Zinc Finger-Homeodomain Transcriptional Factors (ZHDs) in Cucumber (Cucumis sativus L.): Identification, Evolution, Expression Profiles, and Function under Abiotic Stresses
Abstract
1. Introduction
2. Results
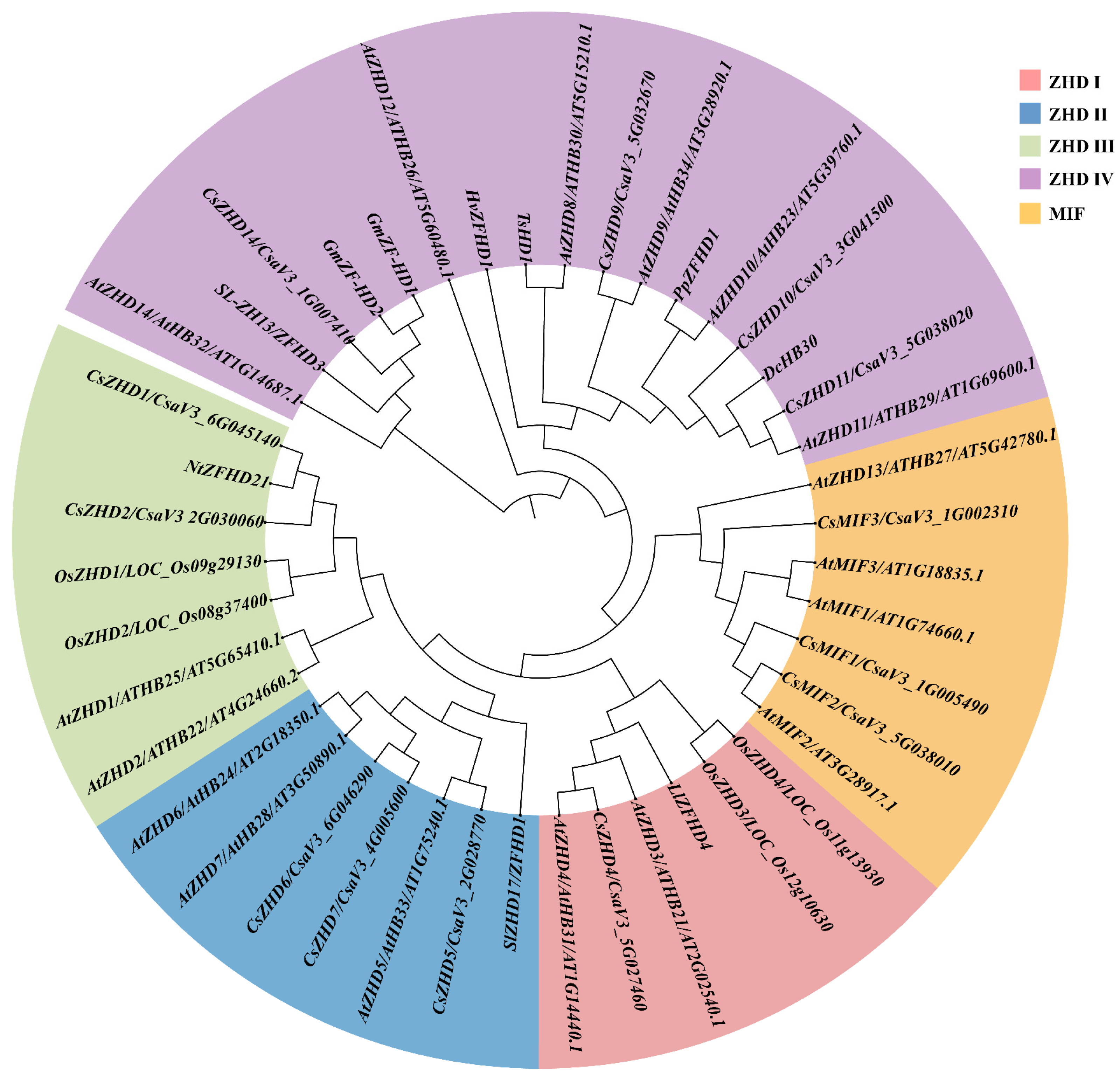
2.1. Systematic Profiles of CsZHD Genes
2.2. Chromosomal Localization and Collinearity Analysis of CsZHD Genes
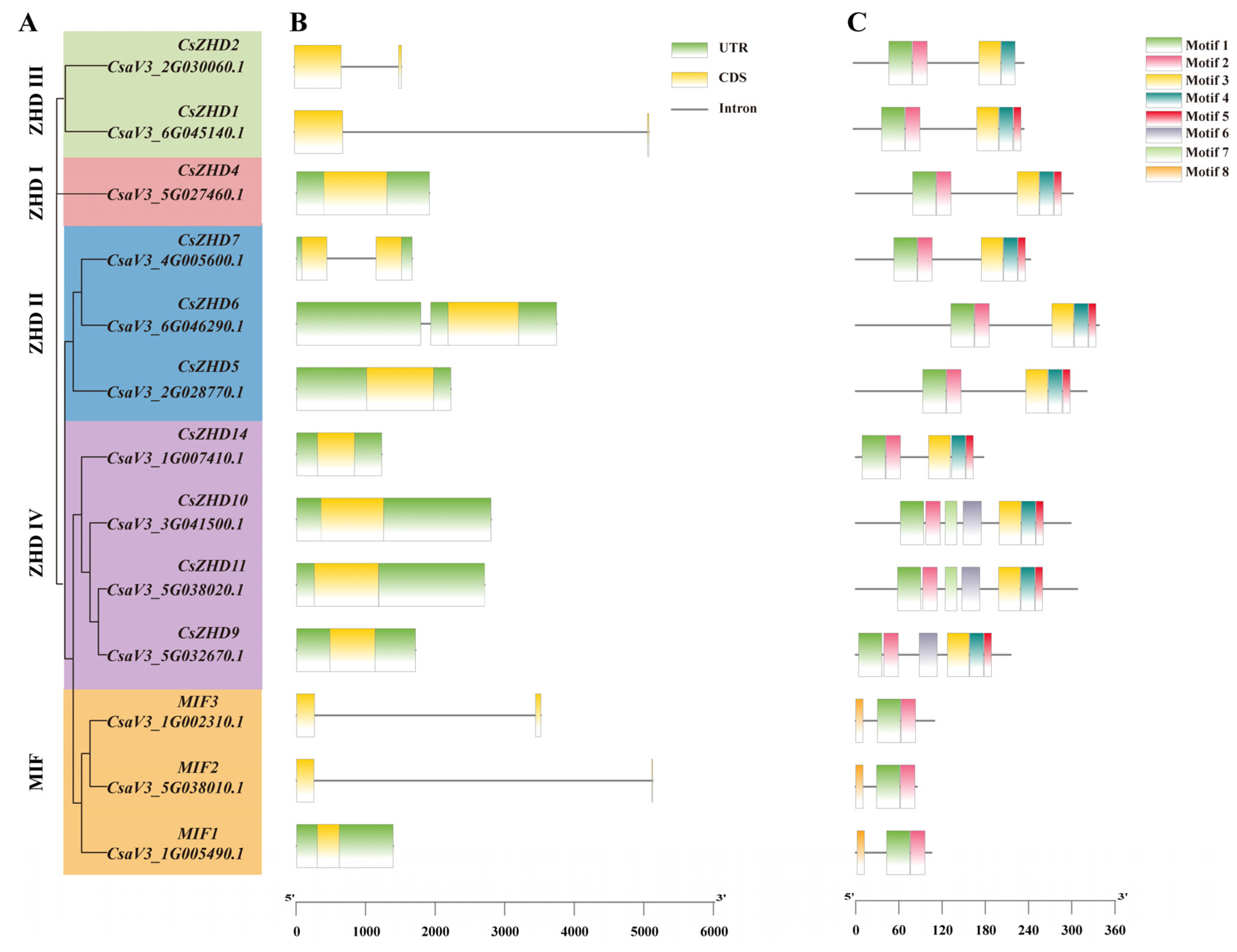
2.3. Gene Structure and Conserved Motifs of CsZHDs
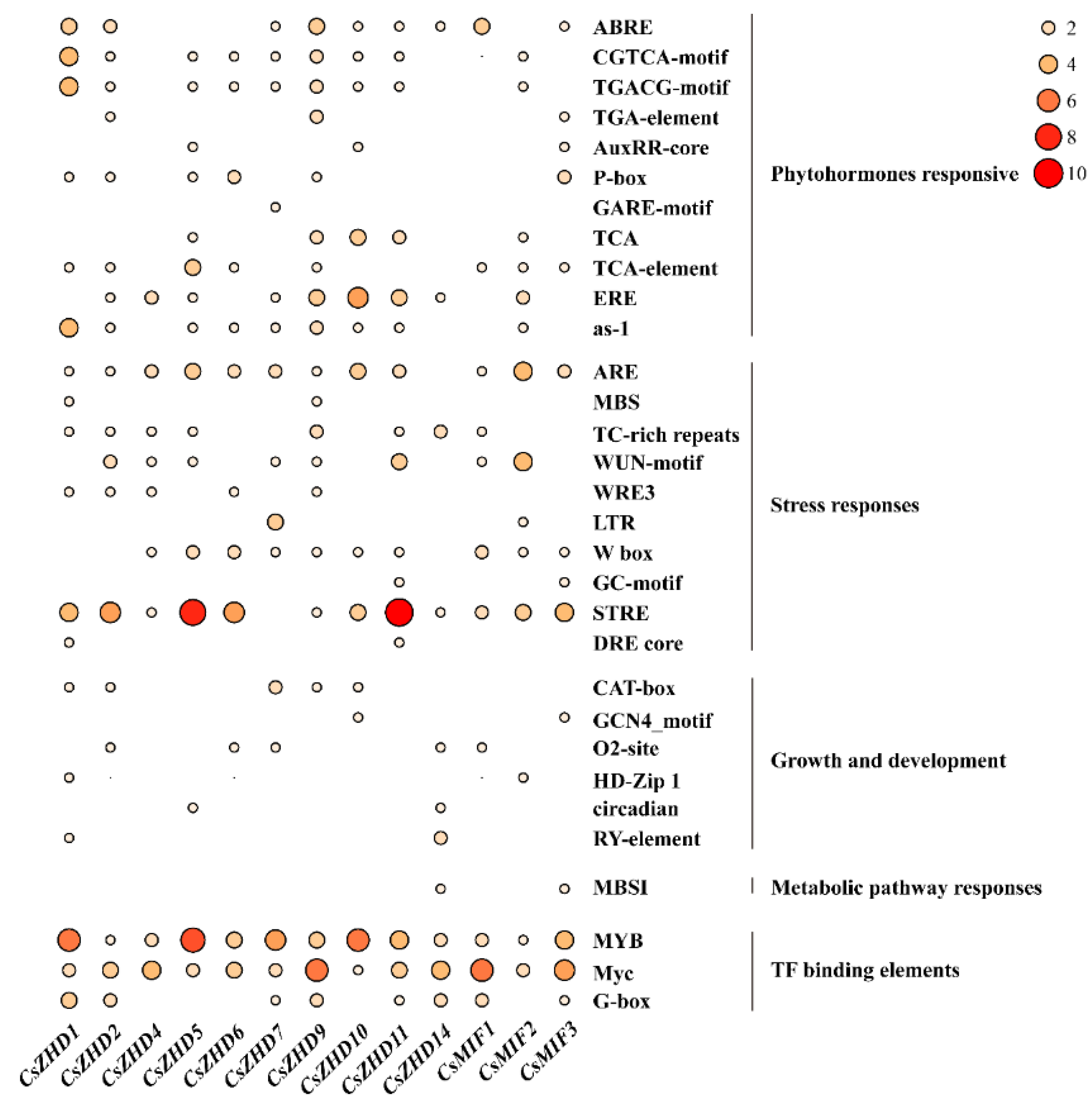
2.4. Analysis of Cis-Acting Elements in CsZHD Promotors
2.5. Tissue-Specific Expression Analysis of CsZHD Genes
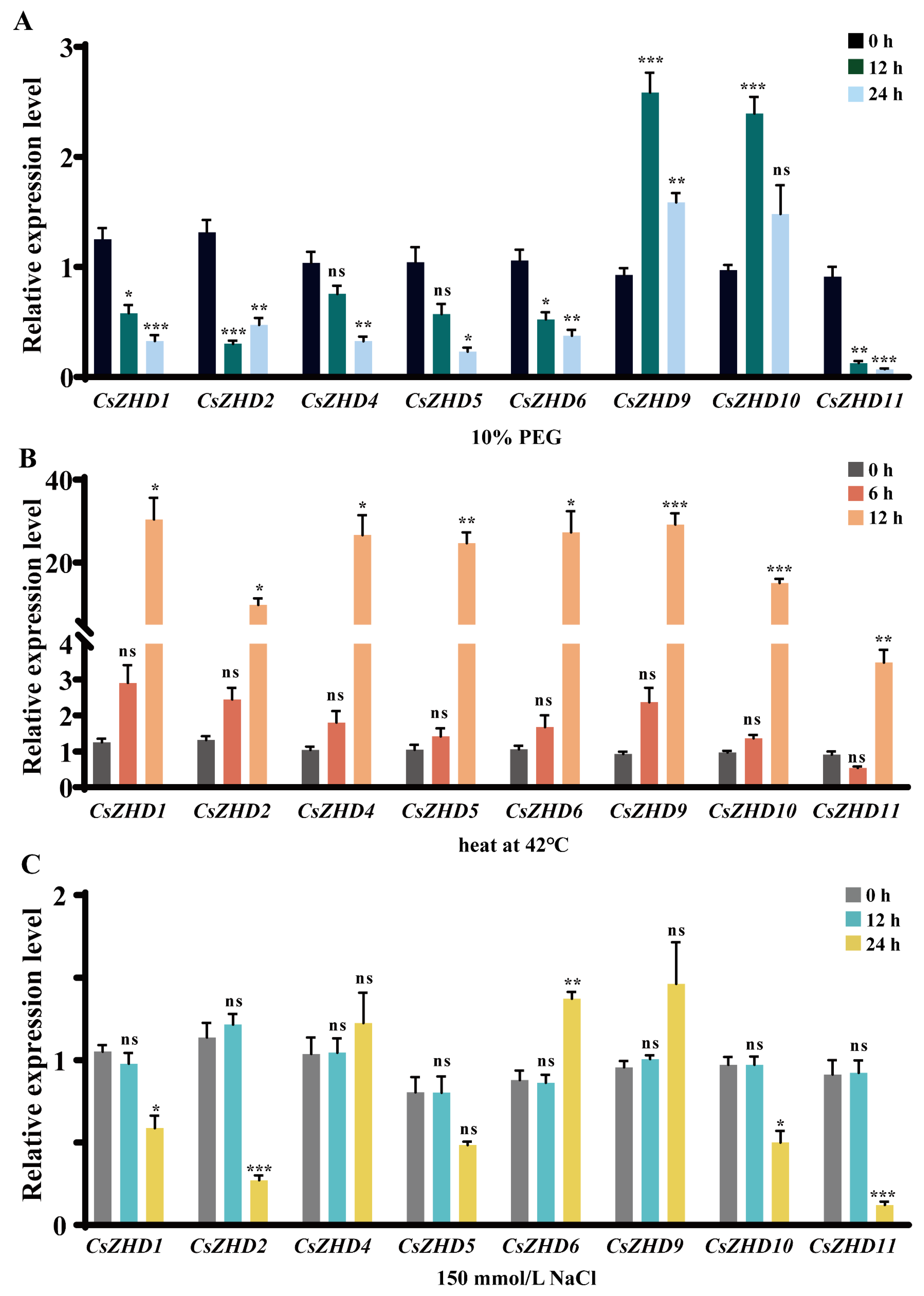
2.6. Expression Patterns of CsZHDs under Drought, Heat and Salt Stresses
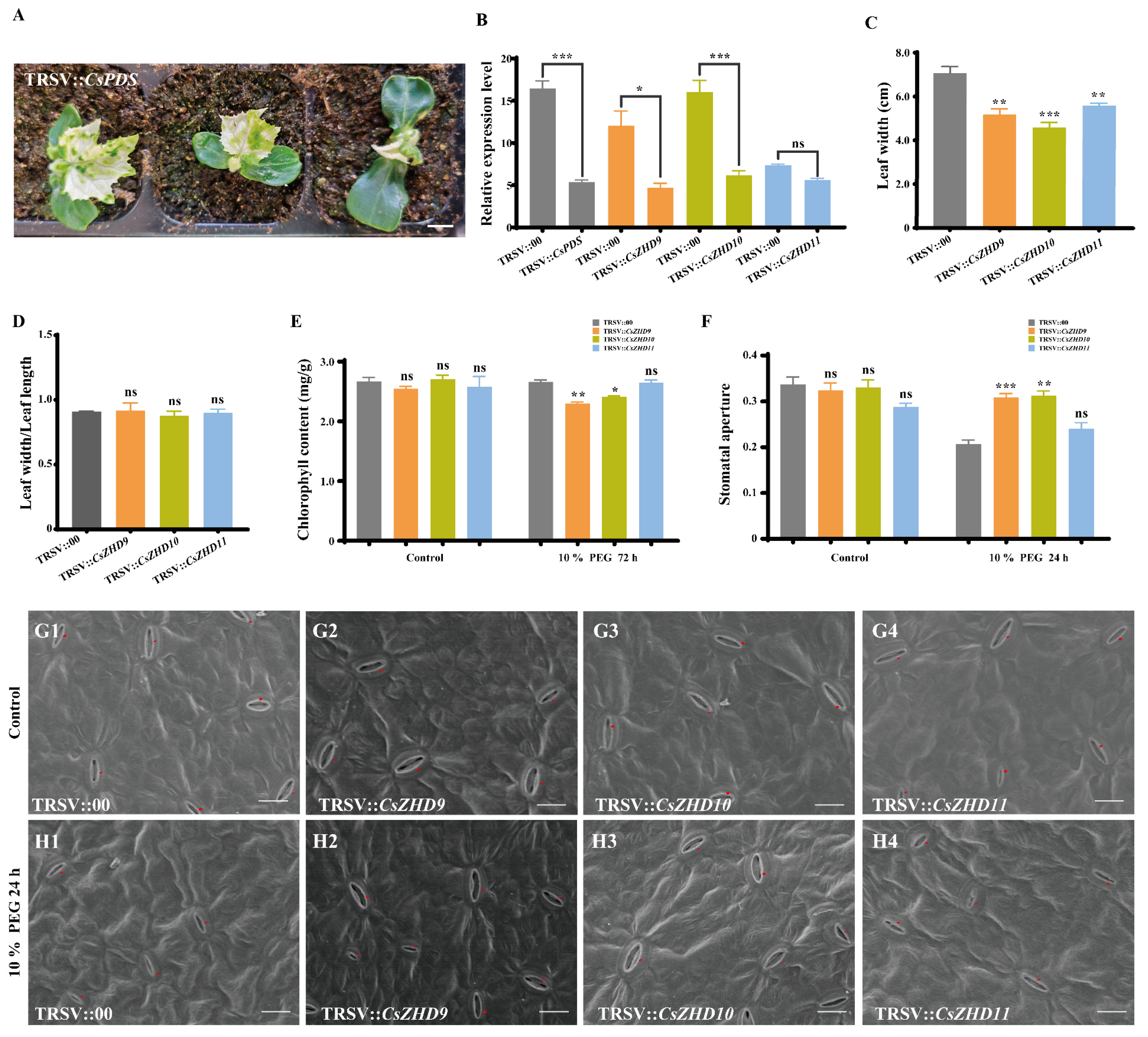
2.7. Silencing of CsZHD9 and CsZHD10 Decreases Drought Tolerance by Regulating Stomatal Movements
3. Discussion
4. Materials and Methods
4.1. Plant Cultivation and Treatment
4.2. Phylogenetic Analysis of ZHD Family
4.3. The Analysis of Chromosomal Location and Synteny
4.4. The Analysis of Gene Structures, Conserved Motifs, and Cis-Elements
4.5. RNA Extraction and qRT-PCR
4.6. VIGS Assay and Phenotypic Observation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Elfving, N.; Davoine, C.; Benlloch, R.; Blomberg, J.; Brannstrom, K.; Muller, D.; Nilsson, A.; Ulfstedt, M.; Ronne, H.; Wingsle, G.; et al. The Arabidopsis thaliana Med25 mediator subunit integrates environmental cues to control plant development. Proc. Natl. Acad. Sci. USA. 2011, 108, 8245–8250. [Google Scholar] [CrossRef] [PubMed]
- Hrmova, M.; Hussain, S.S. Plant transcription factors involved in drought and associated stresses. Int. J. Mol. Sci. 2021, 22, 5662. [Google Scholar] [CrossRef] [PubMed]
- McGinnis, W.; Garber, R.L.; Wirz, J.; Kuroiwa, A.; Gehring, W.J. A homologous protein-coding sequence in Drosophila homeotic genes and its conservation in other metazoans. Cell 1984, 37, 403–408. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, K.; Brocchieri, L.; Bürglin, T.R. A comprehensive classification and evolutionary analysis of plant homeobox genes. Mol. Biol. Evol. 2009, 26, 2775–2794. [Google Scholar] [CrossRef] [PubMed]
- Bürglin, T.R.; Affolter, M. Homeodomain proteins: An update. Chromosoma 2016, 125, 497–521. [Google Scholar] [CrossRef] [PubMed]
- Biggin, M.D.; Tjian, R. A purified Drosophila homeodomain protein represses transcription in vitro. Cell 1989, 58, 433–440. [Google Scholar] [CrossRef] [PubMed]
- Wolberger, C. Homeodomain interactions. Curr. Opin. Struct. Biol. 1996, 6, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Gamsjaeger, R.; Liew, C.K.; Loughlin, F.E.; Crossley, M.; Mackay, J.P. Sticky fingers: Zinc-fingers as protein-recognition motifs. Trends Biochem. Sci. 2007, 32, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Bollier, N.; Gonzalez, N.; Chevalier, C.; Hernould, M. Zinc Finger-Homeodomain and Mini Zinc Finger proteins are key players in plant growth and responses to environmental stresses. J. Exp. Bot. 2022, 73, 4662–4673. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; DePamphilis, C.W.; Ma, H. Phylogenetic analysis of the plant-specific zinc finger-homeobox and mini zinc finger gene families. J. Integr. Plant Biol. 2008, 50, 1031–1045. [Google Scholar] [CrossRef]
- Xu, Y.; Wang, Y.; Long, Q.; Huang, J.; Wang, Y.; Zhou, K.; Zheng, M.; Sun, J.; Chen, H.; Chen, S.; et al. Overexpression of OsZHD1, a zinc finger homeodomain class homeobox transcription factor, induces abaxially curled and drooping leaf in rice. Planta 2014, 239, 803–816. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Yang, Y.; Zhang, L. Zinc Finger-Homeodomain transcriptional factors (ZF-HDs) in wheat (Triticum aestivum L.): Identification, evolution, expression analysis and response to abiotic stresses. Plants 2021, 10, 593. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Wang, X.; Sun, W.; Ma, Z.; Zheng, T.; Huang, L.; Wu, Q.; Tang, Z.; Bu, T.; Li, C.; et al. Genome-wide investigation of the ZF-HD gene family in Tartary buckwheat (Fagopyrum tataricum). BMC Plant Biol. 2019, 19, 248. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wu, P.; Li, Y.; Hou, X. Genome-wide analysis and expression patterns of ZF-HD transcription factors under different developmental tissues and abiotic stresses in Chinese cabbage. Mol. Genet. Genom. 2016, 291, 1451–1464. [Google Scholar] [CrossRef] [PubMed]
- Khatun, K.; Nath, U.K.; Robin AH, K.; Park, J.I.; Lee, D.J.; Kim, M.B.; Kim, C.K.; Lim, K.B.; Nou, I.S.; Chung, M.Y. Genome-wide analysis and expression profiling of zinc finger homeodomain (ZHD) family genes reveal likely roles in organ development and stress responses in tomato. BMC Genom. 2017, 18, 695. [Google Scholar] [CrossRef] [PubMed]
- Islam MA, U.; Nupur, J.A.; Shafiq, M.; Ali, Q.; Sami, A.; Shahid, M.A. In silico and computational analysis of zinc finger motif-associated homeodomain (ZF-HD) family genes in chilli (Capsicum annuum L). BMC Genom. 2023, 24, 603. [Google Scholar] [CrossRef] [PubMed]
- Abdullah, M.; Cheng, X.; Cao, Y.; Su, X.; Manzoor, M.A.; Gao, J.; Cai, Y.; Lin, Y. Zinc Finger-Homeodomain transcriptional factors (ZHDs) in upland Cotton (Gossypium hirsutum): Genome-wide identification and expression analysis in fiber development. Front. Genet. 2018, 9, 357. [Google Scholar] [CrossRef]
- Street, N.R.; Sjodin, A.; Bylesjo, M.; Gustafsson, P.; Trygg, J.; Jansson, S. A cross-species transcriptomics approach to identify genes involved in leaf development. BMC Genom. 2008, 9, 589. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.Y.; Kim, O.K.; Kim, S.G.; Yang, M.S.; Park, C.M. Nuclear import and DNA binding of the ZHD5 transcription factor is modulated by a competitive peptide inhibitor in Arabidopsis. J. Biol. Chem. 2011, 286, 1659–1668. [Google Scholar] [CrossRef] [PubMed]
- Bueso, E.; Munoz-Bertomeu, J.; Campos, F.; Brunaud, V.; Martinez, L.; Sayas, E.; Ballester, P.; Yenush, L.; Serrano, R. ARABIDOPSIS THALIANA HOMEOBOX25 uncovers a role for Gibberellins in seed longevity. Plant Physiol. 2014, 164, 999–1010. [Google Scholar] [CrossRef] [PubMed]
- Perrella, G.; Davidson, M.L.H.; O’Donnell, L.; Nastase, A.M.; Herzyk, P.; Breton, G.; Pruneda-Paz, J.L.; Kay, S.A.; Chory, J.; Kaiserli, E. ZINC-FINGER interactions mediate transcriptional regulation of hypocotyl growth in Arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, E4503–E4511. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Wang, Y.; Zhao, S.; Fu, Y.; Zhu, L. HOMEOBOX PROTEIN 24 mediates the conversion of indole-3-butyric acid to indole-3-acetic acid to promote root hair elongation. New Phytol. 2021, 232, 2057–2070. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhao, H.; Xu, L.; Zhang, H.; Xing, H.; Fu, Y.; Zhu, L. PUB30-mediated downregulation of the HB24-SWEET11 module is involved in root growth inhibition under salt stress by attenuating sucrose supply in Arabidopsis. New Phytol. 2023, 237, 1667–1683. [Google Scholar] [CrossRef]
- Tan, Q.K.; Irish, V.F. The Arabidopsis zinc finger-homeodomain genes encode proteins with unique biochemical properties that are coordinately expressed during floral development. Plant Physiol. 2006, 140, 1095–1108. [Google Scholar] [CrossRef] [PubMed]
- Tran, L.S.; Nakashima, K.; Sakuma, Y.; Osakabe, Y.; Qin, F.; Simpson, S.D.; Maruyama, K.; Fujita, Y.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Co-expression of the stress-inducible zinc finger homeodomain ZFHD1 and NAC transcription factors enhances expression of the ERD1 gene in Arabidopsis. Plant J. 2007, 49, 46–63. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Hua, D.; He, J.; Duan, Y.; Chen, Z.; Hong, X.; Gong, Z. Auxin Response Factor2 (ARF2) and its regulated homeodomain gene HB33 mediate abscisic acid response in Arabidopsis. PLoS Genet. 2011, 7, e1002172. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Hu, J.; Gao, Y.; Wang, Z.; Bao, Y.; Zhang, X.; Yang, H.; Zhang, D.; Jiang, J.; Zhang, H.; et al. Silencing of the SL-ZH13 transcription factor gene decreases the salt stress tolerance of tomato. J. Am. Soc. Hortic. Sci. 2018, 143, 391–396. [Google Scholar] [CrossRef]
- Abu-Romman, S.; Al-Hadid, K. Novel Zinc Finger-Homeodomain gene from barley (HvZFHD1) is differentially regulated during spike development and under hormonal treatments and abiotic stresses. Not. Bot. Horti Agrobot. 2017, 45, 89–96. [Google Scholar] [CrossRef][Green Version]
- Park, H.C.; Kim, M.L.; Lee, S.M.; Bahk, J.D.; Yun, D.J.; Lim, C.O.; Hong, J.C.; Lee, S.Y.; Cho, M.J.; Chung, W.S. Pathogen-induced binding of the soybean zinc finger homeodomain proteins GmZF-HD1 and GmZF-HD2 to two repeats of ATTA homeodomain binding site in the calmodulin isoform 4 (GmCaM4) promoter. Nucleic Acids Res. 2007, 35, 3612–3623. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Zhu, M.; Liu, W.; Jahan, M.S.; Gu, Q.; Shu, S.; Sun, J.; Guo, S. CsPAO2 improves salt tolerance of cucumber through the interaction with CsPSA3 by affecting photosynthesis and polyamine conversion. Int. J. Mol. Sci. 2022, 23, 2413. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; Ming, F.; Liang, Y.; Wang, Y.; Gan, Y.; Qiu, Z.; Yan, S.; Cao, B. Heat stress resistance mechanisms of two cucumber varieties from different regions. Int. J. Mol. Sci. 2022, 23, 1817. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Wan, Z.; Luo, S.; Wei, H.; Zhao, J.; Wang, G.; Yu, J.; Zhang, G. Silencing the CsSnRK2.11 gene decreases drought tolerance of Cucumis sativus L. Int. J. Mol. Sci. 2023, 24, 5761. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.A.U.; Nupur, J.A.; Khalid, M.H.B.; Din, A.M.U.; Shafiq, M.; Alshegaihi, R.M.; Ali, Q.; Ali, Q.; Kamran, Z.; Manzoor, M.; et al. Genome-wide identification and in silico analysis of ZF-HD transcription factor genes in Zea mays L. Genes 2022, 13, 2112. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Yin, X.; Li, X.; Wang, L.; Zheng, Y.; Xu, X.; Zhang, Y.; Wang, X. Genome-wide identification, evolution and expression analysis of the grape (Vitis vinifera L.) zinc finger-homeodomain gene family. Int. J. Mol. Sci. 2014, 15, 5730–5748. [Google Scholar] [CrossRef] [PubMed]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef] [PubMed]
- Verma, V.; Ravindran, P.; Kumar, P.P. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016, 16, 86. [Google Scholar] [CrossRef] [PubMed]
- Larkindale, J.; Hall, J.D.; Knight, M.R.; Vierling, E. Heat stress phenotypes of Arabidopsis mutants implicate multiple signaling pathways in the acquisition of thermotolerance. Plant Physiol. 2005, 138, 882–897. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.; Cho, L.H.; Yang, W.; Pasriga, R.; Wu, Y.; Hong, W.J.; Bureau, C.; Wi, S.J.; Zhang, T.; Wang, R.; et al. Homeobox transcription factor OsZHD2 promotes root meristem activity in rice by inducing ethylene biosynthesis. J. Exp. Bot. 2020, 71, 5348–5364. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.; Baek, G.; Pasriga, R.; Tun, W.; Min, C.W.; Kim, S.T.; Cho, L.H.; An, G. Homeobox transcription factors OsZHD1 and OsZHD2 induce inflorescence meristem activity at floral transition in rice. Plant Cell Environ. 2022, 46, 1327–1339. [Google Scholar] [CrossRef] [PubMed]
- Abu-Romman, S. Molecular cloning and expression analysis of zinc finger-homeodomain transcription factor TaZFHD1 in wheat. S. Afr. J. Bot. 2014, 91, 32–36. [Google Scholar] [CrossRef]
- Zhao, T.; Wang, Z.; Bao, Y.; Zhang, X.; Yang, H.; Zhang, D.; Jiang, J.; Zhang, H.; Li, J.; Chen, Q.; et al. Downregulation of SL-ZH13 transcription factor gene expression decreases drought tolerance of tomato. J. Integr. Agric. 2019, 18, 1579–1586. [Google Scholar] [CrossRef]
- Tan, Q.; Jiang, S.; Wang, N.; Liu, X.; Zhang, X.; Wen, B.; Fang, Y.; He, H.; Chen, X.; Fu, X.; et al. OVATE family protein PpOFP1 physically interacts with PpZFHD1 and confers salt tolerance to tomato and yeast. Front. Plant Sci. 2021, 12, 759955. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Wang, S.; Larkin, R.M.; Zhang, F. DcHB30 and DcWRKY75 transcription factors antagonistically regulate ethylene induced petal senescence in carnation (Dianthus caryophyllus). J. Exp. Bot. 2022, 73, 7326–7343. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Ma, H.; Zhou, J.; Li, Z.; Peng, Z.; Guo, F.; Zhang, J. TsHD1 and TsNAC1 cooperatively play roles in plant growth and abiotic stress resistance of Thellungiella halophile. Plant J. 2019, 99, 81–97. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Wu, M.; Zhang, M.; Jiang, W.; Ren, X.; Liang, E.; Zhang, D.; Zhang, C.; Xiao, N.; Li, Y.; et al. A maize phytochrome-interacting factors protein ZmPIF1 enhances drought tolerance by inducing stomatal closure and improves grain yield in Oryza sativa. Plant Biotechnol. J. 2018, 16, 1375–1387. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Hou, S.; Rodrigues, O.; Wang, P.; Luo, D.; Munemasa, S.; Lei, J.; Liu, J.; Ortiz-Morea, F.A.; Wang, X.; et al. Phytocytokine signalling reopens stomata in plant immunity and water loss. Nature 2022, 605, 332–339. [Google Scholar] [CrossRef] [PubMed]
- Qian, Z.J.; Song, J.J.; Chumont, F.; Ye, Q. Differential responses of plasma membrane aquaporins in mediating water transport of cucumber seedlings under osmotic and salt stresses. Plant Cell Environ. 2015, 38, 461–473. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Hai, Z.; Wang, R.; Yu, Y.; Chen, X.; Liang, W.; Wang, H. Genome-wide analysis of HSP20 gene family and expression patterns under heat stress in cucumber (Cucumis sativus L.). Front. Plant Sci. 2022, 13, 968418. [Google Scholar] [CrossRef] [PubMed]
- Unel, N.M.; Baloglu, M.C.; Altunoglu, Y.Ç. Comprehensive investigation of cucumber heat shock proteins under abiotic stress conditions: A multi-omics survey. J. Biotechnol. 2023, 374, 49–69. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Yang, S.; Sun, L.; Feng, Z.; Gao, Y.; Zhai, X.; Dong, Y.; Wu, H.; Cui, Y.; Li, S.; et al. A CBL4-CIPK6 module confers salt tolerance in cucumber. Veg. Res. 2022, 2, 1–10. [Google Scholar] [CrossRef]
- Shi, Y.; Pang, X.; Liu, W.; Wang, R.; Su, D.; Gao, Y.; Wu, M.; Deng, W.; Liu, Y.; Li, Z. SlZHD17 is involved in the control of chlorophyll and carotenoid metabolism in tomato fruit. Hortic. Res. 2021, 8, 259. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Xie, M.; Li, X.; Li, Z.; Wang, Q.; Ding, A.; Wang, W.; Sun, Y. Systematic investigations of the ZF-HD gene family in tobacco reveal their multiple roles in abiotic stresses. Agronomy 2021, 11, 406. [Google Scholar] [CrossRef]
- Zhang, L.; Wu, P.; Li, W.; Feng, T.; Shockey, J.; Chen, L.; Zhang, L.; Lu, S. Triacylglycerol biosynthesis in shaded seeds of tung tree (Vernicia fordii) is regulated in part by Homeodomain Leucine Zipper 21. Plant J. 2021, 108, 1735–1753. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Wan, H.; Zhao, Z.; Qian, C.; Sui, Y.; Malik, A.A.; Chen, J. Selection of appropriate reference genes for gene expression studies by quantitative real-time polymerase chain reaction in cucumber. Anal. Biochem. 2010, 399, 257–261. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Fang, L.; Wei, X.Y.; Liu, L.Z.; Zhou, L.X.; Tian, Y.P.; Geng, C.; Li, X.D. A tobacco ringspot virus-based vector system for gene and microRNA function studies in cucurbits. Plant Physiol. 2021, 186, 853–864. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Sun, L.; Dong, M.; Fan, S.; Shi, K.; Qu, Y.; Zhu, L.; Shi, J.; Wang, W.; Liu, Y.; et al. Identification and functional characterization of CsMYCs in cucumber glandular trichome development. Int. J. Mol. Sci. 2023, 24, 6435. [Google Scholar] [CrossRef] [PubMed]


| Gene Name | Gene ID | Gene Position | CDS (bp) | AA (aa) | MW (kDa) | pI | |
|---|---|---|---|---|---|---|---|
| Start | End (+/−) | ||||||
| CsZHD1 | CsaV3_6G045140 | 26,710,896 | 26,715,981 (−) | 711 | 236 | 25.57 | 8.83 |
| CsZHD2 | CsaV3_2G030060 | 19,666,125 | 19,667,660 (−) | 711 | 236 | 26.22 | 6.15 |
| CsZHD4 | CsaV3_5G027460 | 22,469,732 | 22,471,636 (−) | 906 | 301 | 33.59 | 8.19 |
| CsZHD5 | CsaV3_2G028770 | 18,872,023 | 18,874,235 (+) | 963 | 320 | 34.06 | 6.52 |
| CsZHD6 | CsaV3_6G046290 | 27,375,407 | 27,379,138 (+) | 1014 | 337 | 37.73 | 8.79 |
| CsZHD7 | CsaV3_4G005600 | 3,724,756 | 3,726,413 (+) | 729 | 242 | 26.86 | 8.94 |
| CsZHD9 | CsaV3_5G032670 | 26,404,212 | 26,405,916 (−) | 648 | 215 | 24.26 | 9.46 |
| CsZHD10 | CsaV3_3G041500 | 33,868,099 | 33,870,888 (+) | 897 | 298 | 31.82 | 8.49 |
| CsZHD11 | CsaV3_5G038020 | 30,174,070 | 30,176,768 (−) | 924 | 307 | 33.29 | 8.19 |
| CsZHD14 | CsaV3_1G007410 | 4,706,303 | 4,707,523 (+) | 534 | 177 | 19.34 | 7.67 |
| CsMIF1 | CsaV3_1G005490 | 3,579,914 | 3,581,299 (+) | 318 | 105 | 11.31 | 8.86 |
| CsMIF2 | CsaV3_5G038010 | 30,158,234 | 30,163,341 (−) | 258 | 85 | 9.23 | 9.04 |
| CsMIF3 | CsaV3_1G002310 | 1,486,596 | 1,490,100 (+) | 330 | 109 | 12.16 | 8.71 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, Y.; Zhu, L.; An, M.; Wang, Y.; Li, S.; Dong, Y.; Yang, S.; Shi, K.; Fan, S.; Chen, X.; et al. Zinc Finger-Homeodomain Transcriptional Factors (ZHDs) in Cucumber (Cucumis sativus L.): Identification, Evolution, Expression Profiles, and Function under Abiotic Stresses. Int. J. Mol. Sci. 2024, 25, 4408. https://doi.org/10.3390/ijms25084408
Gao Y, Zhu L, An M, Wang Y, Li S, Dong Y, Yang S, Shi K, Fan S, Chen X, et al. Zinc Finger-Homeodomain Transcriptional Factors (ZHDs) in Cucumber (Cucumis sativus L.): Identification, Evolution, Expression Profiles, and Function under Abiotic Stresses. International Journal of Molecular Sciences. 2024; 25(8):4408. https://doi.org/10.3390/ijms25084408
Chicago/Turabian StyleGao, Yiming, Liyan Zhu, Menghang An, Yaru Wang, Sen Li, Yuming Dong, Songlin Yang, Kexin Shi, Shanshan Fan, Xiaofeng Chen, and et al. 2024. "Zinc Finger-Homeodomain Transcriptional Factors (ZHDs) in Cucumber (Cucumis sativus L.): Identification, Evolution, Expression Profiles, and Function under Abiotic Stresses" International Journal of Molecular Sciences 25, no. 8: 4408. https://doi.org/10.3390/ijms25084408
APA StyleGao, Y., Zhu, L., An, M., Wang, Y., Li, S., Dong, Y., Yang, S., Shi, K., Fan, S., Chen, X., Ren, H., & Liu, X. (2024). Zinc Finger-Homeodomain Transcriptional Factors (ZHDs) in Cucumber (Cucumis sativus L.): Identification, Evolution, Expression Profiles, and Function under Abiotic Stresses. International Journal of Molecular Sciences, 25(8), 4408. https://doi.org/10.3390/ijms25084408







