High-Resolution Cryo-Electron Microscopy Structure Determination of Haemophilus influenzae Tellurite-Resistance Protein A via 200 kV Transmission Electron Microscopy
Abstract
1. Introduction
2. Results
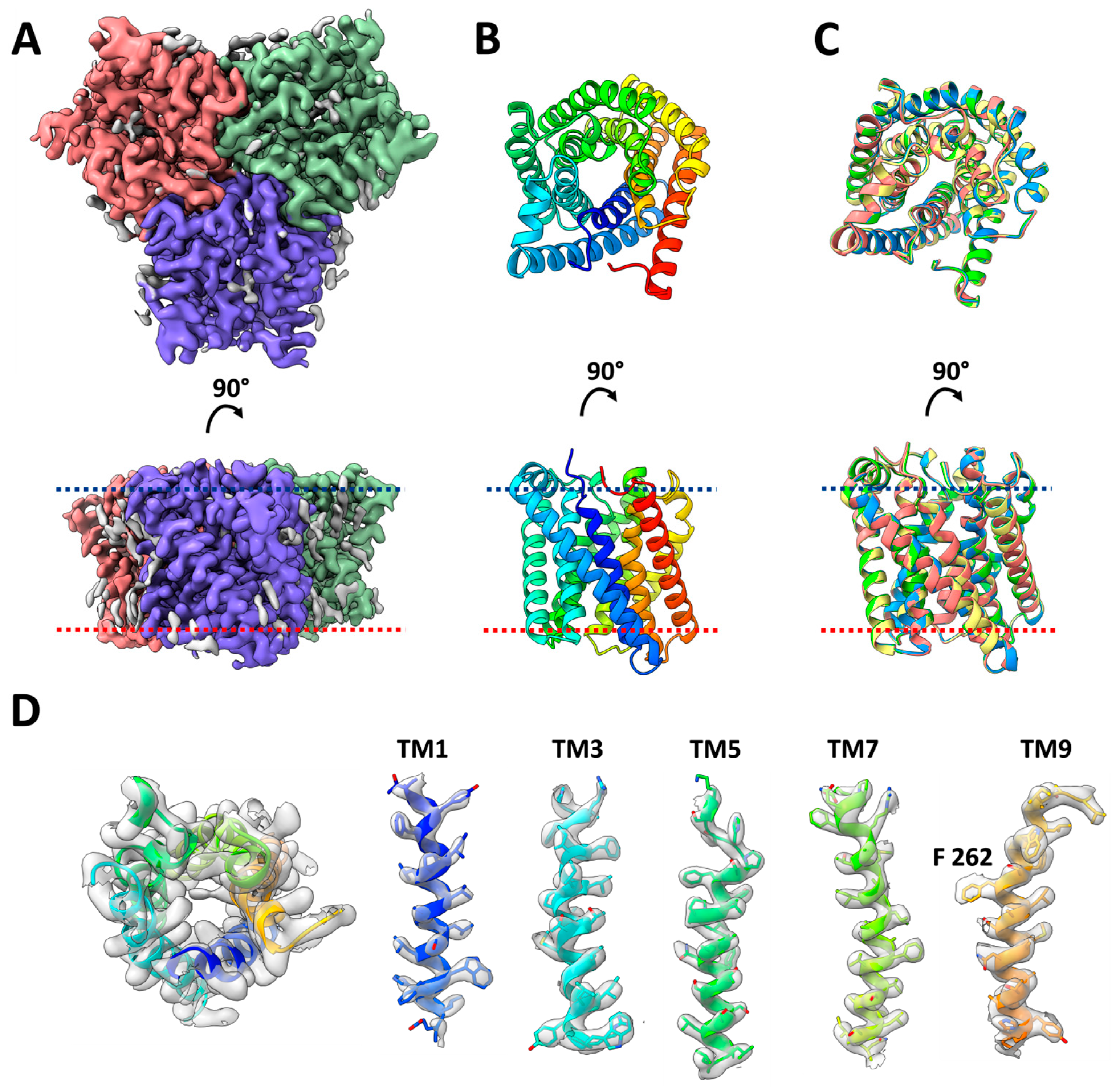
2.1. Structure Determination of HiTehA
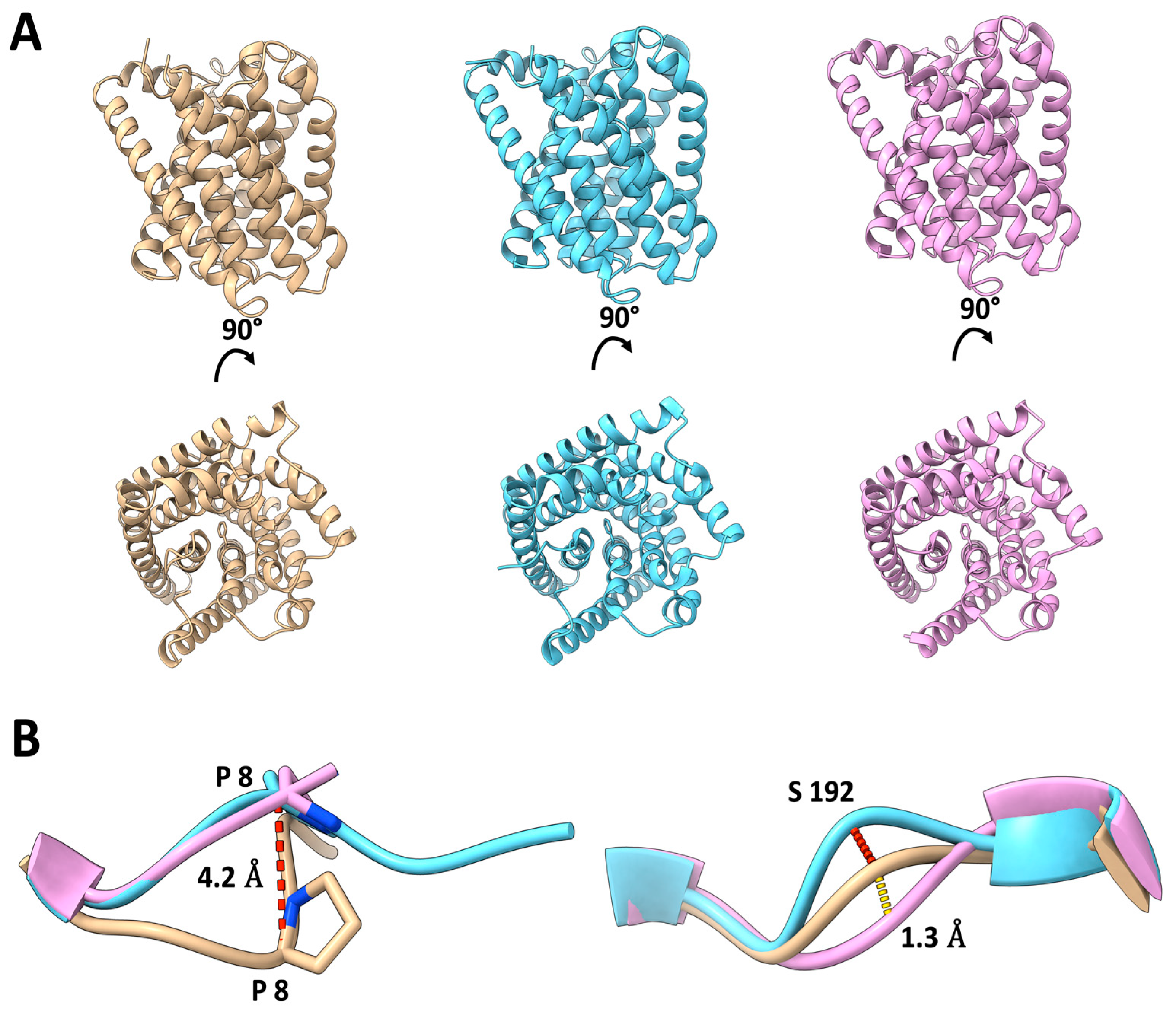
2.2. Comparison of Cryo-EM and X-ray Structures
3. Discussion
4. Materials and Methods
4.1. Protein Expression and Purification
4.2. Sample Preparation
4.3. Data Processing
4.4. Model Building
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fagerberg, L.; Jonasson, K.; von Heijne, G.; Uhlén, M.; Berglund, L. Prediction of the human membrane proteome. Proteomics 2010, 10, 1141–1149. [Google Scholar] [CrossRef] [PubMed]
- Bakheet, T.M.; Doig, A.J. Properties and identification of human protein drug targets. Bioinformatics 2009, 25, 451–457. [Google Scholar] [CrossRef] [PubMed]
- Seddon, A.M.; Curnow, P.; Booth, P.J. Membrane proteins, lipids and detergents: Not just a soap opera. Biochim. Biophys. Acta (BBA) Biomembr. 2004, 1666, 105–117. [Google Scholar] [CrossRef] [PubMed]
- Lichtenberg, D.; Ahyayauch, H.; Goñi, F.M. The Mechanism of Detergent Solubilization of Lipid Bilayers. Biophys. J. 2013, 105, 289–299. [Google Scholar] [CrossRef] [PubMed]
- le Maire, M.; Champeil, P.; Møller, J.V. Interaction of membrane proteins and lipids with solubilizing detergents. Biochim. Biophys. Acta (BBA) Biomembr. 2000, 1508, 86–111. [Google Scholar] [CrossRef]
- Guo, Y. Be Cautious with Crystal Structures of Membrane Proteins or Complexes Prepared in Detergents. Crystals 2020, 10, 86. [Google Scholar] [CrossRef]
- Yang, Z.; Wang, C.; Zhou, Q.; An, J.; Hildebrandt, E.; Aleksandrov, L.A.; Kappes, J.C.; DeLucas, L.J.; Riordan, J.R.; Urbatsch, I.L.; et al. Membrane protein stability can be compromised by detergent interactions with the extramembranous soluble domains. Protein Sci. 2014, 23, 769–789. [Google Scholar] [CrossRef] [PubMed]
- Chipot, C.; Dehez, F.; Schnell, J.R.; Zitzmann, N.; Pebay-Peyroula, E.; Catoire, L.J.; Miroux, B.; Kunji, E.R.S.; Veglia, G.; Cross, T.A. Perturbations of Native Membrane Protein Structure in Alkyl Phosphocholine Detergents: A Critical Assessment of NMR and Biophysical Studies. Chem. Rev. 2018, 118, 3559–3607. [Google Scholar] [CrossRef]
- Singh, S.K.; Sigworth, F.J. Cryo-EM: Spinning the Micelles Away. Structure 2015, 23, 1561. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lipfert, J.; Columbus, L.; Chu, V.B.; Lesley, S.A.; Doniach, S. Size and Shape of Detergent Micelles Determined by Small-Angle X-ray Scattering. J. Phys. Chem. B 2007, 111, 12427–12438. [Google Scholar] [CrossRef]
- Choy, B.C.; Cater, R.J.; Mancia, F.; Pryor, E.E. A 10-year meta-analysis of membrane protein structural biology: Detergents, membrane mimetics, and structure determination techniques. Biochim. Biophys. Acta (BBA) Biomembr. 2021, 1863, 183533. [Google Scholar] [CrossRef] [PubMed]
- Li, S. Detergents and alternatives in cryo-EM studies of membrane proteins. Acta Biochim. Biophys. Sin. 2022, 54, 1049–1056. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.H.; Hu, L.; Punta, M.; Bruni, R.; Hillerich, B.; Kloss, B.; Rost, B.; Love, J.; Siegelbaum, S.A.; Hendrickson, W.A. Homologue structure of the SLAC1 anion channel for closing stomata in leaves. Nature 2010, 467, 1074–1080. [Google Scholar] [CrossRef]
- Tsutsumi, N.; Mukherjee, S.; Waghray, D.; Janda, C.Y.; Jude, K.M.; Miao, Y.; Burg, J.S.; Aduri, N.G.; Kossiakoff, A.A.; Gati, C. Structure of human Frizzled5 by fiducial-assisted cryo-EM supports a heterodimeric mechanism of canonical Wnt signaling. eLife 2020, 9, e58464. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Rapoport, T.A. Cryo-EM structure determination of small proteins by nanobody-binding scaffolds (Legobodies). Proc. Natl. Acad. Sci. USA 2021, 118, e211500111. [Google Scholar] [CrossRef] [PubMed]
- Motiwala, Z.; Aduri, N.G.; Shaye, H.; Han, G.W.; Lam, J.H.; Katritch, V.; Cherezov, V.; Gati, C. Structural basis of GABA reuptake inhibition. Nature 2022, 606, 820–826. [Google Scholar] [CrossRef] [PubMed]
- Giri, N.; Roy, R.S.; Cheng, J. Deep learning for reconstructing protein structures from cryo-EM density maps: Recent advances and future directions. Curr. Opin. Struct. Biol. 2023, 79, 102536. [Google Scholar] [CrossRef] [PubMed]
- Cheng, A.; Yu, Y. Recent advances in data collection for Cryo-EM methods. Curr. Opin. Struct. Biol. 2024, 86, 102795. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Lander, G.C. How low can we go? Structure determination of small biological complexes using single-particle cryo-EM. Curr. Opin. Struct. Biol. 2020, 64, 9–16. [Google Scholar] [CrossRef]
- Chua, E.Y.; Mendez, J.H.; Rapp, M.; Ilca, S.L.; Tan, Y.Z.; Maruthi, K.; Kuang, H.; Zimanyi, C.M.; Cheng, A.; Eng, E.T.; et al. Better, Faster, Cheaper: Recent Advances in Cryo–Electron Microscopy. Annu. Rev. Biochem. 2022, 91, 1–32. [Google Scholar] [CrossRef]
- Axford, D.; Foadi, J.; Hu, N.J.; Choudhury, H.G.; Iwata, S.; Beis, K.; Evans, G.; Alguel, Y. Structure determination of an integral membrane protein at room temperature from crystals in situ. Acta Crystallogr. Sect. D 2015, 71, 1228–1237. [Google Scholar] [CrossRef] [PubMed]
- Suloway, C.; Pulokas, J.; Fellmann, D.; Cheng, A.; Guerra, F.; Quispe, J.; Stagg, S.; Potter, C.S.; Carragher, B. Automated molecular microscopy: The new Leginon system. J. Struct. Biol. 2005, 151, 41–60. [Google Scholar] [CrossRef]
- Cheng, A.; Negro, C.; Bruhn, J.F.; Rice, W.J.; Dallakyan, S.; Eng, E.T.; Waterman, D.G.; Potter, C.S.; Carragher, B. Leginon: New features and applications. Protein Sci. 2021, 30, 136–150. [Google Scholar] [CrossRef]
- Punjani, A.; Rubinstein, J.L.; Fleet, D.J.; Brubaker, M.A. cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 2017, 14, 290–296. [Google Scholar] [CrossRef]
- Rohou, A.; Grigorieff, N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 2015, 192, 216–221. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 2016, 193, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Bepler, T.; Morin, A.; Rapp, M.; Brasch, J.; Shapiro, L.; Noble, A.J.; Berger, B. Positive-unlabeled convolutional neural networks for particle picking in cryo-electron micrographs. Nat. Methods 2019, 16, 1153–1160. [Google Scholar] [CrossRef]
- Goddard, T.D.; Huang, C.C.; Meng, E.C.; Pettersen, E.F.; Couch, G.S.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 2018, 27, 14–25. [Google Scholar] [CrossRef] [PubMed]
- Afonine, P.V.; Poon, B.K.; Read, R.J.; Sobolev, O.V.; Terwilliger, T.C.; Urzhumtsev, A.; Adams, P.D. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D Struct. Biol. 2018, 74, 531–544. [Google Scholar] [CrossRef]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef]
- Chen, V.B.; Arendall, W.B.; Headd, J.J.; Keedy, D.A.; Immormino, R.M.; Kapral, G.J.; Murray, L.W.; Richardson, J.S.; Richardson, D.C. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 12–21. [Google Scholar] [CrossRef] [PubMed]

| Parameter | TehA:GDN | TehA:LMNG | TehA:DDM | TehA:OG |
|---|---|---|---|---|
| Data Collection | ||||
| Blot Time | 10 s | 10 s | 10 s | 3 s |
| Blot Force | 10 | 10 | 10 | 25 |
| Wait Time | 0 s | 20 s | 20 s | 0 s |
| Microscope | Glacios | Glacios | Glacios | Glacios |
| Voltage (kV) | 200 kV | 200 kV | 200 kV | 200 kV |
| Magnification | 240,000 | 240,000 | 240,000 | 240,000 |
| Detector | Falcon 4 | Falcon 4 | Falcon 4 | Falcon 4 |
| Pixel Size (Å/pixel) | 0.566 | 0.566 | 0.566 | 0.566 |
| Total Electron Dose | 36.43 e−/Å2 | 36.43 e−/Å2 | 36.43 e−/Å2 | 36.43 e−/Å2 |
| EER Internal Frames | 840 | 840 | 840 | 840 |
| Defocus Range (μm) | 0.5–1.5 | 0.5–1.5 | 0.5–1.5 | 0.5–1.5 |
| Micrograph Images (no.) | 7352 | 11370 | 3585 | 8354 |
| 3D Reconstruction | ||||
| Software | cryoSPARC 4.4 | cryoSPARC 4.4 | cryoSPARC 4.4 | cryoSPARC 4.4 |
| Final Number of Particles in the Reconstruction | 40,701 | 36,758 | 63,824 | 30,170 |
| Imposed Symmetry | C3 | C3 | C3 | C3 |
| Fourier Shell Correlation (FSC) Cut-Off | 0.143 | 0.143 | 0.143 | 0.143 |
| Map Resolution (Å) | 2.9 | 3.1 | 3.1 | 3.2 |
| Atomic Modelling and Refinement Statistics | ||||
| Software | COOT, Phenix | COOT, Phenix | COOT, Phenix | COOT, Phenix |
| Homology Model (PDB Accession Code) | 3M71 | 3M71 | 3M71 | 3M71 |
| B-factor (Å2) | 82.1 | 87.7 | 106.1 | 104.7 |
| Total Number of Atoms | 7359 | 7359 | 7382 | 7359 |
| Protein Residues | 924 | 924 | 924 | 924 |
| RMSD2 bond length(Å) | 0.002 | 0.003 | 0.002 | 0.003 |
| RMSD2 bond angles (°) | 0.436 | 0.463 | 0.456 | 0.494 |
| Molprobity Score | 1.22 | 1.24 | 1.40 | 1.24 |
| All Atom Clash Score | 4.45 | 4.66 | 3.83 | 4.66 |
| Ramachandran Plot (%) | ||||
| Favored | 98.80 | 98.26 | 96.51 | 98.47 |
| Allowed | 1.20 | 1.74 | 3.49 | 1.53 |
| Outliers | 0.00 | 0.00 | 0.00 | 0.00 |
| Data | ||||
| Box Pixel Size | 440/440/440 | 440/440/440 | 440/440/440 | 440/440/440 |
| Angles (o) | 90.00, 90.00, 90.00 | 90.00, 90.00, 90.00 | 90.00, 90.00, 90.00 | 90.00, 90.00, 90.00 |
| Resolution Estimates (Å) | Masked-Unmasked | Masked-Unmasked | Masked-Unmasked | Masked-Unmasked |
| d FSC (half maps; 0.143) | 3.0–3.2 | 3.1–3.2 | 3.2–3.3 | 3.2–3.4 |
| d 99 (full/half1/half2) | 3.4/1.2/1.2–3.3/1.1/1.1 | 3.4/1.1/1.1–3.4/1.1/1.1 | 3.5/1.2/1.2–3.5/1.1/1.1 | 3.5/1.2/1.2–3.4/1.1/1.1 |
| d Model | 3.3–3.3 | 3.4–3.4 | 3.5–3.5 | 3.5–3.5 |
| d FSC Model (0/0.143/0.5) | 2.9/2.9/3.2–2.9/3.0/3.2 | 3.0/3.0/3.2–3.0/3.1/3.3 | 3.0/3.1/3.3–3.0/3.1/3.4 | 3.1/3.1/3.3–3.1/3.2/3.4 |
| Map min/max/mean | −0.36/0.59/0.02 | −0.34/0.58/0.02 | −0.31/0.53/0.02 | −0.36/0.55/0.01 |
| Model vs. Data | ||||
| CC (Mask) | 0.88 | 0.88 | 0.85 | 0.85 |
| CC (Box) | 0.70 | 0.70 | 0.62 | 0.63 |
| CC (Peaks) | 0.68 | 0.68 | 0.60 | 0.59 |
| CC (Volume) | 0.82 | 0.82 | 0.83 | 0.84 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tran, N.L.; Senko, S.; Lucier, K.W.; Farwell, A.C.; Silva, S.M.; Dip, P.V.; Poweleit, N.; Scapin, G.; Catalano, C. High-Resolution Cryo-Electron Microscopy Structure Determination of Haemophilus influenzae Tellurite-Resistance Protein A via 200 kV Transmission Electron Microscopy. Int. J. Mol. Sci. 2024, 25, 4528. https://doi.org/10.3390/ijms25084528
Tran NL, Senko S, Lucier KW, Farwell AC, Silva SM, Dip PV, Poweleit N, Scapin G, Catalano C. High-Resolution Cryo-Electron Microscopy Structure Determination of Haemophilus influenzae Tellurite-Resistance Protein A via 200 kV Transmission Electron Microscopy. International Journal of Molecular Sciences. 2024; 25(8):4528. https://doi.org/10.3390/ijms25084528
Chicago/Turabian StyleTran, Nhi L., Skerdi Senko, Kyle W. Lucier, Ashlyn C. Farwell, Sabrina M. Silva, Phat V. Dip, Nicole Poweleit, Giovanna Scapin, and Claudio Catalano. 2024. "High-Resolution Cryo-Electron Microscopy Structure Determination of Haemophilus influenzae Tellurite-Resistance Protein A via 200 kV Transmission Electron Microscopy" International Journal of Molecular Sciences 25, no. 8: 4528. https://doi.org/10.3390/ijms25084528
APA StyleTran, N. L., Senko, S., Lucier, K. W., Farwell, A. C., Silva, S. M., Dip, P. V., Poweleit, N., Scapin, G., & Catalano, C. (2024). High-Resolution Cryo-Electron Microscopy Structure Determination of Haemophilus influenzae Tellurite-Resistance Protein A via 200 kV Transmission Electron Microscopy. International Journal of Molecular Sciences, 25(8), 4528. https://doi.org/10.3390/ijms25084528






