Alteration of Gut Microbiota Composition in the Progression of Liver Damage in Patients with Metabolic Dysfunction-Associated Steatotic Liver Disease (MASLD)
Abstract
1. Introduction
2. Results
2.1. Population Description
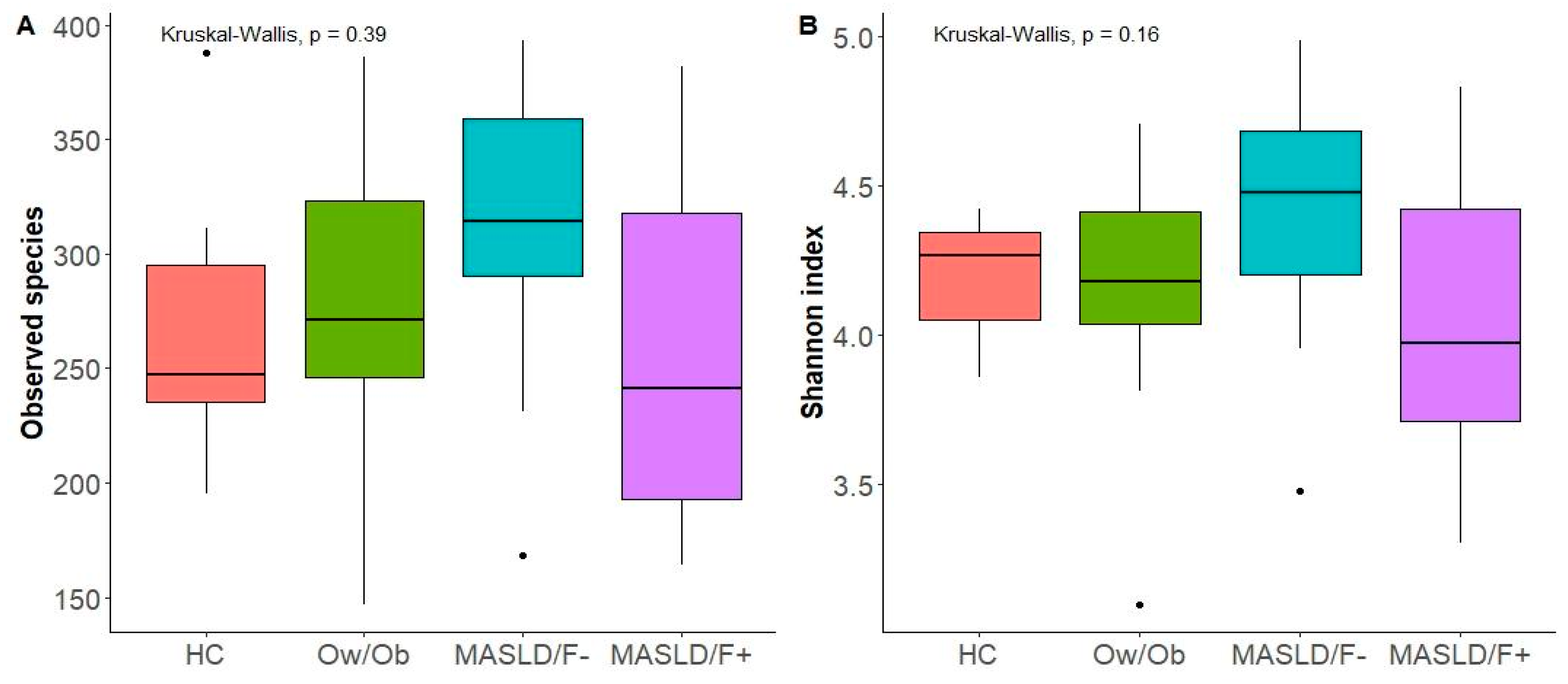
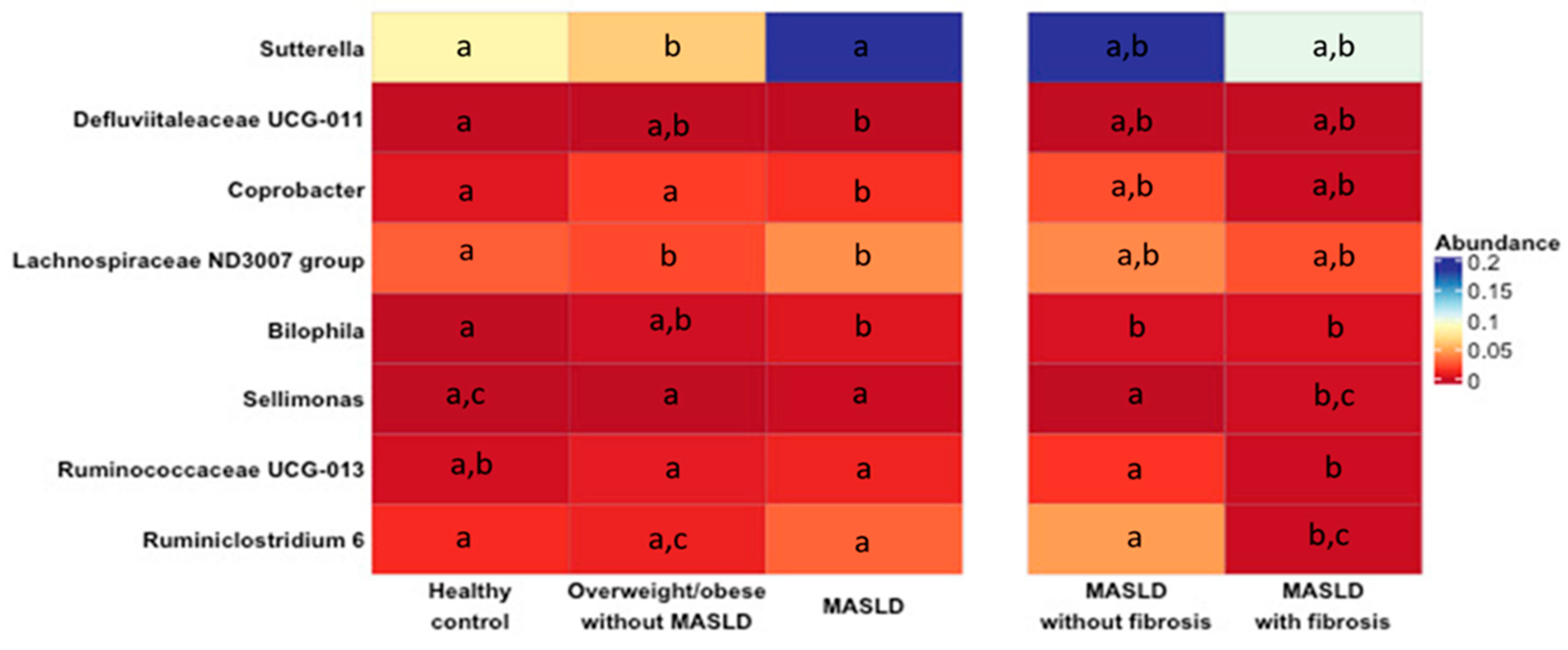
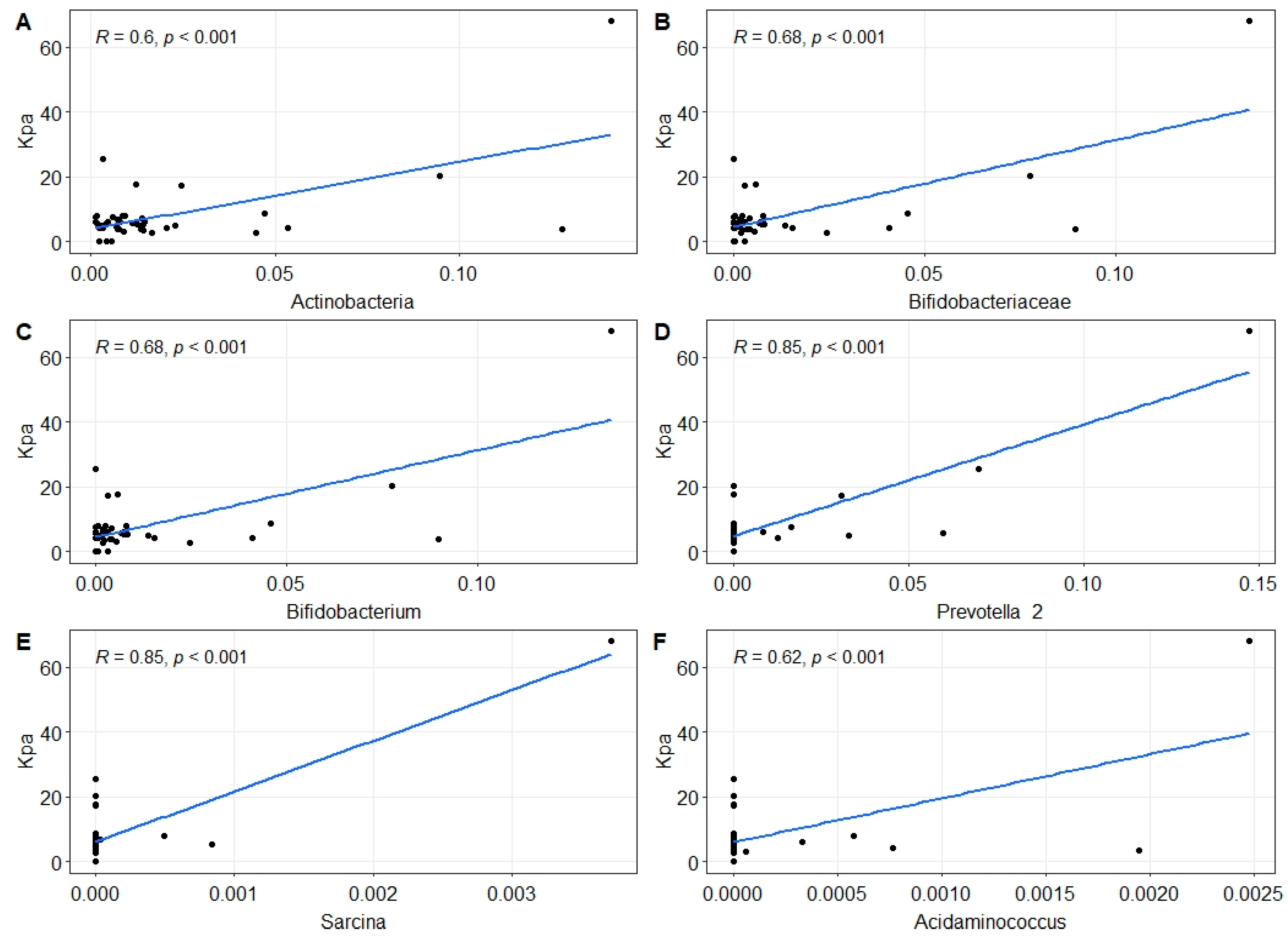
2.2. Changes in Gut Microbiota in MASLD Patients
3. Discussion
4. Materials and Methods
4.1. Study Design
4.2. Ethics Approval and Informed Consent
4.3. Participants
- Healthy control: BMI < 25 Kg/m2, with no MASLD and fibrosis (Fibroscan® CAP < 294 dB/m and stiffness < 8.2 pKa) (HC);
- Overweight/obese subjects: (BMI ≥ 25 kg/m2) with no MASLD and fibrosis (Fibroscan® CAP < 294 dB/m and stiffness < 8.2 pKa) (Ow/Ob);
- Patients with MASLD (Fibroscan® CAP ≥ 294 dB/m or biopsy-proven steatosis or steatohepatitis), without significant fibrosis (stiffness < 8.2 KPa, F < 2) (MASLD/F−);
- Patients with MASLD and significant fibrosis (Fibroscan® CAP ≥ 294 dB/m and stiffness ≥ 8.2 pKa or a biopsy-proven steatosis or steatohepatitis with significant fibrosis ≥ F2) (MASLD/F+).
4.4. Clinical, Anthropometric, and Metabolic Variables
4.5. DNA Extraction, 16S rRNA Amplicon Sequencing, and Bioinformatic Analysis
4.6. Statistical Analyses
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fouda, S.; Jeeyavudeen, M.S.; Pappachan, J.M.; Jayanthi, V. Pathobiology of Metabolic-Associated Fatty Liver Disease. Endocrinol. Metab. Clin. N. Am. 2023, 52, 405–416. [Google Scholar] [CrossRef] [PubMed]
- Ng, C.H.; Huang, D.Q.; Nguyen, M.H. Nonalcoholic fatty liver disease versus metabolic-associated fatty liver disease: Prevalence, outcomes and implications of a change in name. Clin. Mol. Hepatol. 2022, 28, 790–801. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, W.; Morgan, M.P.; Robson, T.; Annett, S. Obesity, non-alcoholic fatty liver disease and hepatocellular carcinoma: Current status and therapeutic targets. Front. Endocrinol. 2023, 14, 1148934. [Google Scholar] [CrossRef] [PubMed]
- Castellanos-Fernandez, M.I.; Pal, S.C.; Arrese, M.; Arab, J.P.; George, J.; Mendez-Sanchez, N. Nonalcoholic Fatty Liver Disease in Latin America and Australia. Clin. Liver Dis. 2023, 27, 301–315. [Google Scholar] [CrossRef] [PubMed]
- Barber, T.M.; Hanson, P.; Weickert, M.O. Metabolic-Associated Fatty Liver Disease and the Gut Microbiota. Endocrinol. Metab. Clin. N. Am. 2023, 52, 485–496. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Lopez, N.; Fuenzalida, C.; Dufeu, M.S.; Pinto-Leon, A.; Escobar, A.; Poniachik, J.; Roblero, J.P.; Valenzuela-Perez, L.; Beltran, C.J. The immune response as a therapeutic target in non-alcoholic fatty liver disease. Front. Immunol. 2022, 13, 954869. [Google Scholar] [CrossRef]
- Liu, L.; Yin, M.; Gao, J.; Yu, C.; Lin, J.; Wu, A.; Zhu, J.; Xu, C.; Liu, X. Intestinal Barrier Function in the Pathogenesis of Nonalcoholic Fatty Liver Disease. J. Clin. Transl. Hepatol. 2023, 11, 452–458. [Google Scholar] [CrossRef] [PubMed]
- Soppert, J.; Brandt, E.F.; Heussen, N.M.; Barzakova, E.; Blank, L.M.; Kuepfer, L.; Hornef, M.W.; Trebicka, J.; Jankowski, J.; Berres, M.L.; et al. Blood Endotoxin Levels as Biomarker of Nonalcoholic Fatty Liver Disease: A Systematic Review and Meta-analysis. Clin. Gastroenterol. Hepatol. 2023, 21, 2746–2758. [Google Scholar] [CrossRef] [PubMed]
- Redondo-Useros, N.; Nova, E.; Gonzalez-Zancada, N.; Diaz, L.E.; Gomez-Martinez, S.; Marcos, A. Microbiota and Lifestyle: A Special Focus on Diet. Nutrients 2020, 12, 1776. [Google Scholar] [CrossRef]
- Salazar, J.; Duran, P.; Diaz, M.P.; Chacin, M.; Santeliz, R.; Mengual, E.; Gutierrez, E.; Leon, X.; Diaz, A.; Bernal, M.; et al. Exploring the Relationship between the Gut Microbiota and Ageing: A Possible Age Modulator. Int. J. Environ. Res. Public Health 2023, 20, 5845. [Google Scholar] [CrossRef]
- Pinart, M.; Dotsch, A.; Schlicht, K.; Laudes, M.; Bouwman, J.; Forslund, S.K.; Pischon, T.; Nimptsch, K. Gut Microbiome Composition in Obese and Non-Obese Persons: A Systematic Review and Meta-Analysis. Nutrients 2021, 14, 12. [Google Scholar] [CrossRef]
- Pitisuttithum, P.; Treeprasertsuk, S. Nonalcoholic fatty liver disease (NAFLD) among older adults. Port. Hypertens. Cirrhos. 2022, 1, 184–191. [Google Scholar] [CrossRef]
- Lake, A.D.; Novak, P.; Shipkova, P.; Aranibar, N.; Robertson, D.; Reily, M.D.; Lu, Z.; Lehman-McKeeman, L.D.; Cherrington, N.J. Decreased hepatotoxic bile acid composition and altered synthesis in progressive human nonalcoholic fatty liver disease. Toxicol. Appl. Pharmacol. 2013, 268, 132–140. [Google Scholar] [CrossRef]
- Kalhan, S.C.; Guo, L.; Edmison, J.; Dasarathy, S.; McCullough, A.J.; Hanson, R.W.; Milburn, M. Plasma metabolomic profile in nonalcoholic fatty liver disease. Metabolism 2011, 60, 404–413. [Google Scholar] [CrossRef]
- Kakiyama, G.; Pandak, W.M.; Gillevet, P.M.; Hylemon, P.B.; Heuman, D.M.; Daita, K.; Takei, H.; Muto, A.; Nittono, H.; Ridlon, J.M.; et al. Modulation of the fecal bile acid profile by gut microbiota in cirrhosis. J. Hepatol. 2013, 58, 949–955. [Google Scholar] [CrossRef] [PubMed]
- Friedman, E.S.; Li, Y.; Shen, T.D.; Jiang, J.; Chau, L.; Adorini, L.; Babakhani, F.; Edwards, J.; Shapiro, D.; Zhao, C.; et al. FXR-Dependent Modulation of the Human Small Intestinal Microbiome by the Bile Acid Derivative Obeticholic Acid. Gastroenterology 2018, 155, 1741–1752.e5. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Luo, Y.; Ranjit, S.; Xie, C.; Libby, A.E.; Orlicky, D.J.; Dvornikov, A.; Wang, X.X.; Myakala, K.; Jones, B.A.; et al. Bile acid sequestration reverses liver injury and prevents progression of nonalcoholic steatohepatitis in Western diet-fed mice. J. Biol. Chem. 2020, 295, 4733–4747. [Google Scholar] [CrossRef] [PubMed]
- Natividad, J.M.; Lamas, B.; Pham, H.P.; Michel, M.L.; Rainteau, D.; Bridonneau, C.; da Costa, G.; van Hylckama Vlieg, J.; Sovran, B.; Chamignon, C.; et al. Bilophila wadsworthia aggravates high fat diet induced metabolic dysfunctions in mice. Nat. Commun. 2018, 9, 2802. [Google Scholar] [CrossRef]
- Liu, X.; Mao, B.; Gu, J.; Wu, J.; Cui, S.; Wang, G.; Zhao, J.; Zhang, H.; Chen, W. Blautia—A new functional genus with potential probiotic properties? Gut Microbes 2021, 13, 1875796. [Google Scholar] [CrossRef]
- Lee, J.Y.; Arai, H.; Nakamura, Y.; Fukiya, S.; Wada, M.; Yokota, A. Contribution of the 7beta-hydroxysteroid dehydrogenase from Ruminococcus gnavus N53 to ursodeoxycholic acid formation in the human colon. J. Lipid Res. 2013, 54, 3062–3069. [Google Scholar] [CrossRef] [PubMed]
- Chiang, J.Y.L.; Ferrell, J.M. Discovery of farnesoid X receptor and its role in bile acid metabolism. Mol. Cell. Endocrinol. 2022, 548, 111618. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, L.E.; Guo, G.L. Role of FXR in Liver Inflammation during Nonalcoholic Steatohepatitis. Curr. Pharmacol. Rep. 2017, 3, 92–100. [Google Scholar] [CrossRef]
- Clifford, B.L.; Sedgeman, L.R.; Williams, K.J.; Morand, P.; Cheng, A.; Jarrett, K.E.; Chan, A.P.; Brearley-Sholto, M.C.; Wahlstrom, A.; Ashby, J.W.; et al. FXR activation protects against NAFLD via bile-acid-dependent reductions in lipid absorption. Cell Metab. 2021, 33, 1671–1684.e4. [Google Scholar] [CrossRef]
- Munoz, M.; Guerrero-Araya, E.; Cortes-Tapia, C.; Plaza-Garrido, A.; Lawley, T.D.; Paredes-Sabja, D. Comprehensive genome analyses of Sellimonas intestinalis, a potential biomarker of homeostasis gut recovery. Microb. Genom. 2020, 6, mgen000476. [Google Scholar] [PubMed]
- Sun, Y.; Chen, Q.; Lin, P.; Xu, R.; He, D.; Ji, W.; Bian, Y.; Shen, Y.; Li, Q.; Liu, C.; et al. Characteristics of Gut Microbiota in Patients with Rheumatoid Arthritis in Shanghai, China. Front. Cell. Infect. Microbiol. 2019, 9, 369. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.Q.; Wang, W.; Li, J.; Ma, M.S.; Zhong, L.Q.; Wei, Q.J.; Song, H.M. Characterization of microbiota in systemic-onset juvenile idiopathic arthritis with different disease severities. World J. Clin. Cases 2019, 7, 2734–2745. [Google Scholar] [CrossRef] [PubMed]
- Lun, H.; Yang, W.; Zhao, S.; Jiang, M.; Xu, M.; Liu, F.; Wang, Y. Altered gut microbiota and microbial biomarkers associated with chronic kidney disease. Microbiologyopen 2019, 8, e00678. [Google Scholar] [CrossRef]
- Juanola, O.; Ferrusquia-Acosta, J.; Garcia-Villalba, R.; Zapater, P.; Magaz, M.; Marin, A.; Olivas, P.; Baiges, A.; Bellot, P.; Turon, F.; et al. Circulating levels of butyrate are inversely related to portal hypertension, endotoxemia, and systemic inflammation in patients with cirrhosis. FASEB J. 2019, 33, 11595–11605. [Google Scholar] [CrossRef]
- Rodriguez-Diaz, C.; Taminiau, B.; Garcia-Garcia, A.; Cueto, A.; Robles-Diaz, M.; Ortega-Alonso, A.; Martin-Reyes, F.; Daube, G.; Sanabria-Cabrera, J.; Jimenez-Perez, M.; et al. Microbiota diversity in nonalcoholic fatty liver disease and in drug-induced liver injury. Pharmacol. Res. 2022, 182, 106348. [Google Scholar] [CrossRef]
- Iino, C.; Endo, T.; Mikami, K.; Hasegawa, T.; Kimura, M.; Sawada, N.; Nakaji, S.; Fukuda, S. Significant decrease in Faecalibacterium among gut microbiota in nonalcoholic fatty liver disease: A large BMI- and sex-matched population study. Hepatol. Int. 2019, 13, 748–756. [Google Scholar] [CrossRef]
- Deleu, S.; Machiels, K.; Raes, J.; Verbeke, K.; Vermeire, S. Short chain fatty acids and its producing organisms: An overlooked therapy for IBD? EBioMedicine 2021, 66, 103293. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Ye, J.; Shao, C.; Zhong, B. Compositional alterations of gut microbiota in nonalcoholic fatty liver disease patients: A systematic review and Meta-analysis. Lipids Health Dis. 2021, 20, 22. [Google Scholar] [CrossRef]
- Tokuhara, D. Role of the Gut Microbiota in Regulating Non-alcoholic Fatty Liver Disease in Children and Adolescents. Front. Nutr. 2021, 8, 700058. [Google Scholar] [CrossRef] [PubMed]
- Ohata, A.; Usami, M.; Miyoshi, M. Short-chain fatty acids alter tight junction permeability in intestinal monolayer cells via lipoxygenase activation. Nutrition 2005, 21, 838–847. [Google Scholar] [CrossRef] [PubMed]
- Sharifnia, T.; Antoun, J.; Verriere, T.G.; Suarez, G.; Wattacheril, J.; Wilson, K.T.; Peek, R.M., Jr.; Abumrad, N.N.; Flynn, C.R. Hepatic TLR4 signaling in obese NAFLD. Am. J. Physiol. Gastrointest. Liver Physiol. 2015, 309, G270–G278. [Google Scholar] [CrossRef]
- De la Cuesta-Zuluaga, J.; Mueller, N.T.; Corrales-Agudelo, V.; Velasquez-Mejia, E.P.; Carmona, J.A.; Abad, J.M.; Escobar, J.S. Metformin Is Associated with Higher Relative Abundance of Mucin-Degrading Akkermansia muciniphila and Several Short-Chain Fatty Acid-Producing Microbiota in the Gut. Diabetes Care 2017, 40, 54–62. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Vitetta, L. Gut Microbiota Metabolites in NAFLD Pathogenesis and Therapeutic Implications. Int. J. Mol. Sci. 2020, 21, 5214. [Google Scholar] [CrossRef] [PubMed]
- Chalasani, N.; Younossi, Z.; Lavine, J.E.; Charlton, M.; Cusi, K.; Rinella, M.; Harrison, S.A.; Brunt, E.M.; Sanyal, A.J. The diagnosis and management of nonalcoholic fatty liver disease: Practice guidance from the American Association for the Study of Liver Diseases. Hepatology 2018, 67, 328–357. [Google Scholar] [CrossRef]
- Rinella, M.E.; Lazarus, J.V.; Ratziu, V.; Francque, S.M.; Sanyal, A.J.; Kanwal, F.; Romero, D.; Abdelmalek, M.F.; Anstee, Q.M.; Arab, J.P.; et al. A multisociety Delphi consensus statement on new fatty liver disease nomenclature. Hepatology 2023, 78, 1966–1986. [Google Scholar] [CrossRef]
- Kleiner, D.E.; Brunt, E.M.; Van Natta, M.; Behling, C.; Contos, M.J.; Cummings, O.W.; Ferrell, L.D.; Liu, Y.C.; Torbenson, M.S.; Unalp-Arida, A.; et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology 2005, 41, 1313–1321. [Google Scholar] [CrossRef]
- Petroff, D.; Blank, V.; Newsome, P.N.; Shalimar; Voican, C.S.; Thiele, M.; de Ledinghen, V.; Baumeler, S.; Chan, W.K.; Perlemuter, G.; et al. Assessment of hepatic steatosis by controlled attenuation parameter using the M and XL probes: An individual patient data meta-analysis. Lancet Gastroenterol. Hepatol. 2021, 6, 185–198. [Google Scholar] [CrossRef] [PubMed]
- Eddowes, P.J.; Sasso, M.; Allison, M.; Tsochatzis, E.; Anstee, Q.M.; Sheridan, D.; Guha, I.N.; Cobbold, J.F.; Deeks, J.J.; Paradis, V.; et al. Accuracy of FibroScan Controlled Attenuation Parameter and Liver Stiffness Measurement in Assessing Steatosis and Fibrosis in Patients with Nonalcoholic Fatty Liver Disease. Gastroenterology 2019, 156, 1717–1730. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]

| Healthy Controls (n = 7) | Overweight/Obese Subjects with No MASLD and Fibrosis (n = 13) | Patients with MASLD and Non-Significant Fibrosis (n = 11) | Patients with MASLD and Significant Fibrosis (n = 9) | p Value | |
|---|---|---|---|---|---|
| % women | 71.4 | 61.5 | 81.8 | 33.3 | 0.1536 |
| Age (years) | 39.9 ± 17.4 (23–63) | 48.9 ± 16.3 (18–73) | 49.0 ± 15.1 (25–67) | 63.2 ± 9.8 Ψ (47–73) | 0.0293 * |
| Weight (Kg) | 60.9 ± 6.3 (53.0–70.0) | 76.2 ± 8.4 Ψ (67.0–98.0) | 75.4 ± 13.8 (57.0–101-0) | 83.3 ± 13.5 † (60.7–103.0) | 0.0047 ** |
| BMI (kg/m2) | 22.3 ± 0.9 (20.8–23.7) | 28.2 ± 2.7 † (25.4–34.7) | 28.4 ± 3.4 † (23.4–34.1) | 31.2 ± 5.1 ƒ (26.0–39.2) | 0.0004 *** |
| Waist circumference (cm) | 79.8 ± 8.8 | 96.4 ± 7.1 | 98.6 ± 9.1 Ψ | 101.8 ± 8.8 † | 0.0103 * |
| Nutritional status (%) | |||||
| Normal weight | 100 | 0 | 18.2 | 0 | |
| Overweight | 0 | 76.9 | 54.5 | 33.3 | |
| Obesity grade 1 | 0 | 23.1 | 27.3 | 55.6 | |
| Obesity grade 2 | 0 | 0 | 0 | 11.1 | |
| Biochemical parameters | |||||
| Cholesterol (mg/dL) | 181.6 ± 50.6 (119–278) | 193.2 ± 42.58 (125–257) | 210.5 ± 63.6 (116–311) | 141.6 ± 24.4 ¥ (103–175) | 0.0247 * |
| Triglycerides (mg/dL) | 87.1 ± 53.5 (35–180) | 145.2 ± 94.3 (48–361) | 154.9 ± 80.7 (69–323) | 109.8 ± 83.8 (41–254) | 0.1654 |
| ALP (UI/L) | 76.7 ± 19.9 (42–97) | 89.44 ± 25.2 (62–121) | 111.1 ± 36.3 (65–173) | 124.0 ± 54.5 (67–200) | 0.2163 |
| GGT (UI/L) | 17.5 ± 5.1 (13 -24) | 28.2 ± 19.0 (16–76) | 113 ± 150 (15–470) | 177 ± 166 ∂,† (35–474) | 0.0031 ** |
| AST (UI/L) | 24.0 ± 5.3 (19.0–31.0) | 34.3 ± 22.3 (21.0–93.0) | 77.6 ± 68.6 Ψ (17.0–231.0) | 47.9 ± 19.7 (21.0–83.0) | 0.0227 * |
| ALT (UI/L) | 15.2 ± 5.4 (11.0–23.0) | 50.0 ± 65.5 (12.0–224.0) | 121.1 ± 105.6 Ψ (14.0–289.0) | 43.6 ± 23.4 (20.0–88.0) | 0.0139 * |
| Albumin (g/dL) | 4.35 ± 0.24 (4.00–4.70) | 4.23 ± 0.28 (3.80–4.70) | 4.62 ± 0.34 (4.20–5.20) | 3.86 ± 0.45 § (3.20–4.60) | 0.0078 ** |
| Tobacco consumption (%) | 1 (14.3) | 3 (23.1) | 1 (9.1) | 1 (11.1) | |
| Alcohol consumption (<20 g/day) (%) | 5 (71.4) | 8 (61.5) | 4 (36.4) | 3 (33.3) | |
| Comorbidities N (%) | |||||
| Diabetes Mellitus | 0 (0) | 1 (7.7) | 2 (18.2) | 5 (55.5) | |
| Hypertension | 2 (28.6) | 1 (7.7) | 4 (36.4) | 3 (33.3) | |
| Dyslipidemia | 2 (28.6) | 4 (30.8) | 3 (27.3) | 2 (22.2) | |
| Atherosclerosis | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zazueta, A.; Valenzuela-Pérez, L.; Ortiz-López, N.; Pinto-León, A.; Torres, V.; Guiñez, D.; Aliaga, N.; Merino, P.; Sandoval, A.; Covarrubias, N.; et al. Alteration of Gut Microbiota Composition in the Progression of Liver Damage in Patients with Metabolic Dysfunction-Associated Steatotic Liver Disease (MASLD). Int. J. Mol. Sci. 2024, 25, 4387. https://doi.org/10.3390/ijms25084387
Zazueta A, Valenzuela-Pérez L, Ortiz-López N, Pinto-León A, Torres V, Guiñez D, Aliaga N, Merino P, Sandoval A, Covarrubias N, et al. Alteration of Gut Microbiota Composition in the Progression of Liver Damage in Patients with Metabolic Dysfunction-Associated Steatotic Liver Disease (MASLD). International Journal of Molecular Sciences. 2024; 25(8):4387. https://doi.org/10.3390/ijms25084387
Chicago/Turabian StyleZazueta, Alejandra, Lucía Valenzuela-Pérez, Nicolás Ortiz-López, Araceli Pinto-León, Verónica Torres, Danette Guiñez, Nicolás Aliaga, Pablo Merino, Alexandra Sandoval, Natalia Covarrubias, and et al. 2024. "Alteration of Gut Microbiota Composition in the Progression of Liver Damage in Patients with Metabolic Dysfunction-Associated Steatotic Liver Disease (MASLD)" International Journal of Molecular Sciences 25, no. 8: 4387. https://doi.org/10.3390/ijms25084387
APA StyleZazueta, A., Valenzuela-Pérez, L., Ortiz-López, N., Pinto-León, A., Torres, V., Guiñez, D., Aliaga, N., Merino, P., Sandoval, A., Covarrubias, N., Pérez de Arce, E., Cattaneo, M., Urzúa, A., Roblero, J. P., Poniachik, J., Gotteland, M., Magne, F., & Beltrán, C. J. (2024). Alteration of Gut Microbiota Composition in the Progression of Liver Damage in Patients with Metabolic Dysfunction-Associated Steatotic Liver Disease (MASLD). International Journal of Molecular Sciences, 25(8), 4387. https://doi.org/10.3390/ijms25084387







