D-Loop Mutations as Prognostic Markers in Glioblastoma—A Pilot Study
Abstract
1. Introduction
2. Results and Discussion
2.1. Results
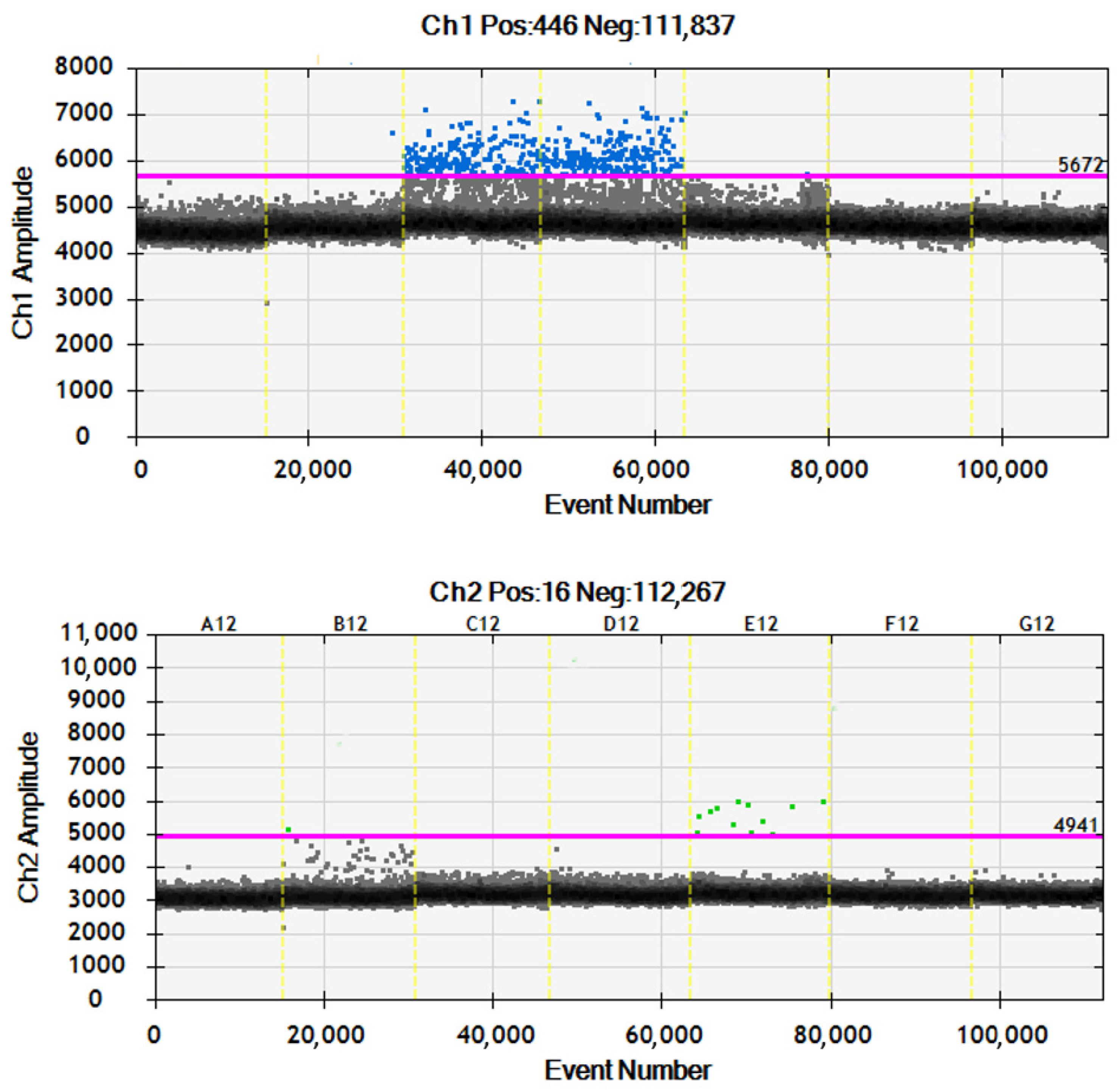
2.1.1. Molecular Marker Analysis
2.1.2. Clinical Outcome
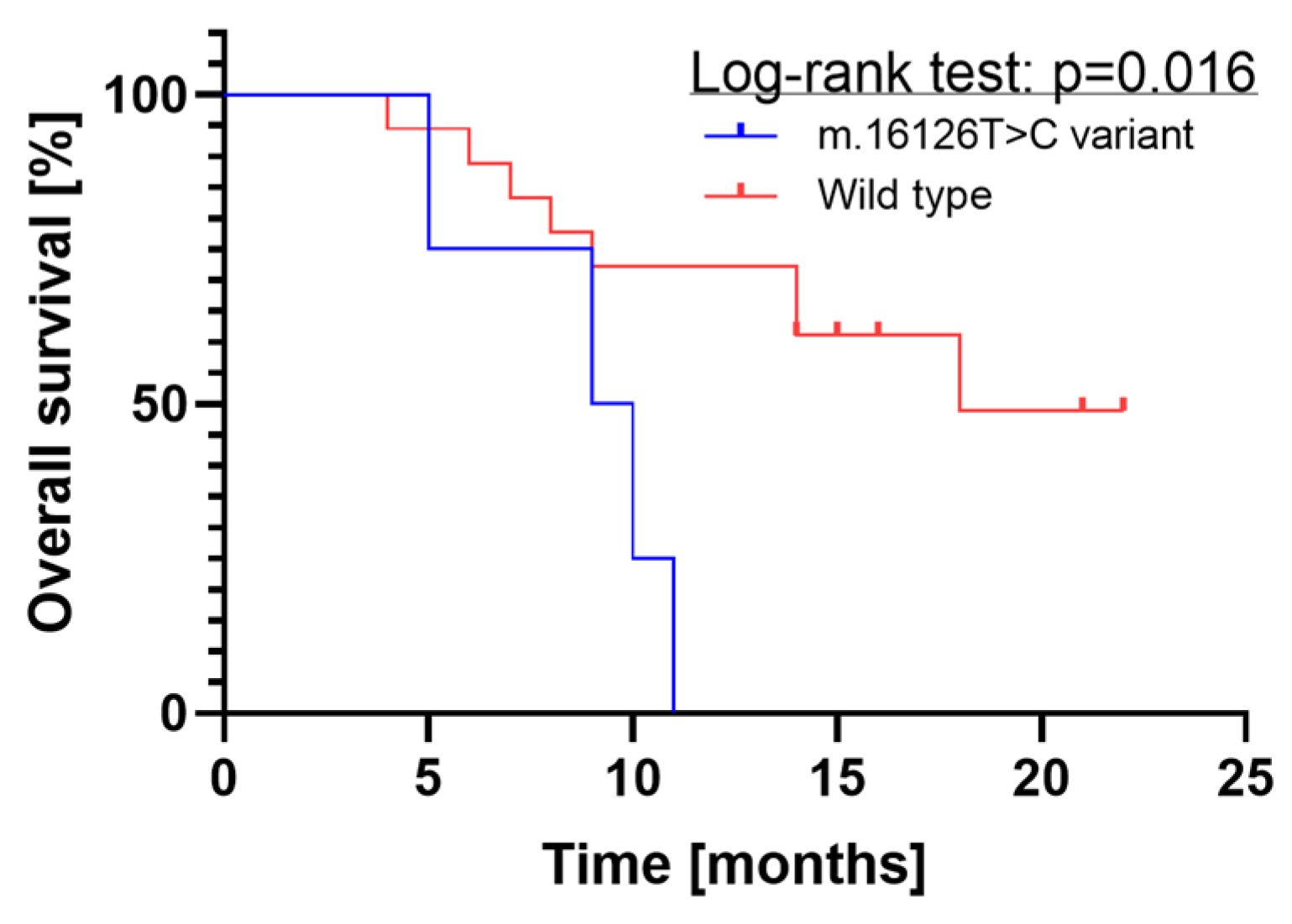
2.1.3. Survival Analysis
2.1.4. Regression Analysis
2.2. Discussion
2.3. Strengths and Limitations
2.4. Conclusions
3. Materials and Methods
3.1. Patients
3.2. Routine Immunohistochemistry Staging
3.3. Material Preparation and DNA Isolation
3.4. Mutation Identification
3.5. Primer Design Strategy
3.6. Statistical Analyses
3.7. Literature Search
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Louis, D.N.; Aldape, K.; Brat, D.J.; Capper, D.; Ellison, D.W.; Hawkins, C.; Paulus, W.; Perry, A.; Reifenberger, G.; Figarella-Branger, D.; et al. Announcing CIMPACT-NOW: The Consortium to Inform Molecular and Practical Approaches to CNS Tumor Taxonomy. Acta Neuropathol. 2017, 133, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Ebner, V.; Gupta, P.; Vibhute, A.K.; Agarwal, R.K.; Rigsby, P.; Brahmbhatt, A.B.; Desai, G.; Bathla, B.A.; Rigsby, R.K.; Brahmbhatt, P.; et al. Newly Recognized CNS Tumors in the 2021 World Health Organization Classification: Imaging Overview with Histopathologic and Genetic Correlation. Am. J. Neuroradiol. 2023, 44, 367–380. [Google Scholar] [CrossRef]
- Poon, M.T.C.; Sudlow, C.L.M.; Figueroa, J.D.; Brennan, P.M. Longer-Term (≥2 Years) Survival in Patients with Glioblastoma in Population-Based Studies Pre- and Post-2005: A Systematic Review and Meta-Analysis. Sci. Rep. 2020, 10, 11622. [Google Scholar] [CrossRef]
- Bigner, S.H.; Mark, J.; Burger, P.C.; Mahaley, M.S.; Bullard, D.E.; Muhlbaier, L.H.; Bigner, D.D. Specific Chromosomal Abnormalities in Malignant Human Gliomas. Cancer Res. 1988, 48, 405–411. [Google Scholar]
- Stoczynska-Fidelus, E.; Szybka, M.; Piaskowski, S.; Bienkowski, M.; Hulas-Bigoszewska, K.; Banaszczyk, M.; Zawlik, I.; Jesionek-Kupnicka, D.; Kordek, R.; Liberski, P.P.; et al. Limited Importance of the Dominant-Negative Effect of TP53missense Mutations. BMC Cancer 2011, 11, 243. [Google Scholar] [CrossRef] [PubMed]
- Kozakiewicz, P.; Grzybowska-Szatkowska, L.; Ciesielka, M.; Całka, P.; Osuchowski, J.; Szmygin, P.; Jarosz, B.; Ostrowska-Leśko, M.; Dudka, J.; Tkaczyk-Wlizło, A.; et al. Mitochondrial DNA Changes in Respiratory Complex I Genes in Brain Gliomas. Biomedicines 2023, 11, 1183. [Google Scholar] [CrossRef] [PubMed]
- Leão Barros, M.B.; Pinheiro, D.D.R.; Borges, B.D.N. Mitochondrial DNA Alterations in Glioblastoma (GBM). Int. J. Mol. Sci. 2021, 22, 5855. [Google Scholar] [CrossRef] [PubMed]
- Su, J.; Li, Y.; Liu, Q.; Peng, G.; Qin, C.; Li, Y. Identification of SSBP1 as a Ferroptosis-Related Biomarker of Glioblastoma Based on a Novel Mitochondria-Related Gene Risk Model and in Vitro Experiments. J. Transl. Med. 2022, 20, 440. [Google Scholar] [CrossRef] [PubMed]
- Kasamatsu, H.; Robberson, D.L.; Vinograd, J. A Novel Closed-Circular Mitochondrial DNA with Properties of a Replicating Intermediate. Proc. Natl. Acad. Sci. USA 1971, 68, 2252–2257. [Google Scholar] [CrossRef]
- McIlwraith, M.J.; West, S.C. DNA Repair Synthesis Facilitates RAD52-Mediated Second-End Capture during DSB Repair. Mol. Cell 2008, 29, 510–516. [Google Scholar] [CrossRef]
- Kirches, E.; Michael, M.; Woy, C.; Schneider, T.; Warich-Kirches, M.; Schneider-Stock, R.; Winkler, K.; Wittig, H.; Dietzmann, K. Loss of Heteroplasmy in the Displacement Loop of Brain Mitochondrial DNA in Astrocytic Tumors. Genes Chromosom. Cancer 1999, 26, 80–83. [Google Scholar] [CrossRef]
- Kirches, E.; Krause, G.; Warich-Kirches, M.; Weis, S.; Schneider, T.; Meyer-Puttlitz, B.; Mawrin, C.; Dietzmann, K. High Frequency of Mitochondrial DNA Mutations in Glioblastoma Multiforme Identified by Direct Sequence Comparison to Blood Samples. Int. J. Cancer 2001, 93, 534–538. [Google Scholar] [CrossRef] [PubMed]
- Altafi, D.; Sadeghi, S.; Hojatian, H.; Afra, M.T.; Kar, S.P.; Gorji, M.; Houshmand, M. Mitochondrial Polymorphisms, in the D-Loop Area, Are Associated with Brain Tumors. Cell J. 2019, 21, 350–356. [Google Scholar] [CrossRef] [PubMed]
- Vega, A.; Salas, A.; Gamborino, E.; Sobrido, M.J.; Macaulay, V.; Carracedo, Á. MtDNA Mutations in Tumors of the Central Nervous System Reflect the Neutral Evolution of MtDNA in Populations. Oncogene 2004, 23, 1314–1320. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Montanini, L.; Regna-Gladin, C.; Eoli, M.; Albarosa, R.; Carrara, F.; Zeviani, M.; Bruzzone, M.G.; Broggi, G.; Boiardi, A.; Finocchiaro, G. Instability of Mitochondrial DNA and MRI and Clinical Correlations in Malignant Gliomas. J. Neurooncol. 2005, 74, 87–90. [Google Scholar] [CrossRef] [PubMed]
- Yeung, K.Y.; Dickinson, A.; Donoghue, J.F.; Polekhina, G.; White, S.J.; Grammatopoulos, D.K.; McKenzie, M.; Johns, T.G.; John, J.C.S. The Identification of Mitochondrial DNA Variants in Glioblastoma Multiforme. Acta Neuropathol. Commun. 2014, 2, 1. [Google Scholar] [CrossRef] [PubMed]
- Yusoff, A.A.M.; Nasir, K.N.M.; Haris, K.; Khair, S.Z.N.M.; Ghani, A.R.I.A.; Idris, Z.; Abdullah, J.M. Detection of Somatic Mutations in the Mitochondrial DNA Control Region D-loop in Brain Tumors: The First Report in Malaysian Patients. Oncol. Lett. 2017, 14, 5179–5188. [Google Scholar] [CrossRef]
- Kilicturgay Yuksel, S.; Ozduman, K.; Yilmaz, E.; Pamir, N.; Boylu Akyerli, C. Analysis of Mitochondrial Dna Control Region D-Loop in Gliomas: Result of 52 Patients. Turk. Neurosurg. 2020, 31, 368–372. [Google Scholar] [CrossRef] [PubMed]
- Abd Radzak, S.M.; Mohd Khair, S.Z.N.; Ahmad, F.; Idris, Z.; Mohamed Yusoff, A.A. Accumulation of Mitochondrial Dna Microsatellite Instability in Malaysian Patients with Primary Central Nervous System Tumors. Turk. Neurosurg. 2020, 31, 99–106. [Google Scholar] [CrossRef]
- Ren, J.; Wu, X.; Shang, F.-F.; Qi, Y.; Tang, Z.; Wen, C.; Cao, W.; Cheng, Q.; Tan, L.; Chen, H.; et al. The TRNA-Cys-GCA Derived TsRNAs Suppress Tumor Progression of Gliomas via Regulating VAV2. Dis. Markers 2022, 2022, 8708312. [Google Scholar] [CrossRef]
- Gao, H.-Z.; Wang, M.-X.; Hu, W.-P.; Chen, J.-Y.; Chen, X.-R.; Lin, L. Complete Mitochondrial Genome Sequence and Mutations of the Glioma Model Inbred C57BL/6 Mice Strain. Mitochondrial DNA 2014, 27, 1852–1853. [Google Scholar] [CrossRef]
- Sharawat, S.K.; Bakhshi, R.; Vishnubhatla, S.; Bakhshi, S. Mitochondrial D-loop Variations in Paediatric Acute Myeloid Leukaemia: A Potential Prognostic Marker. Br. J. Haematol. 2010, 149, 391–398. [Google Scholar] [CrossRef] [PubMed]
- Soffietti, R.; Bettegowda, C.; Mellinghoff, I.K.; Warren, K.E.; Ahluwalia, M.S.; De Groot, J.F.; Galanis, E.; Gilbert, M.R.; Jaeckle, K.A.; Le Rhun, E.; et al. Liquid Biopsy in Gliomas: A RANO Review and Proposals for Clinical Applications. Neuro. Oncol. 2022, 24, 855–871. [Google Scholar] [CrossRef]
- Bobeff, E.J.; Szczesna, D.; Bieńkowski, M.; Janczar, K.; Chmielewska-Kassassir, M.; Wiśniewski, K.; Papierz, W.; Wozniak, L.A.; Jaskólski, D.J. Plasma Amino Acids Indicate Glioblastoma with ATRX Loss. Amino Acids 2021, 53, 119–132. [Google Scholar] [CrossRef]
- Anderson, S.; Bankier, A.T.; Barrell, B.G.; de Bruijn, M.H.L.; Coulson, A.R.; Drouin, J.; Eperon, I.C.; Nierlich, D.P.; Roe, B.A.; Sanger, F.; et al. Sequence and Organization of the Human Mitochondrial Genome. Nature 1981, 290, 457–465. [Google Scholar] [CrossRef]
- Kogelnik, A.M.; Lott, M.T.; Brown, M.D.; Navathe, S.B.; Wallace, D.C. MITOMAP: A Human Mitochondrial Genome Database. Nucleic Acids Res. 1996, 24, 177–179. [Google Scholar] [CrossRef]
- Ronvaux, L.; Riva, M.; Coosemans, A.; Herzog, M.; Rommelaere, G.; Donis, N.; D’Hondt, L.; Douxfils, J. Liquid Biopsy in Glioblastoma. Cancers 2022, 14, 3394. [Google Scholar] [CrossRef] [PubMed]
- Izquierdo, E.; Proszek, P.; Pericoli, G.; Temelso, S.; Clarke, M.; Carvalho, D.M.; MacKay, A.; Marshall, L.V.; Carceller, F.; Hargrave, D.; et al. Droplet Digital PCR-Based Detection of Circulating Tumor DNA from Pediatric High Grade and Diffuse Midline Glioma Patients. Neuro-Oncol. Adv. 2021, 3, vdab013. [Google Scholar] [CrossRef] [PubMed]
- Klekner, Á.; Szivos, L.; Virga, J.; Árkosy, P.; Bognár, L.; Birkó, Z.; Nagy, B. Significance of Liquid Biopsy in Glioblastoma—A Review. J. Biotechnol. 2019, 298, 82–87. [Google Scholar] [CrossRef]
- Auzmendi-iriarte, J.; Carrasco-garcia, E.; Moreno-cugnon, L.; Ruiz, I.; Villanua, J.; Otaegui, D.; Matheu, A. Liquid Biopsy in Glioblastoma: Opportunities, Applications and Challenges. Cancers 2019, 11, 950. [Google Scholar] [CrossRef]
| Characteristic | Study Group (n = 22) | Control Group (n = 25) |
|---|---|---|
| Age (median, IQR) | 64 (45–72) | 58 (48–73) |
| Sex (female/male) | 12/10 | 12/13 |
| DD-PCR | ||
| D-loop Mut. Tumor (%) | 4 (18%) | N/A |
| D-loop Mut. Plasma (%) | 0 | 0 |
| Immunohistochemistry | ||
| IDH1 Mutation (%) | 2 (9%) | N/A |
| GFAP Expression (%) | 22 (100%) | N/A |
| Ki67 (median, IQR) | 30% (19–40) | N/A |
| ATRX Loss (%) | 11 (50%) | N/A |
| P53 Expression (%) | 6 (27%) | N/A |
| 12-month mortality (%) | 9 (41%) | N/A |
| Mortality (%) | 12 (55%) | N/A |
| Survival (median, IQR) | 9 months (6–14) | N/A |
| Follow-up (median, IQR) | 14 months (9–16) 1 | N/A |
| No. | Findings | Reference |
|---|---|---|
| 1 | Loss of D-loop heteroplasmy in glioblastoma. | [11] |
| 2 | The analysis of gliomas and corresponding blood samples revealed instability in mtDNA D-loop repeat regions and identified somatic mutations, suggesting an amplification of mechanisms generating mtDNA polymorphisms in gliomas. | [12] |
| 3 | Most of the mtDNA variants in the D-loop region found in tumors are neutral regarding mitochondrial function and unlikely to impact tumor formations. | [14] |
| 4 | Despite the high frequency of mtDNA mutations in gliomas, they do not directly contribute to increased aggressiveness, potentially due to clonal evolution driven by mutations in nuclear genes that promote rapid growth and infiltration of surrounding tissues. However, they may have diagnostic value as genetic markers for tumors. | [15] |
| 5 | The D-loop region exhibited the highest mutation frequency among 12 glioblastoma cell lines. | [16] |
| 6 | Description of the complete mitochondrial genome sequence of a mouse inbred model. | [21] |
| 7 | Half of the brain tumor patients carried somatic D-loop mtDNA mutations, mostly homoplasmic. Mutations were more common in men and in patients older than 45 years. | [17] |
| 8 | The study revealed an association between mtDNA variants of the D-loop, such as C16069T, T16126C, C16186T, G16274A, C16355T, and T16362C, and brain tumor risk, highlighting the potential role of these variations in the pathogenesis of brain tumors. | [13] |
| 9 | D-loop mtDNA variants C16223T, T16189C, T16311C, and T16126C in gliomas were found to be relevant to WHO classification and morphological grade, highlighting their potential as tumor biomarkers for assessing tumor risk and progression. | [18] |
| 10 | A relatively high prevalence of microsatellite instability (mtMSI) in the D-loop region of mitochondrial DNA (mtDNA) has been demonstrated in brain tumors, with no significant association found with clinical features. | [19] |
| 11 | tRNA-derived small RNAs originating from D-loop and T-loop mtDNA fragments were identified in glioma samples. Some of these tsRNAs were found to be down-regulated in gliomas and correlated with poorer survival outcomes, while others demonstrated the potential to inhibit glioma cell proliferation and tumor growth in vivo. Certain tsRNAs may play a role in glioma progression. | [20] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Szmyd, B.; Stanisławska, P.; Podstawka, M.; Zaczkowski, K.; Izbiński, P.M.; Kulczycka-Wojdala, D.; Stawski, R.; Wiśniewski, K.; Janczar, K.; Braun, M.; et al. D-Loop Mutations as Prognostic Markers in Glioblastoma—A Pilot Study. Int. J. Mol. Sci. 2024, 25, 4334. https://doi.org/10.3390/ijms25084334
Szmyd B, Stanisławska P, Podstawka M, Zaczkowski K, Izbiński PM, Kulczycka-Wojdala D, Stawski R, Wiśniewski K, Janczar K, Braun M, et al. D-Loop Mutations as Prognostic Markers in Glioblastoma—A Pilot Study. International Journal of Molecular Sciences. 2024; 25(8):4334. https://doi.org/10.3390/ijms25084334
Chicago/Turabian StyleSzmyd, Bartosz, Patrycja Stanisławska, Małgorzata Podstawka, Karol Zaczkowski, Patryk M. Izbiński, Dominika Kulczycka-Wojdala, Robert Stawski, Karol Wiśniewski, Karolina Janczar, Marcin Braun, and et al. 2024. "D-Loop Mutations as Prognostic Markers in Glioblastoma—A Pilot Study" International Journal of Molecular Sciences 25, no. 8: 4334. https://doi.org/10.3390/ijms25084334
APA StyleSzmyd, B., Stanisławska, P., Podstawka, M., Zaczkowski, K., Izbiński, P. M., Kulczycka-Wojdala, D., Stawski, R., Wiśniewski, K., Janczar, K., Braun, M., Białasiewicz, P., Jaskólski, D. J., & Bobeff, E. J. (2024). D-Loop Mutations as Prognostic Markers in Glioblastoma—A Pilot Study. International Journal of Molecular Sciences, 25(8), 4334. https://doi.org/10.3390/ijms25084334









