Genital Dysbiosis and Different Systemic Immune Responses Based on the Trimester of Pregnancy in SARS-CoV-2 Infection
Abstract
1. Introduction
2. Results
2.1. Study Cohort
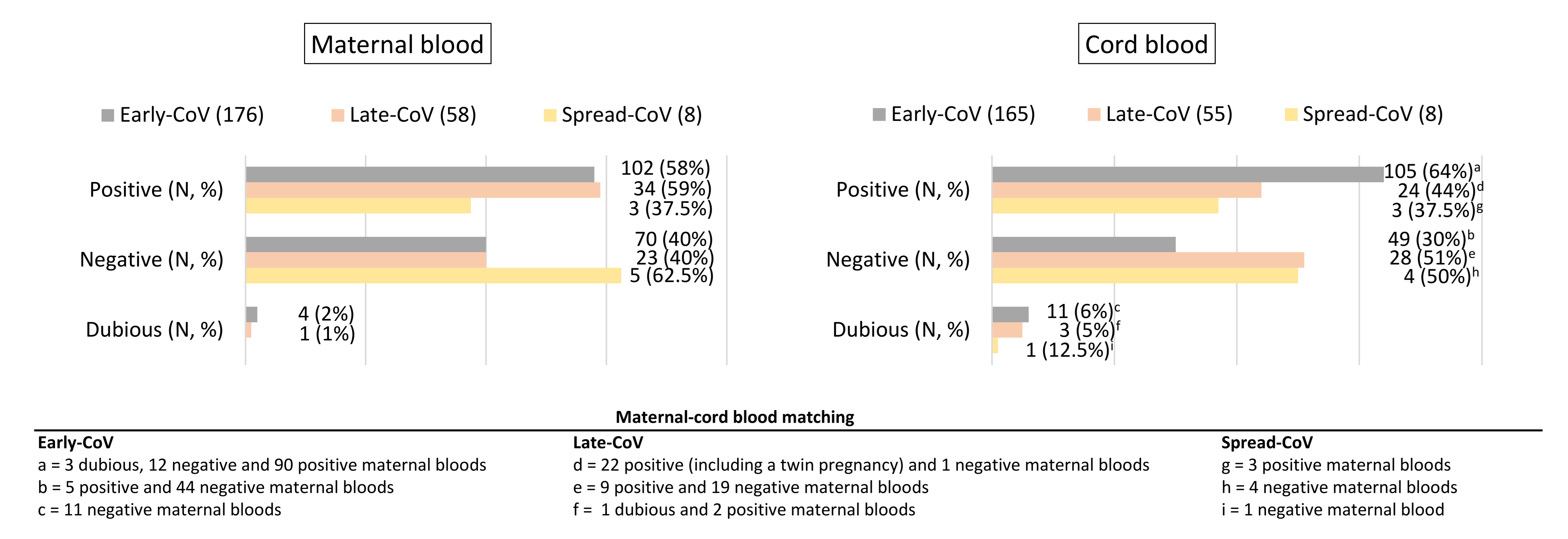
2.2. SARS-CoV-2 Antibody Transplacental Passage
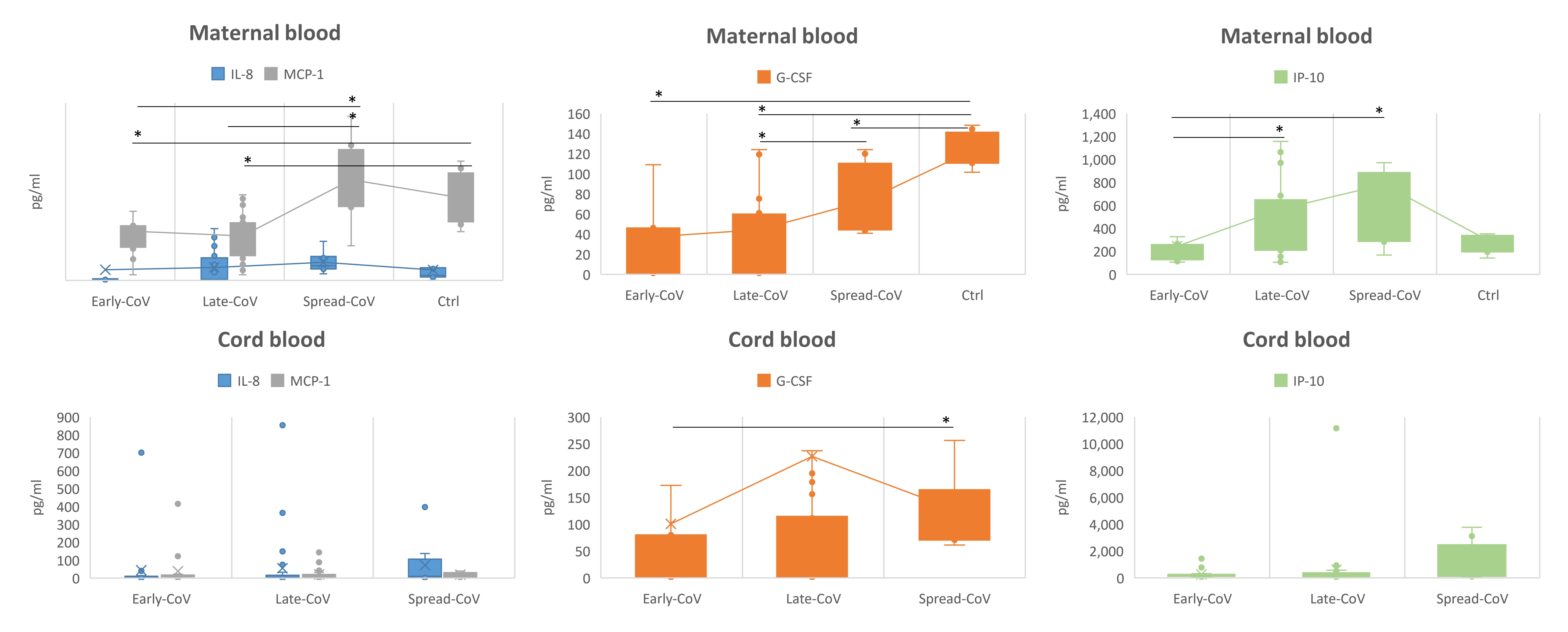
2.3. Inflammatory Response
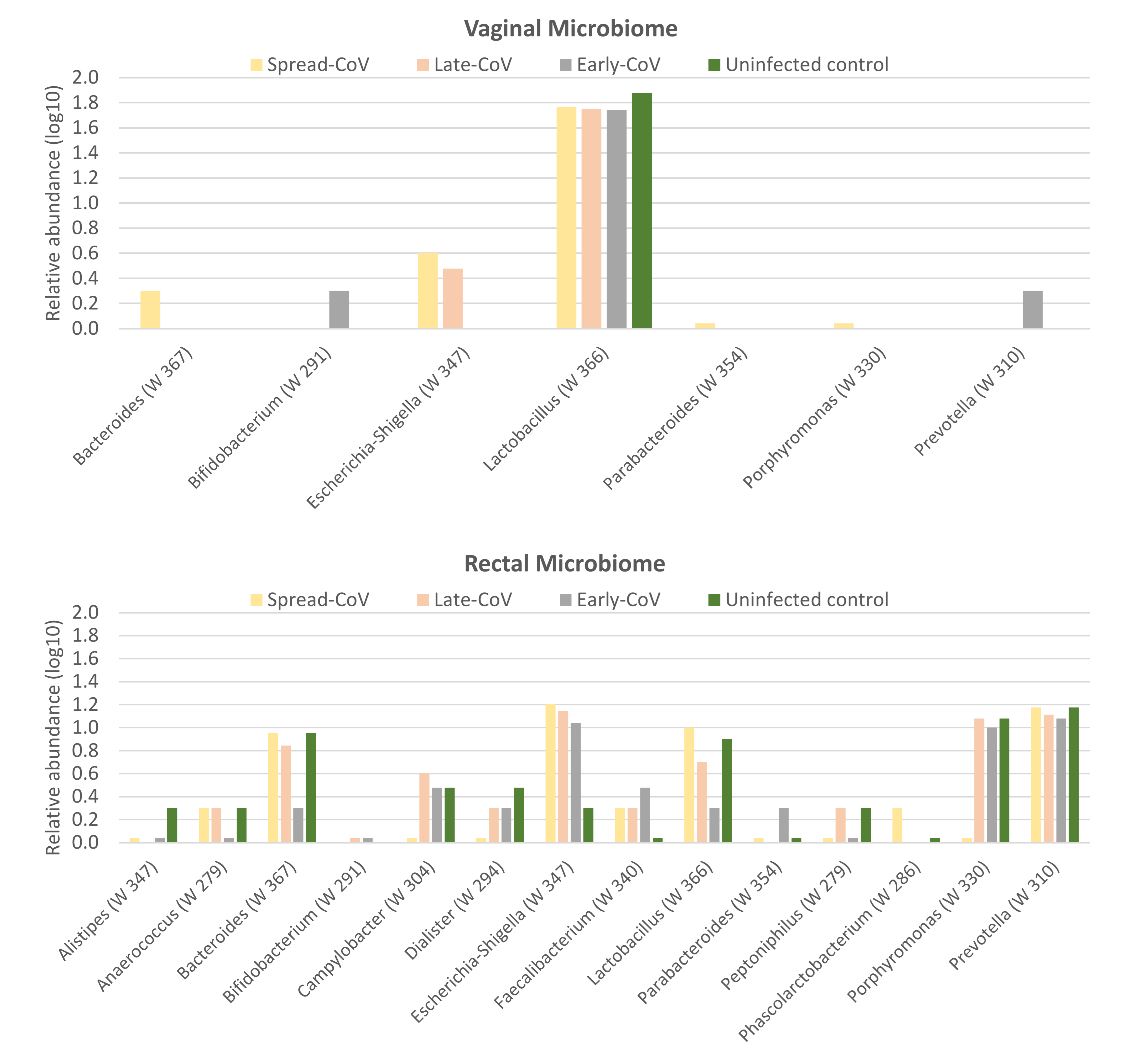
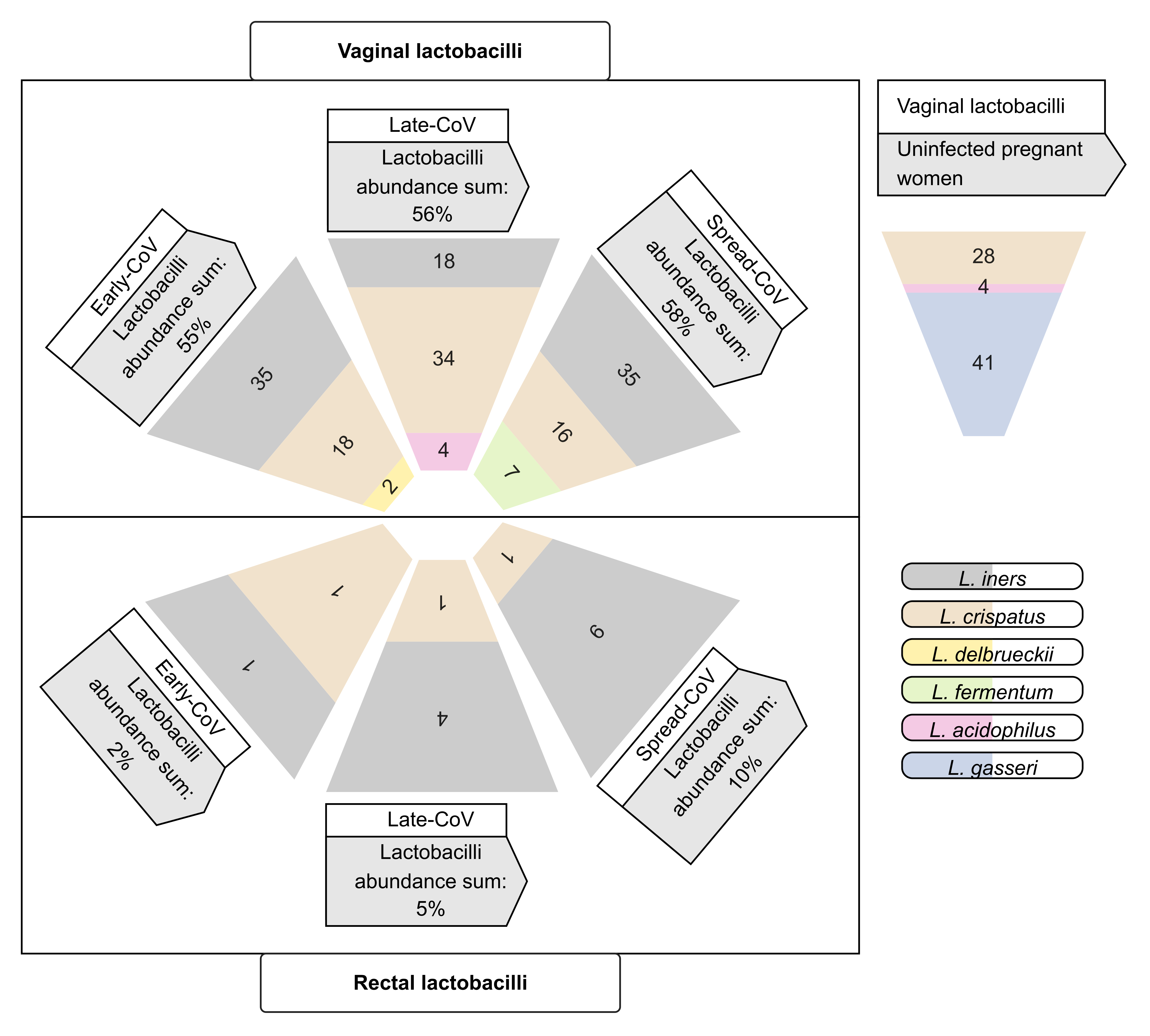
2.4. Maternal Microbiome
3. Discussion
4. Materials and Methods
4.1. Study Cohorts and Workflow of the Study
4.2. Specimen Collection
4.3. Nucleic Acids Extraction
4.4. RT–PCR for SARS-CoV-2
4.5. COVID-19 Serology
4.6. Analyses of the Inflammatory Response
4.7. Microbiome Analysis
4.8. Statistical Analysis
4.9. Compliance with Ethical Standards
4.10. Availability of Data
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dallmann, A.; Ince, I.; Meyer, M.; Willmann, S.; Eissing, T.; Hempel, G. Gestation-Specific Changes in the Anatomy and Physiology of Healthy Pregnant Women: An Extended Repository of Model Parameters for Physiologically Based Pharmacokinetic Modeling in Pregnancy. Clin. Pharmacokinet. 2017, 56, 1303–1330. [Google Scholar] [CrossRef] [PubMed]
- Jahromi, A.S.; Shojaei, M.; Ghobadifar, M.A. Insulin Resistance and Serum Levels of Interleukin-17 and Interleukin-18 in Normal Pregnancy. Immune. Netw. 2014, 14, 149. [Google Scholar] [CrossRef] [PubMed]
- Vivanti, A.J.; Vauloup-Fellous, C.; Prevot, S.; Zupan, V.; Suffee, C.; do Cao, J.; Benachi, A.; de Luca, D. Transplacental Transmission of SARS-CoV-2 Infection. Nat. Commun. 2020, 11, 3572. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Flores, V.; Romero, R.; Xu, Y.; Theis, K.R.; Arenas-Hernandez, M.; Miller, D.; Peyvandipour, A.; Bhatti, G.; Galaz, J.; Gershater, M.; et al. Maternal-Fetal Immune Responses in Pregnant Women Infected with SARS-CoV-2. Nat. Commun. 2022, 13, 320. [Google Scholar] [CrossRef]
- Hu, X.; Gao, J.; Luo, X.; Feng, L.; Liu, W.; Chen, J.; Benachi, A.; de Luca, D.; Chen, L. Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Vertical Transmission in Neonates Born to Mothers with Coronavirus Disease 2019 (COVID-19) Pneumonia. Obstet. Gynecol. 2020, 136, 65–67. [Google Scholar] [CrossRef] [PubMed]
- Norman, M.; Navér, L.; Söderling, J.; Ahlberg, M.; Hervius Askling, H.; Aronsson, B.; Byström, E.; Jonsson, J.; Sengpiel, V.; Ludvigsson, J.F.; et al. Association of Maternal SARS-CoV-2 Infection in Pregnancy with Neonatal Outcomes. JAMA 2021, 325, 2076–2086. [Google Scholar] [CrossRef]
- Gale, C.; Quigley, M.A.; Placzek, A.; Knight, M.; Ladhani, S.; Draper, E.S.; Sharkey, D.; Doherty, C.; Mactier, H.; Kurinczuk, J.J. Characteristics and Outcomes of Neonatal SARS-CoV-2 Infection in the UK: A Prospective National Cohort Study Using Active Surveillance. Lancet Child Adolesc. Health 2021, 5, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Dashraath, P.; Wong, J.L.J.; Lim, M.X.K.; Lim, L.M.; Li, S.; Biswas, A.; Choolani, M.; Mattar, C.; Su, L.L. Coronavirus Disease 2019 (COVID-19) Pandemic and Pregnancy. Am. J. Obs. Gynecol. 2020, 222, 521–531. [Google Scholar] [CrossRef] [PubMed]
- DeBolt, C.A.; Bianco, A.; Limaye, M.A.; Silverstein, J.; Penfield, C.A.; Roman, A.S.; Rosenberg, H.M.; Ferrara, L.; Lambert, C.; Khoury, R.; et al. Pregnant Women with Severe or Critical Coronavirus Disease 2019 Have Increased Composite Morbidity Compared with Nonpregnant Matched Controls. Am. J. Obs. Gynecol. 2021, 224, 510.e1–510.e12. [Google Scholar] [CrossRef] [PubMed]
- Hantoushzadeh, S.; Shamshirsaz, A.A.; Aleyasin, A.; Seferovic, M.D.; Aski, S.K.; Arian, S.E.; Pooransari, P.; Ghotbizadeh, F.; Aalipour, S.; Soleimani, Z.; et al. Maternal Death Due to COVID-19. Am. J. Obs. Gynecol. 2020, 223, 109.e1–109.e16. [Google Scholar] [CrossRef]
- Villar, J.; Ariff, S.; Gunier, R.B.; Thiruvengadam, R.; Rauch, S.; Kholin, A.; Roggero, P.; Prefumo, F.; do Vale, M.S.; Cardona-Perez, J.A.; et al. Maternal and Neonatal Morbidity and Mortality Among Pregnant Women with and Without COVID-19 Infection: The INTERCOVID Multinational Cohort Study. JAMA Pediatr. 2021, 175, 817–826. [Google Scholar] [CrossRef] [PubMed]
- Shook, L.L.; Sullivan, E.L.; Lo, J.O.; Perlis, R.H.; Edlow, A.G. COVID-19 in Pregnancy: Implications for Fetal Brain Development. Trends Mol. Med. 2022, 28, 319–330. [Google Scholar] [CrossRef] [PubMed]
- Han, V.X.; Patel, S.; Jones, H.F.; Dale, R.C. Maternal Immune Activation and Neuroinflammation in Human Neurodevelopmental Disorders. Nat. Rev. Neurol. 2021, 17, 564–579. [Google Scholar] [CrossRef] [PubMed]
- Liong, S.; Oseghale, O.; To, E.E.; Brassington, K.; Erlich, J.R.; Luong, R.; Liong, F.; Brooks, R.; Martin, C.; O’Toole, S.; et al. Influenza A Virus Causes Maternal and Fetal Pathology via Innate and Adaptive Vascular Inflammation in Mice. Proc. Natl. Acad. Sci. USA 2020, 117, 24964–24973. [Google Scholar] [CrossRef] [PubMed]
- Tsatsaris, V.; Mariaggi, A.A.; Launay, O.; Couffignal, C.; Rousseau, J.; Ancel, P.Y.; Marcault, E.; Ville, Y.; Cordier, A.G.; Vivanti, A.; et al. SARS-CoV-2 IgG Antibody Response in Pregnant Women at Delivery. J. Gynecol. Obstet. Hum. Reprod. 2021, 50, 102041. [Google Scholar] [CrossRef] [PubMed]
- Crovetto, F.; Crispi, F.; Llurba, E.; Pascal, R.; Larroya, M.; Trilla, C.; Camacho, M.; Medina, C.; Dobaño, C.; Gomez-Roig, M.D.; et al. Impact of SARS-CoV-2 Infection on Pregnancy Outcomes: A Population-Based Study. Clin. Infect. Dis. 2021, 73, 1768–1775. [Google Scholar] [CrossRef] [PubMed]
- Adhikari, E.H.; Moreno, W.; Zofkie, A.C.; MacDonald, L.; McIntire, D.D.; Collins, R.R.J.; Spong, C.Y. Pregnancy Outcomes Among Women with and without Severe Acute Respiratory Syndrome Coronavirus 2 Infection. JAMA Netw. Open 2020, 3, e2029256. [Google Scholar] [CrossRef] [PubMed]
- Zelini, P.; Perotti, F.; Licia Scatigno, A.; Dominoni, M.; Zavaglio, F.; Arossa, A.; Piccini, S.; Angelini, M.; Ghirardello, S.; Lilleri, D.; et al. Impact of SARS-CoV-2 Infection during Pregnancy and Persistence of Antibody Response. New Microbiol. 2022, 45, 181–189. [Google Scholar] [PubMed]
- Liu, S.; Zhong, J.; Zhang, D. Transplacental Transfer of Maternal Antibody against SARS-CoV-2 and Its Influencing Factors: A Review. Vaccines 2022, 10, 1083. [Google Scholar] [CrossRef] [PubMed]
- Koren, O.; Goodrich, J.K.; Cullender, T.C.; Spor, A.; Laitinen, K.; Kling Bäckhed, H.; Gonzalez, A.; Werner, J.J.; Angenent, L.T.; Knight, R.; et al. Host Remodeling of the Gut Microbiome and Metabolic Changes during Pregnancy. Cell 2012, 150, 470. [Google Scholar] [CrossRef] [PubMed]
- DiGiulio, D.B.; Callahan, B.J.; McMurdie, P.J.; Costello, E.K.; Lyell, D.J.; Robaczewska, A.; Sun, C.L.; Goltsman, D.S.A.; Wong, R.J.; Shawa, G.; et al. Temporal and Spatial Variation of the Human Microbiota during Pregnancy. Proc. Natl. Acad. Sci. USA 2015, 112, 11060–11065. [Google Scholar] [CrossRef] [PubMed]
- Gupta, P.; Singh, M.P.; Goyal, K. Diversity of Vaginal Microbiome in Pregnancy: Deciphering the Obscurity. Front. Public Health 2020, 8, 326. [Google Scholar] [CrossRef] [PubMed]
- Varea-Jiménez, E.; Aznar Cano, E.; Vega-Piris, L.; Martínez Sánchez, E.V.; Mazagatos, C.; García San Miguel Rodríguez-Alarcón, L.; Casas, I.; Sierra Moros, M.J.; Iglesias-Caballero, M.; Vazquez-Morón, S.; et al. Comparative Severity of COVID-19 Cases Caused by Alpha, Delta or Omicron SARS-CoV-2 Variants and Its Association with Vaccination. Enferm. Infecc. Microbiol. Clin. 2022, 42, 187–194. [Google Scholar] [CrossRef] [PubMed]
- Falahi, S.; Abdoli, A.; Kenarkoohi, A. Maternal COVID-19 Infection and the Fetus: Immunological and Neurological Perspectives. New Microbes New Infect. 2023, 53, 101135. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, S.Y.; Petersen, L.H.; Murra, M.; Hvidman, L.; Helmig, R.B.; Møller, J.K.; Khalil, M.R.; Kirkeby, M.; Henriksen, T.B. Transplacental Transfer of SARS-CoV-2 Antibodies: A Cohort Study. Eur. J. Clin. Microbiol. Infect. Dis. 2023, 42, 277–285. [Google Scholar] [CrossRef] [PubMed]
- Edwards, K.M. Maternal Antibodies and Infant Immune Responses to Vaccines. Vaccine 2015, 33, 6469–6472. [Google Scholar] [CrossRef] [PubMed]
- Hussain, T.; Murtaza, G.; Kalhoro, D.H.; Kalhoro, M.S.; Yin, Y.; Chughtai, M.I.; Tan, B.; Yaseen, A.; Rehman, Z.U. Understanding the Immune System in Fetal Protection and Maternal Infections during Pregnancy. J. Immunol. Res. 2022, 2022, 7567708. [Google Scholar] [CrossRef] [PubMed]
- Jenner, A.L.; Aogo, R.A.; Alfonso, S.; Crowe, V.; Deng, X.; Smith, A.P.; Morel, P.A.; Davis, C.L.; Smith, A.M.; Craig, M. COVID-19 Virtual Patient Cohort Suggests Immune Mechanisms Driving Disease Outcomes. PLoS Pathog. 2021, 17, e1009753. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Wang, J.; Cai, X.; Yin, T.; Zhang, Y.; Yang, C.; Yang, J. Granulocyte Colony-Stimulating Factor in Reproductive-Related Disease: Function, Regulation and Therapeutic Effect. Biomed. Pharmacother. 2022, 150, 112903. [Google Scholar] [CrossRef] [PubMed]
- Rutella, S.; Zavala, F.; Danese, S.; Kared, H.; Leone, G. Granulocyte Colony-Stimulating Factor: A Novel Mediator of T Cell Tolerance. J. Immunol. 2005, 175, 7085–7091. [Google Scholar] [CrossRef] [PubMed]
- Denison, F.C.; Kelly, R.W.; Calder, A.A. Differential Secretion of Chemokines from Peripheral Blood in Pregnant Compared with Non-Pregnant Women. J. Reprod. Immunol. 1997, 34, 225–240. [Google Scholar] [CrossRef]
- Bränn, E.; Edvinsson, Å.; Rostedt Punga, A.; Sundström-Poromaa, I.; Skalkidou, A. Inflammatory and Anti-Inflammatory Markers in Plasma: From Late Pregnancy to Early Postpartum. Sci. Rep. 2019, 9, 1863. [Google Scholar] [CrossRef] [PubMed]
- Berhan, Y. What Immunological and Hormonal Protective Factors Lower the Risk of COVID-19 Related Deaths in Pregnant Women? J. Reprod. Immunol. 2020, 142, 103180. [Google Scholar] [CrossRef] [PubMed]
- Carr, R.; Modi, N.; Doré, C.J. G-CSF and GM-CSF for Treating or Preventing Neonatal Infections. Cochrane Database Syst. Rev. 2003, 2003, CD003066. [Google Scholar] [CrossRef]
- Budden, K.F.; Gellatly, S.L.; Wood, D.L.A.; Cooper, M.A.; Morrison, M.; Hugenholtz, P.; Hansbro, P.M. Emerging Pathogenic Links between Microbiota and the Gut–Lung Axis. Nat. Rev. Microbiol. 2016, 15, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, J.M. The Role of Escherichia Coli in Inflammatory Bowel Disease. Gut 2007, 56, 610–612. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, M.; Tanaka, Y.; Benno, Y.; Ohkuma, M. Butyricimonas Faecihominis Sp. Nov. and Butyricimonas Paravirosa Sp. Nov., Isolated from Human Faeces, and Emended Description of the Genus Butyricimonas. Int. J. Syst. Evol. Microbiol. 2014, 64, 2992–2997. [Google Scholar] [CrossRef] [PubMed]
- Juárez-Castelán, C.J.; Vélez-Ixta, J.M.; Corona-Cervantes, K.; Piña-Escobedo, A.; Cruz-Narváez, Y.; Hinojosa-Velasco, A.; Landero-Montes-de-Oca, M.E.; Davila-Gonzalez, E.; González-Del-Olmo, E.; Bastida-Gonzalez, F.; et al. The Entero-Mammary Pathway and Perinatal Transmission of Gut Microbiota and SARS-CoV-2. Int. J. Mol. Sci. 2022, 23, 10306. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.H.; Chen, C.C.; Chiang, H.L.; Liou, J.M.; Chang, C.M.; Lu, T.P.; Chuang, E.Y.; Tai, Y.C.; Cheng, C.; Lin, H.Y.; et al. Altered Gut Microbiota and Inflammatory Cytokine Responses in Patients with Parkinson’s Disease. J. Neuroinflamm. 2019, 16, 129. [Google Scholar] [CrossRef]
- Freitas, A.C.; Hill, J.E. Quantification, Isolation and Characterization of Bifidobacterium from the Vaginal Microbiomes of Reproductive Aged Women. Anaerobe 2017, 47, 145–156. [Google Scholar] [CrossRef] [PubMed]
- Petrova, M.I.; Reid, G.; Vaneechoutte, M.; Lebeer, S. Lactobacillus Iners: Friend or Foe? Trends Microbiol. 2017, 25, 182–191. [Google Scholar] [CrossRef] [PubMed]
- De Seta, F.; Campisciano, G.; Zanotta, N.; Ricci, G.; Comar, M. The Vaginal Community State Types Microbiome-Immune Network as Key Factor for Bacterial Vaginosis and Aerobic Vaginitis. Front Microbiol. 2019, 10, 483585. [Google Scholar] [CrossRef]
- Vásquez, A.; Jakobsson, T.; Ahrné, S.; Forsum, U.; Molin, G. Vaginal Lactobacillus Flora of Healthy Swedish Women. J. Clin. Microbiol. 2002, 40, 2746–2749. [Google Scholar] [CrossRef] [PubMed]
- Chourasia, R.; Padhi, S.; Chiring Phukon, L.; Abedin, M.M.; Singh, S.P.; Rai, A.K. A Potential Peptide from Soy Cheese Produced Using Lactobacillus Delbrueckii WS4 for Effective Inhibition of SARS-CoV-2 Main Protease and S1 Glycoprotein. Front. Mol. Biosci. 2020, 7, 601753. [Google Scholar] [CrossRef] [PubMed]
- Mirashrafi, S.; Moravejolahkami, A.R.; Balouch Zehi, Z.; Hojjati Kermani, M.A.; Bahreini-Esfahani, N.; Haratian, M.; Ganjali Dashti, M.; Pourhossein, M. The Efficacy of Probiotics on Virus Titres and Antibody Production in Virus Diseases: A Systematic Review on Recent Evidence for COVID-19 Treatment. Clin. Nutr. ESPEN 2021, 46, 1–8. [Google Scholar] [CrossRef]
- Reid, G.; Charbonneau, D.; Erb, J.; Kochanowski, B.; Beuerman, D.; Poehner, R.; Bruce, A.W. Oral Use of Lactobacillus Rhamnosus GR-1 and L. Fermentum RC-14 Significantly Alters Vaginal Flora: Randomized, Placebo-Controlled Trial in 64 Healthy Women. FEMS Immunol. Med. Microbiol. 2003, 35, 131–134. [Google Scholar] [CrossRef]
- Gardiner, G.E.; Heinemann, C.; Baroja, M.L.; Bruce, A.W.; Beuerman, D.; Madrenas, J.; Reid, G. Oral Administration of the Probiotic Combination Lactobacillus Rhamnosus GR-1 and L. Fermentum RC-14 for Human Intestinal Applications. Int. Dairy J. 2002, 12, 191–196. [Google Scholar] [CrossRef]
- Yeruva, T.; Rajkumar, H.; Donugama, V. Vaginal Lactobacilli Profile in Pregnant Women with Normal & Abnormal Vaginal Flora. Indian J. Med. Res. 2017, 146, 534–540. [Google Scholar] [PubMed]
- Mousavi, E.; Makvandi, M.; Teimoori, A.; Ataei, A.; Ghafari, S.; Samarbaf-Zadeh, A. Antiviral Effects of Lactobacillus Crispatus against HSV-2 in Mammalian Cell Lines. J. Chin. Med. Assoc. 2018, 81, 262–267. [Google Scholar] [CrossRef]
- Kawahara, T.; Shimizu, I.; Tanaka, Y.; Tobita, K.; Tomokiyo, M.; Watanabe, I. Lactobacillus Crispatus Strain KT-11 S-Layer Protein Inhibits Rotavirus Infection. Front. Microbiol. 2022, 13, 398. [Google Scholar] [CrossRef] [PubMed]
- Campisciano, G.; Iebba, V.; Zito, G.; Luppi, S.; Martinelli, M.; Fischer, L.; De Seta, F.; Basile, G.; Ricci, G.; Comar, M. Lactobacillus Iners and Gasseri, Prevotella Bivia and HPV Belong to the Microbiological Signature Negatively Affecting Human Reproduction. Microorganisms 2020, 9, 39. [Google Scholar] [CrossRef] [PubMed]
- Underwood, M.A.; Mukhopadhyay, S.; Lakshminrusimha, S.; Bevins, C.L. Neonatal Intestinal Dysbiosis. J. Perinatol. 2020, 40, 1597–1608. [Google Scholar] [CrossRef] [PubMed]
- Baldassarre, M.E.; di Mauro, A.; Capozza, M.; Rizzo, V.; Schettini, F.; Panza, R.; Laforgia, N. Dysbiosis and Prematurity: Is There a Role for Probiotics? Nutrients 2019, 11, 1273. [Google Scholar] [CrossRef] [PubMed]

| Demographics | Early-CoV | Late-CoV | Spread-CoV | Uninfected Control |
|---|---|---|---|---|
| Patients | 176 | 58 | 8 | 20 |
| Mean age ± SD | 33 ± 5.3 | 31 ± 5.6 | 35 ± 3.6 | 30 ± 5 |
| Delivery (mean of weeks + days) | 39.1 ± 2.6 | 39 ± 1.6 | 37.9 ± 2 | 39.1 ± 2.2 |
| Vaginal Birth—n (%) | 148 (84.1) | 48 (82.7) | 6 (75) | 10 |
| C-section—n (%) | 26 (14.8) | 10 (17.3) | 2 (25) | 10 |
| Spontaneous Abortion | 2 (1.1) | 0 | 0 | 0 |
| SARS-CoV-2 positive newborns (%) | 0 | 3.3 | 13 | 0 |
| SARS-CoV-2 negative newborns (%) | 100 | 96.7 | 87 | 0 |
| Delivery complications | ||||
| Pathological CTG 1—n (%) | 11 (6.2) | 3 (5.1) | 2 (25) | 0 |
| Prolonged second stage—n (%) | 1 (0.5) | 3 (5.1) | 0 | 0 |
| Gestosis | 2 (1.1) | 0 | 0 | 0 |
| PROM ** | 29 (16.5) | 6 (10.3) | 0 | 0 |
| Maternal risk factors | ||||
| Gestational diabetes—n (%) | 20 (11.4) | 10 (17.2) | 2 (25) | 0 |
| Hypertension—n (%) | 4 (2.2) | 3 (5.1) | 0 | 0 |
| Obesity—n (%) | 9 (5.1) | 2 (3.4) | 0 | 0 |
| COVID-19 pneumonia—n (%) | 2 (1.1) | 4 (6.9) | 0 | 0 |
| Fetal complications | ||||
| Growth restriction—n (%) | 11 (6.2) | 3 (5.1) | 0 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Campisciano, G.; Sorz, A.; Cason, C.; Zanotta, N.; Gionechetti, F.; Piazza, M.; Carli, P.; Uliana, F.M.; Ballaminut, L.; Ricci, G.; et al. Genital Dysbiosis and Different Systemic Immune Responses Based on the Trimester of Pregnancy in SARS-CoV-2 Infection. Int. J. Mol. Sci. 2024, 25, 4298. https://doi.org/10.3390/ijms25084298
Campisciano G, Sorz A, Cason C, Zanotta N, Gionechetti F, Piazza M, Carli P, Uliana FM, Ballaminut L, Ricci G, et al. Genital Dysbiosis and Different Systemic Immune Responses Based on the Trimester of Pregnancy in SARS-CoV-2 Infection. International Journal of Molecular Sciences. 2024; 25(8):4298. https://doi.org/10.3390/ijms25084298
Chicago/Turabian StyleCampisciano, Giuseppina, Alice Sorz, Carolina Cason, Nunzia Zanotta, Fabrizia Gionechetti, Maria Piazza, Petra Carli, Francesca Maria Uliana, Lisa Ballaminut, Giuseppe Ricci, and et al. 2024. "Genital Dysbiosis and Different Systemic Immune Responses Based on the Trimester of Pregnancy in SARS-CoV-2 Infection" International Journal of Molecular Sciences 25, no. 8: 4298. https://doi.org/10.3390/ijms25084298
APA StyleCampisciano, G., Sorz, A., Cason, C., Zanotta, N., Gionechetti, F., Piazza, M., Carli, P., Uliana, F. M., Ballaminut, L., Ricci, G., De Seta, F., Maso, G., & Comar, M. (2024). Genital Dysbiosis and Different Systemic Immune Responses Based on the Trimester of Pregnancy in SARS-CoV-2 Infection. International Journal of Molecular Sciences, 25(8), 4298. https://doi.org/10.3390/ijms25084298







