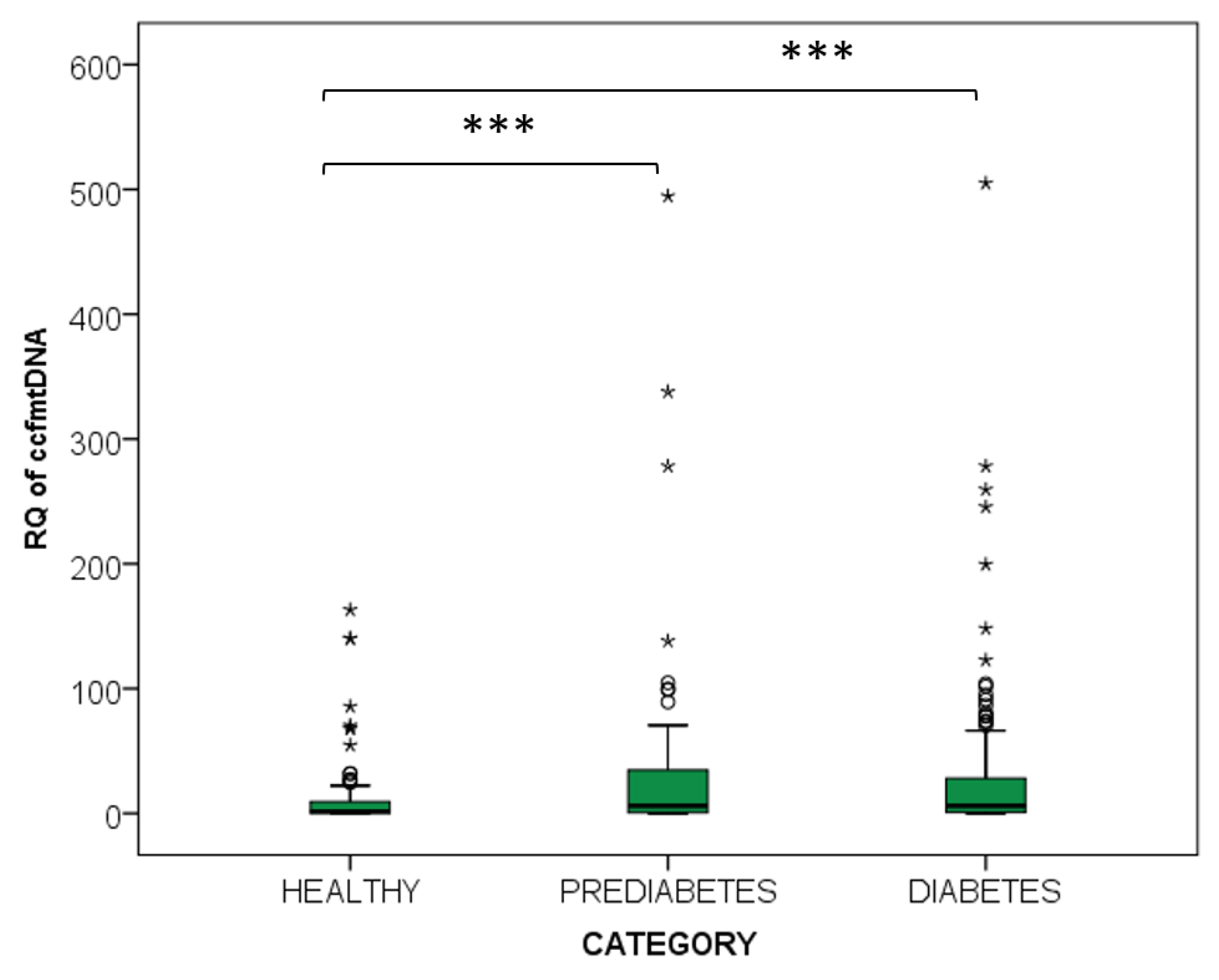
2.2.2. Diabetes and Prediabetes
2.2.3. Osteoarthritis
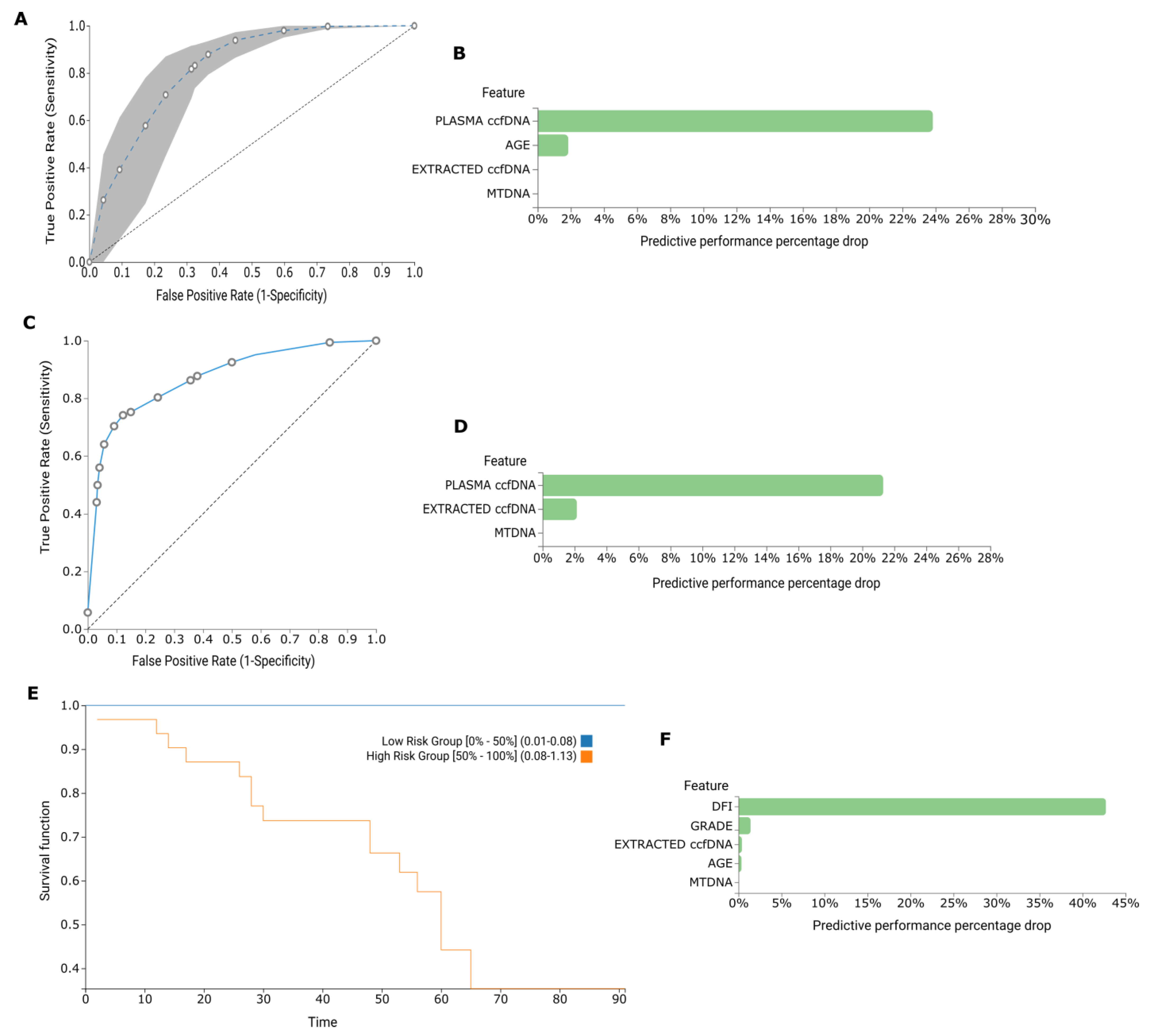
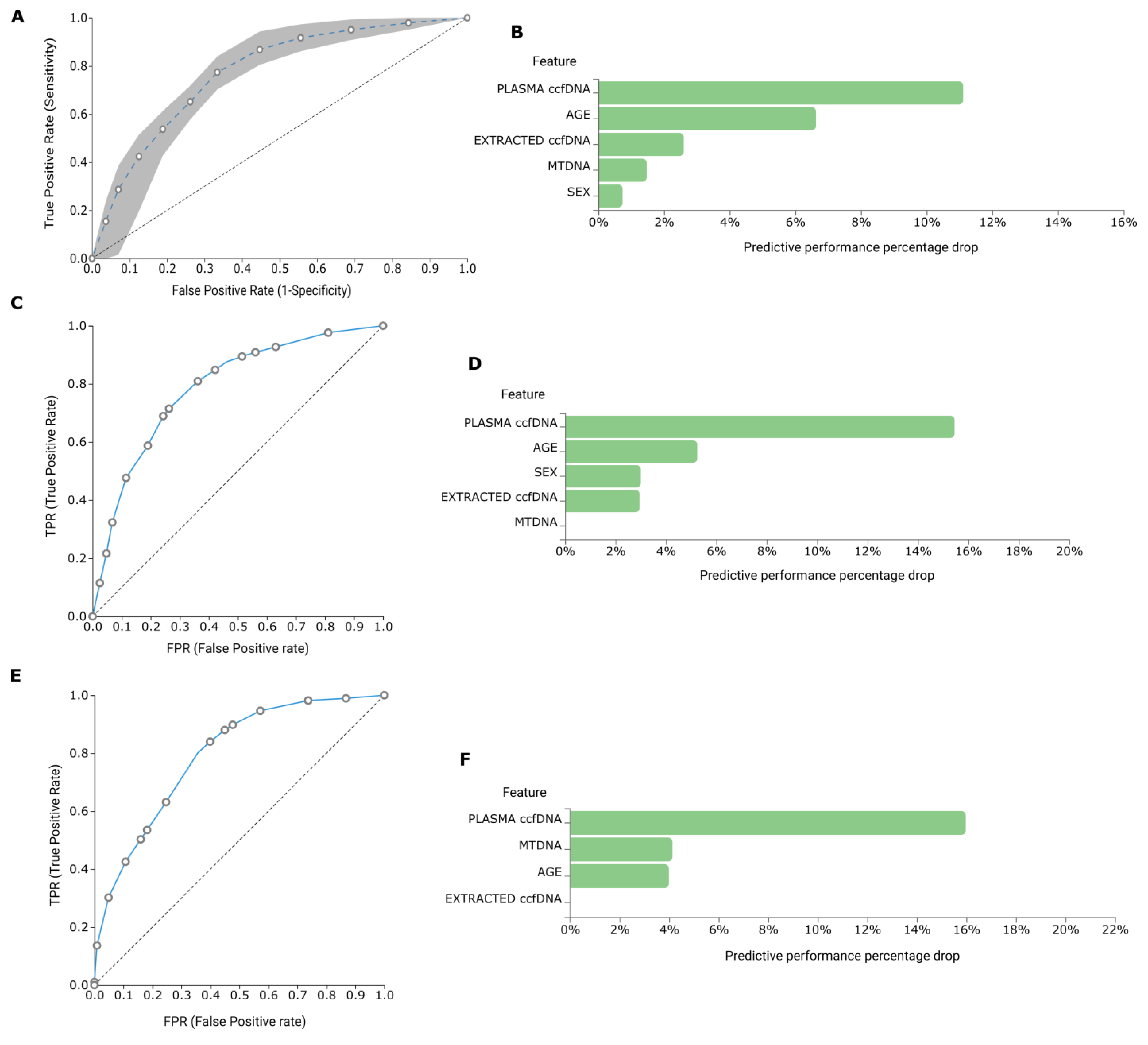
2.3. AutoML Multivariate Analysis
2.3.1. BrCa
2.3.2. Diabetes and Prediabetes
2.3.3. Osteoarthritis
3. Discussion
4. Materials and Methods
4.1. Clinical Samples
4.1.1. BrCa
4.1.2. Diabetes and Pre-Diabetes
4.1.3. Osteoarthritis
4.1.4. Healthy Individuals
4.2. Human Samples Pre-Analytical Procedures
4.3. Quantification of ccf-mtDNA
4.4. Statistical Analysis
4.5. AutoML Multivariate Analysis
5. Conclusions
6. Patents
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mandel, P.; Metais, P. Nuclear Acids in Human Blood Plasma. C. R. Seances Soc. Biol. Fil. 1948, 142, 241–243. [Google Scholar] [PubMed]
- Lu, J.L.; Liang, Z.Y. Circulating free DNA in the era of precision oncology: Pre- and post-analytical concerns. Chronic Dis. Transl. Med. 2016, 2, 223–230. [Google Scholar] [CrossRef]
- Panagopoulou, M.; Karaglani, M.; Balgkouranidou, I.; Pantazi, C.; Kolios, G.; Kakolyris, S.; Chatzaki, E. Circulating cell-free DNA release in vitro: Kinetics, size profiling, and cancer-related gene methylation. J. Cell. Physiol. 2019, 234, 14079–14089. [Google Scholar] [CrossRef]
- Anderson, S.; Bankier, A.T.; Barrell, B.G.; de Bruijn, M.H.; Coulson, A.R.; Drouin, J.; Eperon, I.C.; Nierlich, D.P.; Roe, B.A.; Sanger, F.; et al. Sequence and organization of the human mitochondrial genome. Nature 1981, 290, 457–465. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Nakahira, K.; Guo, X.; Choi, A.M.; Gu, Z. Very Short Mitochondrial DNA Fragments and Heteroplasmy in Human Plasma. Sci. Rep. 2016, 6, 36097. [Google Scholar] [CrossRef] [PubMed]
- Delsite, R.; Kachhap, S.; Anbazhagan, R.; Gabrielson, E.; Singh, K.K. Nuclear genes involved in mitochondria-to-nucleus communication in breast cancer cells. Mol. Cancer 2002, 1, 6. [Google Scholar] [CrossRef] [PubMed]
- Jiang, P.; Chan, C.W.; Chan, K.C.; Cheng, S.H.; Wong, J.; Wong, V.W.; Wong, G.L.; Chan, S.L.; Mok, T.S.; Chan, H.L.; et al. Lengthening and shortening of plasma DNA in hepatocellular carcinoma patients. Proc. Natl. Acad. Sci. USA 2015, 112, E1317–E1325. [Google Scholar] [CrossRef] [PubMed]
- Bonner, M.R.; Shen, M.; Liu, C.S.; Divita, M.; He, X.; Lan, Q. Mitochondrial DNA content and lung cancer risk in Xuan Wei, China. Lung Cancer 2009, 63, 331–334. [Google Scholar] [CrossRef] [PubMed]
- Ellinger, J.; Müller, D.C.; Müller, S.C.; Hauser, S.; Heukamp, L.C.; von Ruecker, A.; Bastian, P.J.; Walgenbach-Brunagel, G. Circulating mitochondrial DNA in serum: A universal diagnostic biomarker for patients with urological malignancies. Urol. Oncol. 2012, 30, 509–515. [Google Scholar] [CrossRef]
- Fernandes, J.; Michel, V.; Camorlinga-Ponce, M.; Gomez, A.; Maldonado, C.; De Reuse, H.; Torres, J.; Touati, E. Circulating mitochondrial DNA level, a noninvasive biomarker for the early detection of gastric cancer. Cancer Epidemiol. Biomark. Prev. A Publ. Am. Assoc. Cancer Res. Cosponsored Am. Soc. Prev. Oncol. 2014, 23, 2430–2438. [Google Scholar] [CrossRef]
- Zachariah, R.R.; Schmid, S.; Buerki, N.; Radpour, R.; Holzgreve, W.; Zhong, X. Levels of circulating cell-free nuclear and mitochondrial DNA in benign and malignant ovarian tumors. Obstet. Gynecol. 2008, 112, 843–850. [Google Scholar] [CrossRef] [PubMed]
- Mahmoud, E.H.; Fawzy, A.; Ahmad, O.K.; Ali, A.M. Plasma Circulating Cell-free Nuclear and Mitochondrial DNA as Potential Biomarkers in the Peripheral Blood of Breast Cancer Patients. Asian Pac. J. Cancer Prev. APJCP 2015, 16, 8299–8305. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Platek, M.; Mahasneh, A.; Ambrosone, C.B.; Zhao, H. Mitochondrial copy number and risk of breast cancer: A pilot study. Mitochondrion 2010, 10, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Xia, P.; An, H.X.; Dang, C.X.; Radpour, R.; Kohler, C.; Fokas, E.; Engenhart-Cabillic, R.; Holzgreve, W.; Zhong, X.Y. Decreased mitochondrial DNA content in blood samples of patients with stage I breast cancer. BMC Cancer 2009, 9, 454. [Google Scholar] [CrossRef] [PubMed]
- Silzer, T.; Barber, R.; Sun, J.; Pathak, G.; Johnson, L.; O’Bryant, S.; Phillips, N. Circulating mitochondrial DNA: New indices of type 2 diabetes-related cognitive impairment in Mexican Americans. PLoS ONE 2019, 14, e0213527. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Cai, X.; Xie, L.; Tang, Y.; Cheng, J.; Wang, J.; Wang, L.; Gong, J. Circulating Cell Free Mitochondrial DNA is a Biomarker in the Development of Coronary Heart Disease in the Patients with Type 2 Diabetes. Clin. Lab. 2015, 61, 661–667. [Google Scholar] [CrossRef] [PubMed]
- Alfatni, A.; Riou, M.; Charles, A.-L.; Meyer, A.; Barnig, C.; Andres, E.; Lejay, A.; Talha, S.; Geny, B. Peripheral Blood Mononuclear Cells and Platelets Mitochondrial Dysfunction, Oxidative Stress, and Circulating mtDNA in Cardiovascular Diseases. J. Clin. Med. 2020, 9, 311. [Google Scholar] [CrossRef] [PubMed]
- Panagopoulou, M.; Karaglani, M.; Balgkouranidou, I.; Biziota, E.; Koukaki, T.; Karamitrousis, E.; Nena, E.; Tsamardinos, I.; Kolios, G.; Lianidou, E.; et al. Circulating cell-free DNA in breast cancer: Size profiling, levels, and methylation patterns lead to prognostic and predictive classifiers. Oncogene 2019, 38, 3387–3401. [Google Scholar] [CrossRef] [PubMed]
- Safi, M.; Najib, A.R. Evaluation of circulating cell-free nuclear and mitochondrial DNA levels in Syrian patients with breast tumor. Exp. Ther. Med. 2021, 21, 65. [Google Scholar] [CrossRef]
- Pasha, H.A.; Rezk, N.A.; Riad, M.A. Circulating Cell Free Nuclear DNA, Mitochondrial DNA and Global DNA Methylation: Potential Noninvasive Biomarkers for Breast Cancer Diagnosis. Cancer Investig. 2019, 37, 432–439. [Google Scholar] [CrossRef]
- Khurram, I.; Khan, M.U.; Ibrahim, S.; Saleem, A.; Khan, Z.; Mubeen, M.; Khawar, A.; Ali, Q. Efficacy of cell-free DNA as a diagnostic biomarker in breast cancer patients. Sci. Rep. 2023, 13, 15347. [Google Scholar] [CrossRef] [PubMed]
- Yuzefovych, L.V.; Pastukh, V.M.; Ruchko, M.V.; Simmons, J.D.; Richards, W.O.; Rachek, L.I. Plasma mitochondrial DNA is elevated in obese type 2 diabetes mellitus patients and correlates positively with insulin resistance. PLoS ONE 2019, 14, e0222278. [Google Scholar] [CrossRef] [PubMed]
- Rosa, H.S.; Ajaz, S.; Gnudi, L.; Malik, A.N. A case for measuring both cellular and cell-free mitochondrial DNA as a disease biomarker in human blood. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2020, 34, 12278–12288. [Google Scholar] [CrossRef] [PubMed]
- Berezina, T.A.; Kopytsya, M.P.; Petyunina, O.V.; Berezin, A.A.; Obradovic, Z.; Schmidbauer, L.; Lichtenauer, M.; Berezin, A.E. Lower Circulating Cell-Free Mitochondrial DNA Is Associated with Heart Failure in Type 2 Diabetes Mellitus Patients. Cardiogenetics 2023, 13, 15–30. [Google Scholar] [CrossRef]
- Dong, C.; Liu, Y.; Sun, C.; Liang, H.; Dai, L.; Shen, J.; Wei, S.; Guo, S.; Leong, K.W.; Chen, Y.; et al. Identification of Specific Joint-Inflammatogenic Cell-Free DNA Molecules From Synovial Fluids of Patients With Rheumatoid Arthritis. Front. Immunol. 2020, 11, 662. [Google Scholar] [CrossRef]
- Hajizadeh, S.; DeGroot, J.; TeKoppele, J.M.; Tarkowski, A.; Collins, L.V. Extracellular mitochondrial DNA and oxidatively damaged DNA in synovial fluid of patients with rheumatoid arthritis. Arthritis Res. Ther. 2003, 5, R234. [Google Scholar] [CrossRef] [PubMed]
- Rykova, E.; Sizikov, A.; Roggenbuck, D.; Antonenko, O.; Bryzgalov, L.; Morozkin, E.; Skvortsova, K.; Vlassov, V.; Laktionov, P.; Kozlov, V. Circulating DNA in rheumatoid arthritis: Pathological changes and association with clinically used serological markers. Arthritis Res. Ther. 2017, 19, 85. [Google Scholar] [CrossRef] [PubMed]
- Seewald, L.A.; Sabino, I.G.; Bohner, A.; Delco, M.L. Synovial fluid mitochondrial DNA as a biomarker after naturally occurring intra-articular fracture. Osteoarthr. Cartil. 2021, 29, S91–S92. [Google Scholar] [CrossRef]
- Nie, S.; Lu, J.; Wang, L.; Gao, M. Pro-inflammatory role of cell-free mitochondrial DNA in cardiovascular diseases. IUBMB Life 2020, 72, 1879–1890. [Google Scholar] [CrossRef]
- Gambardella, S.; Limanaqi, F.; Ferese, R.; Biagioni, F.; Campopiano, R.; Centonze, D.; Fornai, F. ccf-mtDNA as a Potential Link Between the Brain and Immune System in Neuro-Immunological Disorders. Front. Immunol. 2019, 10, 430401. [Google Scholar] [CrossRef]
- Tsamardinos, I.; Charonyktakis, P.; Papoutsoglou, G.; Borboudakis, G.; Lakiotaki, K.; Zenklusen, J.C.; Juhl, H.; Chatzaki, E.; Lagani, V. Just Add Data: Automated predictive modeling for knowledge discovery and feature selection. NPJ Precis. Oncol. 2022, 6, 38. [Google Scholar] [CrossRef] [PubMed]
- Tsamardinos, I.; Greasidou, E.; Borboudakis, G. Bootstrapping the out-of-sample predictions for efficient and accurate cross-validation. Mach. Learn. 2018, 107, 1895–1922. [Google Scholar] [CrossRef] [PubMed]


| All BrCa (n = 119) | Adjuvant (n = 81) | Metastatic (n = 21) | Neo-Adjuvant (n = 17) | Healthy (n = 77) | |
|---|---|---|---|---|---|
| Total plasma ccfDNA (ng/μL) | 1035 (565–2270) | 1045 (565–1800) | 1030 (573–1260) | 951 (667–2270) | 733 (261–3720) |
| RQ of ccf-mtDNA | 3.84 (0.01–982.00) | 6.02 (0.01–982.00) | 2.78 (0.01–398.00) | 2.23 (0.01–962.00) | 0.63 (0.01–24.77) |
| Diabetes Group (n = 158) | Prediabetes Group (n = 78) | Healthy Group (n = 100) | |
|---|---|---|---|
| Total plasma ccfDNA (ng/μL) | 989 (507–1840) | 938 (439–1698) | 886 (261–2800) |
| RQ of ccf-mtDNA | 28.00 (0.01–504.90) | 31.90 (0.01–494.60) | 12.30 (0.01–161.10) |
| Osteoarthritis Group (n = 17) | Healthy Group (n = 17) | |
|---|---|---|
| Total plasma ccfDNA (ng/μL) | 1591 (1100–2440) | 906 (319–2580) |
| RQ of ccf-mtDNA | 6.96 (0.01–29.40) | 6.34 (0.01–24.80) |
| Adjuvant Group (n = 81) | Metastatic Group (n = 21) | Neo-Adjuvant Group (n = 17) | Healthy Group (n = 77) | |
|---|---|---|---|---|
| Age (years) | 59 (±12) | 66 (±14) | 55 (±19) | 56 (±13) |
| Menopause | 44 | 15 | 6 | |
| Stage | * | |||
| Ι | 16 | |||
| ΙΙ | 32 | |||
| ΙΙΙ | 26 | |||
| IV | 21 | |||
| Not available | 7 | |||
| Grade | ||||
| 1 | 5 | |||
| 2 | 34 | 4 | 8 | |
| 3 | 37 | 13 | 5 | |
| Not available | 5 | 4 | 4 | |
| Lymph node status | * | |||
| Negative | 33 | - | - | |
| Positive | 40 | |||
| Not available | 8 | |||
| ER status | ||||
| Positive | 52 | 13 | 13 | |
| Negative | 29 | 8 | 4 | |
| PR status | ||||
| Positive | 48 | 13 | 11 | |
| Negative | 33 | 8 | 6 | |
| Her2 status | ||||
| Positive | 30 | 9 | 7 | |
| Negative | 51 | 12 | 10 |
| Diabetes Group (n = 158) | Prediabetes Group (n = 78) | Healthy Group (n = 100) | |
|---|---|---|---|
| Age (years) | 54 ± 8 | 51 ± 9 | 53 ± 10 |
| Sex (male) | 92 | 43 | 52 |
| HbA1c (%) | 6.9 (5.9–7.4) | 6.1 (5.7–6.3) | 5.5 (5.4–5.7) |
| Fasting glucose (mg/dL) | 131 ± 7 | 117 ± 9 | 86 ± 11 |
| BMI (kg/m2) | 32.5 ± 6.2 | 31.8 ± 5.4 | 32.1 ± 6.0 |
| Treatment | All patients receive oral or insulin medication | - | - |
| Osteoarthritis Group (n = 17) | Healthy Group (n = 17) | |
|---|---|---|
| Age (years) | 72 ± 6 | 67 ± 7 |
| Sex (female) | 17 | 17 |
| BMI (kg/m2) | 32.5 ± 6.2 | 30.2 ± 5.1 |
| Smoking (no) | 17 | 17 |
| Menopause (yes) | 17 | 17 |
| Primer/Probe | Primer/Probe Sequence (5′–3′) |
|---|---|
| MTATP8F | ATCACCCAACTAAAAATATTAAACACAAACTA |
| MTATP8R | ATTTTGGTTCTCAGGGTTTGTTA |
| MTATP8PROBE | FAM-CTACCTCCCTCACCAAACCCATA-BHQ1 |
| GAPDHF | TCCCCACACACATGCACTTA |
| GAPDHR | TAGTCCCAGGGCTTTGATT |
| GAPDHPROBE | CY5-GAGCTAGGAAGGACAGGCAACTT-BHQ2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Panagopoulou, M.; Karaglani, M.; Tzitzikou, K.; Kessari, N.; Arvanitidis, K.; Amarantidis, K.; Drosos, G.I.; Gerou, S.; Papanas, N.; Papazoglou, D.; et al. Mitochondrial Fraction of Circulating Cell-Free DNA as an Indicator of Human Pathology. Int. J. Mol. Sci. 2024, 25, 4199. https://doi.org/10.3390/ijms25084199
Panagopoulou M, Karaglani M, Tzitzikou K, Kessari N, Arvanitidis K, Amarantidis K, Drosos GI, Gerou S, Papanas N, Papazoglou D, et al. Mitochondrial Fraction of Circulating Cell-Free DNA as an Indicator of Human Pathology. International Journal of Molecular Sciences. 2024; 25(8):4199. https://doi.org/10.3390/ijms25084199
Chicago/Turabian StylePanagopoulou, Maria, Makrina Karaglani, Konstantina Tzitzikou, Nikoleta Kessari, Konstantinos Arvanitidis, Kyriakos Amarantidis, George I. Drosos, Spyros Gerou, Nikolaos Papanas, Dimitrios Papazoglou, and et al. 2024. "Mitochondrial Fraction of Circulating Cell-Free DNA as an Indicator of Human Pathology" International Journal of Molecular Sciences 25, no. 8: 4199. https://doi.org/10.3390/ijms25084199
APA StylePanagopoulou, M., Karaglani, M., Tzitzikou, K., Kessari, N., Arvanitidis, K., Amarantidis, K., Drosos, G. I., Gerou, S., Papanas, N., Papazoglou, D., Baritaki, S., Constantinidis, T. C., & Chatzaki, E. (2024). Mitochondrial Fraction of Circulating Cell-Free DNA as an Indicator of Human Pathology. International Journal of Molecular Sciences, 25(8), 4199. https://doi.org/10.3390/ijms25084199










