Quercetin Impairs the Growth of Uveal Melanoma Cells by Interfering with Glucose Uptake and Metabolism
Abstract
1. Introduction
2. Results
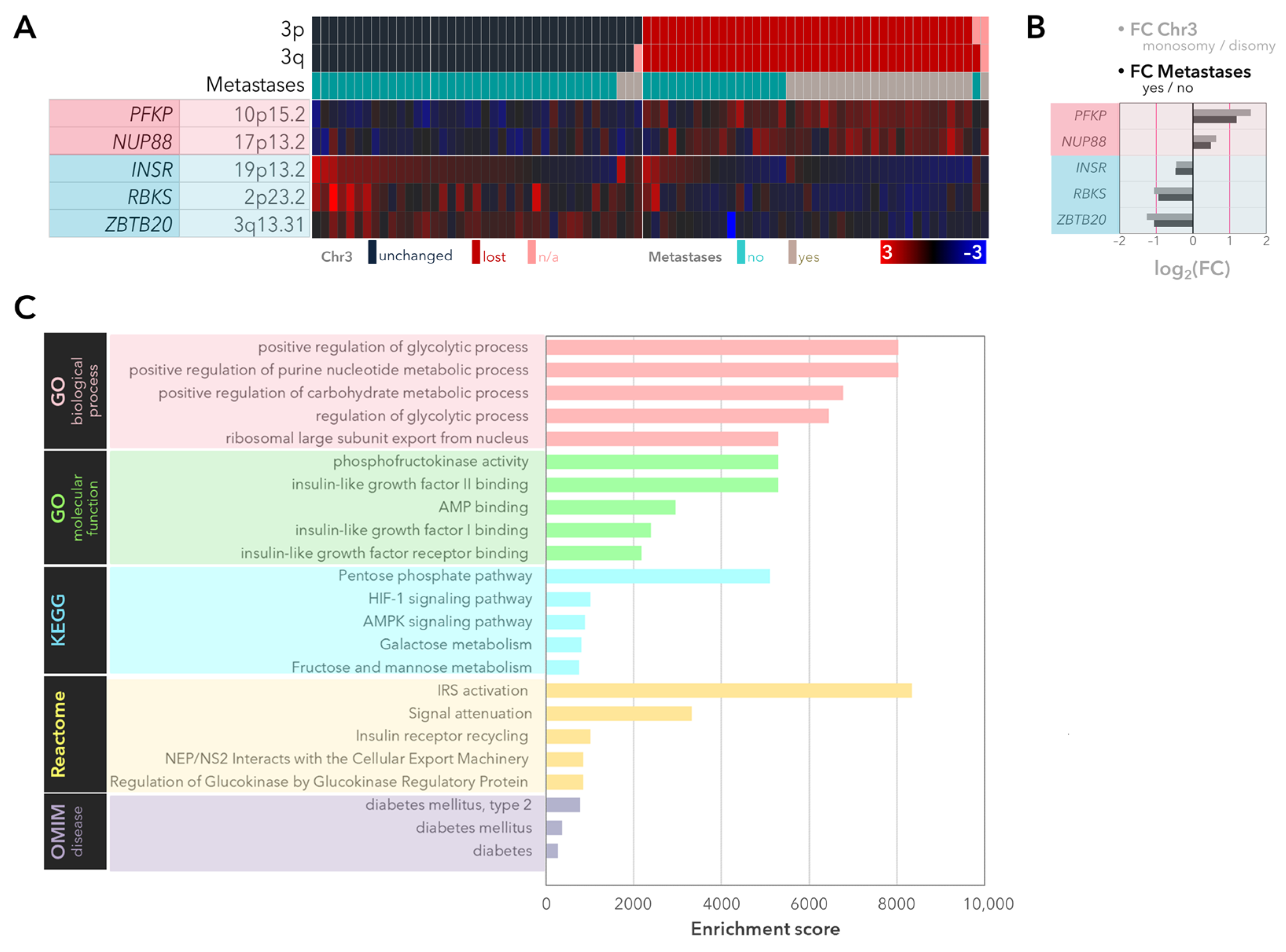
2.1. Expression of the Genes Involved in Glycolysis and the PPP with Regard to Monosomy 3 Status and Metastases in the UM-Cohort of the TCGA Study
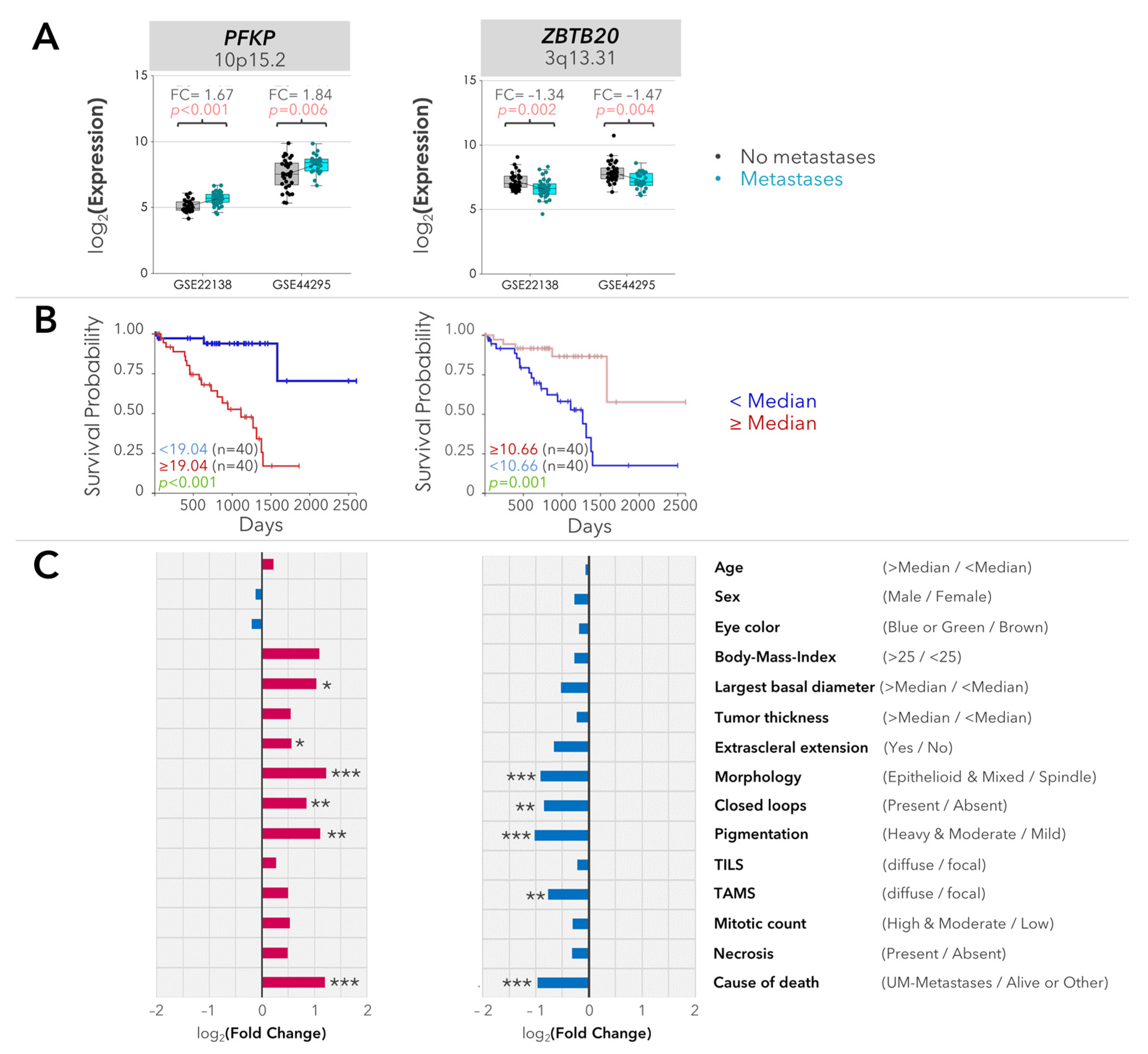
2.2. Validation of the Differentially Expressed Genes
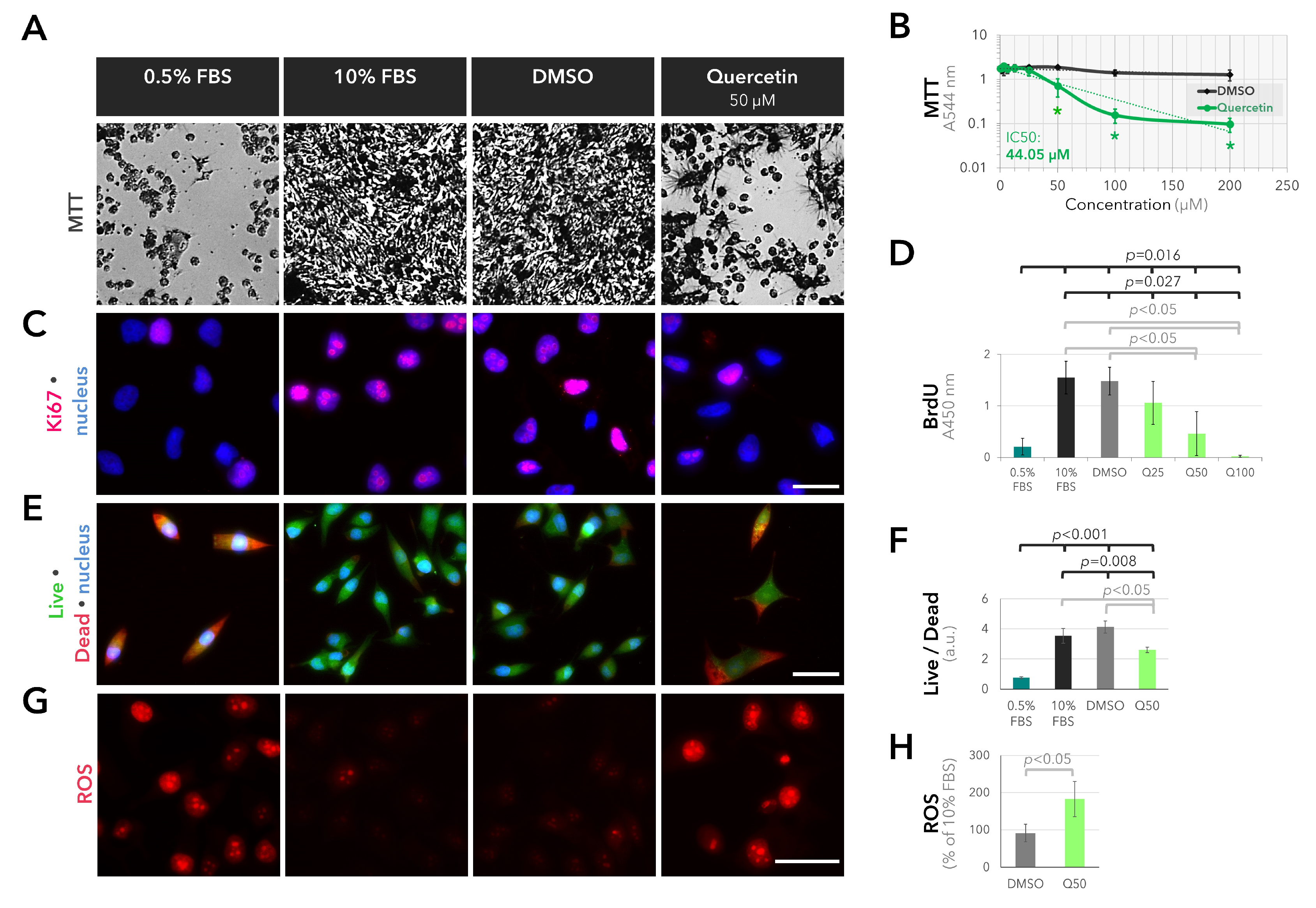
2.3. Anti-Proliferative and Pro-Oxidant Effects of Quercetin on the Cultured UM Cells
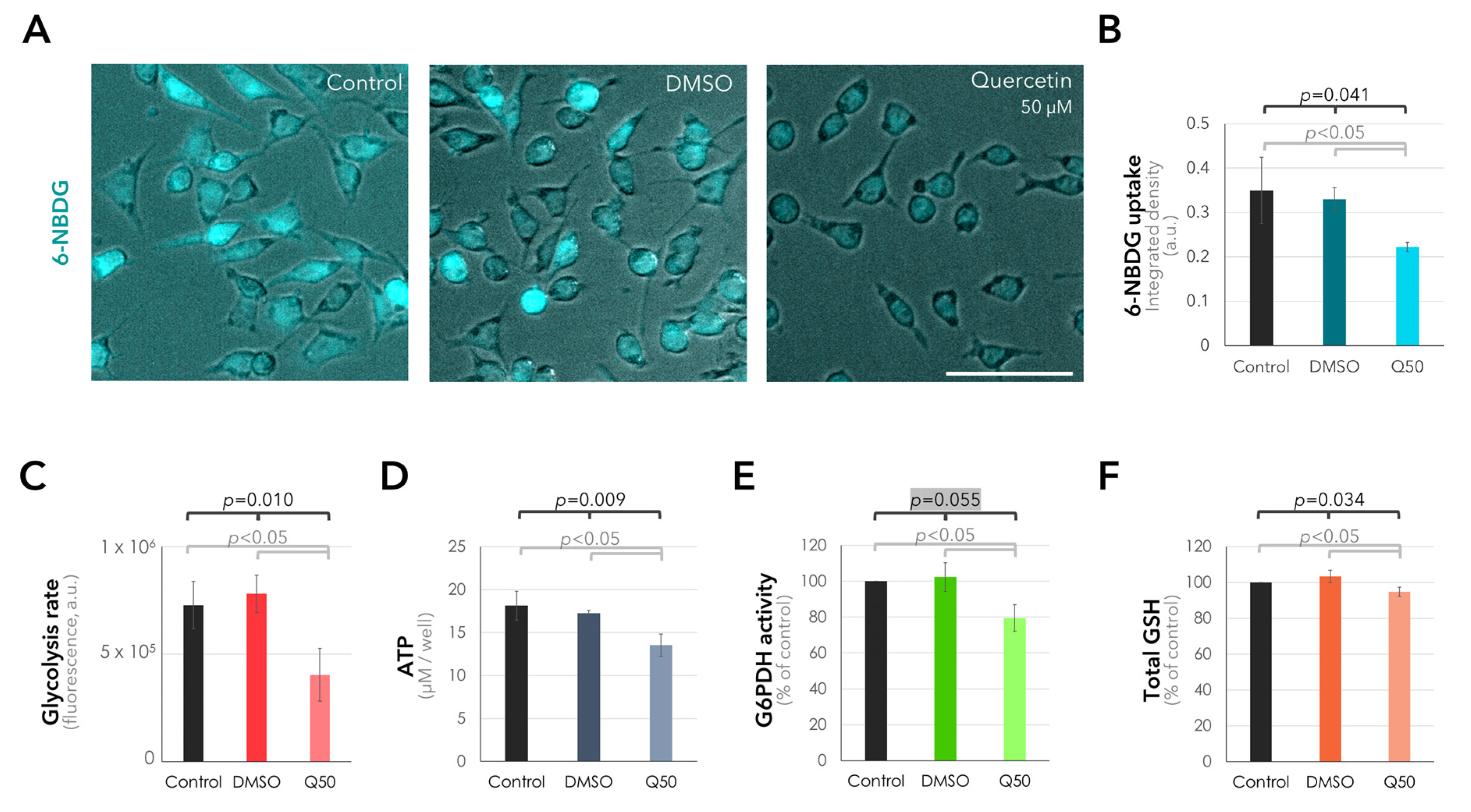
2.4. Suppression of Glucose Uptake and Metabolism in the UM Cells in Response to Quercetin
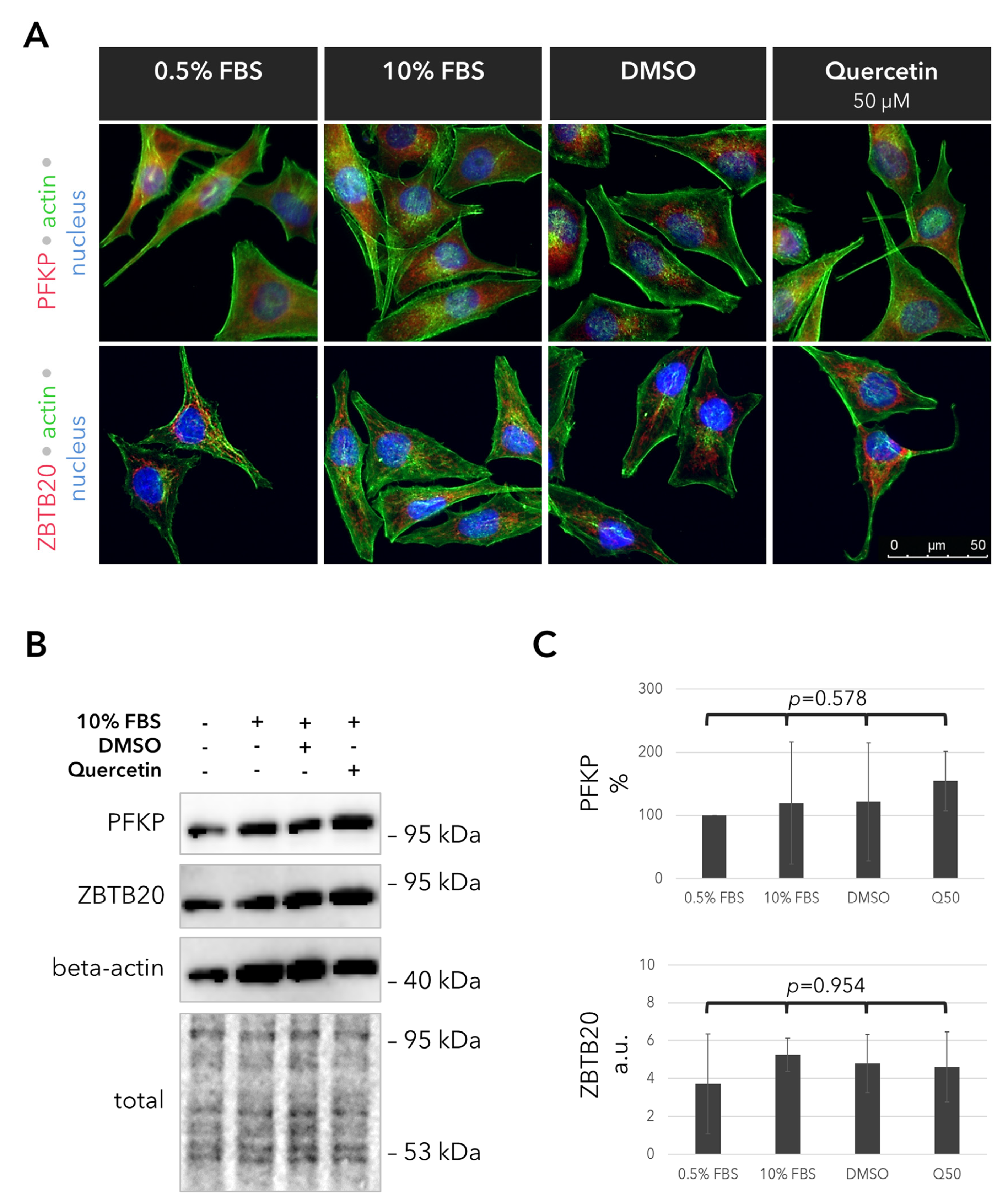
2.5. Outcomes of Quercetin Treatment on the Expression of the PFKP and ZBTB20 Proteins in the UM Cells
3. Discussion
4. Materials and Methods
4.1. Analysis of Gene Expression
4.2. Cell Culture and Test Substances
4.3. Immunocytochemistry
4.4. MTT Assay and IC50 Calculation
4.5. Live/Dead Assay
4.6. BrdU Assay
4.7. Measurement of Reactive Oxygen Species
4.8. Glucose Uptake
4.9. Glycolysis Assay
4.10. ATP Assay
4.11. G6PDH Activity
4.12. Total Glutathione Assay
4.13. Immunoblotting
4.14. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Blum, E.S.; Yang, J.; Komatsubara, K.M.; Carvajal, R.D. Clinical Management of Uveal and Conjunctival Melanoma. Oncology 2016, 30, 29–32, 34–43, 48. [Google Scholar] [PubMed]
- Nichols, E.E.; Richmond, A.; Daniels, A.B. Tumor Characteristics, Genetics, Management, and the Risk of Metastasis in Uveal Melanoma. Semin. Ophthalmol. 2016, 31, 304–309. [Google Scholar] [CrossRef] [PubMed]
- Jager, M.J.; Shields, C.L.; Cebulla, C.M.; Abdel-Rahman, M.H.; Grossniklaus, H.E.; Stern, M.H.; Carvajal, R.D.; Belfort, R.N.; Jia, R.; Shields, J.A.; et al. Uveal Melanoma. Nat. Rev. Dis. Primers 2020, 6, 24. [Google Scholar] [CrossRef] [PubMed]
- Prescher, G.; Bornfeld, N.; Hirche, H.; Horsthemke, B.; Jöckel, K.H.; Becher, R. Prognostic Implications of Monosomy 3 in Uveal Melanoma. Lancet 1996, 347, 1222–1225. [Google Scholar] [PubMed]
- Kaliki, S.; Shields, C.L.; Shields, J.A. Uveal Melanoma: Estimating Prognosis. Indian J. Ophthalmol. 2015, 63, 93–102. [Google Scholar] [CrossRef]
- Abdel-Rahman, M.H.; Cebulla, C.M.; Verma, V.; Christopher, B.N.; Carson, W.E., 3rd; Olencki, T.; Davidorf, F.H. Monosomy 3 Status of Uveal Melanoma Metastases Is Associated with Rapidly Progressive Tumors and Short Survival. Exp. Eye Res. 2012, 100, 26–31. [Google Scholar] [CrossRef] [PubMed]
- Sevim, D.G.; Kiratli, H. Serum Adiponectin, Insulin Resistance, and Uveal Melanoma: Clinicopathological Correlations. Melanoma Res. 2016, 26, 164–172. [Google Scholar] [CrossRef] [PubMed]
- McCannel, T.A.; Reddy, S.; Burgess, B.L.; Auerbach, M. Association of Positive Dual-Modality Positron Emission Tomography/Computed Tomography Imaging of Primary Choroidal Melanoma with Chromosome 3 Loss and Tumor Size. Retina 2010, 30, 146–151. [Google Scholar] [CrossRef] [PubMed]
- Papastefanou, V.P.; Islam, S.; Szyszko, T.; Grantham, M.; Sagoo, M.S.; Cohen, V.M. Metabolic Activity of Primary Uveal Melanoma on PET/CT Scan and Its Relationship with Monosomy 3 and Other Prognostic Factors. Br. J. Ophthalmol. 2014, 98, 1659–1665. [Google Scholar] [CrossRef][Green Version]
- Maaßen, T.; Vardanyan, S.; Brosig, A.; Merz, H.; Ranjbar, M.; Kakkassery, V.; Grisanti, S.; Tura, A. Monosomy-3 Alters the Expression Profile of the Glucose Transporters GLUT1-3 in Uveal Melanoma. Int. J. Mol. Sci. 2020, 21, 9345. [Google Scholar] [CrossRef]
- Mueckler, M.; Thorens, B. The SLC2 (GLUT) Family of Membrane Transporters. Mol. Asp. Med. 2013, 34, 121–138. [Google Scholar] [CrossRef] [PubMed]
- Ward, P.S.; Thompson, C.B. Metabolic Reprogramming: A Cancer Hallmark Even Warburg Did Not Anticipate. Cancer Cell 2012, 21, 297–308. [Google Scholar] [CrossRef] [PubMed]
- Bronkhorst, I.H.; Jager, M.J. Inflammation in Uveal Melanoma. Eye 2013, 27, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Kozal, K.; Jóźwiak, P.; Krześlak, A. Contemporary Perspectives on the Warburg Effect Inhibition in Cancer Therapy. Cancer Control. 2021, 28, 10732748211041243. [Google Scholar] [CrossRef] [PubMed]
- Kubicka, A.; Matczak, K.; Łabieniec-Watała, M. More Than Meets the Eye Regarding Cancer Metabolism. Int. J. Mol. Sci. 2021, 22, 9507. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Jin, Y.; Fan, Z. The Mechanism of Warburg Effect-Induced Chemoresistance in Cancer. Front. Oncol. 2021, 11, 698023. [Google Scholar] [CrossRef] [PubMed]
- Jiang, P.; Du, W.; Wu, M. Regulation of the Pentose Phosphate Pathway in Cancer. Protein Cell 2014, 5, 592–602. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Farias, M.; Carrasco-Pozo, C. The Anti-Cancer Effect of Quercetin: Molecular Implications in Cancer Metabolism. Int. J. Mol. Sci. 2019, 20, 3177. [Google Scholar] [CrossRef] [PubMed]
- Liberti, M.V.; Locasale, J.W. The Warburg Effect: How Does It Benefit Cancer Cells? Trends Biochem. Sci. 2016, 41, 211–218. [Google Scholar] [CrossRef]
- Ge, T.; Yang, J.; Zhou, S.; Wang, Y.; Li, Y.; Tong, X. The Role of the Pentose Phosphate Pathway in Diabetes and Cancer. Front. Endocrinol. 2020, 11, 365. [Google Scholar] [CrossRef]
- Stincone, A.; Prigione, A.; Cramer, T.; Wamelink, M.M.; Campbell, K.; Cheung, E.; Olin-Sandoval, V.; Grüning, N.M.; Krüger, A.; Tauqeer Alam, M.; et al. The Return of Metabolism: Biochemistry and Physiology of the Pentose Phosphate Pathway. Biol. Rev. Camb. Philos. Soc. 2015, 90, 927–963. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Huo, Y.; Wang, D.; Tai, Y.; Li, J.; Pang, D.; Zhang, Y.; Zhao, W.; Du, N.; Huang, Y. MiR-216a-5p/Hexokinase 2 Axis Regulates Uveal Melanoma Growth through Modulation of Warburg Effect. Biochem. Biophys. Res. Commun. 2018, 501, 885–892. [Google Scholar] [CrossRef]
- Bao, R.; Surriga, O.; Olson, D.J.; Allred, J.B.; Strand, C.A.; Zha, Y.; Carll, T.; Labadie, B.W.; Bastos, B.R.; Butler, M.; et al. Transcriptional Analysis of Metastatic Uveal Melanoma Survival Nominates NRP1 as a Therapeutic Target. Melanoma Res. 2021, 31, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Lu, J.; Li, W. A Comprehensive Prognostic and Immunological Analysis of a Six-Gene Signature Associated with Glycolysis and Immune Response in Uveal Melanoma. Front. Immunol. 2021, 12, 738068. [Google Scholar] [CrossRef] [PubMed]
- Kashyap, D.; Mittal, S.; Sak, K.; Singhal, P.; Tuli, H.S. Molecular Mechanisms of Action of Quercetin in Cancer: Recent Advances. Tumour Biol. 2016, 37, 12927–12939. [Google Scholar] [CrossRef] [PubMed]
- Khan, F.; Niaz, K.; Maqbool, F.; Ismail Hassan, F.; Abdollahi, M.; Nagulapalli Venkata, K.C.; Nabavi, S.M.; Bishayee, A. Molecular Targets Underlying the Anticancer Effects of Quercetin: An Update. Nutrients 2016, 8, 529. [Google Scholar] [CrossRef]
- Yi, H.; Peng, H.; Wu, X.; Xu, X.; Kuang, T.; Zhang, J.; Du, L.; Fan, G. The Therapeutic Effects and Mechanisms of Quercetin on Metabolic Diseases: Pharmacological Data and Clinical Evidence. Oxid. Med. Cell. Longev. 2021, 2021, 6678662. [Google Scholar] [CrossRef] [PubMed]
- Hosseini, A.; Razavi, B.M.; Banach, M.; Hosseinzadeh, H. Quercetin and Metabolic Syndrome: A Review. Phytother. Res. 2021, 35, 5352–5364. [Google Scholar] [CrossRef] [PubMed]
- Brito, A.F.; Ribeiro, M.; Abrantes, A.M.; Pires, A.S.; Teixo, R.J.; Tralhão, J.G.; Botelho, M.F. Quercetin in Cancer Treatment, Alone or in Combination with Conventional Therapeutics? Curr. Med. Chem. 2015, 22, 3025–3039. [Google Scholar] [CrossRef]
- Sak, K. Site-Specific Anticancer Effects of Dietary Flavonoid Quercetin. Nutr. Cancer 2014, 66, 177–193. [Google Scholar] [CrossRef]
- Sak, K.; Everaus, H. Role of Flavonoids in Future Anticancer Therapy by Eliminating the Cancer Stem Cells. Curr. Stem Cell Res. Ther. 2015, 10, 271–282. [Google Scholar] [CrossRef] [PubMed]
- Umar, S.M.; Kashyap, A.; Kahol, S.; Mathur, S.R.; Gogia, A.; Deo, S.V.S.; Prasad, C.P. Prognostic and Therapeutic Relevance of Phosphofructokinase Platelet-Type (PFKP) in Breast Cancer. Exp. Cell Res. 2020, 396, 112282. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Pan, L.; Gao, C.; Xu, H.; Li, Y.; Zhang, L.; Ma, L.; Meng, L.; Sun, X.; Qin, H. Quercetin Inhibits the Proliferation of Glycolysis-Addicted HCC Cells by Reducing Hexokinase 2 and Akt-MTOR Pathway. Molecules 2019, 24, 1993. [Google Scholar] [CrossRef] [PubMed]
- Gibellini, L.; Pinti, M.; Nasi, M.; De Biasi, S.; Roat, E.; Bertoncelli, L.; Cossarizza, A. Interfering with ROS Metabolism in Cancer Cells: The Potential Role of Quercetin. Cancers 2010, 2, 1288–1311. [Google Scholar] [CrossRef] [PubMed]
- Li, X.B.; Gu, J.D.; Zhou, Q.H. Review of Aerobic Glycolysis and Its Key Enzymes—New Targets for Lung Cancer Therapy. Thorac Cancer 2015, 6, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, P.; Zhong, J.; Tan, M.; Ge, J.; Tao, L.; Li, Y.; Zhu, Y.; Wu, L.; Qiu, J.; et al. The Platelet Isoform of Phosphofructokinase Contributes to Metabolic Reprogramming and Maintains Cell Proliferation in Clear Cell Renal Cell Carcinoma. Oncotarget 2016, 7, 27142–27157. [Google Scholar] [CrossRef] [PubMed]
- Al Hasawi, N.; Alkandari, M.F.; Luqmani, Y.A. Phosphofructokinase: A Mediator of Glycolytic Flux in Cancer Progression. Crit. Rev. Oncol. Hematol. 2014, 92, 312–321. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Preiss, N.K.; Valenteros, K.B.; Kamal, Y.; Usherwood, Y.K.; Frost, H.R.; Usherwood, E.J. Zbtb20 Restrains CD8 T Cell Immunometabolism and Restricts Memory Differentiation and Antitumor Immunity. J. Immunol. 2020, 205, 2649–2666. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Tan, Y.X.; Ren, Y.B.; Dong, L.W.; Xie, Z.F.; Tang, L.; Cao, D.; Zhang, W.P.; Hu, H.P.; Wang, H.Y. Zinc Finger Protein ZBTB20 Expression Is Increased in Hepatocellular Carcinoma and Associated with Poor Prognosis. BMC Cancer 2011, 11, 271. [Google Scholar] [CrossRef]
- Liu, J.; Jiang, J.; Hui, X.; Wang, W.; Fang, D.; Ding, L. Mir-758-5p Suppresses Glioblastoma Proliferation, Migration and Invasion by Targeting ZBTB20. Cell. Physiol. Biochem. 2018, 48, 2074–2083. [Google Scholar] [CrossRef]
- Fan, D.; Qiu, B.; Yang, X.J.; Tang, H.L.; Peng, S.J.; Yang, P.; Dong, Y.M.; Yang, L.; Bao, G.Q.; Zhao, H.D. LncRNA SNHG8 Promotes Cell Migration and Invasion in Breast Cancer Cell through MiR-634/ZBTB20 Axis. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 11639–11649. [Google Scholar] [PubMed]
- Zhang, Y.; Zhou, X.; Zhang, M.; Cheng, L.; Zhang, Y.; Wang, X. ZBTB20 Promotes Cell Migration and Invasion of Gastric Cancer by Inhibiting IκBα to Induce NF-ΚB Activation. Artif. Cells Nanomed. Biotechnol. 2019, 47, 3862–3872. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Zhou, L.; Zhang, H.; Chen, R.; Zhang, Y.; Li, L.; Lu, J.Y.; Jiang, H.; Liu, D.; Qi, S.; et al. Regulation of Hepatic Lipogenesis by the Zinc Finger Protein Zbtb20. Nat. Commun. 2017, 8, 14824. [Google Scholar] [CrossRef] [PubMed]
- Silva-Rodríguez, P.; Fernández-Díaz, D.; Bande, M.; Pardo, M.; Loidi, L.; Blanco-Teijeiro, M.J. GNAQ and GNA11 Genes: A Comprehensive Review on Oncogenesis, Prognosis and Therapeutic Opportunities in Uveal Melanoma. Cancers 2022, 14, 3066. [Google Scholar] [CrossRef] [PubMed]
- Onken, M.D.; Noda, S.E.; Kaltenbronn, K.M.; Frankfater, C.; Makepeace, C.M.; Fettig, N.; Piggott, K.D.; Custer, P.L.; Ippolito, J.E.; Blumer, K.J. Oncogenic Gq/11 Signaling Acutely Drives and Chronically Sustains Metabolic Reprogramming in Uveal Melanoma. J. Biol. Chem. 2022, 298, 101495. [Google Scholar] [CrossRef] [PubMed]
- Griewank, K.G.; Yu, X.; Khalili, J.; Sozen, M.M.; Stempke-Hale, K.; Bernatchez, C.; Wardell, S.; Bastian, B.C.; Woodman, S.E. Genetic and Molecular Characterization of Uveal Melanoma Cell Lines. Pigment. Cell Melanoma Res. 2012, 25, 182–187. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhang, W.J.; Choi, J.; Frei, B. Quercetin Affects Glutathione Levels and Redox Ratio in Human Aortic Endothelial Cells not Through Oxidation but Formation and Cellular Export of Quercetin-Glutathione Conjugates and Upregulation of Glutamate-Cysteine Ligase. Redox Biol. 2016, 9, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Ader, P.; Blöck, M.; Pietzsch, S.; Wolffram, S. Interaction of Quercetin Glucosides with the Intestinal Sodium/Glucose Co-Transporter (SGLT-1). Cancer Lett. 2001, 162, 175–180. [Google Scholar] [CrossRef] [PubMed]
- Strobel, P.; Allard, C.; Perez-Acle, T.; Calderon, R.; Aldunate, R.; Leighton, F. Myricetin, Quercetin and Catechin-Gallate Inhibit Glucose Uptake in Isolated Rat Adipocytes. Biochem. J. 2005, 386 Pt 3, 471–478. [Google Scholar] [CrossRef]
- Zhang, H.; Hassan, Y.I.; Liu, R.; Mats, L.; Yang, C.; Liu, C.; Tsao, R. Molecular Mechanisms Underlying the Absorption of Aglycone and Glycosidic Flavonoids in a Caco-2 BBe1 Cell Model. ACS Omega 2020, 5, 10782–10793. [Google Scholar] [CrossRef]
- Ismail, A.; Tanasova, M. Importance of GLUT Transporters in Disease Diagnosis and Treatment. Int. J. Mol. Sci. 2022, 23, 8698. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, C.; Oba, J.; Roszik, J.; Marszalek, J.R.; Chen, K.; Qi, Y.; Eterovic, K.; Robertson, A.G.; Burks, J.K.; McCannel, T.A.; et al. Elevated Endogenous SDHA Drives Pathological Metabolism in Highly Metastatic Uveal Melanoma. Investig. Ophthalmol. Vis. Sci. 2019, 60, 4187–4195. [Google Scholar] [CrossRef] [PubMed]
- Giallongo, S.; Di Rosa, M.; Caltabiano, R.; Longhitano, L.; Reibaldi, M.; Distefano, A.; Lo Re, O.; Amorini, A.M.; Puzzo, L.; Salvatorelli, L.; et al. Loss of MacroH2A1 Decreases Mitochondrial Metabolism and Reduces the Aggressiveness of Uveal Melanoma Cells. Aging 2020, 12, 9745–9760. [Google Scholar] [CrossRef] [PubMed]
- Mor, D.E.; Murphy, C.T. Mitochondrial Hyperactivity as a Potential Therapeutic Target in Parkinson’s Disease. Transl. Med. Aging 2020, 4, 117–120. [Google Scholar] [CrossRef] [PubMed]
- Day, R.M.; Suzuki, Y.J. Cell proliferation, reactive oxygen and cellular glutathione. Dose Response 2006, 3, 425–442. [Google Scholar] [CrossRef] [PubMed]
- Hebert, L.; Bellanger, D.; Guillas, C.; Campagne, A.; Dingli, F.; Loew, D.; Fievet, A.; Jacquemin, V.; Popova, T.; Jean, D.; et al. Modulating BAP1 expression affects ROS homeostasis, cell motility and mitochondrial function. Oncotarget 2017, 8, 72513–72527. [Google Scholar] [CrossRef] [PubMed]
- Bononi, A.; Yang, H.; Giorgi, C.; Patergnani, S.; Pellegrini, L.; Su, M.; Xie, G.; Signorato, V.; Pastorino, S.; Morris, P.; et al. Germline BAP1 Mutations Induce a Warburg Effect. Cell Death Differ. 2017, 24, 1694–1704. [Google Scholar] [CrossRef] [PubMed]
- Patra, K.C.; Hay, N. The Pentose Phosphate Pathway and Cancer. Trends Biochem. Sci. 2014, 39, 347–354. [Google Scholar] [CrossRef] [PubMed]
- Lewis, C.A.; Parker, S.J.; Fiske, B.P.; McCloskey, D.; Gui, D.Y.; Green, C.R.; Vokes, N.I.; Feist, A.M.; Vander Heiden, M.G.; Metallo, C.M. Tracing Compartmentalized NADPH Metabolism in the Cytosol and Mitochondria of Mammalian Cells. Mol. Cell 2014, 55, 253–263. [Google Scholar] [CrossRef]
- Vafadar, A.; Shabaninejad, Z.; Movahedpour, A.; Fallahi, F.; Taghavipour, M.; Ghasemi, Y.; Akbari, M.; Shafiee, A.; Hajighadimi, S.; Moradizarmehri, S.; et al. Quercetin and Cancer: New Insights into Its Therapeutic Effects on Ovarian Cancer Cells. Cell Biosci. 2020, 10, 32. [Google Scholar] [CrossRef]
- Srivastava, S.; Somasagara, R.R.; Hegde, M.; Nishana, M.; Tadi, S.K.; Srivastava, M.; Choudhary, B.; Raghavan, S.C. Quercetin, a Natural Flavonoid Interacts with DNA, Arrests Cell Cycle and Causes Tumor Regression by Activating Mitochondrial Pathway of Apoptosis. Sci. Rep. 2016, 6, 24049. [Google Scholar] [CrossRef]
- Wu, L.; Li, J.; Liu, T.; Li, S.; Feng, J.; Yu, Q.; Zhang, J.; Chen, J.; Zhou, Y.; Ji, J.; et al. Quercetin Shows Anti-Tumor Effect in Hepatocellular Carcinoma LM3 Cells by Abrogating JAK2/STAT3 Signaling Pathway. Cancer Med. 2019, 8, 4806–4820. [Google Scholar] [CrossRef]
- Dabeek, W.M.; Marra, M.V. Dietary Quercetin and Kaempferol: Bioavailability and Potential Cardiovascular-Related Bioactivity in Humans. Nutrients 2019, 11, 2288. [Google Scholar] [CrossRef]
- Eipel, C.; Abshagen, K.; Vollmar, B. Regulation of Hepatic Blood Flow: The Hepatic Arterial Buffer Response Revisited. World J. Gastroenterol. 2010, 16, 6046–6057. [Google Scholar] [CrossRef] [PubMed]
- Egert, S.; Bosy-Westphal, A.; Seiberl, J.; Kürbitz, C.; Settler, U.; Plachta-Danielzik, S.; Wagner, A.E.; Frank, J.; Schrezenmeir, J.; Rimbach, G.; et al. Quercetin Reduces Systolic Blood Pressure and Plasma Oxidised Low-Density Lipoprotein Concentrations in Overweight Subjects with a High-Cardiovascular Disease Risk Phenotype: A Double-Blinded, Placebo-Controlled Cross-over Study. Br. J. Nutr. 2009, 102, 1065–1074. [Google Scholar] [CrossRef] [PubMed]
- Lu, N.T.; Crespi, C.M.; Liu, N.M.; Vu, J.Q.; Ahmadieh, Y.; Wu, S.; Lin, S.; McClune, A.; Durazo, F.; Saab, S.; et al. A Phase I Dose Escalation Study Demonstrates Quercetin Safety and Explores Potential for Bioflavonoid Antivirals in Patients with Chronic Hepatitis C. Phytother. Res. 2016, 30, 160–168. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and Collaborative HTML5 Gene List Enrichment Analysis Tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef]
- De Waard-Siebinga, I.; Blom, D.J.; Griffioen, M.; Schrier, P.I.; Hoogendoorn, E.; Beverstock, G.; Danen, E.H.; Jager, M.J. Establishment and Characterization of an Uveal-Melanoma Cell Line. Int. J. Cancer 1995, 62, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Jager, M.J.; Magner, J.A.; Ksander, B.R.; Dubovy, S.R. Uveal Melanoma Cell Lines: Where do they come from? (An American Ophthalmological Society Thesis). Trans. Am. Ophthalmol. Soc. 2016, 114, T5. [Google Scholar]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An Open-Source Platform for Biological-Image Analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tura, A.; Herfs, V.; Maaßen, T.; Zuo, H.; Vardanyan, S.; Prasuhn, M.; Ranjbar, M.; Kakkassery, V.; Grisanti, S. Quercetin Impairs the Growth of Uveal Melanoma Cells by Interfering with Glucose Uptake and Metabolism. Int. J. Mol. Sci. 2024, 25, 4292. https://doi.org/10.3390/ijms25084292
Tura A, Herfs V, Maaßen T, Zuo H, Vardanyan S, Prasuhn M, Ranjbar M, Kakkassery V, Grisanti S. Quercetin Impairs the Growth of Uveal Melanoma Cells by Interfering with Glucose Uptake and Metabolism. International Journal of Molecular Sciences. 2024; 25(8):4292. https://doi.org/10.3390/ijms25084292
Chicago/Turabian StyleTura, Aysegül, Viktoria Herfs, Tjorge Maaßen, Huaxin Zuo, Siranush Vardanyan, Michelle Prasuhn, Mahdy Ranjbar, Vinodh Kakkassery, and Salvatore Grisanti. 2024. "Quercetin Impairs the Growth of Uveal Melanoma Cells by Interfering with Glucose Uptake and Metabolism" International Journal of Molecular Sciences 25, no. 8: 4292. https://doi.org/10.3390/ijms25084292
APA StyleTura, A., Herfs, V., Maaßen, T., Zuo, H., Vardanyan, S., Prasuhn, M., Ranjbar, M., Kakkassery, V., & Grisanti, S. (2024). Quercetin Impairs the Growth of Uveal Melanoma Cells by Interfering with Glucose Uptake and Metabolism. International Journal of Molecular Sciences, 25(8), 4292. https://doi.org/10.3390/ijms25084292





