Abstract
Aberrant expression of miR-145-5p has been observed in prostate cancer where is has been suggested to play a tumor suppressor role. In other cancers, miR-145-5p acts as an inhibitor of epithelial-to-mesenchymal transition (EMT), a key molecular process for tumor progression. However, the interaction between miR-145-5p and EMT remains to be elucidated in prostate cancer. In this paper the link between miR-145-5p and EMT in prostate cancer was investigated using a combination of in silico and in vitro analyses. miR-145-5p expression was significantly lower in prostate cancer cell lines compared to normal prostate cells. Bioinformatic analysis of The Cancer Genome Atlas prostate adenocarcinoma (TCGA PRAD) data showed significant downregulation of miR-145-5p in prostate cancer, correlating with disease progression. Functional enrichment analysis significantly associated miR-145-5p and its target genes with EMT. MYO6, an EMT-associated gene, was identified and validated as a novel target of miR-145-5p in prostate cancer cells. In vitro manipulation of miR-145-5p levels significantly altered cell proliferation, clonogenicity, migration and expression of EMT-associated markers. Additional TCGA PRAD analysis suggested miR-145-5p tumor expression may be useful predictor of disease recurrence. In summary, this is the first study to report that miR-145-5p may inhibit EMT by targeting MYO6 in prostate cancer cells. The findings suggest miR-145-5p could be a useful diagnostic and prognostic biomarker for prostate cancer.
1. Introduction
Prostate cancer is a heterogeneous disease affecting millions of men worldwide, predominantly in regions with a high human development index [1]. Epithelial-to-mesenchymal transition (EMT) is a key biological process required for the progression and metastasis of prostate cancer [2]. EMT is a dynamic process in which epithelial cells are reprogrammed to exhibit a mesenchymal phenotype. The loss of cell polarity and cell–cell adhesion leads to enhanced migratory capacity, invasiveness, and resistance to apoptosis [3]. Multiple factors regulate the EMT process, including several miRNAs [4]. MicroRNAs (miRNAs) are short, non-coding RNA molecules that play an important role in regulating gene expression at the post-transcriptional level.
miRNAs bind to specific sequences in the 3′ untranslated region (3′ UTR) of their target messenger RNAs (mRNAs), resulting in either mRNA degradation or translational repression [5]. By inhibiting the translation of target mRNAs or causing their degradation, miRNAs can effectively silence the expression of specific genes [6]. The small size of miRNAs, typically only 18–25 nucleotides long, allows them to recognize related but not identical target sites, so each miRNA can regulate multiple target mRNAs [7]. Through this sequence-specific interaction, miRNAs provide an important mechanism for fine-tuning the levels of proteins encoded by mRNAs in the cell [8]. The ability of miRNAs to modulate gene expression has implicated them in many biological processes and human diseases, including prostate cancer [9].
Our laboratory has previously demonstrated the role of several miRNAs in prostate cancer, including miR-200c [10], miR-24 [11], miR-210 [12], miR-21 [13], miR-182 [14] and miR-143 [15]. However, there remains a need for further investigation into miRNAs that are specifically associated with the EMT process in prostate cancer [16]. With this objective in mind, miR-145-5p is promising miRNA for further investigation.
The role of miR-145-5p has been studied in various cancers and is generally characterized as a tumor suppressor (reviewed in [17]). Expression of miR-145-5p is downregulated in several cancer types, including bladder [18], breast [19], gastric [20], cholangiocarcinoma [21], clear cell renal cell carcinoma (ccRCC) [22], colorectal [23], oesophageal squamous cell carcinoma (ESCC) [24], oesophageal carcinoma [25], gallbladder carcinoma (GBC) [26], gastric cancer [27], glioma [28], hepatocellular carcinoma (HCC) [29], laryngeal squamous cell carcinoma (LSCC) [30], chronic lymphocytic leukemia (CLL) [31], lung adenocarcinoma (LUAC) [32], lung squamous cell carcinoma (Lung SCC) [33], melanoma [34], non-small cell lung cancer (NSCLC) [35], osteosarcoma [36], retinoblastoma [37], and papillary thyroid carcinoma (PTC) [38].
Given this body of evidence, it is not surprising that emerging research has suggested miR-145-5p also acts as a tumor suppressor in prostate cancer [39]. Various studies have validated multiple gene targets of miR-145-5p in prostate cancer cells, demonstrating its capacity to modulate cell behavior through various signaling pathways [40,41,42]. Previous studies have also demonstrated that overexpression of miR-145-5p can increase the chemosensitivity of prostate cancer cells to first-line chemotherapeutic agents [43,44]. However, the link between miR-145-5p and EMT remains poorly characterized in prostate cancer. Some studies have identified that overexpression of miR-145-5p decreases migration of prostate cells, but have not directly linked miR-145-5p and EMT [45,46,47,48]. The studies that have examined a role for miR-145-5p in EMT have produced contradicting evidence, so there is a clear need for further work in this area [49,50].
Hence, although miR-145-5p has been implicated in EMT and prostate cancer, its precise mechanistic contributions and relevant molecular targets in this context remain poorly defined. Therefore, this study aims to elucidate the functional role of miR-145-5p associated with EMT in prostate cancer by examining the expression profile, downstream effects on putative target genes, and impacts on cell behavior. Additionally, complementary in silico analyses are used to evaluate the clinical significance of miR-145-5p in prostate cancer progression and determine its potential as a prognostic or diagnostic biomarker.
2. Results
2.1. Reduced miR-145-5p Expression Correlates with Prostate Cancer Progression
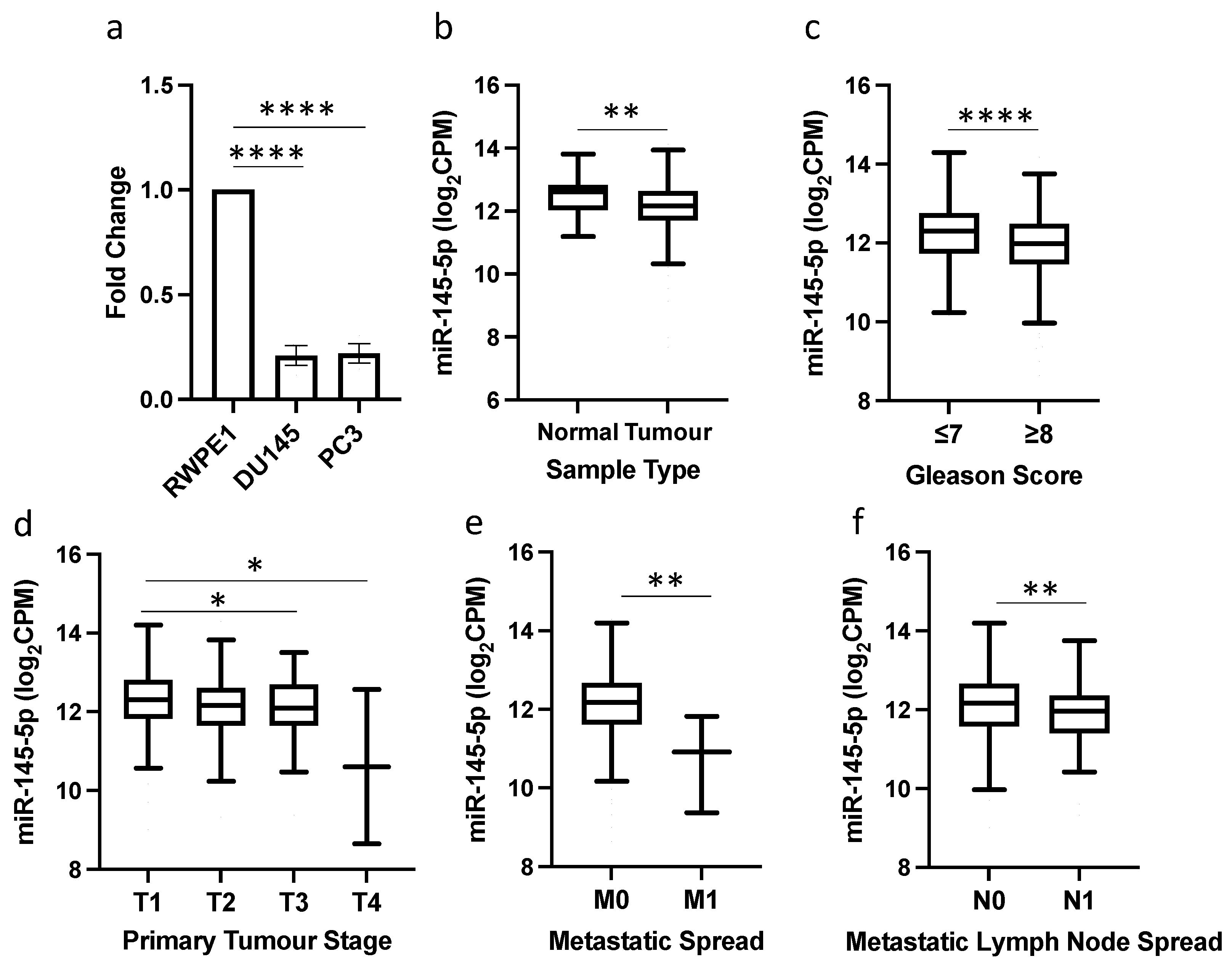
qRT-PCR analysis revealed significantly reduced expression of miR-145-5p in the prostate cancer cell lines DU145 and PC3 compared to normal prostate RWPE1 cells (Figure 1a). This reduction in miR-145-5p levels was validated in TCGA PRAD clinical samples, with miR-145-5p expression significantly lower in prostate tumor tissue than normal prostate tissue (Figure 1b). Further in silico analysis of the TCGA PRAD dataset demonstrated an association between decreased miR-145-5p expression and established clinicopathological markers of prostate cancer progression, including increased Gleason score, higher pathological T stage, presence of distant metastasis, and lymph node involvement (Figure 1c–f).
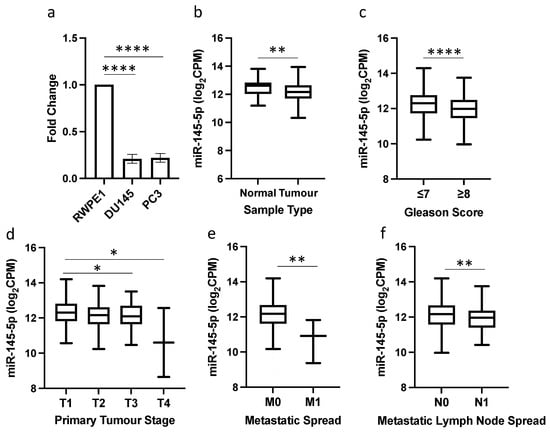
Figure 1.
miR-145-5p downregulation is associated with prostate cancer and disease progression. (a) qRT-PCR shows miR-145-5p expression is significantly lower in DU145 and PC3 prostate cancer cell lines compared to normal prostate cell line, RWPE1 (n = 3, housekeeping: snRNA U6). (b) UCSC Xena analysis of TCGA PRAD samples shows miR-145-5p expression is significantly decreased in prostate tumor tissue (n = 494) compared to normal prostate tissue (n = 52). UCSC Xena analysis of TCGA PRAD samples shows expression of miR-145-5p is significantly lower in patients with (c) Gleason score ≥ 8 (n = 210) compared to those scored ≤ 7 (n = 336), (d) pathological stage T3 (n = 55) and T4 (n = 2) compared to T1 (n = 197) (e) pathological stage M1 (n = 3) compared to M0 (n = 542) and (f) pathological stage N1 (n = 80) compared to N0 (n = 385). All p-values generated by unpaired two-tailed t-test (* p < 0.05, ** p < 0.01, **** p < 0.0001). CPM = Copies per million; n = number.
Functional enrichment analysis confirmed that miR-145-5p targets several gene sets that are significantly associated with prostate cancer (Table 1, Supplementary Figure S1). Additional functional enrichment analysis to help identify the key biological mechanisms involved revealed several significant gene networks associated with EMT and extracellular matrix remodeling. (Table 2, Supplementary Figure S2).

Table 1.
Functional enrichment analysis of miR-145-5p in prostate cancer. Table shows the significant association of miR-145-5p target genes with Gene Set descriptions related to prostate cancer. Analysis performed using cluster Profiler in CancerMIRNome.

Table 2.
Functional enrichment analysis of EMT-related functions associated with miR-145-5p. Table shows the significant association of miR-145-5p target genes with Gene Set descriptions related to EMT. Analysis performed using cluster Profiler in CancerMIRNome.
2.2. MYO6 Is a Novel Target of miR-145-5p in Prostate Cancer
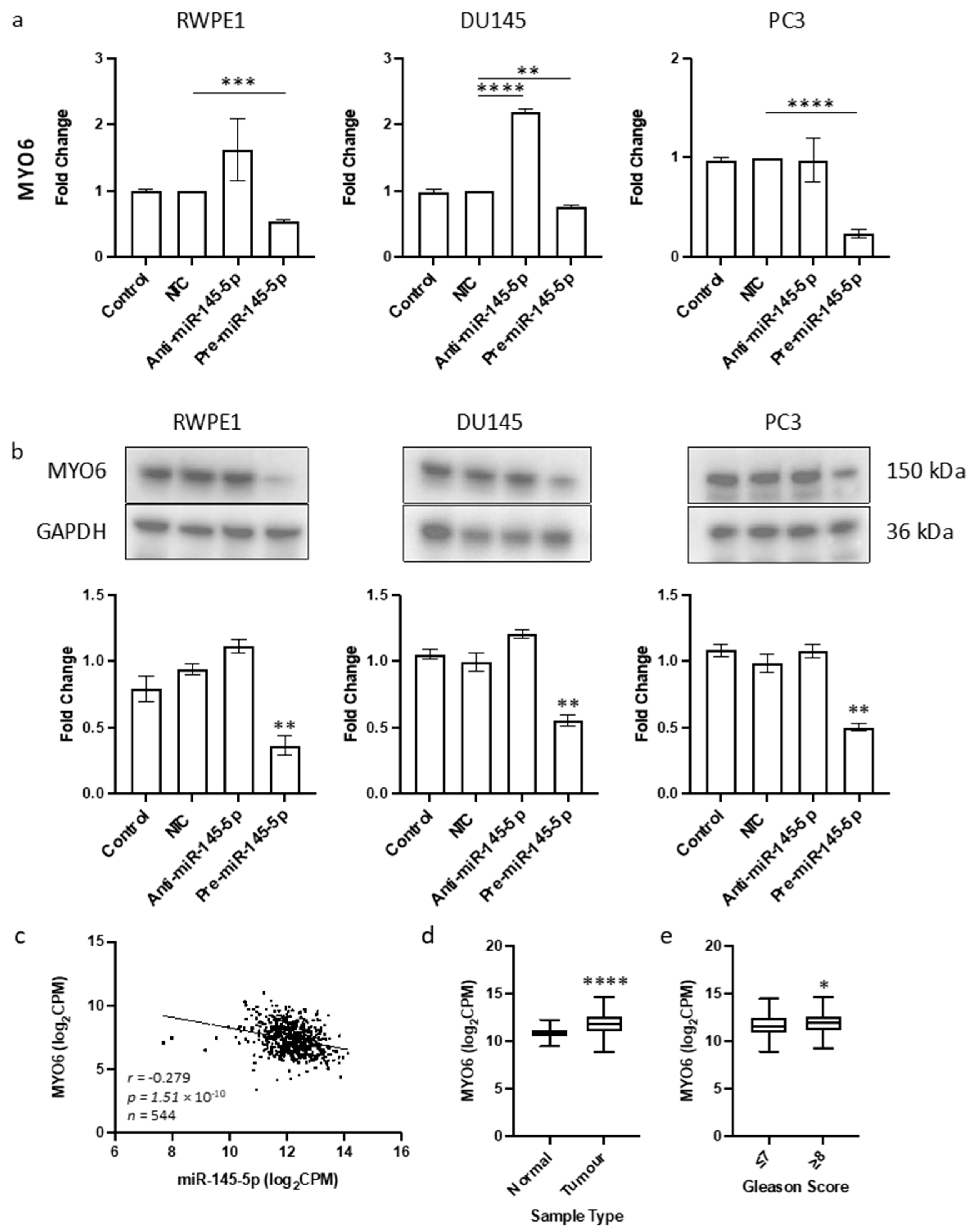
The objective of this study was to identify a novel target of miR-145-5p in prostate cancer. Given the tumor suppressive role of miR-145-5p and its association with EMT in other cancers, we aimed to find a target gene known to promote EMT that would be upregulated when miR-145-5p levels are decreased. By cross-referencing known EMT genes, validated miR-145-5p targets, and genes negatively correlated with miR-145-5p expression in TCGA PRAD database, we identified MYO6 as a likely candidate (Supplementary Figure S3). MYO6 was selected for further analysis due to its previously established link with prostate cancer [51] which we also observed in the functional enrichment analysis performed here (Table 1). The predicted base-pairing between miR-145-5p and the MYO6 transcript was confirmed (Supplementary Figure S3). Transient overexpression of miR-145-5p in RWPE1, DU145 and PC3 prostate cancer cell lines (Supplementary Figure S4) consistently led to decreased MYO6 mRNA and protein levels, which was validated by a significant negative correlation between miR-145-5p and MYO6 expression in TCGA PRAD samples (Figure 2). We also demonstrated that MYO6 expression is upregulated in prostate tumors compared to normal tissue, and in higher Gleason grade tumors (Figure 2c). Taken together, these data provide evidence that MYO6 is a direct target of miR-145-5p in prostate cancer. The loss of miR-145-5p expression may therefore promote cancer progression through disinhibition of MYO6 and subsequent induction of EMT.
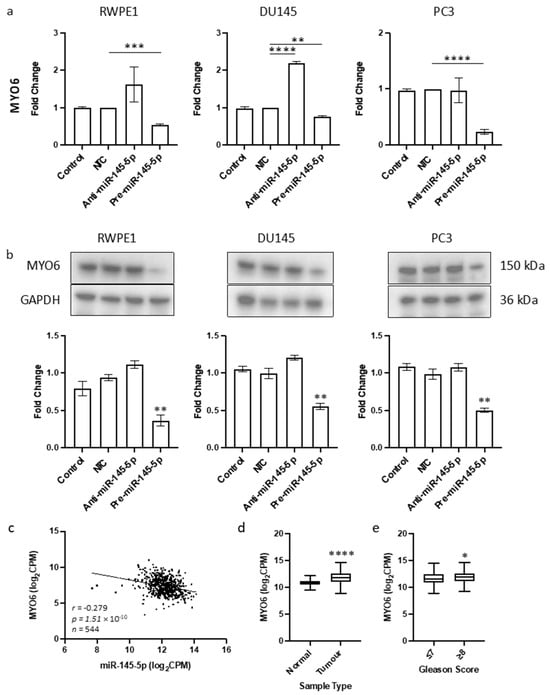
Figure 2.
Identification of MYO6 as novel target of miR-145-5p in prostate cancer. (a) Transient overexpression of miR-145-5p significantly reduces MYO6 gene expression in RWPE1, DU145 and PC3 cell lines. (n = 3, expression values normalized to GAPDH) (b) Quantified Western blotting and representative images (n = 3) shows transient overexpression of miR-145-5p causes significant downregulation of MYO6 protein in RWPE1, DU145 and PC3 cell lines. Bar graphs show mean ± SEM. p-values generated by unpaired two-tailed t-test, relative to NTC (* p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001). (c) CancerMIRNome analysis of the TCGA PRAD specimens (n = 543) shows the expression of miR-145-5p and MYO6 are significantly negatively correlated (Pearson correlation, **** p < 0.0001). UCSC Xena analysis of TCGA PRAD samples shows MYO6 expression is significantly elevated in (d) tumor tissue (n = 497) relative to normal (n = 52) tissue and in (e) samples with Gleason score ≥ 8 (n = 154) compared to those scored ≤ 7 (n = 213). (both Welch’s t-test, (* p < 0.05, **** p < 0.0001). NTC = Non-Targeting Control; n = number.
2.3. miR-145-5p Alters Key EMT Markers in Prostate Cell Lines
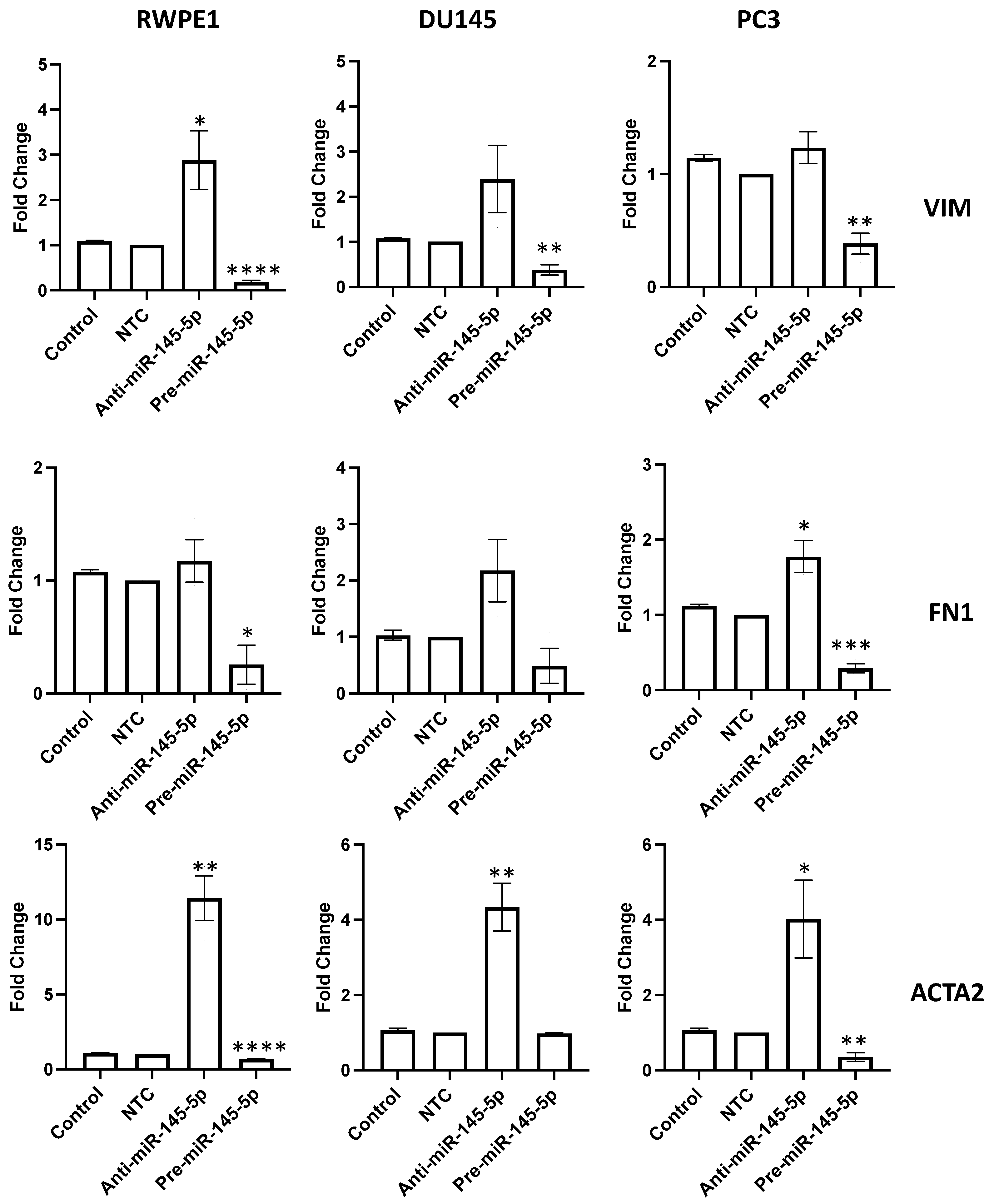
To validate the role of miR-145-5p in regulating EMT in prostate cells, the expression of several EMT markers were examined following transient knockdown or overexpression of miR-145-5p in RWPE1, DU145, and PC3 prostate cell lines. qRT-PCR analysis revealed that mRNA levels of mesenchymal markers vimentin (VIM), fibronectin 1 (FN1), and actin alpha 2 (ACTA2) were downregulated with miR-145-5p overexpression (Figure 3). Taken together, these results confirm that miR-145-5p modulates the expression of multiple EMT marker genes in prostate cell lines.
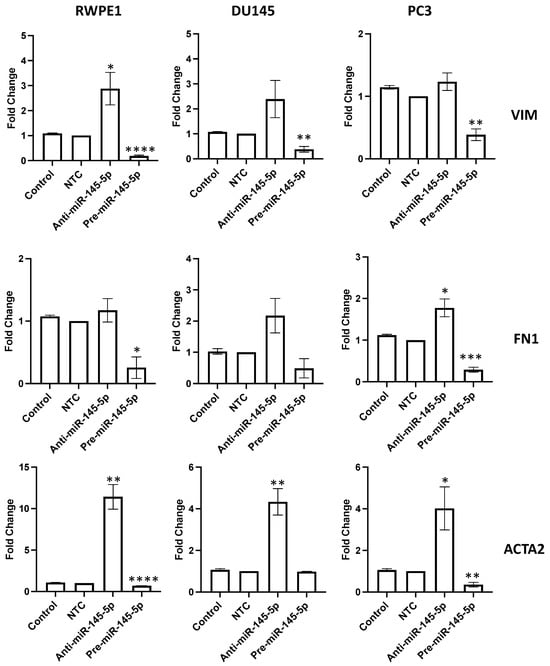
Figure 3.
miR-145-5p alters the expression of key EMT markers. VIM, FN1 and ACTA2 quantified 72 h post transfection using qRT-PCR. Bars show mean ± SEM (n = 3, expression values normalized to GAPDH). p-values generated by unpaired two-tailed t-test, relative to NTC (* p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001). NTC = Non-Targeting Control; n = number.
2.4. miR-145-5p Regulates Proliferation, Migration, and Clonogenic POTENTIAL in Prostate Cell Lines
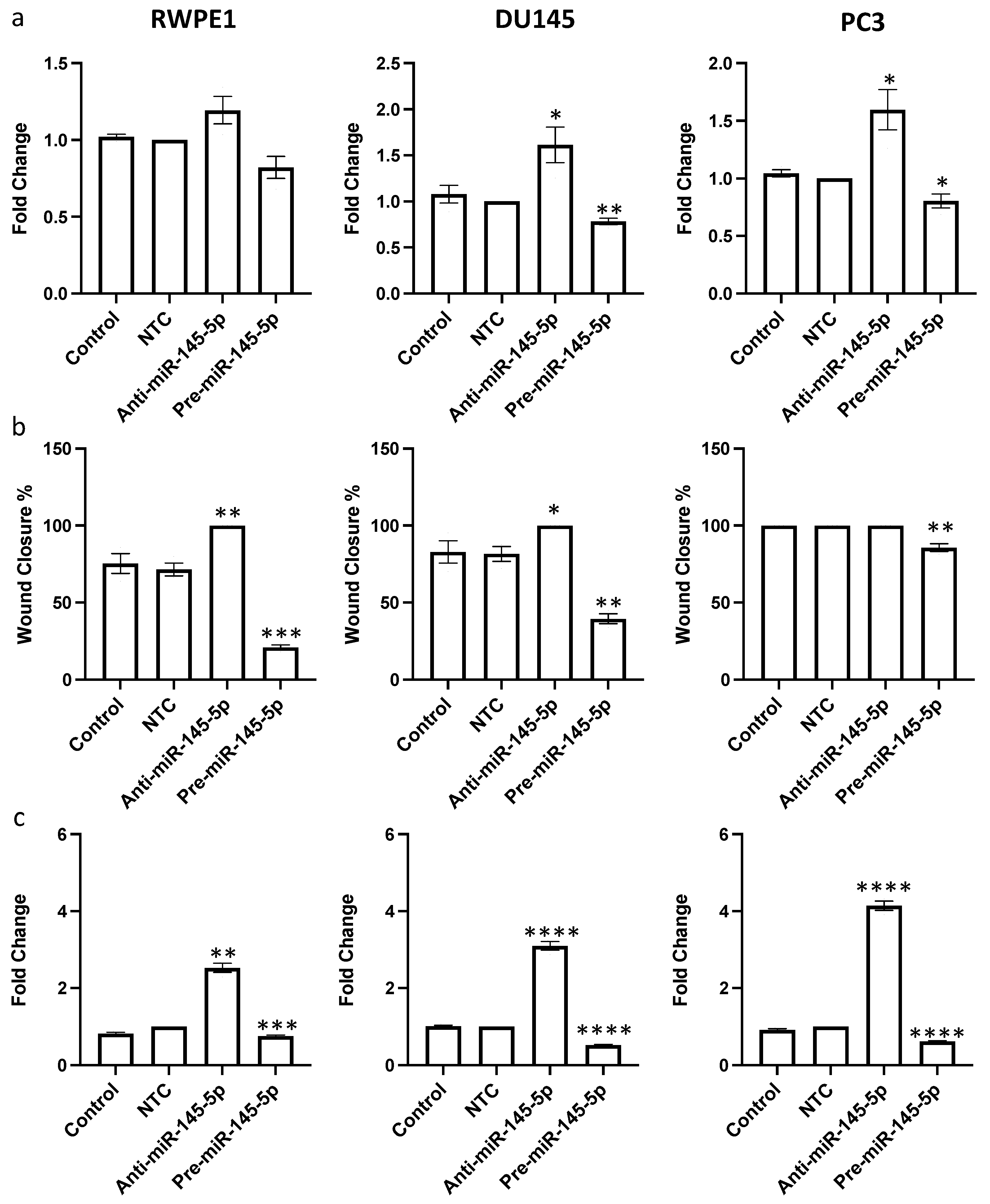
After confirming that miR-145-5p regulates expression of multiple EMT marker genes, we hypothesized that miR-145-5p would consequently impact prostate cell behaviors relevant to tumor progression. Since miR-145-5p regulates multiple oncogenic targets that drive cell proliferation, we first examined the effects of miR-145-5p modulation on proliferation rates. miR-145-5p inhibition significantly increased proliferation, while miR-145-5p overexpression decreased proliferation in RWPE1, DU145, and PC3 prostate cell lines, as quantified by cell counting assay (Figure 4a).
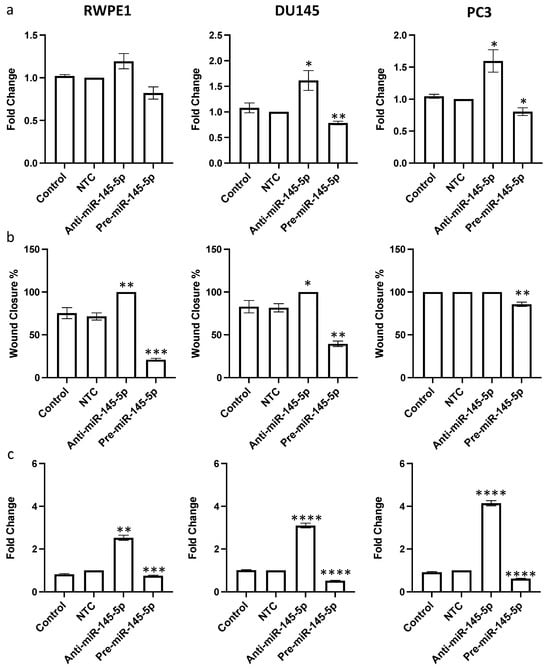
Figure 4.
miR-145-5p alters (a) proliferation, (b) migration and (c) clonogenicity of RWPE1, DU145 and PC3 cells. Proliferation measured by Alamar blue absorbance (600 nm) 72 h post transfection (n = 5). Migration measured by scratch assay performed at 72 h post transfection using ImageJ to quantify wound closure (n = 3). Clonogenicity measured by colony counting performed at 72 h post transfection using ImageJ (Version 1.54h) to quantify. Bar graphs show mean ± SEM. p-values generated by unpaired two-tailed t-test, relative to NTC (* p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001). NTC = Non-Targeting Control; n = number.
Given the established role of MYO6 signaling in promoting cell migration, we next assessed if miR-145-5p impacts prostate cell motility. As hypothesized, miR-145-5p overexpression reduced migration of RWPE1, DU145, and PC3 cells, whereas miR-145-5p knockdown enhanced migration of RWPE1, DU145 and PC3 cells, as determined by wound healing assay (Figure 4b, Supplementary Figure S5). Lastly, we evaluated the effects of miR-145-5p on clonogenic capacity using clonogenic assays. Overexpression of miR-145-5p significantly reduced colony formation, while miR-145-5p inhibition increased colony formation across the tested prostate cell lines (Figure 4c, Supplementary Figure S6).
2.5. Mapping the Functional Network of the miR-145-5p/MYO6 Axis
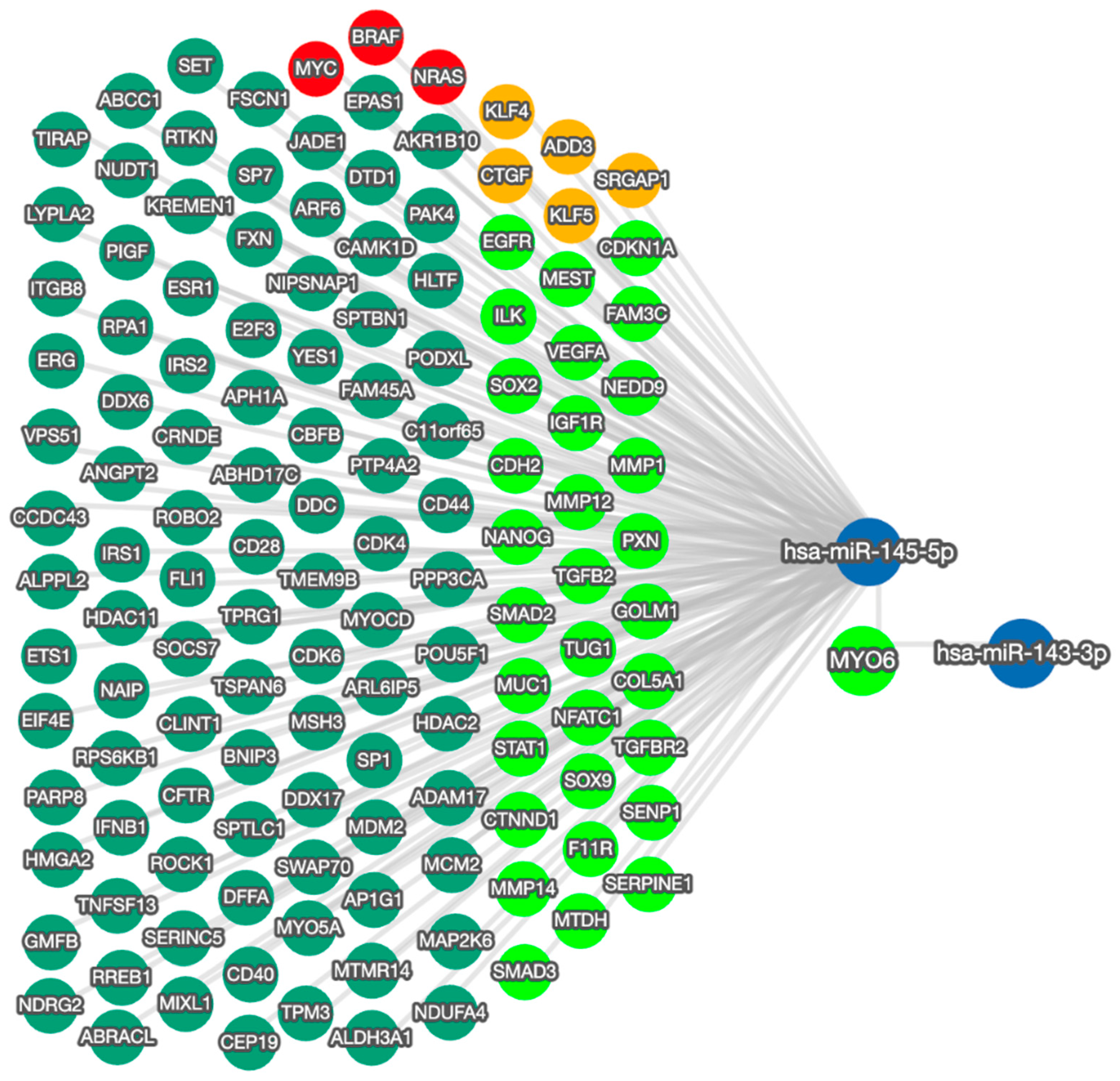
The effects on cell behavior caused by the manipulation of miR-145-5p reflect the function of MYO6 and the wider regulatory network (Figure 5). The bidirectional network highlights the capacity of miR-145-5p to regulate multiple EMT-associated targets. By modulating EMT through multiple molecular targets and pathways, miR-145-5p can clearly exert greater overall impact than through MYO6 alone. Moreover, miR-145-5p regulates several critical oncogenes including MYC, BRAF, and NRAS, which are known promoters of prostate cancer cell proliferation and clonogenic potential when aberrantly activated.
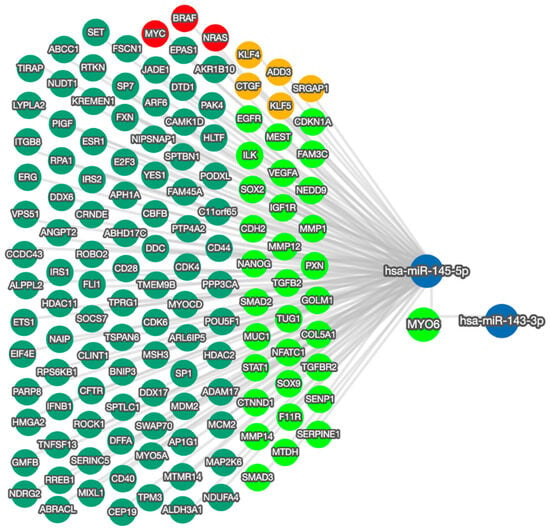
Figure 5.
miRTargetLink 2.0 visualization of miR-145-5p and MYO6 bidirectional network interactions. miR-145-5p and MYO6 were input items, while the connected nodes are gene/miRNA interactions validated by strong experimental evidence (qRT-PCR, Western blot, Luciferase reporter assay). Blue nodes: miRNAs; green nodes: genes; bright green nodes: EMT-associated genes; red nodes: genes significantly associated with prostate cancer; orange nodes: actin cytoskeleton modulators.
2.6. Potential of miR-145-5p as a Biomarker of Prostate Cancer
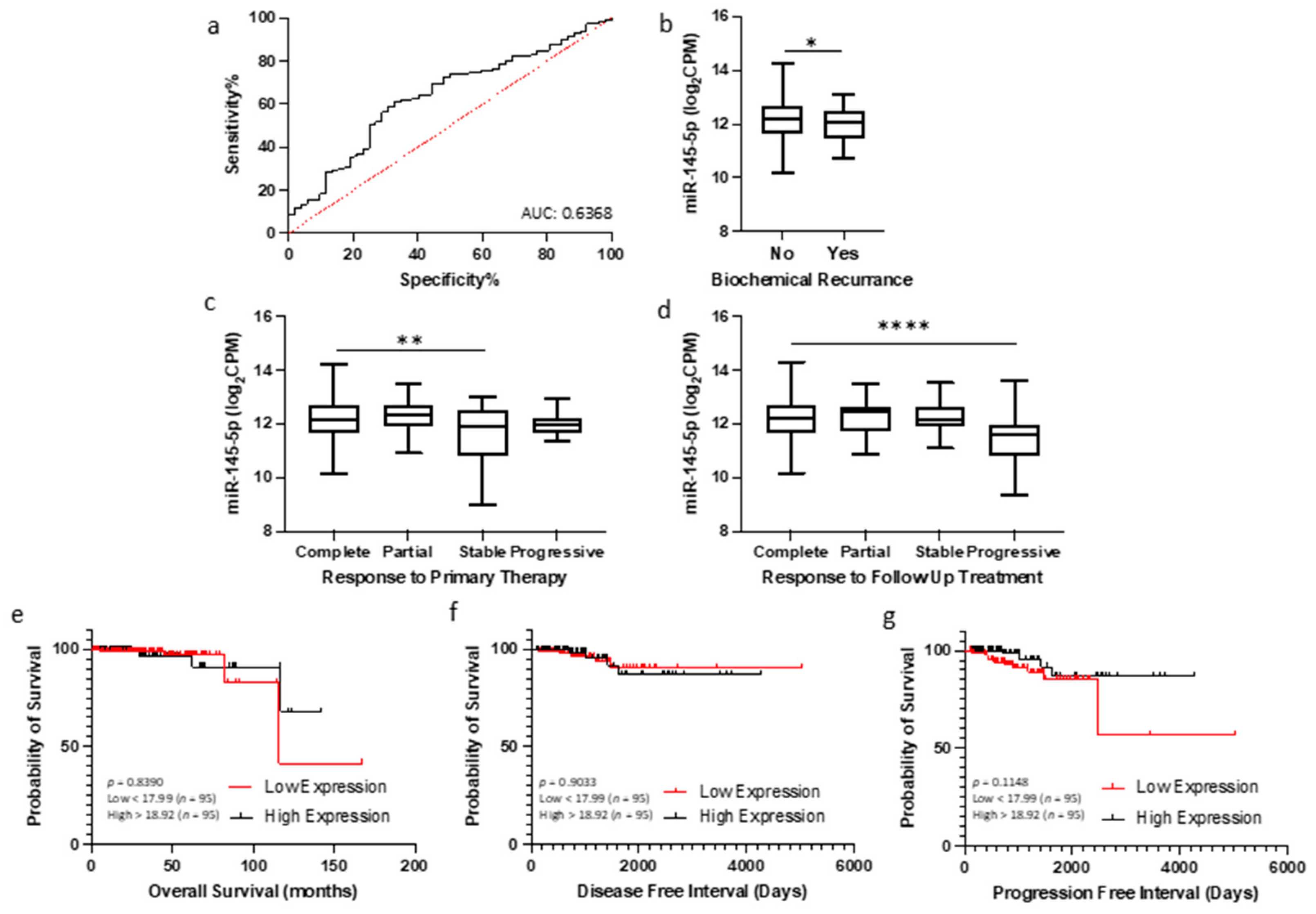
Given the association between miR-145-5p levels and prostate cancer progression observed in clinical datasets, we hypothesized that quantification of miR-145-5p may have utility as a diagnostic or prognostic biomarker for prostate cancer. To investigate this, we analyzed miR-145-5p expression and clinical outcome data from TCGA PRAD cohort. Receiver operating characteristic (ROC) curve analysis demonstrated that miR-145-5p expression can effectively discriminate between tumor and normal prostate tissue samples in this cohort (Figure 6a). A combination panel of five miRNAs (miR-221-3p, miR-222-3p, miR-133b, miR-143-3p, and miR-145-5p) improved AUC to 0.977 (Supplemental Figure S9). Further, patients exhibiting lower miR-145-5p expression showed significantly shorter biochemical recurrence-free survival after initial prostate cancer treatment, as assessed by log-rank test (Figure 6b). Conversely, higher miR-145-5p levels correlated with improved initial and follow-up therapeutic response (Figure 6c,d). For survival analyses, the TCGA PRAD cohort was stratified into quartiles by miR-145-5p expression level (Low < 17.99, High > 18.92 log2CPM). Kaplan–Meier plots revealed no significant difference in overall survival, disease-free and progression free intervals (Figure 6e–g), likely due to the limited number of deaths in this cohort. High expression of the miR-145-5p target MYO6 was associated with reduced disease-free survival (Supplementary Figure S7) but did not meet significance. In summary, our analyses indicate miR-145-5p is a potential diagnostic marker and prognostic indicator of biochemical recurrence in prostate cancer.
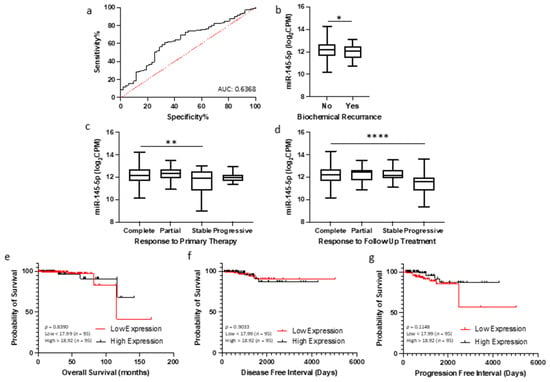
Figure 6.
miR-145-5p as a prognostic biomarker for prostate cancer. (a) ROC curve analysis demonstrating that miR-145-5p shows potential for distinguishing between tumor and normal tissue in TCGA PRAD patient cohort. (b) Patients experiencing biochemical recurrence (BCR) showed significantly lower levels of miR-145-5p compared to those with no recurrence (p-values generated by unpaired two-tailed t-test; n, no recurrence = 407, recurrence = 61). (c) Significant difference in miR-145-5p levels between patient remission response after primary therapy (n, complete = 330, partial = 30, stable = 29, progressive = 23) and (d) follow up therapy (n, complete = 340, partial = 17, stable = 42, progressive = 24)(* p < 0.05, ** p < 0.01, **** p < 0.0001). For KM plots, patients were divided into quartiles based on miR-145-5p expression for (e) Overall survival, (f) Disease-free interval and (g) Progression-free interval. (p-values generated by log-rank (Mantel–Cox) test). ROC = Receiver operating characteristic. AUC = Area Under Curve. n = number.
Furthermore, since loss of miR-145-5p expression has been consistently linked with many other cancers, it was no surprise to find high AUC values for miR-145-5p in multiple TCGA patient cohorts, suggesting the diagnostic potential of miR-145-5p for detecting cancer (Supplementary Figure S8). Moreover, combination of miR-145-5p with other miRNAs increased the discriminatory power for detecting prostate cancer (Supplementary Figure S9). Similarly, the expression levels of both miR-145-5p and MYO6 in tumor tissue is significantly correlated with survival outcomes in several other TCGA patient cohorts, indicating that they could together be useful prognostic biomarkers for different cancers (Supplementary Table S1).
3. Discussion
Previous studies have explored the function of miR-145-5p in various cancer types and signaling pathways [52,53,54,55,56], but its contribution to EMT in prostate cancer has not been thoroughly characterized. Therefore, we aimed to further investigate the relationship between miR-145-5p and EMT in prostate cancer in this study. This is the first report demonstrating that downregulation of miR-145-5p can promote prostate cancer development through targeting MYO6 and regulating EMT.
We first established that miR-145-5p expression was significantly reduced in prostate cancer cell lines and clinical samples compared to normal controls (Figure 1a,b). Further analysis of clinical samples showed that lower miR-145-5p levels were associated with more advanced cancer stage, as determined by clinicopathological parameters (Figure 1c–f). These findings are consistent with previous studies which found that lower levels of miR-145-5p expression are associated with poorer patient prognosis and treatment response [57,58].
Functional enrichment analysis further validated the link between miR-145-5p and target genes associated with prostate cancer development (Table 1) and EMT (Table 2). Together, these results provide evidence that loss of miR-145-5p is detrimental in prostate cancer, as it leads to reduced regulation of EMT and other critical cellular networks [42,48,49]. However, very few studies have demonstrated a functional connection between miR-145-5p and EMT in prostate cancer thus far. Therefore, we aimed to identify a previously unreported EMT-related target of miR-145-5p in this disease.
By cross-referencing three databases, we strategically filtered potential gene targets for further study. We identified MYO6 as the most promising candidate to investigate, since it appeared in our functional enrichment analyses and appears in multiple EMT gene databases, such as dbEMT2 and EMTome. We experimentally confirmed MYO6 as a miR-145-5p target in vitro in three prostate cell lines, showing that miR-145-5p overexpression significantly reduced MYO6 protein and mRNA levels (Figure 2). We corroborated this in vitro data by analyzing TCGA PRAD data, which revealed a significant negative correlation between miR-145-5p and MYO6 levels, as expected for a direct target interaction. We propose that the upregulation of MYO6 expression observed in tumor versus normal prostate tissue is due, at least in part, to downregulation of miR-145-5p.
Taken together, these results suggest that MYO6 is a direct target of miR-145-5p in prostate cells, as previously shown in gastric cancer [57] and 293T kidney cells [58]. This is significant because others have demonstrated how loss of miR-145-5p leading to increased MYO6 expression can promote EMT and uncontrolled cell growth [52,59]. The MYO6 gene encodes a protein called myosin VI, which is a motor protein involved in intracellular transport and actin cytoskeleton organization [60] Myosin VI regulates the cellular trafficking of cell surface receptors, as well as the adhesion molecules involved in cell–cell and cell-matrix interactions, both of which are critical for maintaining the integrity of epithelial cell layers [61]. Disruption of these interactions is a hallmark of EMT, which may be influenced by over-expression of myosin VI. Myosin VI has also been implicated in the regulation of EMT-associated signaling pathways, such as the TGF-β pathway, thereby presenting another mechanism for myosin VI expression to influence the development of EMT [62].
We confirmed this link by showing that modulating miR-145-5p levels significantly altered expression of EMT marker genes in prostate cells (Figure 3). Furthermore, we demonstrated that inhibiting miR-145-5p markedly increased proliferation, migration, and clonogenicity of prostate cells, while overexpression had the opposite effects (Figure 4). These results further illustrate why miR-145-5p loss is deleterious in prostate cancer, as it enhances the capacity for uncontrolled cell growth. Regarding potential therapeutic applications, our data also show that restoring miR-145-5p levels in prostate cancer cells, thereby reducing MYO6 levels, could be a viable strategy to suppress EMT and inhibit cell growth. Targeting MYO6 has been proposed by others as a potential therapeutic approach in gastric [63] and breast [64] cancers to suppress tumor proliferation and metastasis. A prior study revealed that concurrent overexpression of miR-145-5p and miR-143-3p, a member of the miR-145-5p family, synergistically suppressed MYO6 expression in gastric cancer, exhibiting greater inhibitory effects than miR-145-5p alone [57]. Others have shown that modulation of miR-145-5p expression in prostate cancer cells may be a strategy to confer chemosensitivity [43,44]. However, the effective delivery of small RNA-delivering techniques to tumors remains a considerable barrier to clinical use of this type of targeted therapeutic strategy.
However, it would be overly simplistic to attribute the effects on cell behavior solely to the miR-145-5p/MYO6 interaction. This axis resides at the center of an intricate network of interactions (Figure 5), so the combined effects of these connections ultimately determine miR-145-5p impact on cell phenotype. Prior prostate cancer studies have also shown miR-145-5p can inhibit cell growth via other targets, including WIP1 [45], PLD5 [65], and SOX2 [39]. Other validated oncogenic targets of miR-145-5p include MYC [66,67], BRAF [68] and NRAS [69,70]. While our data suggests miR-145-5p regulation of MYO6 exerts a similar effect in prostate cancer cells, its effect on other targets is also important to acknowledge.
For example, other studies have identified different miR-145-5p targets mediating various cellular effects in different cancer types. A range of in vitro experiments have consistently demonstrated that miR-145-5p can reduce proliferation, invasion, and migration in thyroid cancer [71], non-small-cell lung cancer [72], lung adenocarcinoma [73], liver cancer [74], pancreatic cancer [75], colorectal cancer [76], gastric cancer [77], bladder cancer [78], renal cell carcinoma [79], urothelial cancer [80], breast cancer [81], ovarian cancer [82], chondrosarcoma [83], pleural mesothelioma [84], glioma [85] and invasive glioblastoma [86]. All this evidence points towards the crucial role of miR-145-5p in cancer development and progression, throughout its network of targets.
Given the consistent downregulation of miR-145-5p in tumor tissues, we were interested in its potential utility as a clinically useful biomarker. In general, miRNAs are attractive biomarker candidates because they are much more stably preserved in clinical samples compared to mRNAs and can be readily detected using sensitive and specific PCR-based assays [87]. However, identifying the optimal miRNAs to use for a given disease remains challenging. Our data suggest miR-145-5p expression profiling may be a valuable diagnostic biomarker, as it showed significant ability to distinguish between normal and tumor prostate tissues (Figure 6a). Additionally, a biomarker predicting biochemical recurrence (BCR) following prostate cancer treatment would be clinically useful. BCR is typically defined as a rise in prostate-specific antigen after surgery or radiation, indicating potential tumor regrowth. Here, we found patients experiencing BCR had significantly lower miR-145-5p levels, suggesting it may help predict treatment response (Figure 6b). This corroborates previous work proposing miR-145-5p as a potential prostate cancer biomarker. One study evaluated plasma miR-145-5p as a noninvasive biomarker for prostate cancer screening and diagnosis [88]. The expression of miR-145-5p was measured in 170 patients undergoing prostate biopsy. miR-145-5p levels significantly differed between benign, precancerous, and cancerous prostate pathologies. Multivariate analysis showed miR-145-5p could distinguish between patient groups when combined with clinical assessments. Given our analyses, we agree with the authors’ conclusion that miR-145-5p is a promising plasma biomarker for prostate cancer detection and diagnosis when used with other clinical information. However, further validation on different sample types (e.g., tissue, plasma, serum, urine), is needed in larger, independent patient cohorts.
Others have evaluated miR-145-5p as a valuable adjunct to the Gleason grading system [56]. Due to the heterogeneous and often limited nature of prostate biopsy samples, the Gleason scoring system has inherent variability and limitations in predicting prognosis, even among patients receiving the same Gleason score [89,90]. This study found prostate cancer biopsies that were initially scored Gleason 6 and later upgraded to Gleason 7 after surgery had lower expression of miR-145-5p. The differences in miR-145-5p expression suggests an underlying biological distinction between indolent versus aggressive cancer that appear identical on standard histopathology. Again, this demonstrates the potential value of adding new measurements to traditional disease parameters which could improve clinical decision-making about individual patients.
Stronger evidence from other cancers also shows how measuring miR-145-5p expression in various sample types holds promise as a diagnostic and/or prognostic biomarker for breast [91], lung [92], endometrial [93], glioblastoma [94], gastric [95], cervical [96] and urothelial [97] carcinomas. Together with our data, this evidence base warrants further investigation of miR-145-5p as a standalone or adjuvant biomarker for cancer, including prostate cancer. Critically, standardized approaches are fundamental for robust clinical evaluation of miRNA biomarkers [13].
Nonetheless, miR-145-5p alone likely lacks sufficient specificity and sensitivity to serve as a clinically useful biomarker. Instead, it would likely be included in a multivariate biomarker panel with other carefully selected genomic and proteomic markers. We have previously demonstrated the superiority of multivariate biomarker panels over individual markers for prostate cancer diagnosis and prognosis [98,99]. The synergistic combination of multiple miRNAs into a biomarker panel provides greater sensitivity and specificity than any individual miRNA biomarker alone (Supplementary Figure S9). Given the miR-145-5p network highlighted here (Figure 5), a panel emphasizing EMT prediction could incorporate additional related genes or proteins, especially MYO6. In fact, current prostate cancer risk prediction models like STHLM3 incorporate various genomic, proteomic and clinical variables [100]. Our findings suggest miR-145-5p may be a valuable addition to such models. Considering its importance across cancers, miR-145-5p could have broad utility as a cancer biomarker [101].
Ultimately, clinical acceptance of miRNA biomarkers requires evidence that they improve patient management. The work described here is in part due to the need to have further understanding of miRNA biological functions in order to identify the most useful candidates, individually and in combination. Further obstacles to clinical adoption of miRNA biomarkers include concerns around reproducibility, lack of standardization across studies, and insufficient validation in large patient cohorts. Overcoming these barriers necessitates robust research, stringent validation protocols, and close collaboration between researchers, clinicians, and regulatory bodies. It is also important to acknowledge prostate tumor heterogeneity, as profiling is not cell-specific and the miR-145-5p/MYO6 relationship may be more significant in certain cell populations. Therefore, further work in this area is likely to incorporate single-cell analysis and advanced proteomics could provide insights into the cell-specific role of miR-145-5p to inform more precise diagnostics and targeted therapies [102,103].
4. Materials and Methods
4.1. Cell Culture and Transfections
All cell lines were acquired from the American Type Culture Collection (ATCC, Rockville, MD, USA). An in-house genotyping service authenticated the cells, and they were verified to be mycoplasma-free (InvivoGen, Toulouse, France). Cells utilized at a low passage number ranging from 3 to 6. RWPE-1, a normal prostate epithelial cell line, was cultured in keratinocyte growth medium, supplemented with 5 ng/mL of human recombinant epidermal growth factor and 0.05 mg/mL of bovine pituitary extract. (Life Technologies, Paisley, UK). The human prostate cancer cell lines, DU145 and PC3, were cultivated in RPMI-1640 medium, enriched with 10% fetal bovine serum and L-glutamine (Life Technologies). The cells were maintained at 37 °C under a humidified atmosphere of 95% air and 5% CO2. For miRNA transfections, in a 6-well plate, 1 × 105 cells were seeded per well to ensure ~80% confluency at collection. Cells were transfected with 25 nM miR-145-5p precursor (hsa-miR-145-5p miRCURY LNA miRNA Mimic; ID YM0047001), miR-145-5p inhibitor (anti-miR-145-5p; ID YI04102423), or a 25 nM non-targeting negative control (Negative Control A; ID YI00199006) (Qiagen, Manchester, UK) using Lipofectamine 2000 (Life Technologies) for 4 to 6 h then replaced with culture media. After 72 h, cells were collected for RNA or protein extraction. Pre-miR-145-5p (double-stranded RNA molecule designed to mimic endogenous mature miR-145-5p) and anti-miR-145-5p (single stranded oligonucleotide designed to specifically bind and inhibit endogenous miR-145-5p) are chemically modified molecules designed upon the mature miR-145-5p sequence GUCCAGUUUUCCCAGGAAUCCCU.
4.2. Quantitative Real-Time PCR (qRT-PCR)
Total RNA was isolated from cell lines using the miRNeasy Tissue/Cells Advanced Mini Kit (Qiagen, Manchester, UK) following the manufacturer’s protocol. RNA integrity was verified by a NanoDropTM 2000 spectrophotometer (ThermoFisher Scientific, Waltham, MA, USA). First strand cDNA was synthesized from 500 ng total RNA using the Transcriptor First Strand cDNA Synthesis Kit (Roche, Sussex, UK) and random primers according to the manufacturer’s instructions. Quantitative PCR was performed using FastStart SYBR Green Master (Roche) on a Roche LC480 LightCycler with primer sets for:
- MYO6 (fw: GGATCTGTCCGAGCAGGAAG, rv: CTGTACGGGTGAAGCTGGAG),
- ACTA2 (fw: GTTCCGCTCCTCTCTCCAAC, rv: GTGCGGACAGGAATTGAAGC),
- VIM (fw: GGACCAGCTAACCAACGACA, rv: AAGGTCAAGACGTGCCAGAG),
- FN1 (fw: TCAGCTTCCTGGCACTTCTG, rv: TCCCTGGGGATGTGACCAAT), and housekeeping gene GAPDH (fw: GACAGTCAGCCGCATCTTCT, rv: GCGCCCAATACGACCAAATC).
Gene expression was normalized to GAPDH. The data presented were generated from at least three independent biological replicates. Quantitative reverse transcription PCR (qRT-PCR) for miR-145-5p was performed using the miRCURY LNA miRNA PCR Assays system (Qiagen). A total of 20 ng of template RNA was used in each first strand cDNA synthesis reaction. PCR amplification was carried out for 40 cycles and fluorescence was monitored using the Roche LC480 LightCycler. Normalization was performed against the snRNA U6 housekeeping gene. All qRT-PCR miRNA data were generated from a minimum of three independent biological replicates.
4.3. Protein Analysis
Protein extraction was performed using Cell Lysis Buffer (Abcam, Cambridge, UK) supplemented with 2% v/v HaltTM Protease Inhibitor Cocktail (ThermoFisher Scientific). Western blotting was conducted using Bio-Rad mini-Protean TGX gels and Trans-Blot® Turbo Transfer System with associated reagents (Bio-Rad, Watford, UK). Primary antibodies used were rabbit anti-MYO6 and mouse anti-GAPDH as a loading control (both from Proteintech, Manchester, UK). Membranes were blocked with 5% milk in TBS-T (0.05%), followed by incubation with the appropriate horseradish peroxidase (HRP)-conjugated secondary antibody (goat anti-rabbit IgG-HRP at 1:5000 dilution or goat anti-mouse IgG-HRP at 1:5000 dilution, both from Proteintech). Chemiluminescent signal was detected using enhanced chemiluminescent reagent (ThermoFisher Scientific) and imaged on a G:BOX F3 system (Syngene, Cambridge, UK). A minimum of three biological replicates were performed for each experiment.
4.4. Bioassays
For proliferation assays, transfected cells were replated at 0.01 × 106 cells per well. After 24 h, 10% v/v alamarBlueTM Cell Viability Reagent was added and incubated at 37 °C in a humidified atmosphere of 95% air and 5% CO2 for 4 h. Following incubation, absorbance was measured at 570 nm and 600 nm using a FLUOstar Omega plate reader (BMG LabTech, Aylesbury, UK). For migration assays, transfected cells were replated at 0.25 × 106 cells per well and allowed to form a confluent monolayer. A 200 μL pipette tip was used to create an artificial “wound”. Wound images were captured at 0 and 72 h. Percentage closure was calculated as ((initial gap area − remaining gap area)/initial gap area) × 100. For colony formation assays, transfected cells were replated at 0.03 × 106 cells per well and incubated for 72 h at 37 °C in a humidified atmosphere of 95% air and 5% CO2. Cells were quantified using the cell counting function in ImageJ Version 1.54h software. Fold change was calculated relative to the non-transfected control (NTC).
4.5. Databases and Analysis
The prostate adenocarcinoma (PRAD) data from The Cancer Genome Atlas (TCGA) repository was obtained from the Genomic Data Commons Data Portal v39.0 (https://portal.gdc.cancer.gov/, accessed on 10 January 2024). Expression analysis of miR-145-5p and correlation with clinical parameters were performed using the University of California Santa Cruz (UCSC) Xena Functional Genomics Explorer (https://xenabrowser.net/, accessed on 12 January 2024) [104]. Identification of negatively correlated targets and functional enrichment analysis were conducted using CancerMIRNome incorporating clusterProfiler 1.0 (http://bioinfo.jialab-ucr.org/CancerMIRNome/, accessed on 28 July 2023) [105,106]. Epithelial-mesenchymal transition (EMT)-associated targets of miR-145-5p were determined by integrating data from miRTarBase v9.0 (http://mirtarbase.cuhk.edu.cn/, accessed on 14 February 2023) [107], the EMT gene database dbEMT 2.0 (http://dbemt.bioinfo-minzhao.org/, accessed on 16 February 2023) [108], EMTome (http://www.emtome.org/, accessed on 28 July 2023) [109], and Venny 2.0 (https://bioinfogp.cnb.csic.es/tools/venny/, accessed on 28 July 2023) [110] tools. The protein–protein interaction network of MYO6 was generated using STRING v12.0 (https://string-db.org/, accessed on 24 March 2023) [111] and functionally annotated using Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. Additional survival analyses were performed with the Kaplan–Meier Plotter (KM-Plotter) (http://kmplot.com/analysis/, accessed on 25 June 2023) [112]. Network analysis and visualization were conducted using GeneMANIA (https://genemania.org/, accessed on 21 April 2023) [113] and miRTargetLink 2.0 (http://ccbcompute.cs.uni-saarland.de/mirtargetlink2, accessed on 22 April 2023) [114]. Combination biomarker panels accessed using CombiROC (http://CombiROC.eu, accessed on 21 April 2023) [115]. Hallmark enrichment analysis performed with CancerHallmark Tool (https://cancerhallmarks.com/, accessed on 10 March 2024).
4.6. Statistics
Graphs were generated using GraphPad PRISM version 9. Unless otherwise stated, bar graphs show mean ± standard error of at least three biological replicates, with statistical significance evaluated by paired t-test. All boxplots show mean and Tukey whiskers, with statistical significance determined by unpaired t-test with Welch’s correction or nonparametric Kruskal–Wallis one-way ANOVA with Dunn’s multiple comparisons test. Statistical significance for scatterplots was evaluated by Pearson’s correlation with p-values adjusted for multiple hypothesis testing. For Kaplan–Meier graphs, statistical significance was determined by log-rank (Mantel-Cox) test. To correct for multiple hypotheses in functional enrichment tables, adjusted p-values were calculated using the Benjamini-Hochberg procedure. For all analyses, data were considered statistically significant at * p < 0.05, ** p < 0.01, *** p < 0.001, and **** p < 0.0001.
5. Conclusions
Our findings demonstrate that miR-145-5p expression is decreased in prostate cancer and correlates with indicators of disease advancement. To our knowledge, this is the first evidence that miR-145-5p directly regulates MYO6 in prostate cancer cells. We hypothesize that declining miR-145-5p levels may promote EMT in prostate cancer. Further research should explore the miR-145-5p/MYO6 interaction and its role in EMT. MiR-145-5p has potential as a diagnostic or prognostic biomarker for prostate cancer, warranting additional investigation.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms25084301/s1.
Author Contributions
Conceptualization, L.A. and D.J.M.; methodology, L.A. and D.J.M.; formal analysis, L.A. and D.J.M.; data curation, L.A. and D.J.M.; writing—original draft preparation, L.A.; writing—review and editing, L.A., C.E.W. and D.J.M.; visualization, L.A. and D.J.M.; supervision, C.E.W. and D.J.M.; project administration, C.E.W. and D.J.M.; funding acquisition, C.E.W. and D.J.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Department for the Economy (DfE), Northern Ireland as a PhD studentship secured by D.J.M. and C.E.W., and awarded to L.A.
Institutional Review Board Statement
Ethical review and approval were waived for this study due to the fact that only publicly available data and materials were used in this study.
Informed Consent Statement
Patient consent was waived due to the retrospective nature of this study and the use of de-identified data.
Data Availability Statement
The genotypic and phenotypic data for Prostate adenocarcinoma (PRAD) cohort are available at The Cancer Genome Atlas (TCGA) portal. Analysis tools are listed in Methods and other datasets analyzed in the present study are available from the published papers that have been cited in this manuscript.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Rebello, R.J.; Oing, C.; Knudsen, K.E.; Loeb, S.; Johnson, D.C.; Reiter, R.E.; Gillessen, S.; Van der Kwast, T.; Bristow, R.G. Prostate cancer. Nat. Rev. Dis. Primer 2021, 7, 9. [Google Scholar] [CrossRef]
- Khan, M.I.; Hamid, A.; Adhami, V.M.; Lall, R.K.; Mukhtar, H. Role of Epithelial Mesenchymal Transition in Prostate Tumorigenesis. Curr. Pharm. Des. 2015, 21, 1240–1248. [Google Scholar] [CrossRef]
- Kalluri, R.; Weinberg, R.A. The basics of epithelial-mesenchymal transition. J. Clin. Investig. 2009, 119, 1420–1428. [Google Scholar] [CrossRef]
- Hussen, B.M.; Shoorei, H.; Mohaqiq, M.; Dinger, M.E.; Hidayat, H.J.; Taheri, M.; Ghafouri-Fard, S. The Impact of Non-coding RNAs in the Epithelial to Mesenchymal Transition. Front. Mol. Biosci. 2021, 8, 665199. Available online: https://www.frontiersin.org/articles/10.3389/fmolb.2021.665199 (accessed on 29 June 2023). [CrossRef] [PubMed]
- Shang, R.; Lee, S.; Senavirathne, G.; Lai, E.C. microRNAs in action: Biogenesis, function and regulation. Nat. Rev. Genet. 2023, 24, 816–833. [Google Scholar] [CrossRef]
- Dexheimer, P.J.; Cochella, L. MicroRNAs: From Mechanism to Organism. Front. Cell Dev. Biol. 2020, 8, 409. Available online: https://www.frontiersin.org/articles/10.3389/fcell.2020.00409 (accessed on 24 January 2024). [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 388354. Available online: https://www.frontiersin.org/articles/10.3389/fendo.2018.00402 (accessed on 29 June 2023). [CrossRef]
- Orang, A.V.; Safaralizadeh, R.; Kazemzadeh-Bavili, M. Mechanisms of miRNA-Mediated Gene Regulation from Common Downregulation to mRNA-Specific Upregulation. Int. J. Genom. 2014, 2014, 970607. [Google Scholar] [CrossRef]
- Schitcu, V.H.; Raduly, L.; Nutu, A.; Zanoaga, O.; Ciocan, C.; Munteanu, V.C.; Cojocneanu, R.; Petrut, B.; Coman, I.; Braicu, C.; et al. MicroRNA Dysregulation in Prostate Cancer. Pharmacogenom. Pers. Med. 2022, 15, 177–193. [Google Scholar] [CrossRef]
- Lynch, S.M.; O’Neill, K.M.; McKenna, M.M.; Walsh, C.P.; McKenna, D.J. Regulation of miR-200c and miR-141 by Methylation in Prostate Cancer. Prostate 2016, 76, 1146–1159. [Google Scholar] [CrossRef]
- Lynch, S.M.; McKenna, M.M.; Walsh, C.P.; McKenna, D.J. miR-24 regulates CDKN1B/p27 expression in prostate cancer. Prostate 2016, 76, 637–648. [Google Scholar] [CrossRef] [PubMed]
- Angel, C.Z.; Lynch, S.M.; Nesbitt, H.; McKenna, M.M.; Walsh, C.P.; McKenna, D.J. miR-210 is induced by hypoxia and regulates neural cell adhesion molecule in prostate cells. J. Cell. Physiol. 2020, 235, 6194–6203. [Google Scholar] [CrossRef] [PubMed]
- Stafford, M.Y.C.; Willoughby, C.E.; Walsh, C.P.; McKenna, D.J. Prognostic value of miR-21 for prostate cancer: A systematic review and meta-analysis. Biosci. Rep. 2022, 42, BSR20211972. [Google Scholar] [CrossRef] [PubMed]
- Stafford, M.Y.C.; McKenna, D.J. MiR-182 Is Upregulated in Prostate Cancer and Contributes to Tumor Progression by Targeting MITF. Int. J. Mol. Sci. 2023, 24, 1824. [Google Scholar] [CrossRef]
- Armstrong, L.; Willoughby, C.E.; McKenna, D.J. Targeting of AKT1 by miR-143-3p Suppresses Epithelial-to-Mesenchymal Transition in Prostate Cancer. Cells 2023, 12, 2207. [Google Scholar] [CrossRef] [PubMed]
- Sekhon, K.; Bucay, N.; Majid, S.; Dahiya, R.; Saini, S. MicroRNAs and epithelial-mesenchymal transition in prostate cancer. Oncotarget 2016, 7, 67597–67611. [Google Scholar] [CrossRef]
- Kadkhoda, S.; Ghafouri-Fard, S. Function of miRNA-145–5p in the pathogenesis of human disorders. Pathol.-Res. Pract. 2022, 231, 153780. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, H.; Situ, J.; Li, M.; Sun, H. KCNQ1OT1 aggravates cell proliferation and migration in bladder cancer through modulating miR-145-5p/PCBP2 axis. Cancer Cell Int. 2019, 19, 325. [Google Scholar] [CrossRef]
- Dong, M.; Xu, T.; Li, H.; Li, X. LINC00052 promotes breast cancer cell progression and metastasis by sponging miR-145-5p to modulate TGFBR2 expression. Oncol. Lett. 2021, 21, 368. [Google Scholar] [CrossRef]
- He, W.; Liang, B.; Wang, C.; Li, S.; Zhao, Y.; Huang, Q.; Liu, Z.; Yao, Z.; Wu, Q.; Liao, W.; et al. MSC-regulated lncRNA MACC1-AS1 promotes stemness and chemoresistance through fatty acid oxidation in gastric cancer. Oncogene 2019, 38, 4637–4654. [Google Scholar] [CrossRef]
- He, J.; Yan, H.; Wei, S.; Chen, G. LncRNA ST8SIA6-AS1 Promotes Cholangiocarcinoma Progression by Suppressing the miR-145-5p/MAL2 Axis. OncoTargets Ther. 2021, 14, 3209–3223. [Google Scholar] [CrossRef] [PubMed]
- Liep, J.; Kilic, E.; Meyer, H.A.; Busch, J.; Jung, K.; Rabien, A. Cooperative Effect of miR-141-3p and miR-145-5p in the Regulation of Targets in Clear Cell Renal Cell Carcinoma. PLoS ONE 2016, 11, e0157801. [Google Scholar] [CrossRef] [PubMed]
- Kadkhoda, S.; Taslimi, R.; Noorbakhsh, F.; Darbeheshti, F.; Bazzaz, J.T.; Ghafouri-Fard, S.; Shakoori, A. Importance of Circ0009910 in colorectal cancer pathogenesis as a possible regulator of miR-145 and PEAK1. World J. Surg. Oncol. 2021, 19, 265. [Google Scholar] [CrossRef] [PubMed]
- Mei, L.-L.; Wang, W.-J.; Qiu, Y.-T.; Xie, X.-F.; Bai, J.; Shi, Z.-Z. miR-145-5p Suppresses Tumor Cell Migration, Invasion and Epithelial to Mesenchymal Transition by Regulating the Sp1/NF-κB Signaling Pathway in Esophageal Squamous Cell Carcinoma. Int. J. Mol. Sci. 2017, 18, 1833. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.; Chen, P.; Li, S. miR-145-5p Inhibits the Proliferation, Migration, and Invasion of Esophageal Carcinoma Cells by Targeting ABRACL. BioMed Res. Int. 2021, 2021, e6692544. [Google Scholar] [CrossRef] [PubMed]
- Goeppert, B.; Truckenmueller, F.; Ori, A.; Fritz, V.; Albrecht, T.; Fraas, A.; Scherer, D.; Silos, R.G.; Sticht, C.; Gretz, N.; et al. Profiling of gallbladder carcinoma reveals distinct miRNA profiles and activation of STAT1 by the tumor suppressive miRNA-145-5p. Sci. Rep. 2019, 9, 4796. [Google Scholar] [CrossRef] [PubMed]
- Zhou, K.; Song, B.; Wei, M.; Fang, J.; Xu, Y. MiR-145-5p suppresses the proliferation, migration and invasion of gastric cancer epithelial cells via the ANGPT2/NOD_LIKE_RECEPTOR axis. Cancer Cell Int. 2020, 20, 416. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Chen, T.; Zhu, Y.; Li, Y.; Zhang, Y.; Wang, Y.; Li, X.; Xie, X.; Wang, J.; Huang, M.; et al. circPTN sponges miR-145-5p/miR-330-5p to promote proliferation and stemness in glioma. J. Exp. Clin. Cancer Res. 2019, 38, 398. [Google Scholar] [CrossRef] [PubMed]
- Ding, B.; Fan, W.; Lou, W. hsa_circ_0001955 Enhances In Vitro Proliferation, Migration, and Invasion of HCC Cells through miR-145-5p/NRAS Axis. Mol. Ther.-Nucleic Acids 2020, 22, 445–455. [Google Scholar] [CrossRef]
- Gao, W.; Zhang, C.; Li, W.; Li, H.; Sang, J.; Zhao, Q.; Bo, Y.; Luo, H.; Zheng, X.; Lu, Y.; et al. Promoter Methylation-Regulated miR-145-5p Inhibits Laryngeal Squamous Cell Carcinoma Progression by Targeting FSCN1. Mol. Ther. 2019, 27, 365–379. [Google Scholar] [CrossRef]
- Bagheri, M.; Khansarinejad, B.; Mosayebi, G.; Moradabadi, A.; Mondanizadeh, M. Diagnostic Value of Plasma miR-145 and miR-185 as Targeting of the APRIL Oncogene in the B-cell Chronic Lymphocytic Leukemia. Asian Pac. J. Cancer Prev. APJCP 2021, 22, 111–117. [Google Scholar] [CrossRef]
- Jiang, W.; Zhang, C.; Kang, Y.; Li, G.; Feng, Y.; Ma, H. The roles and mechanisms of the circular RNA circ_104640 in early-stage lung adenocarcinoma: A potential diagnostic and therapeutic target. Ann. Transl. Med. 2021, 9, 138. [Google Scholar] [CrossRef] [PubMed]
- Mataki, H.; Seki, N.; Mizuno, K.; Nohata, N.; Kamikawaji, K.; Kumamoto, T.; Koshizuka, K.; Goto, Y.; Inoue, H. Dual-strand tumor-suppressor microRNA-145 (miR-145-5p and miR-145-3p) coordinately targeted MTDH in lung squamous cell carcinoma. Oncotarget 2016, 7, 72084. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.J.; Liu, S.; Han, D.M.; Han, D.Z.; Sun, W.J.; Zhao, X.C.; Liang, J.Q.; Yu, L. FUT8-AS1 Inhibits the Malignancy of Melanoma Through Promoting miR-145-5p Biogenesis and Suppressing NRAS/MAPK Signaling. Front. Oncol. 2021, 10, 586085. Available online: https://www.frontiersin.org/articles/10.3389/fonc.2020.586085 (accessed on 24 January 2024). [CrossRef] [PubMed]
- Zhu, Z.; Wu, Q.; Zhang, M.; Tong, J.; Zhong, B.; Yuan, K. Hsa_circ_0016760 exacerbates the malignant development of non-small cell lung cancer by sponging miR-145-5p/FGF5. Oncol. Rep. 2021, 45, 501–512. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Pan, R.; Lu, Q.; Ren, C.; Sun, J.; Wu, H.; Wen, J.; Chen, H. MicroRNA-145-5p inhibits osteosarcoma cell proliferation by targeting E2F transcription factor 3. Int. J. Mol. Med. 2020, 45, 1317–1326. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Yang, J.; Gong, F.; Li, L.; Li, A. Long non-coding RNA CASC9 promotes the progression of retinoblastoma via interacting with miR-145-5p. Cell Cycle 2020, 19, 2270–2280. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Zhou, Q.; Yi, H.; Ma, S.; Li, D.; Xu, Y.; Wang, J.; Yin, S. A novel lncRNA n384546 promotes thyroid papillary cancer progression and metastasis by acting as a competing endogenous RNA of miR-145-5p to regulate AKT3. Cell Death Dis. 2019, 10, 433. [Google Scholar] [CrossRef] [PubMed]
- Ozen, M.; Karatas, O.F.; Gulluoglu, S.; Bayrak, O.F.; Sevli, S.; Guzel, E.; Ekici, I.D.; Caskurlu, T.; Solak, M.; Creighton, C.J.; et al. Overexpression of miR-145–5p Inhibits Proliferation of Prostate Cancer Cells and Reduces SOX2 Expression. Cancer Investig. 2015, 33, 251–258. [Google Scholar] [CrossRef]
- Ji, S.; Shi, Y.; Yang, L.; Zhang, F.; Li, Y.; Xu, F. miR-145-5p Inhibits Neuroendocrine Differentiation and Tumor Growth by Regulating the SOX11/MYCN Axis in Prostate cancer. Front. Genet. 2022, 13, 790621. Available online: https://www.frontiersin.org/articles/10.3389/fgene.2022.790621 (accessed on 24 January 2024). [CrossRef]
- Guo, H.; Zhao, J.; Li, X.; Sun, F.; Qin, Y.; Yang, X.; Xiong, X.; Yin, Q.; Wang, X.; Gao, L.; et al. Identification of miR-1-3p, miR-143–3p and miR-145–5p association with bone metastasis of Gleason 3+4 prostate cancer and involvement of LASP1 regulation. Mol. Cell. Probes 2023, 68, 101901. [Google Scholar] [CrossRef] [PubMed]
- Le Hars, M.; Castro-Vega, L.J.; Rajabi, F.; Tabatadze, D.; Romero, M.; Pinskaya, M.; Groisman, I. Pro-tumorigenic role of lnc-ZNF30-3 as a sponge counteracting miR-145-5p in prostate cancer. Biol. Direct 2023, 18, 38. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Qin, P.; Cao, C.; Dai, G.; Xu, L.; Yang, D. Use of miR-145 and testicular nuclear receptor 4 inhibition to reduce chemoresistance to docetaxel in prostate cancer. Oncol. Rep. 2021, 45, 963–974. [Google Scholar] [CrossRef] [PubMed]
- Tohidast, M.; Memari, N.; Amini, M.; Hosseini, S.S.; Jebelli, A.; Doustvandi, M.A.; Baradaran, B.; Mokhtarzadeh, A. MiR-145 inhibits cell migration and increases paclitaxel chemosensitivity in prostate cancer cells. Iran. J. Basic Med. Sci. 2023, 26, 1350–1359. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Deng, L.; Gong, Y. MiR-145-5p Inhibits the Invasion of Prostate Cancer and Induces Apoptosis by Inhibiting WIP1. J. Oncol. 2021, 2021, 4412705. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.; Huang, Y.; Liu, Q.; Liu, H.; Long, T.; Zhu, C.; Wu, X. MiR-145 suppresses the motility of prostate cancer cells by targeting cadherin-2. Mol. Cell. Biochem. 2021, 476, 3635–3646. [Google Scholar] [CrossRef] [PubMed]
- Ren, D.; Wang, M.; Guo, W.; Zhao, X.; Tu, X.; Huang, S.; Zou, X.; Peng, X. Wild-type p53 suppresses the epithelial-mesenchymal transition and stemness in PC-3 prostate cancer cells by modulating miR-145. Int. J. Oncol. 2013, 42, 1473–1481. [Google Scholar] [CrossRef] [PubMed]
- Ren, D.; Wang, M.; Guo, W.; Huang, S.; Wang, Z.; Zhao, X.; Du, H.; Song, L.; Peng, X. Double-negative feedback loop between ZEB2 and miR-145 regulates epithelial-mesenchymal transition and stem cell properties in prostate cancer cells. Cell Tissue Res. 2014, 358, 763–778. [Google Scholar] [CrossRef] [PubMed]
- Luo, B.; Yuan, Y.; Zhu, Y.; Liang, S.; Dong, R.; Hou, J.; Li, P.; Xing, Y.; Lu, Z.; Lo, R.; et al. microRNA-145-5p inhibits prostate cancer bone metastatic by modulating the epithelial-mesenchymal transition. Front. Oncol. 2022, 12, 988794. Available online: https://www.frontiersin.org/journals/oncology/articles/10.3389/fonc.2022.988794 (accessed on 31 January 2024). [CrossRef]
- Chen, Q.; Zhou, L.; Ye, X.; Tao, M.; Wu, J. miR-145-5p suppresses proliferation, metastasis and EMT of colorectal cancer by targeting CDCA3. Pathol.-Res. Pract. 2020, 216, 152872. [Google Scholar] [CrossRef]
- Wang, D.; Zhu, L.; Liao, M.; Zeng, T.; Zhuo, W.; Yang, S.; Wu, W. MYO6 knockdown inhibits the growth and induces the apoptosis of prostate cancer cells by decreasing the phosphorylation of ERK1/2 and PRAS40. Oncol. Rep. 2016, 36, 1285–1292. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Zhang, X.; Li, W.; Chen, Y. MicroRNA-145-5p regulates the proliferation of epithelial ovarian cancer cells via targeting SMAD4. J. Ovarian Res. 2020, 13, 54. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Wu, S.; Zhu, K.; Zhang, J.; Shi, X.; Shen, J.; Xu, J. The role of miR-145-5p in esophageal squamous cell carcinoma tumor-associated macrophages and selection of immunochemotherapy. J. Thorac. Dis. 2022, 14, 2493–2510. [Google Scholar] [CrossRef] [PubMed]
- Mozammel, N.; Amini, M.; Baradaran, B.; Mahdavi, S.Z.B.; Hosseini, S.S.; Mokhtarzadeh, A. The function of miR-145 in colorectal cancer progression; an updated review on related signaling pathways. Pathol.-Res. Pract. 2023, 242, 154290. [Google Scholar] [CrossRef] [PubMed]
- Avgeris, M.; Stravodimos, K.; Fragoulis, E.G.; Scorilas, A. The loss of the tumour-suppressor miR-145 results in the shorter disease-free survival of prostate cancer patients. Br. J. Cancer 2013, 108, 2573–2581. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Dong, L.; Sun, J.; Shao, J.; Zhang, J.; Chen, S.; Wang, C.; Wu, G.; Wang, X. miR-145-5p: A Potential Biomarker in Predicting Gleason Upgrading of Prostate Biopsy Samples Scored 3+3=6. Cancer Manag. Res. 2021, 13, 9095–9106. [Google Scholar] [CrossRef] [PubMed]
- Lei, C.; Du, F.; Sun, L.; Li, T.; Li, T.; Min, Y.; Nie, A.; Wang, X.; Geng, L.; Lu, Y.; et al. miR-143 and miR-145 inhibit gastric cancer cell migration and metastasis by suppressing MYO6. Cell Death Dis. 2017, 8, e3101. [Google Scholar] [CrossRef] [PubMed]
- Szczyrba, J.; Löprich, E.; Wach, S.; Jung, V.; Unteregger, G.; Barth, S.; Grobholz, R.; Wieland, W.; Stöhr, R.; Hartmann, A.; et al. The microRNA profile of prostate carcinoma obtained by deep sequencing. Mol. Cancer Res. MCR 2010, 8, 529–538. [Google Scholar] [CrossRef]
- Zhang, L.; Yu, R.; Li, C.; Dang, Y.; Yi, X.; Wang, L. Circ_0026416 downregulation blocks the development of colorectal cancer through depleting MYO6 expression by enriching miR-545-3p. World J. Surg. Oncol. 2021, 19, 299. [Google Scholar] [CrossRef]
- Buss, F.; Kendrick-Jones, J. How are the cellular functions of myosin VI regulated within the cell? Biochem. Biophys. Res. Commun. 2008, 369, 165–175. [Google Scholar] [CrossRef]
- Maddugoda, M.P.; Crampton, M.S.; Shewan, A.M.; Yap, A.S. Myosin VI and vinculin cooperate during the morphogenesis of cadherin cell–cell contacts in mammalian epithelial cells. J. Cell Biol. 2007, 178, 529–540. [Google Scholar] [CrossRef] [PubMed]
- Chung, C.-L.; Tai, S.-B.; Hu, T.-H.; Chen, J.-J.; Chen, C.-L. Roles of Myosin-Mediated Membrane Trafficking in TGF-β Signaling. Int. J. Mol. Sci. 2019, 20, 3913. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Ying, M.; Wu, Q.; Wang, R.; Li, Y. Overexpression of myosin VI regulates gastric cancer cell progression. Gene 2016, 593, 100–109. [Google Scholar] [CrossRef]
- Zhan, X.-J.; Wang, R.; Kuang, X.-R.; Zhou, J.-Y.; Hu, X.-L. Elevated expression of myosin VI contributes to breast cancer progression via MAPK/ERK signaling pathway. Cell. Signal. 2023, 106, 110633. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, J.; Ma, Y.; Xu, C.; Wang, Y.; He, Y. MicroRNA miR-145-5p inhibits Phospholipase D 5 (PLD5) to downregulate cell proliferation and metastasis to mitigate prostate cancer. Bioengineered 2021, 12, 3240–3251. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Zeng, H.; Guo, Y.; Liu, P.; Pan, H.; Deng, A.; Hu, J. miRNA-145 inhibits non-small cell lung cancer cell proliferation by targeting c-Myc. J. Exp. Clin. Cancer Res. CR 2010, 29, 151. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Qu, Y.; Dang, S.; Yao, B.; Ji, M. MiR-145 inhibits oral squamous cell carcinoma (OSCC) cell growth by targeting c-Myc and Cdk6. Cancer Cell Int. 2013, 13, 51. [Google Scholar] [CrossRef]
- Pagliuca, A.; Valvo, C.; Fabrizi, E.; di Martino, S.; Biffoni, M.; Runci, D.; Forte, S.; De Maria, R.; Ricci-Vitiani, L. Analysis of the combined action of miR-143 and miR-145 on oncogenic pathways in colorectal cancer cells reveals a coordinate program of gene repression. Oncogene 2013, 32, 4806–4813. [Google Scholar] [CrossRef] [PubMed]
- Zou, C.; Xu, Q.; Mao, F.; Li, D.; Bian, C.; Liu, L.-Z.; Jiang, Y.; Chen, X.; Qi, Y.; Zhang, X.; et al. MiR-145 inhibits tumor angiogenesis and growth by N-RAS and VEGF. Cell Cycle Georget. Tex 2012, 11, 2137–2145. [Google Scholar] [CrossRef]
- Yin, Y.; Yan, Z.-P.; Lu, N.-N.; Xu, Q.; He, J.; Qian, X.; Yu, J.; Guan, X.; Jiang, B.-H.; Liu, L.-Z. Downregulation of miR-145 associated with cancer progression and VEGF transcriptional activation by targeting N-RAS and IRS1. Biochim. Biophys. Acta 2013, 1829, 239–247. [Google Scholar] [CrossRef]
- Boufraqech, M.; Zhang, L.; Jain, M.; Patel, D.; Ellis, R.; Xiong, Y.; He, M.; Nilubol, N.; Merino, M.J.; Kebebew, E. miR-145 suppresses thyroid cancer growth and metastasis and targets AKT3. Endocr. Relat. Cancer 2014, 21, 517–531. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Wang, J.; Deng, J.; Li, X.; Long, W.; Chang, Y. MiR-145 acts as a metastasis suppressor by targeting metadherin in lung cancer. Med. Oncol. Northwood Lond. Engl. 2015, 32, 344. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Qiu, M.; Jiang, F.; Zhang, S.; Yang, X.; Wang, J.; Xu, L.; Yin, R. MiR-145 regulates cancer stem-like properties and epithelial-to-mesenchymal transition in lung adenocarcinoma-initiating cells. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2014, 35, 8953–8961. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wu, C.; Wang, Y.; Wen, S.; Wang, J.; Chen, Z.; He, Q.; Feng, D. MicroRNA-145 inhibits cell proliferation by directly targeting ADAM17 in hepatocellular carcinoma. Oncol. Rep. 2014, 32, 1923–1930. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Ebeling, M.C.; Zaman, M.S.; Sikander, M.; Yallapu, M.M.; Chauhan, N.; Yacoubian, A.M.; Behrman, S.W.; Zafar, N.; Kumar, D.; et al. MicroRNA-145 targets MUC13 and suppresses growth and invasion of pancreatic cancer. Oncotarget 2014, 5, 7599–7609. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Guo, H.; Zhang, H.; Wang, H.; Qian, G.; Fan, X.; Hoffman, A.R.; Hu, J.; Ge, S. Putative tumor suppressor miR-145 inhibits colon cancer cell growth by targeting oncogene Friend leukemia virus integration 1. Cancer 2011, 117, 86–95. [Google Scholar] [CrossRef] [PubMed]
- Takagi, T.; Iio, A.; Nakagawa, Y.; Naoe, T.; Tanigawa, N.; Akao, Y. Decreased expression of microRNA-143 and -145 in human gastric cancers. Oncology 2009, 77, 12–21. [Google Scholar] [CrossRef]
- Kou, B.; Gao, Y.; Du, C.; Shi, Q.; Xu, S.; Wang, C.Q.; Wang, X.; He, D.; Guo, P. miR-145 inhibits invasion of bladder cancer cells by targeting PAK1. Urol. Oncol. 2014, 32, 846–854. [Google Scholar] [CrossRef]
- Wu, D.; Li, M.; Wang, L.; Zhou, Y.; Zhou, J.; Pan, H.; Qu, P. microRNA-145 inhibits cell proliferation, migration and invasion by targeting matrix metallopeptidase-11 in renal cell carcinoma. Mol. Med. Rep. 2014, 10, 393–398. [Google Scholar] [CrossRef]
- Ostenfeld, M.S.; Bramsen, J.B.; Lamy, P.; Villadsen, S.B.; Fristrup, N.; Sørensen, K.D.; Ulhøi, B.; Borre, M.; Kjems, J.; Dyrskjøt, L.; et al. miR-145 induces caspase-dependent and -independent cell death in urothelial cancer cell lines with targeting of an expression signature present in Ta bladder tumors. Oncogene 2010, 29, 1073–1084. [Google Scholar] [CrossRef]
- Wang, S.; Bian, C.; Yang, Z.; Bo, Y.; Li, J.; Zeng, L.; Zhou, H.; Zhao, R.C. miR-145 inhibits breast cancer cell growth through RTKN. Int. J. Oncol. 2009, 34, 1461–1466. [Google Scholar] [PubMed]
- Dong, R.; Liu, X.; Zhang, Q.; Jiang, Z.; Li, Y.; Wei, Y.; Li, Y.; Yang, Q.; Liu, J.; Wei, J.-J.; et al. miR-145 inhibits tumor growth and metastasis by targeting metadherin in high-grade serous ovarian carcinoma. Oncotarget 2014, 5, 10816–10829. [Google Scholar] [CrossRef]
- Mak, I.W.Y.; Singh, S.; Turcotte, R.; Ghert, M. The epigenetic regulation of SOX9 by miR-145 in human chondrosarcoma. J. Cell. Biochem. 2015, 116, 37–44. [Google Scholar] [CrossRef]
- Cioce, M.; Ganci, F.; Canu, V.; Sacconi, A.; Mori, F.; Canino, C.; Korita, E.; Casini, B.; Alessandrini, G.; Cambria, A.; et al. Protumorigenic effects of mir-145 loss in malignant pleural mesothelioma. Oncogene 2014, 33, 5319–5331. [Google Scholar] [CrossRef]
- Wan, X.; Cheng, Q.; Peng, R.; Ma, Z.; Chen, Z.; Cao, Y.; Jiang, B. ROCK1, a novel target of miR-145, promotes glioma cell invasion. Mol. Med. Rep. 2014, 9, 1877–1882. [Google Scholar] [CrossRef] [PubMed]
- Koo, S.; Martin, G.; Toussaint, L.G. MicroRNA-145 Promotes the Phenotype of Human Glioblastoma Cells Selected for Invasion. Anticancer Res. 2015, 35, 3209–3215. [Google Scholar]
- Condrat, C.E.; Thompson, D.C.; Barbu, M.G.; Bugnar, O.L.; Boboc, A.; Cretoiu, D.; Suciu, N.; Cretoiu, S.M.; Voinea, S.C. miRNAs as Biomarkers in Disease: Latest Findings Regarding Their Role in Diagnosis and Prognosis. Cells 2020, 9, 276. [Google Scholar] [CrossRef] [PubMed]
- Coradduzza, D.; Solinas, T.; Balzano, F.; Culeddu, N.; Rossi, N.; Cruciani, S.; Azara, E.; Maioli, M.; Zinellu, A.; De Miglio, M.R.; et al. miRNAs as Molecular Biomarkers for Prostate Cancer. J. Mol. Diagn. 2022, 24, 1171–1180. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Jung, G.; Kim, J.H.; Byun, S.-S.; Hong, S.K. Role of prostate health index to predict Gleason score upgrading and high-risk prostate cancer in radical prostatectomy specimens. Sci. Rep. 2021, 11, 17447. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, Y.; Zhang, F.; Ji, Z.; Yang, P.; Tian, Y. Predicting Gleason sum upgrading from biopsy to radical prostatectomy pathology: A new nomogram and its internal validation. BMC Urol. 2021, 21, 3. [Google Scholar] [CrossRef]
- Rajarajan, D.; Kaur, B.; Penta, D.; Natesh, J.; Meeran, S.M. miR-145-5p as a predictive biomarker for breast cancer stemness by computational clinical investigation. Comput. Biol. Med. 2021, 135, 104601. [Google Scholar] [CrossRef] [PubMed]
- Cho, W.C.; Wong, C.F.; Li, K.P.; Fong, A.H.; Fung, K.Y.; Au, J.S. miR-145 as a Potential Biomarker and Therapeutic Target in Patients with Non-Small Cell Lung Cancer. Int. J. Mol. Sci. 2023, 24, 10022. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Han, Y.; Liu, F.; Ruan, L. Downregulations of miR-449a and miR-145-5p Act as Prognostic Biomarkers for Endometrial Cancer. J. Comput. Biol. 2020, 27, 834–844. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Ta, W.-W.; Sun, P.-F.; Meng, Y.-F.; Zhao, C.-Z. Diagnostic and prognostic significance of serum miR-145-5p expression in glioblastoma. Int. J. Clin. Exp. Pathol. 2019, 12, 2536–2543. [Google Scholar] [PubMed]
- Zhang, Y.; Wen, X.; Hu, X.-L.; Cheng, L.-Z.; Yu, J.-Y.; Wei, Z.-B. Downregulation of miR-145-5p correlates with poor prognosis in gastric cancer. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 3026–3030. [Google Scholar]
- Hu, W.; Chen, W.-M.; Jiao, J.-B. Plasma miR-145 as a novel biomarker for the diagnosis and radiosensitivity prediction of human cervical cancer. J. Int. Med. Res. 2017, 45, 1054–1060. [Google Scholar] [CrossRef]
- Li, H.; Huang, G.; Lai, Y.; Ni, L.; Lai, Y. A Panel of Three Serum MicroRNAs as a Potential Diagnostic Biomarker for Urothelial Carcinoma. Oncol. Res. Treat. 2022, 45, 344–352. [Google Scholar] [CrossRef] [PubMed]
- McNally, C.J.; Ruddock, M.W.; Moore, T.; McKenna, D.J. Biomarkers That Differentiate Benign Prostatic Hyperplasia from Prostate Cancer: A Literature Review. Cancer Manag. Res. 2020, 12, 5225–5241. [Google Scholar] [CrossRef] [PubMed]
- McNally, C.J.; Watt, J.; Kurth, M.J.; Lamont, J.V.; Moore, T.; Fitzgerald, P.; Pandha, H.; McKenna, D.J.; Ruddock, M.W. A Novel Combination of Serum Markers in a Multivariate Model to Help Triage Patients Into “Low-” and “High-Risk” Categories for Prostate Cancer. Front. Oncol. 2022, 12, 837127. [Google Scholar] [CrossRef]
- Eklund, M.; Nordström, T.; Aly, M.; Adolfsson, J.; Wiklund, P.; Brandberg, Y.; Thompson, J.; Wiklund, F.; Lindberg, J.; Presti, J.C.; et al. The Stockholm-3 (STHLM3) Model can Improve Prostate Cancer Diagnostics in Men Aged 50-69 yr Compared with Current Prostate Cancer Testing. Eur. Urol. Focus 2018, 4, 707–710. [Google Scholar] [CrossRef]
- Ye, D.; Shen, Z.; Zhou, S. Function of microRNA-145 and mechanisms underlying its role in malignant tumor diagnosis and treatment. Cancer Manag. Res. 2019, 11, 969–979. [Google Scholar] [CrossRef] [PubMed]
- Kwon, Y.W.; Jo, H.-S.; Bae, S.; Seo, Y.; Song, P.; Song, M.; Yoon, J.H. Application of Proteomics in Cancer: Recent Trends and Approaches for Biomarkers Discovery. Front. Med. 2021, 8, 747333. [Google Scholar] [CrossRef] [PubMed]
- De Vargas Roditi, L.; Jacobs, A.; Rueschoff, J.H.; Bankhead, P.; Chevrier, S.; Jackson, H.W.; Hermanns, T.; Fankhauser, C.D.; Poyet, C.; Chun, F.; et al. Single-cell proteomics defines the cellular heterogeneity of localized prostate cancer. Cell Rep. Med. 2022, 3, 100604. [Google Scholar] [CrossRef] [PubMed]
- Goldman, M.J.; Craft, B.; Hastie, M.; Repečka, K.; McDade, F.; Kamath, A.; Banerjee, A.; Luo, Y.; Rogers, D.; Brooks, A.N.; et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 2020, 38, 675–678. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Qu, H.; Wang, S.; Chater, J.M.; Wang, X.; Cui, Y.; Yu, L.; Zhou, R.; Jia, Q.; Traband, R.; et al. CancerMIRNome: An interactive analysis and visualization database for miRNome profiles of human cancer. Nucleic Acids Res. 2022, 50, D1139–D1146. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.-Y.; Lin, Y.-C.; Cui, S.; Huang, Y.; Tang, Y.; Xu, J.; Bao, J.; Li, Y.; Wen, J.; Zuo, H.; et al. miRTarBase update 2022: An informative resource for experimentally validated miRNA-target interactions. Nucleic Acids Res. 2022, 50, D222–D230. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Liu, Y.; Zheng, C.; Qu, H. dbEMT 2.0: An updated database for epithelial-mesenchymal transition genes with experimentally verified information and precalculated regulation information for cancer metastasis. J. Genet. Genom. Yi Chuan Xue Bao 2019, 46, 595–597. [Google Scholar] [CrossRef]
- Vasaikar, S.V.; Deshmukh, A.P.; Hollander, P.D.; Addanki, S.; Kuburich, N.A.; Kudaravalli, S.; Joseph, R.; Chang, J.T.; Soundararajan, R.; Mani, S.A. EMTome: A resource for pan-cancer analysis of epithelial-mesenchymal transition genes and signatures. Br. J. Cancer 2021, 124, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Venny 2.1.0. Available online: https://bioinfogp.cnb.csic.es/tools/venny/ (accessed on 29 June 2023).
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Lánczky, A.; Győrffy, B. Web-Based Survival Analysis Tool Tailored for Medical Research (KMplot): Development and Implementation. J. Med. Internet Res. 2021, 23, e27633. [Google Scholar] [CrossRef] [PubMed]
- Warde-Farley, D.; Donaldson, S.L.; Comes, O.; Zuberi, K.; Badrawi, R.; Chao, P.; Franz, M.; Grouios, C.; Kazi, F.; Lopes, C.T.; et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010, 38 (Suppl. 2), W214–W220. [Google Scholar] [CrossRef] [PubMed]
- Kern, F.; Aparicio-Puerta, E.; Li, Y.; Fehlmann, T.; Kehl, T.; Wagner, V.; Ray, K.; Ludwig, N.; Lenhof, H.-P.; Meese, E.; et al. miRTargetLink 2.0—Interactive miRNA target gene and target pathway networks. Nucleic Acids Res. 2021, 49, W409–W416. [Google Scholar] [CrossRef] [PubMed]
- Mazzara, S.; Rossi, R.L.; Grifantini, R.; Donizetti, S.; Abrignani, S.; Bombaci, M. CombiROC: An interactive web tool for selecting accurate marker combinations of omics data. Sci. Rep. 2017, 7, 45477. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).