Investigating the Impact of Electrostatic Interactions on Calmodulin Binding and Ca2+-Dependent Activation of the Calcium-Gated Potassium SK4 Channel
Abstract
1. Introduction
2. Results
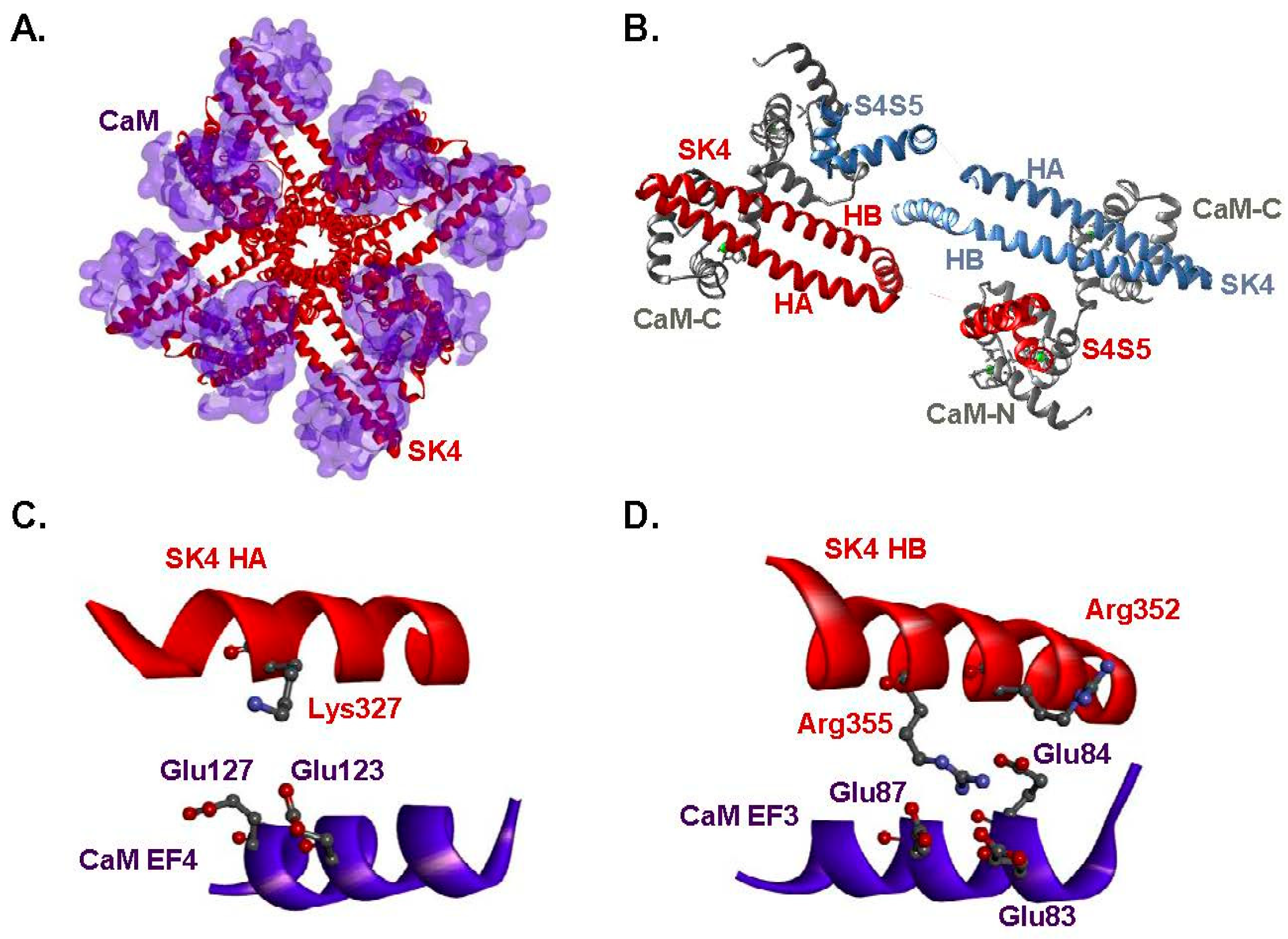
2.1. Two Hydrophilic Pockets Are Predicted between CaM and SK4 HA-HB Helices
2.2. Endogenous CaM Supports Function of Recombinant SK4 in HEK293T Cells
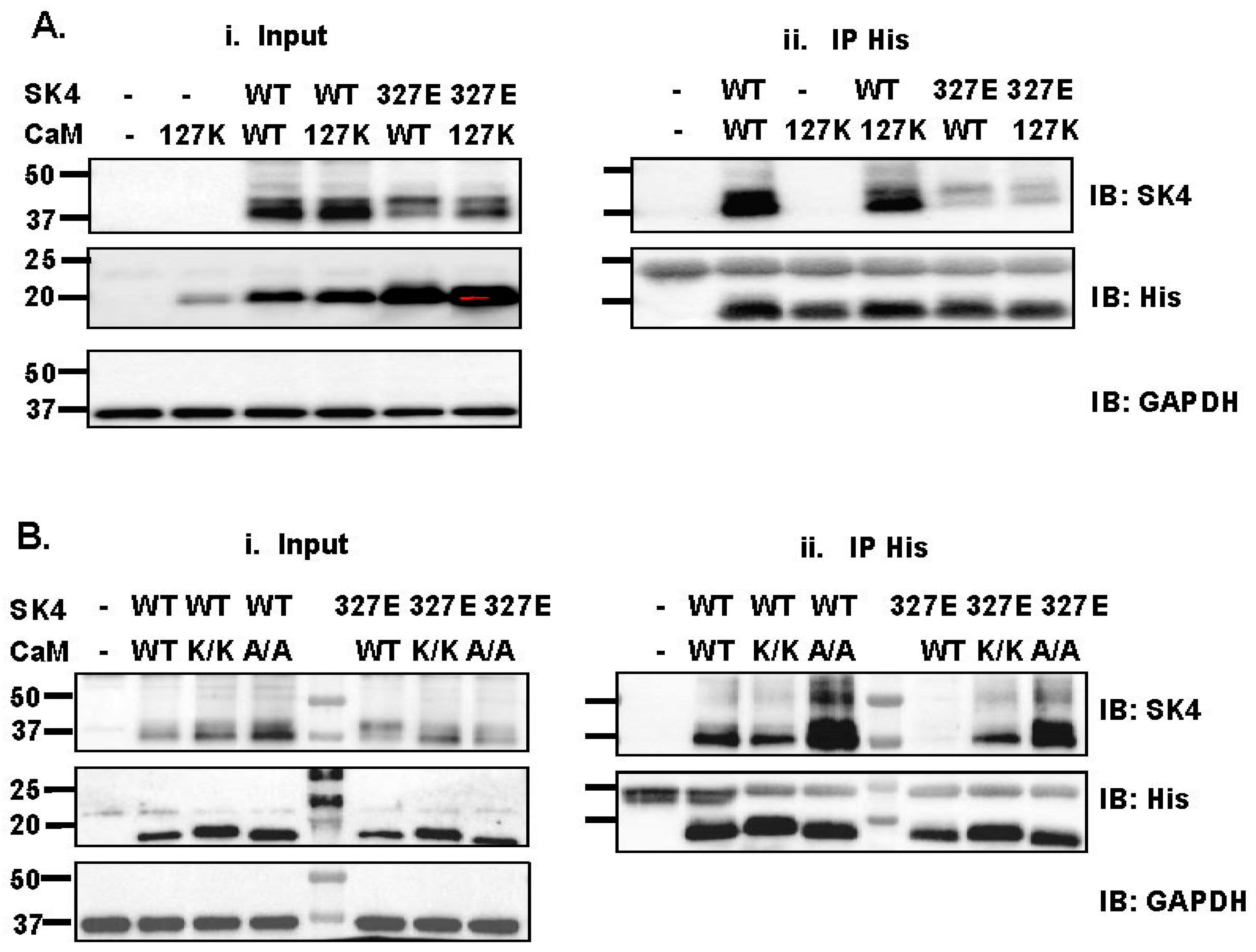
2.3. Single Substitution at Lys327 in the SK4 HA Helix Decrease Interaction with CaM
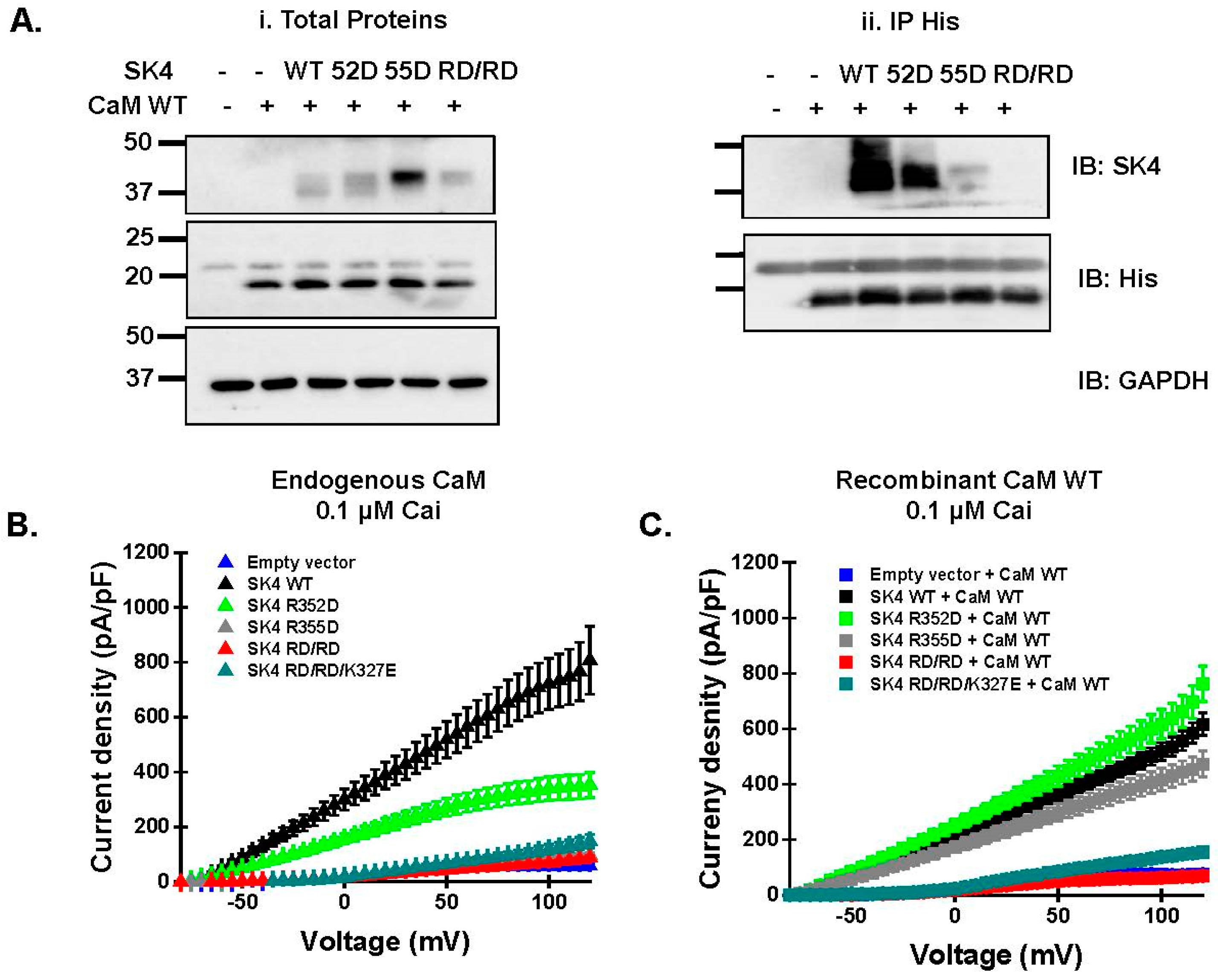
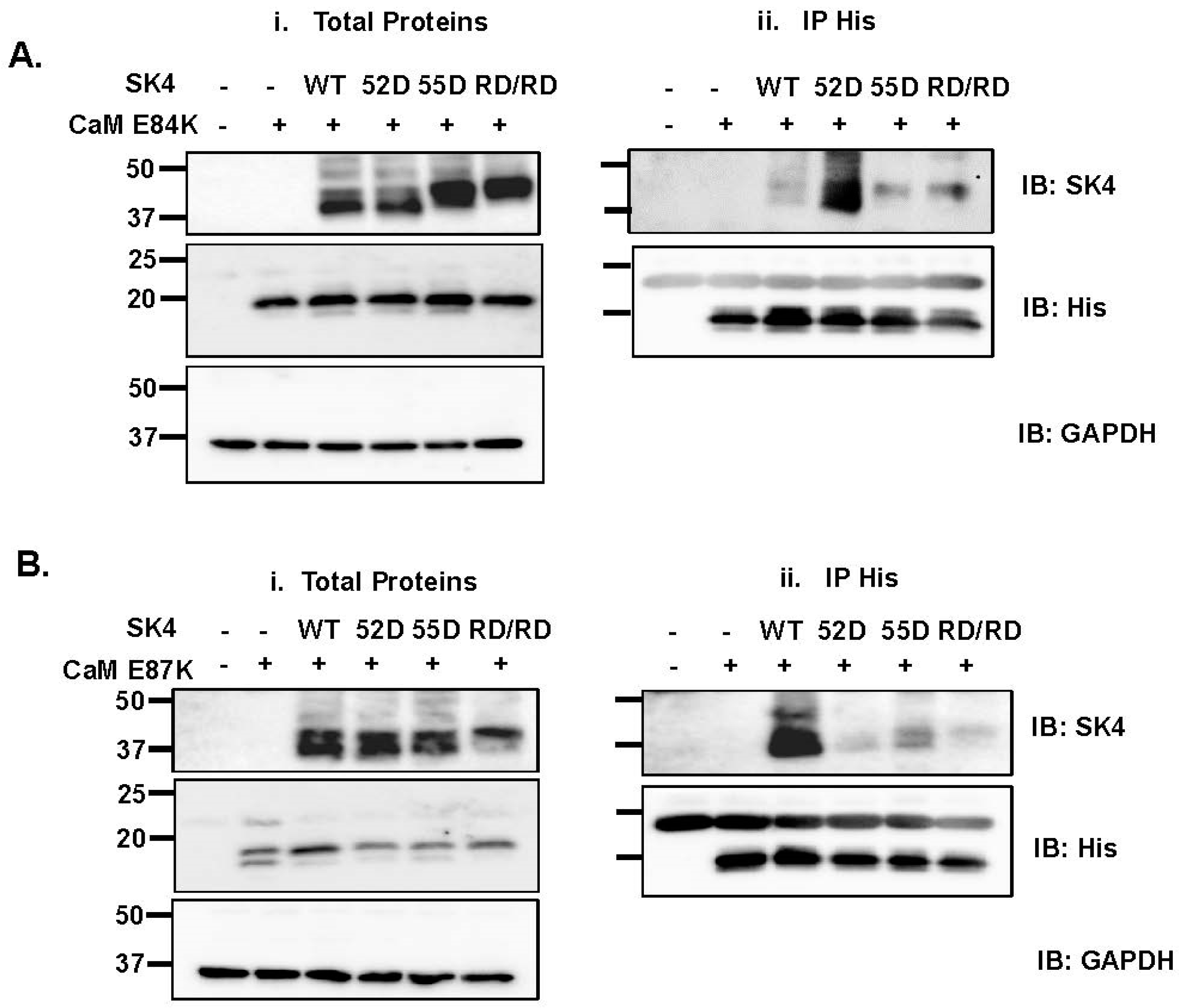
2.4. Double Charge Substitution in the SK4 HB Helix Disrupts Protein Interaction and Channel Function
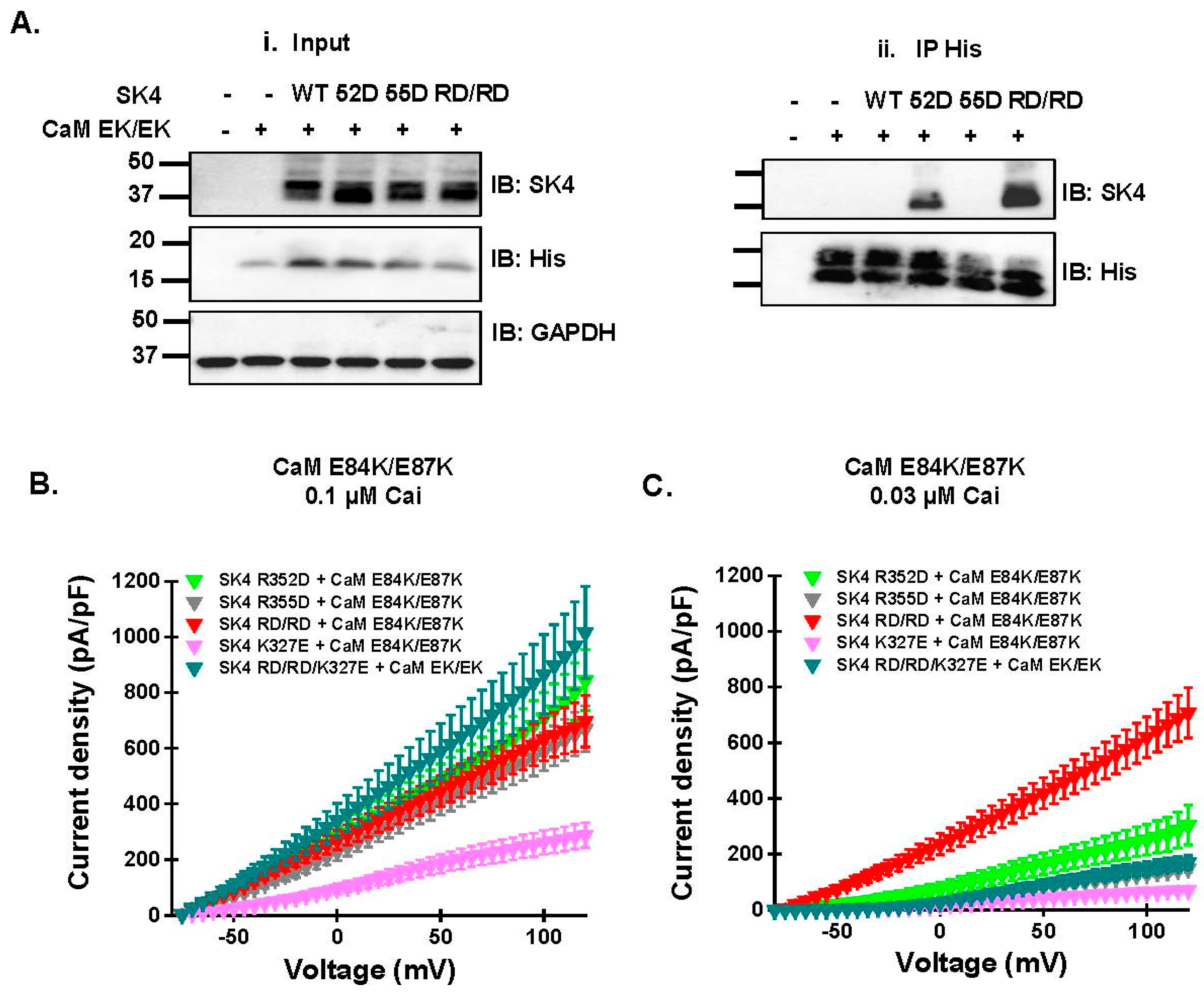
2.5. Electrostatic Interactions Promote CaM Binding onto the SK4 HB Helix
3. Discussion
3.1. Electrostatic Interactions Underlie CaM Binding on the HB Helix of SK4
3.2. A Role for the Intracellular C-Terminal of SK4 in the Channel Activation Gating
4. Materials and Methods
4.1. Computer-Based Imaging and Molecular Dynamics Simulation
4.2. Recombinant DNA Techniques
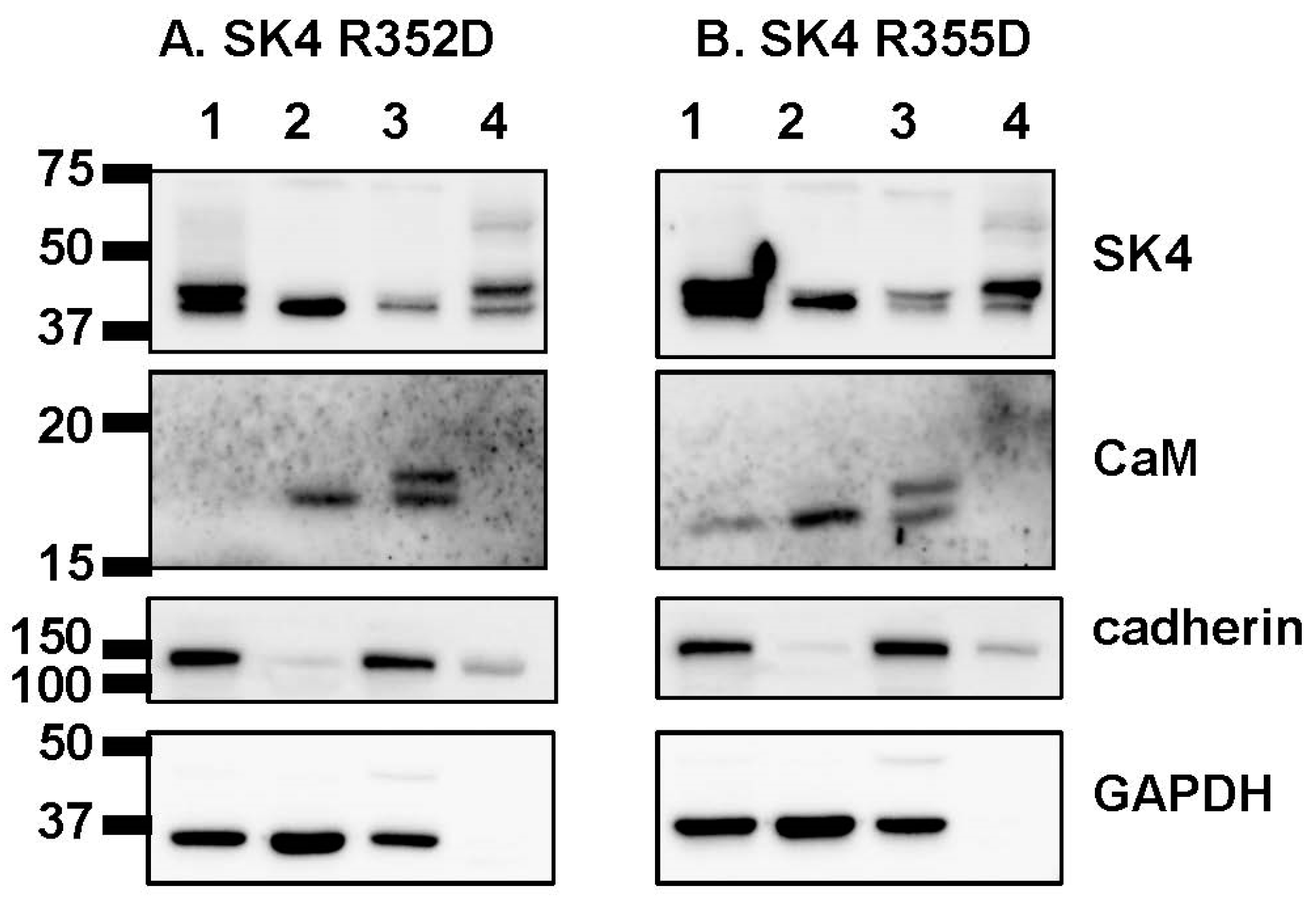
4.3. Cell Surface Fractionation Assays
4.4. Co-Immunoprecipitation Assays
4.5. Whole-Cell Patch-Clamp Experiments
4.6. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Meech, R. Intracellular calcium injection causes increased potassium conductance in Aplysia. Comp. Biochem. Physiol. A Comp. Physiol. 1972, 42, 493–499. [Google Scholar] [CrossRef] [PubMed]
- Gardos, G. Effect of ethylenediaminetetraacetate on the permeability of human erythrocytes. Acta Physiol. Acad. Sci. Hung. 1958, 14, 1–5. [Google Scholar] [PubMed]
- Klein, H.; Abu-Arish, A.; Trinh, N.T.N.; Luo, Y.; Wiseman, P.W.; Hanrahan, J.W.; Brochiero, E.; Sauvé, R. Investigating CFTR and KCa3.1 Protein/Protein Interactions. PLoS ONE 2016, 11, e0153665. [Google Scholar] [CrossRef] [PubMed]
- Kshatri, A.S.; Gonzalez-Hernandez, A.; Giraldez, T. Physiological Roles and Therapeutic Potential of Ca2+ Activated Potassium Channels in the Nervous System. Front. Mol. Neurosci. 2018, 11, 258. [Google Scholar] [CrossRef] [PubMed]
- Skibsbye, L.; Poulet, C.; Diness, J.G.; Bentzen, B.H.; Yuan, L.; Kappert, U.; Matschke, K.; Wettwer, E.; Ravens, U.; Grunnet, M.; et al. Small-conductance calcium-activated potassium (SK) channels contribute to action potential repolarization in human atria. Cardiovasc. Res. 2014, 103, 156–167. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-D.; Thai, P.N.; Lieu, D.K.; Chiamvimonvat, N. Cardiac small-conductance calcium-activated potassium channels in health and disease. Pflügers Arch. Eur. J. Physiol. 2021, 473, 477–489. [Google Scholar] [CrossRef]
- Liu, T.; Li, T.; Xu, D.; Wang, Y.; Zhou, Y.; Wan, J.; Huang, C.L.H.; Tan, X. Small-conductance calcium-activated potassium channels in the heart: Expression, regulation and pathological implications. Philos. Trans. R. Soc. B Biol. Sci. 2023, 378, 20220171. [Google Scholar] [CrossRef]
- Burg, S.; Shapiro, S.; Peretz, A.; Haimov, E.; Redko, B.; Yeheskel, A.; Simhaev, L.; Engel, H.; Raveh, A.; Ben-Bassat, A.; et al. Allosteric inhibitors targeting the calmodulin-PIP2 interface of SK4 K+ channels for atrial fibrillation treatment. Proc. Natl. Acad. Sci. USA 2022, 119, e2202926119. [Google Scholar] [CrossRef] [PubMed]
- Adelman, J.P.; Maylie, J.; Sah, P. Small-Conductance Ca2+-Activated K+ Channels: Form and Function. Ann. Rev. Physiol. 2012, 74, 245–269. [Google Scholar] [CrossRef]
- Xia, X.M.; Fakler, B.; Rivard, A.; Wayman, G.; Johnson-Pais, T.; Keen, J.E.; Ishii, T.; Hirschberg, B.; Bond, C.T.; Lutsenko, S.; et al. Mechanism of calcium gating in small-conductance calcium-activated potassium channels. Nature 1998, 395, 503–507. [Google Scholar] [CrossRef] [PubMed]
- Kretsinger, R.H. Gene Triplication deduced from the Tertiary Structure of a Muscle Calcium Binding Protein. Nat. New Biol. 1972, 240, 85–88. [Google Scholar] [CrossRef]
- Kretsinger, R.H. EF-hands embrace [news; comment]. Nat. Struct. Biol. 1997, 4, 514–516. [Google Scholar] [CrossRef] [PubMed]
- Moncrief, N.D.; Kretsinger, R.H.; Goodman, M. Evolution of EF-hand calcium-modulated proteins. I. Relationships based on amino acid sequences. J. Mol. Evol. 1990, 30, 522–562. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, H.; Nakayama, S.; Kretsinger, R.H. Classification and evolution of EF-hand proteins. Biometals 1998, 11, 277–295. [Google Scholar] [CrossRef] [PubMed]
- Persechini, A.; Moncrief, N.D.; Kretsinger, R.H. The EF-hand family of calcium-modulated proteins. Trends Neurosci. 1989, 12, 462–467. [Google Scholar] [CrossRef]
- Lee, C.-H.; MacKinnon, R. Activation mechanism of a human SK-calmodulin channel complex elucidated by cryo-EM structures. Science 2018, 360, 508–513. [Google Scholar] [CrossRef] [PubMed]
- Wissmann, R.; Bildl, W.; Neumann, H.; Rivard, A.F.; Klöcker, N.; Weitz, D.; Schulte, U.; Adelman, J.P.; Bentrop, D.; Fakler, B. A Helical Region in the C Terminus of Small-conductance Ca2+-activated K+ Channels Controls Assembly with Apo-calmodulin. J. Biol. Chem. 2002, 277, 4558–4564. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.Y.; Rumpf, C.H.; Fujiwara, Y.; Cooley, E.S.; Van Petegem, F.; Minor, J. Structures of CaV2 Ca2+/CaM-IQ Domain Complexes Reveal Binding Modes that Underlie Calcium-Dependent Inactivation and Facilitation. Structure 2008, 16, 1455–1467. [Google Scholar] [CrossRef] [PubMed]
- Ramis, R.; Ballesteros, Ó.R.; Muguruza-Montero, A.; M-Alicante, S.; Núñez, E.; Villarroel, Á.; Leonardo, A.; Bergara, A. Molecular dynamics simulations of the calmodulin-induced α-helix in the SK2 calcium-gated potassium ion channel. J. Biol. Chem. 2023, 299, 102850. [Google Scholar] [CrossRef]
- Lee, W.-S.; Ngo-Anh, T.J.; Bruening-Wright, A.; Maylie, J.; Adelman, J.P. Small Conductance Ca2+-activated K+ Channels and Calmodulin: Cell Surface Expression and Gating. J. Biol. Chem. 2003, 278, 25940–25946. [Google Scholar] [CrossRef] [PubMed]
- Weisbrod, D. Small and Intermediate Calcium Activated Potassium Channels in the Heart: Role and Strategies in the Treatment of Cardiovascular Diseases. Front. Physiol. 2020, 11, 590534. [Google Scholar] [CrossRef]
- Westerlund, A.M.; Delemotte, L. Effect of Ca2+ on the promiscuous target-protein binding of calmodulin. PLoS Comput. Biol. 2018, 14, e1006072. [Google Scholar] [CrossRef]
- Persechini, A.; Cronk, B. The Relationship between the Free Concentrations of Ca2+ and Ca2+-calmodulin in Intact Cells. J. Biol. Chem. 1999, 274, 6827–6830. [Google Scholar] [CrossRef] [PubMed]
- Persechini, A.; Stemmer, P.M. Calmodulin Is a Limiting Factor in the Cell. Trends Cardiovasc. Med. 2002, 12, 32–37. [Google Scholar] [CrossRef] [PubMed]
- Allen, D.; Fakler, B.; Maylie, J.; Adelman, J.P. Organization and regulation of small conductance Calcium -activated potassium channel multiprotein complexes. J. Neurosci. 2007, 27, 2369–2376. [Google Scholar] [CrossRef]
- Nam, Y.-W.; Kong, D.; Wang, D.; Orfali, R.; Sherpa, R.T.; Totonchy, J.; Nauli, S.M.; Zhang, M. Differential modulation of SK channel subtypes by phosphorylation. Cell Calcium 2021, 94, 102346. [Google Scholar] [CrossRef]
- Halling, D.B.; Kenrick, S.A.; Riggs, A.F.; Aldrich, R.W. Calcium-dependent stoichiometries of the KCa2.2 (SK) intracellular domain/calmodulin complex in solution. J. Gen. Physiol. 2014, 143, 231. [Google Scholar] [CrossRef]
- Schumacher, M.A.; Rivard, A.F.; Bachinger, H.P.; Adelman, J.P. Structure of the gating domain of a Ca2+-activated K+ channel complexed with Ca2+/calmodulin. Nature 2001, 410, 1120. [Google Scholar] [CrossRef]
- Halling, D.B.; Philpo, A.E.; Aldrich, R.W. Calcium dependence of both lobes of calmodulin is involved in binding to a cytoplasmic domain of SK channels. eLife 2022, 11, e81303. [Google Scholar] [CrossRef] [PubMed]
- Šali, A.; Blundell, T.L. Comparative Protein Modelling by Satisfaction of Spatial Restraints. J. Mol. Biol. 1993, 234, 779–815. [Google Scholar] [CrossRef] [PubMed]
- Klein, H.; Garneau, L.; Banderali, U.; Simoes, M.; Parent, L.; Sauvé, R. Structural Determinants of the Closed KCa3.1 Channel Pore in Relation to Channel Gating: Results from a Substituted Cysteine Accessibility Analysis. J. Gen. Physiol. 2007, 129, 299–315. [Google Scholar] [CrossRef] [PubMed]
- Garneau, L.; Klein, H.; Banderali, U.; Longpre-Lauzon, A.; Parent, L.; Sauve, R. Hydrophobic Interactions as Key Determinants to the KCa3.1 Channel Closed Configuration: An analysis of KCa3.1 mutants constitutively active in zero calcium. J. Biol. Chem. 2009, 284, 389–403. [Google Scholar] [CrossRef] [PubMed]
- Phillips, J.C.; Hardy, D.J.; Maia, J.D.C.; Stone, J.E.; Ribeiro, J.V.; Bernardi, R.C.; Buch, R.; Fiorin, G.; Hénin, J.; Jiang, W.; et al. Scalable molecular dynamics on CPU and GPU architectures with NAMD. J. Chem. Phys. 2020, 153, 044130. [Google Scholar] [CrossRef] [PubMed]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef]
- Klein, H.; Garneau, L.; Coady, M.; Lemay, G.; Lapointe, J.Y.; Sauvé, R. Molecular Characterization of an Inwardly Rectifying K+ Channel from HeLa Cells. J. Membr. Biol. 1999, 167, 43–52. [Google Scholar] [CrossRef]
- Simoes, M.; Garneau, L.; Klein, H.; Banderali, U.; Hobeila, F.; Roux, B.; Parent, L.; Sauve, R. Cysteine Mutagenesis and Computer Modeling of the S6 Region of an Intermediate Conductance IKCa Channel. J. Gen. Physiol. 2002, 120, 99–116. [Google Scholar] [CrossRef] [PubMed]
- Briot, J.; Methot, O.; Bourdin, B.; Tetreault, M.P.; Najmanovich, R.; Parent, L. A three-way inter-molecular network accounts for the CaVa2d1-induced functional modulation of the pore-forming CaV1.2 subunit. J. Biol. Chem. 2018, 293, 7176–7188. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Segura, E.; Marsolais, M.; Parent, L. A CACNA1C variant associated with cardiac arrhythmias provides mechanistic insights in the calmodulation of L-type Ca2+ channels. J. Biol. Chem. 2022, 298, 102632. [Google Scholar] [CrossRef]
- Bourdin, B.; Briot, J.; Tetreault, M.P.; Sauve, R.; Parent, L. Negatively charged residues in the first extracellular loop of the L-type CaV1.2 channel anchor the interaction with the CaValpha2delta1 auxiliary subunit. J. Biol. Chem. 2017, 292, 17236–17249. [Google Scholar] [CrossRef]
- Segura, E.; Bourdin, B.; Tétreault, M.-P.; Briot, J.; Allen, B.G.; Mayer, G.; Parent, L. Proteolytic cleavage of the hydrophobic domain in the CaVα2δ1 subunit improves assembly and activity of cardiac CaV1.2 channels. J. Biol. Chem. 2017, 292, 11109–11124. [Google Scholar] [CrossRef]
- Lizotte, E.; Tremblay, A.; Allen, B.G.; Fiset, C. Isolation and characterization of subcellular protein fractions from mouse heart. Anal. Biochem. 2005, 345, 47–54. [Google Scholar] [CrossRef] [PubMed]

| SK4 | Co-IP | Native CaM | CaM WT | ||
|---|---|---|---|---|---|
| n/N | Mean ± SD (pA/pF) | n/N | Mean ± SD (pA/pF) | ||
| Empty vector | - | 6/2 | 55 ± 16 | 5/1 | 77 ± 20 |
| WT | ++ | 10/2 | 563 ± 199 p < 0.001 v. Empty vector | 33/9 | 401 ± 135 p < 0.001 v. Empty vector + CaM WT p = 0.03 v. SK4 WT + native CaM |
| K327E | + | 14/2 | 183 ± 50 p < 0.001 v. Empty vector p < 0.001 v. SK4 WT + native CaM | 27/5 | 405 ± 193 p < 0.001 v. Empty vector p < 0.001 v. SK4 K327E + native CaM |
| R352D | + | 18/2 | 289 ± 148 p < 0.001 v. Empty vector p = 0.001 v. SK4 WT + native CaM | 21/4 | 430 ± 163 p < 0.001 v. Empty vector + CaM WT p = 0.008 v. SK4 R352D + native CaM |
| R355D | + | 20/3 | 59 ± 23 p < 0.001 v. SK4 WT + native CaM | 37/5 | 330 ± 172 p < 0.001 v. Empty vector + CaM WT p < 0.001 v. SK4 R355D + native CaM |
| RD/RD | - | 15/3 | 49 ± 22 p < 0.001 v. SK4 WT + native CaM | 11/2 | 54 ± 19 p < 0.001 v. SK4 WT + CaM WT |
| RD/RD/K327E | N/A | 9/1 | 75 ± 30 p < 0.001 v. SK4 WT + native CaM | 10/1 | 97 ± 31 p < 0.001 v. SK4 WT + CaM WT |
| SK4 | CaM | Co-IP | Cai 0.1 μM | Cai 0.03 μM | ||
|---|---|---|---|---|---|---|
| n/N | Mean ± SD (pA/pF) | n/N | Mean ± SD (pA/pF) | |||
| R352D | E84K/E87K | ++ | 19/3 | 525 ± 172 | 8/1 | 182 ± 73 p < 0.001 v. Cai 0.1 μM |
| R355D | E84K/E87K | – | 26/3 | 468 ± 136 | 12/2 | 92 ± 30 p < 0.001 v. Cai 0.1 μM |
| RD/RD | E84K/E87K | ++ | 26/5 | 511 ± 205 | 10/2 | 457 ± 187 |
| K327E | E84K/E87K | N/A | 7/1 | 202 ± 61 | 7/1 | 48 ± 13 p < 0.001 v. Cai 0.1 μM |
| RD/RD/K327E | E84K/E87K | N/A | 8/1 | 642 ± 275 | 6/1 | 106 ± 42 p < 0.001 v. Cai 0.1 μM |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Segura, É.; Zhao, J.; Broszczak, M.; Audet, F.; Sauvé, R.; Parent, L. Investigating the Impact of Electrostatic Interactions on Calmodulin Binding and Ca2+-Dependent Activation of the Calcium-Gated Potassium SK4 Channel. Int. J. Mol. Sci. 2024, 25, 4255. https://doi.org/10.3390/ijms25084255
Segura É, Zhao J, Broszczak M, Audet F, Sauvé R, Parent L. Investigating the Impact of Electrostatic Interactions on Calmodulin Binding and Ca2+-Dependent Activation of the Calcium-Gated Potassium SK4 Channel. International Journal of Molecular Sciences. 2024; 25(8):4255. https://doi.org/10.3390/ijms25084255
Chicago/Turabian StyleSegura, Émilie, Juan Zhao, Marlena Broszczak, Frédéric Audet, Rémy Sauvé, and Lucie Parent. 2024. "Investigating the Impact of Electrostatic Interactions on Calmodulin Binding and Ca2+-Dependent Activation of the Calcium-Gated Potassium SK4 Channel" International Journal of Molecular Sciences 25, no. 8: 4255. https://doi.org/10.3390/ijms25084255
APA StyleSegura, É., Zhao, J., Broszczak, M., Audet, F., Sauvé, R., & Parent, L. (2024). Investigating the Impact of Electrostatic Interactions on Calmodulin Binding and Ca2+-Dependent Activation of the Calcium-Gated Potassium SK4 Channel. International Journal of Molecular Sciences, 25(8), 4255. https://doi.org/10.3390/ijms25084255







