Circulating microRNA Profiles Identify a Patient Subgroup with High Inflammation and Severe Symptoms in Schizophrenia Experiencing Acute Psychosis
Abstract
1. Introduction
2. Results
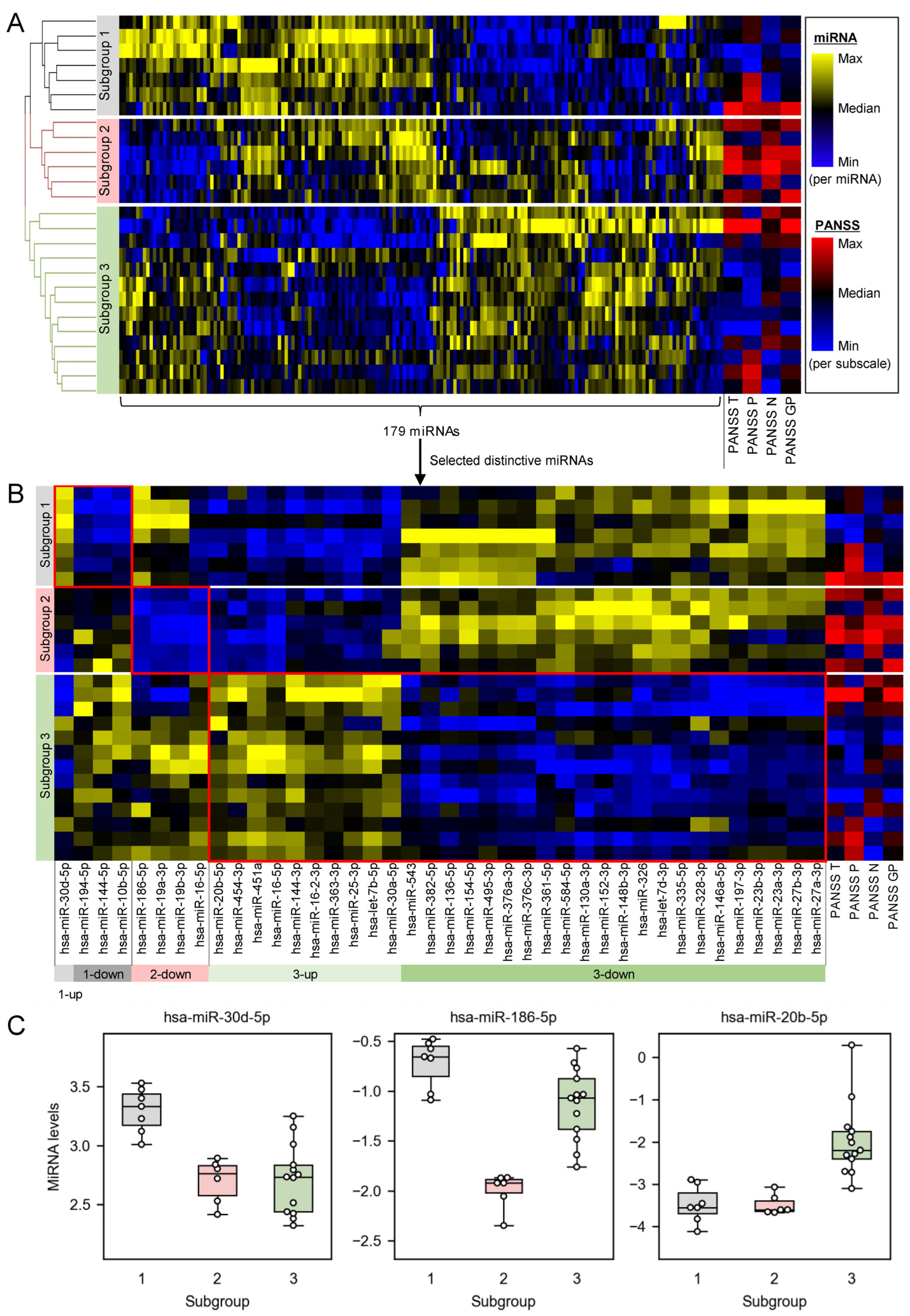
2.1. Plasma miRNA Profiles Revealed Three Subgroups of Schizophrenia Patients
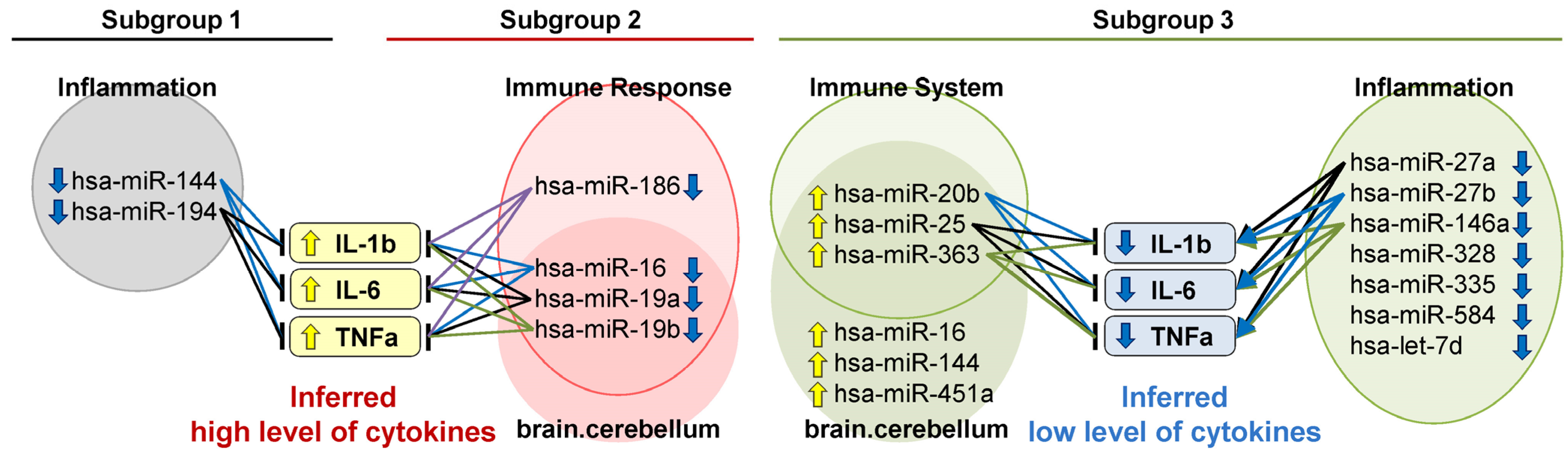
2.2. Distinctively Downregulated miRNAs in the Subgroup with High PANSS Scores Enriched ‘Immune Response’, and Were Reported to Negatively Regulate IL-1β, IL-6, and TNFα
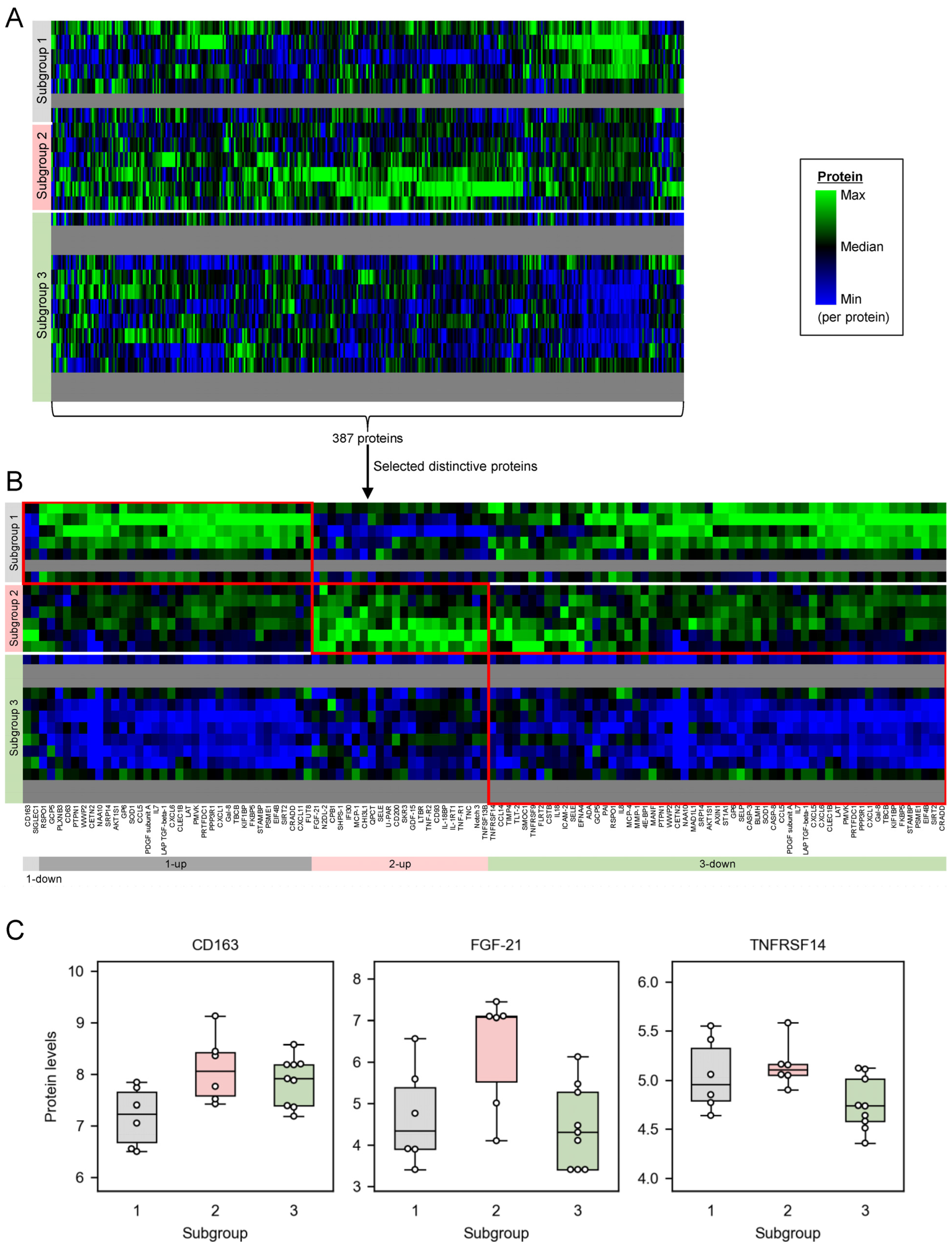
2.3. Distinctively Upregulated Proteins in the Subgroups with High PANSS Scores Enriched ‘Cytokine-Cytokine Receptor Interaction’, and All the Mapped Proteins Were Pro-Inflammatory Cytokines
3. Discussion
3.1. Plasma miRNAs Are a Potential Biomarker to Stratify Schizophrenia Patients into Subgroups of Different Pathophysiological Backgrounds
3.2. The Subgroup with High PANSS Scores Is Associated with High Inflammation
3.3. Anti-Inflammatory Treatments Are Potentially Effective for the Schizophrenia Subgroup with High Inflammation
3.4. This Study Has Limitations Regarding the Data Set and Methods
4. Materials and Methods
4.1. Study Population
4.2. Plasma Sample Preparation
4.3. MicroRNA Measurement
4.4. MicroRNA Set Enrichment Analysis for Distinctive miRNAs in Each Subgroup
4.5. Literature-Based Inference of Regulatory Functions of miRNAs on Pro-Inflammatory Cytokines
4.6. Protein Measurement
4.7. Protein Set Enrichment Analysis for Distinctive Proteins in Each Subgroup
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Charlson, F.J.; Ferrari, A.J.; Santomauro, D.F.; Diminic, S.; Stockings, E.; Scott, J.G.; McGrath, J.J.; Whiteford, H.A. Global Epidemiology and Burden of Schizophrenia: Findings from the Global Burden of Disease Study 2016. Schizophr. Bull. 2018, 44, 1195–1203. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Chant, D.; Welham, J.; McGrath, J. A systematic review of the prevalence of schizophrenia. PLoS Med. 2005, 2, e141. [Google Scholar] [CrossRef]
- Fountoulakis, K.N.; Dragioti, E.; Theofilidis, A.T.; Wikilund, T.; Atmatzidis, X.; Nimatoudis, I.; Thys, E.; Wampers, M.; Hranov, L.; Hristova, T.; et al. Staging of Schizophrenia with the Use of PANSS: An International Multi-Center Study. Int. J. Neuropsychopharmacol. 2019, 22, 681–697. [Google Scholar] [CrossRef] [PubMed]
- Correll, C.U.; Schooler, N.R. Negative Symptoms in Schizophrenia: A Review and Clinical Guide for Recognition, Assessment, and Treatment. Neuropsychiatr. Dis. Treat. 2020, 16, 519–534. [Google Scholar] [CrossRef] [PubMed]
- van Os, J.; Kapur, S. Schizophrenia. Lancet 2009, 374, 635–645. [Google Scholar] [CrossRef] [PubMed]
- Buckley, P.F.; Miller, B.J.; Lehrer, D.S.; Castle, D.J. Psychiatric comorbidities and schizophrenia. Schizophr. Bull. 2009, 35, 383–402. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, J.L.; Altar, C.A.; Taylor, D.L.; Degtiar, I.; Hornberger, J.C. The social and economic burden of treatment-resistant schizophrenia: A systematic literature review. Int. Clin. Psychopharmacol. 2014, 29, 63–76. [Google Scholar] [CrossRef] [PubMed]
- Kay, S.R.; Fiszbein, A.; Opler, L.A. The positive and negative syndrome scale (PANSS) for schizophrenia. Schizophr. Bull. 1987, 13, 261–276. [Google Scholar] [CrossRef] [PubMed]
- Lefort-Besnard, J.; Varoquaux, G.; Derntl, B.; Gruber, O.; Aleman, A.; Jardri, R.; Sommer, I.; Thirion, B.; Bzdok, D. Patterns of schizophrenia symptoms: Hidden structure in the PANSS questionnaire. Transl. Psychiatry 2018, 8, 237. [Google Scholar] [CrossRef]
- Bowen, E.F.W.; Burgess, J.L.; Granger, R.; Kleinman, J.E.; Rhodes, C.H. DLPFC transcriptome defines two molecular subtypes of schizophrenia. Transl. Psychiatry 2019, 9, 147. [Google Scholar] [CrossRef] [PubMed]
- Childers, E.; Bowen, E.F.W.; Rhodes, C.H.; Granger, R. Immune-Related Genomic Schizophrenic Subtyping Identified in DLPFC Transcriptome. Genes 2022, 13, 1200. [Google Scholar] [CrossRef] [PubMed]
- Hertzberg, L.; Maggio, N.; Muler, I.; Yitzhaky, A.; Majer, M.; Haroutunian, V.; Zuk, O.; Katsel, P.; Domany, E.; Weiser, M. Comprehensive Gene Expression Analysis Detects Global Reduction of Proteasome Subunits in Schizophrenia. Schizophr. Bull. 2021, 47, 785–795. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, E.; van Beveren, N.J.; Ramsey, J.; Leweke, F.M.; Rothermundt, M.; Bogerts, B.; Steiner, J.; Guest, P.C.; Bahn, S. Identification of subgroups of schizophrenia patients with changes in either immune or growth factor and hormonal pathways. Schizophr. Bull. 2014, 40, 787–795. [Google Scholar] [CrossRef] [PubMed]
- Rifai, N.; Gillette, M.A.; Carr, S.A. Protein biomarker discovery and validation: The long and uncertain path to clinical utility. Nat. Biotechnol. 2006, 24, 971–983. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.N.; Masud, M.K.; Haque, M.H.; Hossain, M.S.A.; Yamauchi, Y.; Nguyen, N.-T.; Shiddiky, M.J.A. RNA Biomarkers: Diagnostic and Prognostic Potentials and Recent Developments of Electrochemical Biosensors. Small Methods 2017, 1, 1700131. [Google Scholar] [CrossRef]
- Lim, M.; Carollo, A.; Neoh, M.J.Y.; Esposito, G. Mapping miRNA Research in Schizophrenia: A Scientometric Review. Int. J. Mol. Sci. 2022, 24, 436. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, A.; Filant, J.; Moxley, K.M.; Sood, A.; McMeekin, S.; Ramesh, R. Exosomes: A role for naturally occurring nanovesicles in cancer growth, diagnosis and treatment. Curr. Gene Ther. 2015, 15, 182–192. [Google Scholar] [CrossRef]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef] [PubMed]
- Brase, J.C.; Wuttig, D.; Kuner, R.; Sultmann, H. Serum microRNAs as non-invasive biomarkers for cancer. Mol. Cancer 2010, 9, 306. [Google Scholar] [CrossRef] [PubMed]
- Perkins, D.O.; Jeffries, C.D.; Jarskog, L.F.; Thomson, J.M.; Woods, K.; Newman, M.A.; Parker, J.S.; Jin, J.; Hammond, S.M. microRNA expression in the prefrontal cortex of individuals with schizophrenia and schizoaffective disorder. Genome Biol. 2007, 8, R27. [Google Scholar] [CrossRef] [PubMed]
- Beveridge, N.J.; Gardiner, E.; Carroll, A.P.; Tooney, P.A.; Cairns, M.J. Schizophrenia is associated with an increase in cortical microRNA biogenesis. Mol. Psychiatry 2010, 15, 1176–1189. [Google Scholar] [CrossRef] [PubMed]
- Santarelli, D.M.; Carroll, A.P.; Cairns, H.M.; Tooney, P.A.; Cairns, M.J. Schizophrenia-associated MicroRNA-Gene Interactions in the Dorsolateral Prefrontal Cortex. Genom. Proteom. Bioinform. 2019, 17, 623–634. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Du, J.; Qi, Y.; Liang, G.; Wang, T.; Li, S.; Xie, S.; Zeshan, B.; Xiao, Z. Aberrant expression of serum miRNAs in schizophrenia. J. Psychiatr. Res. 2012, 46, 198–204. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Guo, C.; Guo, M.; Tong, S.; Zhang, Q.; Sun, H.; He, L.; Shi, Y. Identification of serum microRNAs as diagnostic biomarkers for schizophrenia. Hereditas 2019, 156, 23. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.Y.; Zhang, J.; Niu, W.; Guo, W.; Song, H.T.; Li, H.Y.; Fan, H.M.; Zhao, L.; Zhong, A.F.; Dai, Y.H.; et al. A preliminary analysis of microRNA as potential clinical biomarker for schizophrenia. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2015, 168B, 170–178. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Jinde, S.; Koike, S.; Tada, M.; Satomura, Y.; Yoshikawa, A.; Nishimura, Y.; Takizawa, R.; Kinoshita, A.; Sakakibara, E.; et al. Altered expression of microRNA-223 in the plasma of patients with first-episode schizophrenia and its possible relation to neuronal migration-related genes. Transl. Psychiatry 2019, 9, 289. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Zhang, F.; Wang, X.; Shugart, Y.Y.; Zhao, Y.; Li, X.; Liu, Z.; Sun, N.; Yang, C.; Zhang, K.; et al. Diagnostic value of blood-derived microRNAs for schizophrenia: Results of a meta-analysis and validation. Sci. Rep. 2017, 7, 15328. [Google Scholar] [CrossRef] [PubMed]
- Geaghan, M.P.; Atkins, J.R.; Brichta, A.M.; Tooney, P.A.; Scott, R.J.; Carr, V.J.; Cairns, M.J. Alteration of miRNA-mRNA interactions in lymphocytes of individuals with schizophrenia. J. Psychiatr. Res. 2019, 112, 89–98. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Xu, Y.; Shugart, Y.Y.; Yue, W.; Qi, G.; Yuan, G.; Cheng, Z.; Yao, J.; Wang, J.; Wang, G.; et al. Converging evidence implicates the abnormal microRNA system in schizophrenia. Schizophr. Bull. 2015, 41, 728–735. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Shang, S.; Wang, J.; Zhang, T.; Nie, F.; Song, X.; Heping Zhao Zhu, C.; Zhang, R.; Hao, D. Identification of miR-22-3p, miR-92a-3p, and miR-137 in peripheral blood as biomarker for schizophrenia. Psychiatry Res. 2018, 265, 70–76. [Google Scholar] [CrossRef]
- Pérez-Rodríguez, D.; Penedo, M.A.; Rivera-Baltanás, T.; Peña-Centeno, T.; Burkhardt, S.; Fischer, A.; Prieto-González, J.M.; Olivares, J.M.; López-Fernández, H.; Agís-Balboa, R.C. MiRNA Differences Related to Treatment-Resistant Schizophrenia. Int. J. Mol. Sci. 2023, 24, 1891. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Li, X.; Li, Z.; Qiang, Z.; Ma, H. Altered expression of MiR-186-5p and its target genes after spinal cord ischemia-reperfusion injury in rats. Neurosci. Lett. 2020, 718, 134669. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.L.; Lin, K.; Lin, S.K. NLRP3 inflammasome signaling as an early molecular response is negatively controlled by miR-186 in CFA-induced prosopalgia mice. Braz. J. Med. Biol. Res. 2018, 51, e7602. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Huang, T.; Qin, L.; Yin, L. Circ_0068087 Silencing Ameliorates Oxidized Low-Density Lipoprotein-Induced Dysfunction in Vascular Endothelial Cells Depending on miR-186-5p-Mediated Regulation of Roundabout Guidance Receptor 1. Front. Cardiovasc. Med. 2021, 8, 650374. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Sun, H.; Ji, Z. Downregulating lncRNA PVT1 Relieves Astrocyte Overactivation Induced Neuropathic Pain Through Targeting miR-186-5p/CXCL13/CXCR5 Axis. Neurochem. Res. 2021, 46, 1457–1469. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Wu, Q.; Shi, Y.; Luo, A.; Lin, S.; Feng, X.; Jiang, J.; Zhang, M.; Wang, F.; Tan, W. MicroRNA-15a/16/SOX5 axis promotes migration, invasion and inflammatory response in rheumatoid arthritis fibroblast-like synoviocytes. Aging 2020, 12, 14376–14390. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Li, J.; Cai, J.; Cheng, L.; Wang, X.; Xu, P.; Li, G.; Liang, X. Overexpression of MicroRNA-16 Alleviates Atherosclerosis by Inhibition of Inflammatory Pathways. BioMed Res. Int. 2020, 2020, 8504238. [Google Scholar] [CrossRef]
- Liang, X.; Xu, Z.; Yuan, M.; Zhang, Y.; Zhao, B.; Wang, J.; Zhang, A.; Li, G. MicroRNA-16 suppresses the activation of inflammatory macrophages in atherosclerosis by targeting PDCD4. Int. J. Mol. Med. 2016, 37, 967–975. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, T.; Li, Y.; Xu, Y. miR-19a/19b improves the therapeutic potential of mesenchymal stem cells in a mouse model of myocardial infarction. Gene Ther. 2021, 28, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Lin, T.Y.; Chen, L.; Liu, Y.; Dian, M.J.; Hao, W.C.; Lin, X.L.; Li, X.Y.; Li, Y.L.; Lian, M.; et al. miR-19 regulates the expression of interferon-induced genes and MHC class I genes in human cancer cells. Int. J. Med. Sci. 2020, 17, 953–964. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; She, S.; Li, D.; Liu, Z.; Yang, X.; Zeng, Z.; Liu, F. Role of miR-19a targeting TNF-α in mediating ulcerative colitis. Scand. J. Gastroenterol. 2013, 48, 815–824. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Liu, X.; Ni, H. Clinical significance of miR-19b-3p in patients with sepsis and its regulatory role in the LPS-induced inflammatory response. Eur. J. Med. Res. 2020, 25, 9. [Google Scholar] [CrossRef] [PubMed]
- Liang, G.H.; Luo, Y.N.; Wei, R.Z.; Yin, J.Y.; Qin, Z.L.; Lu, L.L.; Ma, W.H. CircZNF532 knockdown protects retinal pigment epithelial cells against high glucose-induced apoptosis and pyroptosis by regulating the miR-20b-5p/STAT3 axis. J. Diabetes Investig. 2022, 13, 781–795. [Google Scholar] [CrossRef]
- You, H.; Zhang, L.; Chen, Z.; Liu, W.; Wang, H.; He, H. MiR-20b-5p relieves neuropathic pain by targeting Akt3 in a chronic constriction injury rat model. Synapse 2019, 73, e22125. [Google Scholar] [CrossRef] [PubMed]
- Tang, G.X.; Yang, M.S.; Xiang, K.M.; Yang, B.C.; Liu, Z.L.; Zhao, S.P. MiR-20b-5p modulates inflammation, apoptosis and angiogenesis in severe acute pancreatitis through autophagy by targeting AKT3. Autoimmunity 2021, 54, 460–470. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Duan, J.; Gong, H.; Pang, Y.; Wang, L.; Yan, Y. Exosomes from miR-20b-3p-overexpressing stromal cells ameliorate calcium oxalate deposition in rat kidney. J. Cell. Mol. Med. 2019, 23, 7268–7278. [Google Scholar] [CrossRef] [PubMed]
- Che, Z.; Xueqin, J.; Zhang, Z. LncRNA OIP5-AS1 accelerates intervertebral disc degeneration by targeting miR-25-3p. Bioengineered 2021, 12, 11201–11212. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Zhao, J.L.; Peng, Z.Y.; Xu, W.F.; Yu, G.L. Exosomal miR-25-3p from mesenchymal stem cells alleviates myocardial infarction by targeting pro-apoptotic proteins and EZH2. Cell Death Dis. 2020, 11, 317. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Ye, Z.; Chen, Y.; Qiao, X.; Jin, Y. MicroRNA-25-5p negatively regulates TXNIP expression and relieves inflammatory responses of brain induced by lipopolysaccharide. Sci. Rep. 2022, 12, 17915. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Qian, H.; Zou, H. Suppression of lncRNA OIP5-AS1 Attenuates Apoptosis and Inflammation, and Promotes Proliferation by Mediating miR-25-3p Expression in Lipopolysaccharide-Induced Myocardial Injury. Anal. Cell. Pathol. 2023, 2023, 3154223. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Jin, J.; Xia, Z.; Chen, H. miR-363-3p attenuates the oxygen-glucose deprivation/reoxygenation-induced neuronal injury in vitro by targeting PDCD6IP. Mol. Med. Rep. 2022, 26, 322. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Li, S.; Yang, L.; Xiang, D. microRNA-363-3p reduces endothelial cell inflammatory responses in coronary heart disease via inactivation of the NOX4-dependent p38 MAPK axis. Aging 2021, 13, 11061–11082. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Guo, H.; Xiao, L.; Qing, Z.; Ma, J. MiR-363 downregulates in CD4 (+) T cells from arthritis. Int. J. Clin. Exp. Pathol. 2017, 10, 2581–2588. [Google Scholar]
- Li, R.D.; Shen, C.H.; Tao, Y.F.; Zhang, X.F.; Zhang, Q.B.; Ma, Z.Y.; Wang, Z.X. MicroRNA-144 suppresses the expression of cytokines through targeting RANKL in the matured immune cells. Cytokine 2018, 108, 197–204. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Hao, Y.; Yang, A.; Xie, L.; Ding, N.; Xu, L.; Wang, Y.; Yang, Y.; Bai, Y.; Zhang, H.; et al. TGFB3-AS1 promotes Hcy-induced inflammationof macrophages via inhibiting the maturityof miR-144 and upregulating Rap1a. Mol. Ther. Nucleic Acids 2021, 26, 1318–1335. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Xie, F.; Zhao, J.; Yue, B. Suppressed nuclear factor-kappa B alleviates lipopolysaccharide-induced acute lung injury through downregulation of CXCR4 mediated by microRNA-194. Respir. Res. 2020, 21, 144. [Google Scholar] [CrossRef]
- Wang, M.; Li, Z.; Zuo, Q. miR-194-5p inhibits LPS-induced astrocytes activation by directly targeting neurexophilin 1. Mol. Cell. Biochem. 2020, 471, 203–213. [Google Scholar] [CrossRef]
- Luo, Y.H.; Huang, Z.T.; Zong, K.Z.; Cao, Z.R.; Peng, D.D.; Zhou, B.Y.; Shen, A.; Yan, P.; Wu, Z.J. miR-194 ameliorates hepatic ischemia/reperfusion injury via targeting PHLDA1 in a TRAF6-dependent manner. Int. Immunopharmacol. 2021, 96, 107604. [Google Scholar] [CrossRef]
- Cao, Z.; Chen, L. Inhibition of miR-27a suppresses the inflammatory response via the p38/MAPK pathway in intervertebral disc cells. Exp. Ther. Med. 2017, 14, 4572–4578. [Google Scholar] [CrossRef]
- Xie, N.; Cui, H.; Banerjee, S.; Tan, Z.; Salomao, R.; Fu, M.; Abraham, E.; Thannickal, V.J.; Liu, G. miR-27a regulates inflammatory response of macrophages by targeting IL-10. J. Immunol. 2014, 193, 327–334. [Google Scholar] [CrossRef]
- Li, L.; Qi, C.; Liu, Y.; Shen, Y.; Zhao, X.; Qin, H.; Zhang, Y.; Yu, T. MicroRNA miR-27b-3p regulate microglial inflammation response and cell apoptosis by inhibiting A20 (TNF-α-induced protein 3). Bioengineered 2021, 12, 9902–9913. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Yang, L.J.; Liu, H.; Song, Y.J.; Yang, Q.Q.; Liu, Y.; Qian, S.W.; Tang, Q.Q. Exosomal miR-27b-3p secreted by visceral adipocytes contributes to endothelial inflammation and atherogenesis. Cell Rep. 2023, 42, 111948. [Google Scholar] [CrossRef] [PubMed]
- Jennewein, C.; von Knethen, A.; Schmid, T.; Brüne, B. MicroRNA-27b contributes to lipopolysaccharide-mediated peroxisome proliferator-activated receptor gamma (PPARgamma) mRNA destabilization. J. Biol. Chem. 2010, 285, 11846–11853. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Hu, X.; Liu, S.; Zou, Y.; Wang, Z.; Chu, Y. The clinical significance and function of miR-146 in the promotion of epidural fibrosis. Genet. Mol. Biol. 2021, 44, e20200447. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, S.; Zhang, C.; Wang, M. LncRNA MEG3 inhibits the inflammatory response of ankylosing spondylitis by targeting miR-146a. Mol. Cell. Biochem. 2020, 466, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Yoshimura, A.; Wakabayashi, Y.; Mori, T. Cellular and molecular basis for the regulation of inflammation by TGF-beta. J. Biochem. 2010, 147, 781–792. [Google Scholar] [CrossRef]
- Chen, D.; Tang, T.X.; Deng, H.; Yang, X.P.; Tang, Z.H. Interleukin-7 Biology and Its Effects on Immune Cells: Mediator of Generation, Differentiation, Survival, and Homeostasis. Front. Immunol. 2021, 12, 747324. [Google Scholar] [CrossRef] [PubMed]
- Marques, R.E.; Guabiraba, R.; Russo, R.C.; Teixeira, M.M. Targeting CCL5 in inflammation. Expert Opin. Ther. Targets 2013, 17, 1439–1460. [Google Scholar] [CrossRef] [PubMed]
- Mittal, P.; Romero, R.; Kusanovic, J.P.; Edwin, S.S.; Gotsch, F.; Mazaki-Tovi, S.; Espinoza, J.; Erez, O.; Nhan-Chang, C.L.; Than, N.G.; et al. CXCL6 (granulocyte chemotactic protein-2): A novel chemokine involved in the innate immune response of the amniotic cavity. Am. J. Reprod. Immunol. 2008, 60, 246–257. [Google Scholar] [CrossRef] [PubMed]
- Korbecki, J.; Gąssowska-Dobrowolska, M.; Wójcik, J.; Szatkowska, I.; Barczak, K.; Chlubek, M.; Baranowska-Bosiacka, I. The Importance of CXCL1 in Physiology and Noncancerous Diseases of Bone, Bone Marrow, Muscle and the Nervous System. Int. J. Mol. Sci. 2022, 23, 4205. [Google Scholar] [CrossRef]
- Callahan, V.; Hawks, S.; Crawford, M.A.; Lehman, C.W.; Morrison, H.A.; Ivester, H.M.; Akhrymuk, I.; Boghdeh, N.; Flor, R.; Finkielstein, C.V.; et al. The Pro-Inflammatory Chemokines CXCL9, CXCL10 and CXCL11 Are Upregulated Following SARS-CoV-2 Infection in an AKT-Dependent Manner. Viruses 2021, 13, 1062. [Google Scholar] [CrossRef] [PubMed]
- Dinarello, C.A. Interleukin-1, interleukin-1 receptors and interleukin-1 receptor antagonist. Int. Rev. Immunol. 1998, 16, 457–499. [Google Scholar] [CrossRef] [PubMed]
- Wajant, H.; Siegmund, D. TNFR1 and TNFR2 in the Control of the Life and Death Balance of Macrophages. Front. Cell Dev. Biol. 2019, 7, 91. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.; Lu, B.; Cao, C.; Li, H.; Yang, D.; Huang, L.; Ding, T.; Wu, M.; Lu, G. Plasma TNFSF13B and TNFSF14 Function as Inflammatory Indicators of Severe Adenovirus Pneumonia in Pediatric Patients. Front. Immunol. 2021, 11, 614781. [Google Scholar] [CrossRef]
- Deshmane, S.L.; Kremlev, S.; Amini, S.; Sawaya, B.E. Monocyte chemoattractant protein-1 (MCP-1): An overview. J. Interferon Cytokine Res. 2009, 29, 313–326. [Google Scholar] [CrossRef] [PubMed]
- Parchwani, D.; Dholariya, S.; Katoch, C.; Singh, R. Growth differentiation factor 15 as an emerging novel biomarker in SARS-CoV-2 infection. World J. Methodol. 2022, 12, 438–447. [Google Scholar] [CrossRef] [PubMed]
- Grandoch, M.; Feldmann, K.; Göthert, J.R.; Dick, L.S.; Homann, S.; Klatt, C.; Bayer, J.K.; Waldheim, J.N.; Rabausch, B.; Nagy, N.; et al. Deficiency in lymphotoxin β receptor protects from atherosclerosis in apoE-deficient mice. Circ. Res. 2015, 116, e57–e68. [Google Scholar] [CrossRef] [PubMed]
- Mukaida, N. Pathophysiological roles of interleukin-8/CXCL8 in pulmonary diseases. Am. J. Physiol. Lung Cell Mol. Physiol. 2003, 284, L566–L577. [Google Scholar] [CrossRef] [PubMed]
- Ihim, S.A.; Abubakar, S.D.; Zian, Z.; Sasaki, T.; Saffarioun, M.; Maleknia, S.; Azizi, G. Interleukin-18 cytokine in immunity, inflammation, and autoimmunity: Biological role in induction, regulation, and treatment. Front. Immunol. 2022, 13, 919973. [Google Scholar] [CrossRef]
- Deng, J.; Jiang, R.; Meng, E.; Wu, H. CXCL5: A coachman to drive cancer progression. Front. Oncol. 2022, 12, 944494. [Google Scholar] [CrossRef]
- Mortier, A.; Van Damme, J.; Proost, P. Overview of the mechanisms regulating chemokine activity and availability. Immunol. Lett. 2012, 145, 2–9. [Google Scholar] [CrossRef]
- Lee, W.H.; Kim, S.H.; Lee, Y.; Lee, B.B.; Kwon, B.; Song, H.; Kwon, B.S.; Park, J.E. Tumor necrosis factor receptor superfamily 14 is involved in atherogenesis by inducing proinflammatory cytokines and matrix metalloproteinases. Arter. Thromb. Vasc. Biol. 2001, 21, 2004–2010. [Google Scholar] [CrossRef] [PubMed]
- Ward-Kavanagh, L.K.; Lin, W.W.; Šedý, J.R.; Ware, C.F. The TNF Receptor Superfamily in Co-stimulating and Co-inhibitory Responses. Immunity 2016, 44, 1005–1019. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Dai, F.; Wang, L.; Sun, Y.; Mei, L.; Ran, Y.; Ye, F. CCL13 and human diseases. Front. Immunol. 2023, 14, 1176639. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Sung, Y.M.; Park, J.; Kim, S.; Kim, J.; Park, J.; Ha, H.; Bae, J.Y.; Kim, S.; Baek, D. General rules for functional microRNA targeting. Nat. Genet. 2016, 48, 1517–1526. [Google Scholar] [CrossRef] [PubMed]
- Cui, C.; Cui, Q. The relationship of human tissue microRNAs with those from body fluids. Sci. Rep. 2020, 10, 5644. [Google Scholar] [CrossRef] [PubMed]
- Overall, J.E.; Gorham, D.R. The brief psychiatric rating scale. Psychol. Rep. 1962, 10, 799–812. [Google Scholar] [CrossRef]
- Horai, T.; Boku, S.; Okazaki, S.; Otsuka, I.; Ratta-Apha, W.; Mouri, K.; Yamaki, N.; Hirata, T.; Hishimoto, A. miR-19b is elevated in peripheral blood of schizophrenic patients and attenuates proliferation of hippocampal neural progenitor cells. J. Psychiatr. Res. 2020, 131, 102–107. [Google Scholar] [CrossRef]
- Flace, P.; Livrea, P.; Basile, G.A.; Galletta, D.; Bizzoca, A.; Gennarini, G.; Bertino, S.; Branca, J.J.V.; Gulisano, M.; Bianconi, S.; et al. The Cerebellar Dopaminergic System. Front. Syst. Neurosci. 2021, 15, 650614. [Google Scholar] [CrossRef] [PubMed]
- Pinacho, R.; Villalmanzo, N.; Roca, M.; Iniesta, R.; Monje, A.; Haro, J.M.; Meana, J.J.; Ferrer, I.; Gill, G.; Ramos, B. Analysis of Sp transcription factors in the postmortem brain of chronic schizophrenia: A pilot study of relationship to negative symptoms. J. Psychiatr. Res. 2013, 47, 926–934. [Google Scholar] [CrossRef]
- Kos, M.Z.; Puppala, S.; Cruz, D.; Neary, J.L.; Kumar, A.; Dalan, E.; Li, C.; Nathanielsz, P.; Carless, M.A. Blood-Based miRNA Biomarkers as Correlates of Brain-Based miRNA Expression. Front. Mol. Neurosci. 2022, 15, 817290. [Google Scholar] [CrossRef] [PubMed]
- MacDowell, K.S.; Pinacho, R.; Leza, J.C.; Costa, J.; Ramos, B.; García-Bueno, B. Differential regulation of the TLR4 signalling pathway in post-mortem prefrontal cortex and cerebellum in chronic schizophrenia: Relationship with SP transcription factors. Prog. Neuropsychopharmacol. Biol. Psychiatry 2017, 79 Pt B, 481–492. [Google Scholar] [CrossRef]
- Goldsmith, D.R.; Rapaport, M.H.; Miller, B.J. A meta-analysis of blood cytokine network alterations in psychiatric patients: Comparisons between schizophrenia, bipolar disorder and depression. Mol. Psychiatry 2016, 21, 1696–1709. [Google Scholar] [CrossRef] [PubMed]
- Gallego, J.A.; Blanco, E.A.; Husain-Krautter, S.; Madeline Fagen, E.; Moreno-Merino, P.; Del Ojo-Jiménez, J.A.; Ahmed, A.; Rothstein, T.L.; Lencz, T.; Malhotra, A.K. Cytokines in cerebrospinal fluid of patients with schizophrenia spectrum disorders: New data and an updated meta-analysis. Schizophr. Res. 2018, 202, 64–71. [Google Scholar] [CrossRef] [PubMed]
- van Kesteren, C.F.; Gremmels, H.; de Witte, L.D.; Hol, E.M.; Van Gool, A.R.; Falkai, P.G.; Kahn, R.S.; Sommer, I.E. Immune involvement in the pathogenesis of schizophrenia: A meta-analysis on postmortem brain studies. Transl. Psychiatry 2017, 7, e1075. [Google Scholar] [CrossRef] [PubMed]
- Akhondzadeh, S.; Tabatabaee, M.; Amini, H.; Ahmadi Abhari, S.A.; Abbasi, S.H.; Behnam, B. Celecoxib as adjunctive therapy in schizophrenia: A double-blind, randomized and placebo-controlled trial. Schizophr. Res. 2007, 90, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Müller, N.; Riedel, M.; Scheppach, C.; Brandstätter, B.; Sokullu, S.; Krampe, K.; Ulmschneider, M.; Engel, R.R.; Möller, H.J.; Schwarz, M.J. Beneficial antipsychotic effects of celecoxib add-on therapy compared to risperidone alone in schizophrenia. Am. J. Psychiatry 2002, 159, 1029–1034. [Google Scholar] [CrossRef] [PubMed]
- Müller, N.; Krause, D.; Dehning, S.; Musil, R.; Schennach-Wolff, R.; Obermeier, M.; Möller, H.J.; Klauss, V.; Schwarz, M.J.; Riedel, M. Celecoxib treatment in an early stage of schizophrenia: Results of a randomized, double-blind, placebo-controlled trial of celecoxib augmentation of amisulpride treatment. Schizophr. Res. 2010, 121, 118–124. [Google Scholar] [CrossRef]
- Rapaport, M.H.; Delrahim, K.K.; Bresee, C.J.; Maddux, R.E.; Ahmadpour, O.; Dolnak, D. Celecoxib augmentation of continuously ill patients with schizophrenia. Biol. Psychiatry 2005, 57, 1594–1596. [Google Scholar] [CrossRef] [PubMed]
- Cho, M.; Lee, T.Y.; Kwak, Y.B.; Yoon, Y.B.; Kim, M.; Kwon, J.S. Adjunctive use of anti-inflammatory drugs for schizophrenia: A meta-analytic investigation of randomized controlled trials. Aust. N. Z. J. Psychiatry 2019, 53, 742–759. [Google Scholar] [CrossRef] [PubMed]
- Attari, A.; Mojdeh, A.; Khalifeh Soltani FA, S.; Najarzadegan, M.R. Aspirin Inclusion in Antipsychotic Treatment on Severity of Symptoms in Schizophrenia: A Randimized Clinical Trial. Iran. J. Psychiatry Behav. Sci. 2017, 11, e5848. [Google Scholar] [CrossRef]
- Laan, W.; Grobbee, D.E.; Selten, J.P.; Heijnen, C.J.; Kahn, R.S.; Burger, H. Adjuvant aspirin therapy reduces symptoms of schizophrenia spectrum disorders: Results from a randomized, double-blind, placebo-controlled trial. J. Clin. Psychiatry 2010, 71, 520–527. [Google Scholar] [CrossRef] [PubMed]
- Fond, G.; Lançon, C.; Korchia, T.; Auquier, P.; Boyer, L. The Role of Inflammation in the Treatment of Schizophrenia. Front. Psychiatry 2020, 11, 160. [Google Scholar] [CrossRef] [PubMed]
- Noto, C.; Gadelha, A.; Belangero, S.I.; Spindola, L.M.; Rocha, N.P.; de Miranda, A.S.; Teixeira, A.L.; Cardoso Smith, M.A.; de Jesus Mari, J.; Bressan, R.A.; et al. Circulating levels of sTNFR1 as a marker of severe clinical course in schizophrenia. J. Psychiatr. Res. 2013, 47, 467–471. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Su, H.; Li, J.; Lyon, C.; Tang, W.; Wan, M.; Hu, T.Y. Clinical applications of exosome membrane proteins. Precis. Clin. Med. 2020, 3, 54–66. [Google Scholar] [CrossRef]
- Xia, X.; Wang, Y.; Huang, Y.; Zhang, H.; Lu, H.; Zheng, J.C. Exosomal miRNAs in central nervous system diseases: Biomarkers, pathological mediators, protective factors and therapeutic agents. Prog. Neurobiol. 2019, 183, 101694. [Google Scholar] [CrossRef] [PubMed]
- Lei, J.; Fu, Y.; Zhuang, Y.; Zhang, K.; Lu, D. LncRNA SNHG1 alleviates IL-1β-induced osteoarthritis by inhibiting miR-16-5p-mediated p38 MAPK and NF-κB signaling pathways. Biosci. Rep. 2019, 39, BSR20191523. [Google Scholar] [CrossRef] [PubMed]
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders: DSM-5; American Psychiatric Association: Washington, DC, USA, 2013; Volume 5, p. 5. [Google Scholar]
- Sheehan, D.V.; Lecrubier, Y.; Sheehan, K.H.; Amorim, P.; Janavs, J.; Weiller, E.; Hergueta, T.; Baker, R.; Dunbar, G.C. The Mini-International Neuropsychiatric Interview (M.I.N.I.): The development and validation of a structured diagnostic psychiatric interview for DSM-IV and ICD-10. J. Clin. Psychiatry 1998, 59 (Suppl. 20), 22–33. [Google Scholar] [PubMed]
- Mestdagh, P.; Van Vlierberghe, P.; De Weer, A.; Muth, D.; Westermann, F.; Speleman, F.; Vandesompele, J. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol. 2009, 10, R64. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Han, X.; Wan, Y.; Zhang, S.; Zhao, Y.; Fan, R.; Cui, Q.; Zhou, Y. TAM 2.0: Tool for MicroRNA set analysis. Nucleic Acids Res. 2018, 46, W180–W185. [Google Scholar] [CrossRef] [PubMed]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef]
| Characteristics | Total n = 26 |
|---|---|
| Age, mean ± S.D. | 34.5 ± 8.6 |
| Male, n (%) | 23 (88.5) |
| Race, n (%) | |
| Black or African–American | 20 (76.9) |
| White | 6 (23.1) |
| PANSS total score, mean ± S.D. | 93.3 ± 9.5 |
| PANSS positive symptom subscale score, mean ± S.D. | 24.6 ± 4.0 |
| PANSS negative symptom subscale score, mean ± S.D. | 22.7 ± 3.9 |
| PANSS general psychopathy subscale score, mean ± S.D. | 46.0 ± 5.6 |
| miRNA Sets | Enriched Pathways/Tissues | Nmapped/Npredefined | q-Values |
|---|---|---|---|
| Upregulated 1 miRNA in subgroup 1 | No item was significantly enriched | - | - |
| Downregulated 3 miRNAs in subgroup 1 | Aging | 3/63 | 2.49 × 103 |
| Epithelial-to-Mesenchymal Transition | 3/83 | 3.81 × 103 | |
| Inflammation | 3/112 | 4.00 × 103 | |
| Downregulated 4 miRNAs in subgroup 2 | brain.cerebellum | 5/21 | 3.24 × 108 |
| Immune Response | 6/92 | 3.27 × 107 | |
| Angiogenesis | 5/65 | 4.29 × 106 | |
| Cell Death | 5/78 | 8.18 × 106 | |
| Cell Cycle | 5/83 | 8.95 × 106 | |
| Apoptosis | 5/106 | 2.58 × 105 | |
| Neurotoxicity | 3/20 | 1.31 × 104 | |
| Regulation of Akt Pathway | 3/26 | 2.59 × 104 | |
| Hormone-mediated Signaling Pathway | 3/58 | 1.97 × 103 | |
| Upregulated 10 miRNAs in subgroup 3 | brain.cerebellum | 7/21 | 2.29 × 1010 |
| Aging | 6/63 | 2.09 × 105 | |
| Angiogenesis | 5/65 | 4.79 × 104 | |
| T-Cell Differentiation | 3/16 | 1.48 × 103 | |
| Cell Division | 3/17 | 1.48 × 103 | |
| Neurotoxicity | 3/20 | 2.03 × 103 | |
| Immune System | 3/21 | 2.03 × 103 | |
| Hematopoiesis | 4/57 | 2.15 × 103 | |
| Downregulated 22 miRNAs in subgroup 3 | kidney.cortex_renalis | 7/41 | 2.20 × 105 |
| Neuron Apoptosis | 4/15 | 9.75 × 104 | |
| DNA Damage Response | 4/16 | 9.75 × 104 | |
| Adipocyte Differentiation | 5/41 | 1.98 × 103 | |
| Cholesterol Hydrolysis | 2/2 | 1.98 × 103 | |
| Cholesterol Influx | 2/2 | 1.98 × 103 | |
| Cholesterol Esterification | 2/2 | 1.98 × 103 | |
| Peritoneal Cavity Homeostasis | 4/23 | 1.98 × 103 | |
| Placenta | 3/11 | 3.21 × 103 | |
| Inflammation | 7/112 | 3.58 × 103 |
| miRNAs | Functions | Experimental Materials/Conditions | Ref. |
|---|---|---|---|
| miR-186 | Downregulate IL-1β | Spinal cord after miR-186-5p mimic was injected in rats | [32] |
| Trigeminal ganglions after miR-186 mimic was injected in mice | [33] | ||
| HUVEC (human) after transfected with miR-186 mimic and inhibitor | [34] | ||
| Downregulate IL-6 | Spinal cord after miR-186-5p mimic was injected in rats | [32] | |
| Spinal cord after miR-186-5p was injected via lentiviral vector in rats | [35] | ||
| Downregulate TNFα | Spinal cord after miR-186-5p mimic was injected in rats | [32] | |
| HUVEC (human) after transfected with miR-186 mimic and inhibitor | [34] | ||
| Spinal cord after miR-186-5p was injected via lentiviral vector in rats | [35] | ||
| miR-16 | Downregulate IL-1β | MH7A cells (human) after transfected with miR-16 mimic | [36] |
| Thoracic aorta after agomiR was injected in ApoE-/- mice | [37] | ||
| Downregulate IL-6 | Thoracic aorta after agomiR was injected in ApoE-/- mice | [37] | |
| RAW 264.7 (mouse) after transfected with miR-16 mimic and inhibitor | [38] | ||
| Downregulate TNFα | MH7A cells (human) after transfected with miR-16 mimic | [36] | |
| Thoracic aorta after agomiR was injected in ApoE-/- mice | [37] | ||
| RAW 264.7 (mouse) after transfected with miR-16 mimic and inhibitor | [38] | ||
| miR-19a | Downregulate IL-1β | Mesenchymal stem cells (mouse) after overexpressed with miR-19a/19b | [39] |
| CNE2, HONE2, A549 and HCC827 (human) after transfected with miR-19a mimic | [40] | ||
| Downregulate IL-6 | Mesenchymal stem cells (mouse) after overexpressed with miR-19a/19b | [39] | |
| CNE2, HONE2, A549 and HCC827 (human) after transfected with miR-19a mimic | [40] | ||
| Downregulate TNFα | Mesenchymal stem cells (mouse) after overexpressed with miR-19a/19b | [39] | |
| HT-29 (human) after transfected with miR-19a mimic and inhibitor | [41] | ||
| miR-19b | Downregulate IL-1β | Mesenchymal stem cells (mouse) after overexpressed with miR-19a/19b | [39] |
| CNE2, HONE2, A549 and HCC827 (human) after transfected with miR-19b-1 mimic | [40] | ||
| Downregulate IL-6 | Mesenchymal stem cells (mouse) after overexpressed with miR-19a/19b | [39] | |
| CNE2, HONE2, A549 and HCC827 (human) after transfected with miR-19b-1 mimic | [40] | ||
| Downregulate TNFα | Mesenchymal stem cells (mouse) after overexpressed with miR-19a/19b | [39] | |
| HUVEC (human) with LPS treatment after transfected with miR-19b-3p mimic and inhibitor | [42] | ||
| miR-20b | Downregulate IL-1β | ARPE-19 cell (human) under high glucose conditions after transfected with miR-20b-5p mimic | [43] |
| Spinal dorsal horn after miR-20b mimic was injected in chronic constriction injury model rats | [44] | ||
| Pancreatic acinar cells (rat) with cerulean + LPS treatment after transfected with miR-20b-5p | [45] | ||
| NRK-52E cells (rat) treated with oxalate after incubated with miR-20b-3p-enriched exosomes | [46] | ||
| Downregulate IL-6 | ARPE-19 cell (human) under high glucose conditions after transfected with miR-20b-5p mimic | [43] | |
| Spinal dorsal horn after miR-20b mimic was injected in chronic constriction injury model rats | [44] | ||
| Pancreatic acinar cells (rat) with cerulean + LPS treatment after transfected with miR-20b-5p | [45] | ||
| NRK-52E cells (rat) treated with oxalate after incubated with miR-20b-3p-enriched exosomes | [46] | ||
| Downregulate TNFα | ARPE-19 cell (human) under high glucose conditions after transfected with miR-20b-5p mimic | [43] | |
| Spinal dorsal horn after miR-20b mimic was injected in chronic constriction injury model rats | [44] | ||
| Pancreatic acinar cells (rat) with cerulean + LPS treatment after transfected with miR-20b-5p | [45] | ||
| NRK-52E cells (rat) treated with oxalate after incubated with miR-20b-3p-enriched exosomes | [46] | ||
| miR-25 | Downregulate IL-1β | Nucleus pulposus cells (human) treated with LPS and miR-25b-3p inhibitor | [47] |
| Mesenchymal stem cell (mouse) treated with oxygen-glucose deprivation and miR-25-3p inhibitor | [48] | ||
| CTX TNA2 and serum (mouse) treated with LPS after transfected with miR-25-5p mimic | [49] | ||
| Downregulate IL-6 | Nucleus pulposus cells (human) treated with LPS and miR-25b-3p inhibitor | [47] | |
| Mesenchymal stem cell (mouse) treated with oxygen-glucose deprivation and miR-25-3p inhibitor | [48] | ||
| CTX TNA2 and serum (mouse) treated with LPS after transfected with miR-25-5p mimic | [49] | ||
| H9C2 cells (rat) treated with LPS after transfected with miR-25-3p mimic | [50] | ||
| Downregulate TNFα | Nucleus pulposus cells (human) treated with LPS and miR-25b-3p inhibitor | [47] | |
| Mesenchymal stem cell (mouse) treated with oxygen-glucose deprivation and miR-25-3p inhibitor | [48] | ||
| CTX TNA2 and serum (mouse) treated with LPS after transfected with miR-25-5p mimic | [49] | ||
| H9C2 cells (rat) treated with LPS after transfected with miR-25-3p mimic | [50] | ||
| miR-363 | Downregulate IL-1β | SH-SY5Y cells (human) under oxygen and glucose deprivation/reperfusion after transfected with miR-363-3p mimics | [51] |
| Coronary arterial endothelial cells (mouse) after transfected with miR-363-3p mimic and inhibitor | [52] | ||
| Downregulate IL-6 | SH-SY5Y cells (human) under oxygen and glucose deprivation/reperfusion after transfected with miR-363-3p mimics | [51] | |
| Coronary arterial endothelial cells (mouse) after transfected with miR-363-3p mimic and inhibitor | [52] | ||
| Downregulate TNFα | SH-SY5Y cells (human) under oxygen and glucose deprivation/reperfusion after transfected with miR-363-3p mimics | [51] | |
| Jurkat cells (human) after transfected with miR-363 mimic and inhibitor | [53] | ||
| miR-144 | Downregulate IL-1β | Macrophages (mouse) after transfected with miR-144 mimic and inhibitor | [54] |
| macrophages from THP-1 cell line (human) after transfected with miR-144 mimic and inhibitor | [55] | ||
| Downregulate IL-6 | Macrophages (mouse) after transfected with miR-144 mimic and inhibitor | [54] | |
| Macrophages from THP-1 cell line (human) after transfected with miR-144 mimic and inhibitor | [55] | ||
| Downregulate TNFα | Macrophages (mouse) after transfected with miR-144 mimic and inhibitor | [54] | |
| Macrophages from THP-1 cell line (human) after transfected with miR-144 mimic and inhibitor | [55] | ||
| miR-194 | Downregulate IL-1β | RAW264.7 cell line (mouse) after transfected with miR-194 mimic | [56] |
| Astrocytes (human) after transfected with miR-194-5p mimic or inhibitor | [57] | ||
| Serum after miR-194 agomiR was injected in mice | [58] | ||
| Downregulate IL-6 | RAW264.7 cell line (mouse) after transfected with miR-194 mimic | [56] | |
| Astrocytes (human) after transfected with miR-194-5p mimic or inhibitor | [57] | ||
| Serum after miR-194 agomiR was injected in mice | [58] | ||
| Downregulate TNFα | RAW264.7 cell line (mouse) after transfected with miR-194 mimic | [56] | |
| Astrocytes (human) after transfected with miR-194-5p mimic or inhibitor | [57] | ||
| Serum after miR-194 agomiR was injected in mice | [58] | ||
| miR-27a | Upregulate IL-1β | Nucleus pulposus cells (human) after transfected with miR-27a inhibitor | [59] |
| Macrophages (mouse) after transfected with miR-27a mimics | [60] | ||
| Upregulate IL-6 | Nucleus pulposus cells (human) after transfected with miR-27a inhibitor | [59] | |
| Macrophages (mouse) after transfected with miR-27a mimics | [60] | ||
| Upregulate TNFα | Nucleus pulposus cells (human) after transfected with miR-27a inhibitor | [59] | |
| Macrophages (mouse) after transfected with miR-27a mimics | [60] | ||
| miR-27b | Upregulate IL-1β | Microglial cells (mouse) after transfected with miR-27b-3p mimic | [61] |
| 3T3-L1 cells (mouse) after transfected with miR-27b-3p mimic | [62] | ||
| Upregulate IL-6 | Microglial cells (mouse) after transfected with miR-27b-3p mimic | [61] | |
| 3T3-L1 cells (mouse) after transfected with miR-27b-3p mimic | [62] | ||
| Macrophages (human) after transfected with miR-27b-3p mimic | [63] | ||
| Upregulate TNFα | Microglial cells (mouse) after transfected with miR-27b-3p mimic | [61] | |
| Macrophages (human) after transfected with miR-27b-3p mimic | [63] | ||
| miR-146a | Upregulate IL-1β | Epidural fibroblasts (human) after transfected with miR-146 mimic | [64] |
| Fibroblast-like synovial cells (human) after transfected with miR-146a mimic and inhibitor | [65] | ||
| Upregulate IL-6 | Epidural fibroblasts (human) after transfected with miR-146 mimic | [64] | |
| Fibroblast-like synovial cells (human) after transfected with miR-146a mimic and inhibitor | [65] | ||
| Upregulate TNFα | Epidural fibroblasts (human) after transfected with miR-146 mimic | [64] | |
| Fibroblast-like synovial cells (human) after transfected with miR-146a mimic and inhibitor | [65] |
| Protein Sets | Enriched Pathways | Nmapped/Npredefined | q-Values |
|---|---|---|---|
| Downregulated 2 proteins in subgroup 1 | No item was significantly enriched | - | - |
| Upregulated 34 proteins in subgroup 1 | Cytokine-cytokine receptor interaction | 6/297 | 4.61 × 102 |
| Viral protein interaction with cytokine and cytokine receptor | 4/100 | 4.61 × 102 | |
| Upregulated 22 proteins in subgroup 2 | Cytokine-cytokine receptor interaction | 8/29 | 5.32 × 106 |
| Viral protein interaction with cytokine and cytokine receptor | 74/100 | 2.93 × 103 | |
| NF-kappa B signaling pathway | 4/104 | 2.93 × 103 | |
| TNF signaling pathway | 4/114 | 2.93 × 103 | |
| Osteoclast differentiation | 3/128 | 4.74 × 102 | |
| Downregulated 57 proteins in subgroup 3 | Viral protein interaction with cytokine and cytokine receptor | 9/100 | 8.91 × 107 |
| Cytokine-cytokine receptor interaction | 12/297 | 2.29 × 106 | |
| IL-17 signaling pathway | 7/94 | 7.87 × 105 | |
| TNF signaling pathway | 7/114 | 1.80 × 104 | |
| Chemokine signaling pathway | 7/192 | 2.63 × 103 | |
| AGE-RAGE signaling pathway in diabetic complications | 5/100 | 9.96 × 103 | |
| Cytosolic DNA-sensing pathway | 4/75 | 3.60 × 102 | |
| Cellular senescence | 5/156 | 3.72 × 102 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miyano, T.; Mikkaichi, T.; Nakamura, K.; Yoshigae, Y.; Abernathy, K.; Ogura, Y.; Kiyosawa, N. Circulating microRNA Profiles Identify a Patient Subgroup with High Inflammation and Severe Symptoms in Schizophrenia Experiencing Acute Psychosis. Int. J. Mol. Sci. 2024, 25, 4291. https://doi.org/10.3390/ijms25084291
Miyano T, Mikkaichi T, Nakamura K, Yoshigae Y, Abernathy K, Ogura Y, Kiyosawa N. Circulating microRNA Profiles Identify a Patient Subgroup with High Inflammation and Severe Symptoms in Schizophrenia Experiencing Acute Psychosis. International Journal of Molecular Sciences. 2024; 25(8):4291. https://doi.org/10.3390/ijms25084291
Chicago/Turabian StyleMiyano, Takuya, Tsuyoshi Mikkaichi, Kouichi Nakamura, Yasushi Yoshigae, Kelly Abernathy, Yuji Ogura, and Naoki Kiyosawa. 2024. "Circulating microRNA Profiles Identify a Patient Subgroup with High Inflammation and Severe Symptoms in Schizophrenia Experiencing Acute Psychosis" International Journal of Molecular Sciences 25, no. 8: 4291. https://doi.org/10.3390/ijms25084291
APA StyleMiyano, T., Mikkaichi, T., Nakamura, K., Yoshigae, Y., Abernathy, K., Ogura, Y., & Kiyosawa, N. (2024). Circulating microRNA Profiles Identify a Patient Subgroup with High Inflammation and Severe Symptoms in Schizophrenia Experiencing Acute Psychosis. International Journal of Molecular Sciences, 25(8), 4291. https://doi.org/10.3390/ijms25084291






