Anti-Obesity Therapeutic Targets Studied In Silico and In Vivo: A Systematic Review
Abstract
1. Introduction
2. Methods
2.1. Protocol and Registration
2.2. Search Question
2.3. Inclusion Criteria
2.4. Exclusion Criteria
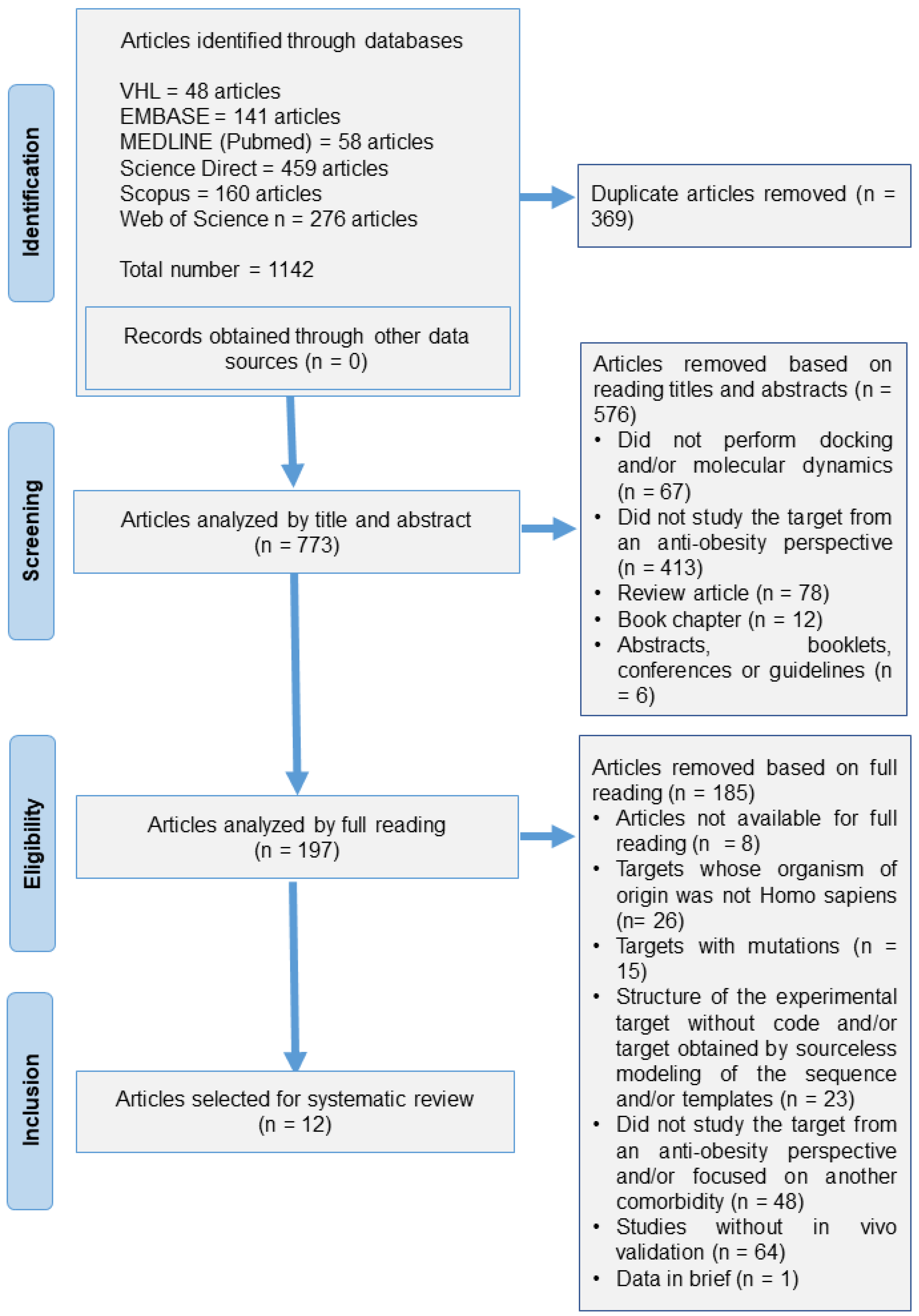
2.5. Search Strategy
2.6. Data Selection and Extraction
2.7. Data Analysis and Synthesis
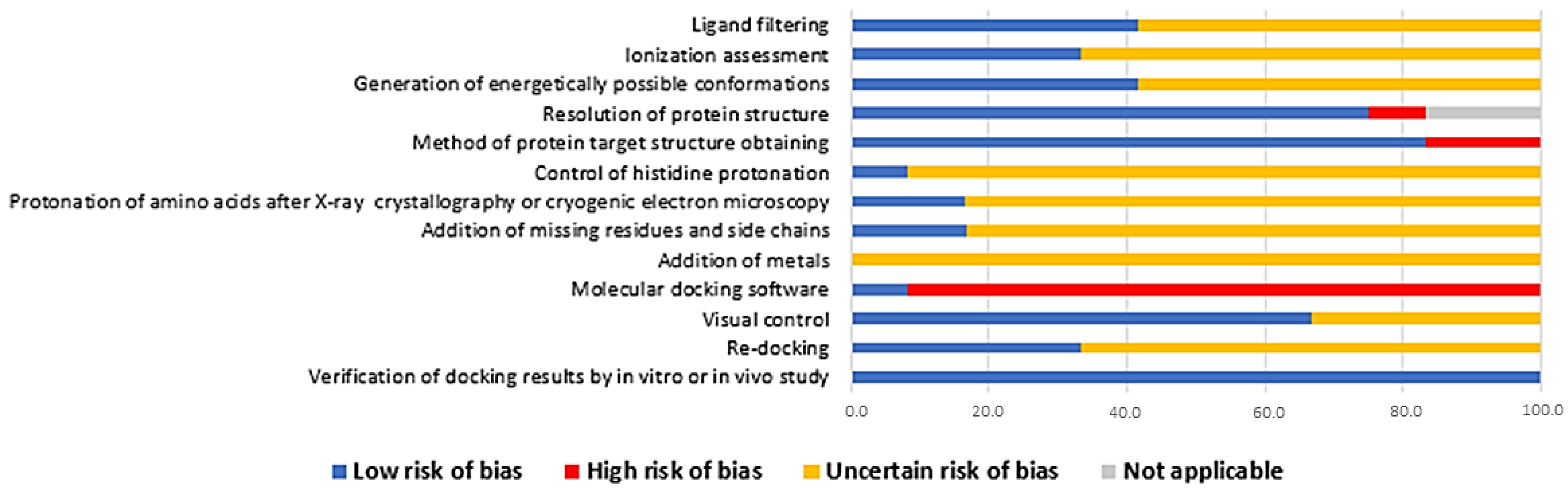
2.8. Assessment of the Risk of Bias in the Included Studies
3. Results and Discussion
3.1. Articles Included in Search
3.2. Risk of Bias Assessment
3.3. Theoretical and Modeling Structures in In Silico Studies
3.4. Animal Models Employed in the Validation of In Silico Studies
3.5. In Silico-Studied Therapeutic Targets in Obesity
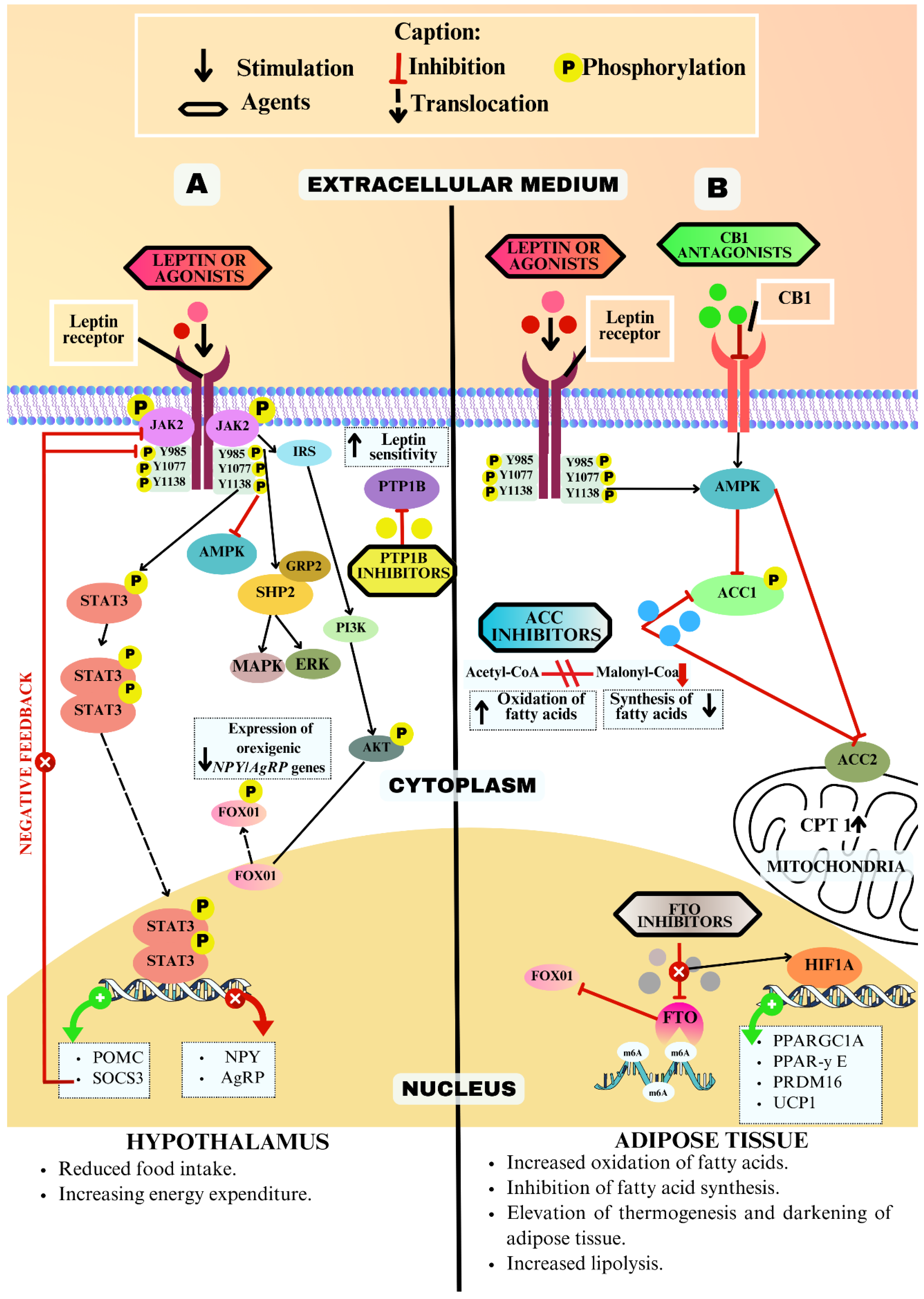
3.5.1. Leptin Receptor (LEPR)
3.5.2. Protein Tyrosine Phosphatase 1B (PTP1B)
3.5.3. Fat Mass and Obesity-Associated Protein (FTO)
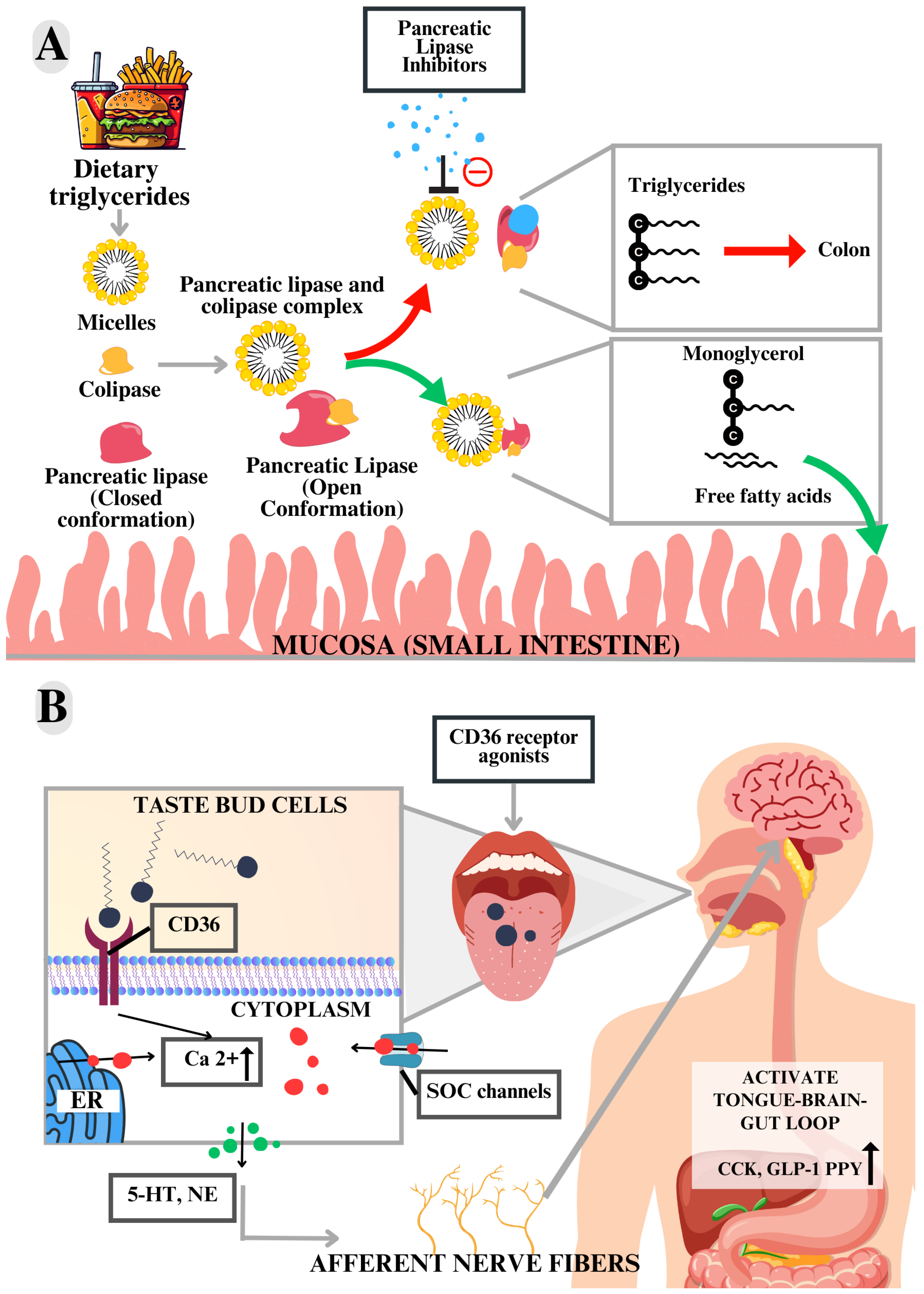
3.5.4. Lingual Cluster of Differentiation 36 (CD36)
3.5.5. Acetyl-CoA Carboxylase (ACC)
3.5.6. Type 1 Cannabinoid Receptor (CB1)
3.5.7. Pancreatic Lipase (PL)
3.6. Limitations of the Study
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Lobstein, T.; Jackson-Leach, R.; Powis, J. Obesity Atlas 2023; World Obesity Federation: London, UK, 2023. [Google Scholar]
- Lobstein, T.; Brinsden, H.; Neveux, M. World Obesity Atlas 2024; World Obesity Federation: London, UK, 2024. [Google Scholar]
- Jackson, S.; Llewellyn, C.; Smith, L. The obesity epidemic—Nature via nurture: A narrative review of high-income countries. SAGE Open Med. 2020, 8, 205031212091826. [Google Scholar] [CrossRef] [PubMed]
- Lobstein, T.; Brinsden, H.; Neveux, M. World Obesity Atlas 2022; World Obesity Federation: London, UK, 2022. [Google Scholar]
- Tajik, S.; Mirzababaei, A.; Ghaedi, E.; Kord-Varkaneh, H.; Mirzaei, K. Risk of type 2 diabetes in metabolically healthy people in different categories of body mass index: An updated network meta-analysis of prospective cohort studies. J. Cardiovasc. Thorac. Res. 2019, 11, 254. [Google Scholar] [CrossRef] [PubMed]
- Barroso, W.K.S.; Rodrigues, C.I.S.; Bortolotto, L.A.; Mota-Gomes, M.A.; Brandão, A.A.; Feitosa, A.D.d.M.; Machado, C.A.; Poli-De-Figueiredo, C.E.; Amodeo, C.; Mion, D.; et al. Diretrizes Brasileiras de Hipertensão Arterial-2020. Arq. Bras. Cardiol. 2021, 116, 516–658. [Google Scholar] [CrossRef] [PubMed]
- Avgerinos, K.; Spyrou, N.; Mantzoros, C.; Dalamaga, M. Obesity and cancer risk: Emerging biological mechanisms and perspectives. Metabolism 2019, 92, 121–135. [Google Scholar] [CrossRef] [PubMed]
- Bays, H.E.; Fitch, A.; Christensen, S.; Burridge, K.; Tondt, J. Anti-Obesity Medications and Investigational Agents: An Obesity Medicine Association (OMA) Clinical Practice Statement (CPS) 2022. Obes. Pillars 2022, 2, 100018. [Google Scholar] [CrossRef] [PubMed]
- Wharton, S.; Lau, D.C.; Vallis, M.; Sharma, A.M.; Biertho, L.; Campbell-Scherer, D.; Adamo, K.; Alberga, A.; Bell, R.; Boulé, N.; et al. Obesity in adults: A clinical practice guideline. Can. Med. Assoc. J. 2020, 192, E875–E891. [Google Scholar] [CrossRef] [PubMed]
- ClinicalTrials.gov. A Study of IBI362 9 mg in Chinese Adults with Obesity. NCT06164873, Phase III Trial. 2023. Available online: https://clinicaltrials.gov/study/NCT06164873?term=NCT06164873&rank=1 (accessed on 11 April 2024).
- ClinicalTrials.gov. A Study to Test Whether BI 456906 Helps Japanese People Living with Obesity Disease (SYNCHRONIZE™JP). NCT06176365, Phase III Trial. 2023. Available online: https://clinicaltrials.gov/study/NCT06176365?term=NCT06176365&rank=1 (accessed on 11 April 2024).
- ClinicalTrials.gov. A Study of Once-Daily Oral Orforglipron (LY3502970) in Japanese Adult Participants with Obesity Disease (ATTAIN-J). NCT05931380. Phase III Trial. 2023. Available online: https://clinicaltrials.gov/study/NCT05931380?term=NCT05931380&rank=1 (accessed on 11 April 2024).
- Rosa-Gonçalves, P.; Majerowicz, D. Pharmacotherapy of Obesity: Limits and Perspectives. Am. J. Cardiovasc. Drugs 2019, 19, 349–364. [Google Scholar] [CrossRef]
- Rehman, K.; Munawar, S.M.; Akash, M.S.H.; Buabeid, M.A.; Chohan, T.A.; Tariq, M.; Jabeen, K.; Arafa, E.-S.A. Hesperidin improves insulin resistance via down-regulation of inflammatory responses: Biochemical analysis and in silico validation. PLoS ONE 2020, 15, e0227637. [Google Scholar] [CrossRef]
- Prabhakar, L.; John, D.; Singh, S.; Murali, A. Computational analysis of marine algal compounds for obesity management against pancreatic lipase. J. Biomol. Struct. Dyn. 2022, 41, 4863–4872. [Google Scholar] [CrossRef] [PubMed]
- Glykofrydi, S.; Kokkinos, A.; Barber, T.; Mastorakos, G.; Valsamakis, G. Existing and Emerging Molecular Targets for the Pharmacotherapy of Obesity; Endotext; MDText.com, Inc.: South Dartmouth, MA, USA, 2020. [Google Scholar]
- Maia, E.; Assis, L.; Oliveira, T.; Silva, A.; Taranto, A. Structure-Based Virtual Screening: From Classical to Artificial Intelligence. Front. Chem. 2020, 8, 343. [Google Scholar] [CrossRef]
- Burley, S.K.; Bhikadiya, C.; Bi, C.; Bittrich, S.; Chen, L.; Crichlow, G.V.; Duarte, J.M.; Dutta, S.; Fayazi, M.; Feng, Z.; et al. RCSBProtein Data Bank: Celebrating 50 years of the PDB with new tools for understanding visualizing biological macromolecules in 3D. Protein Sci. 2022, 31, 187–208. [Google Scholar] [CrossRef] [PubMed]
- Turdzo, S.; Hantz, E.; Lindert, S. Applications of machine learning in computer-aided drug Discovery QRB. Discovery 2022, 3, e14. [Google Scholar] [CrossRef]
- Batool, M.; Ahmad, B.; Choi, S. A Structure-Based Drug Discovery Paradigm. Int. J. Mol. Sci. 2019, 20, 2783. [Google Scholar] [CrossRef]
- Korkmaz, T.; Ayaz, F. Molecular docking: A powerful tool for predicting protein-ligand interactions. In Proceedings of the 7th Advanced Engineering Days (AED), Mersin, Turkey, 9 July 2023. [Google Scholar]
- Ricci-Lopez, J.; Aguila, S.; Gilson, M.; Brizuela, A. Improving Structure-Based Virtual Screening with Ensemble Docking and Machine Learning. J. Chem. Inf. Model. 2021, 61, 5362–5376. [Google Scholar] [CrossRef]
- Ferreira, L.; Santos, R.; Olivia, G.; Andricopulo, A. Molecular Docking and Structure-Based Drug Design Strategies. Molecules 2015, 20, 13384–13421. [Google Scholar] [CrossRef]
- Hu, X.; Zeng, Z.; Zhang, J.; Wu, D.; Li, H.; Geng, F. Molecular dynamics simulation of the interaction of food proteins with small molecules. Food Chem. 2023, 405, 134824. [Google Scholar] [CrossRef] [PubMed]
- Senol, H.; Ghaffari-Moghaddam, M.; Toraman, G.; Guller, U. Novel chalcone derivatives of ursolic acid as acetylcholinesterase inhibitors: Synthesis, characterization, biological activity, ADME prediction, molecular docking and molecular dynamics studies. J. Mol. Struct. 2024, 1295, 136804. [Google Scholar] [CrossRef]
- Westbrook, J.D.; Burley, S.K. How Structural Biologists and the Protein Data Bank Contributed to Recent US FDA New Drug Approvals. Structure 2019, 27, 211. [Google Scholar] [CrossRef] [PubMed]
- Jayarathne, S.; Koboziev, I.; Park, O.; Oldewage-Theron, W.; Shen, C.; Moustaid-Moussa, N. Anti-Inflammatory and Anti-Obesity Properties of Food Bioactive Components: Effects on Adipose Tissue. Prev. Nutr. Food Sci. 2017, 22, 251–262. [Google Scholar] [CrossRef]
- Müller, T.; Blüher, M.; Tschöp, M.; DiMarchi, R. Anti-obesity drug discovery: Advances and challenges. Nat. Rev. Drug Discov. 2021, 21, 201–223. [Google Scholar] [CrossRef]
- Gomes, A.F.T.; de Medeiros, W.F.; de Oliveira, G.S.; Medeiros, I.; Maia, J.K.d.S.; Bezerra, I.W.L.; Piuvezam, G.; Morais, A.H.d.A. In silico structure-based designers of therapeutic targets for diabetes mellitus or obesity: A protocol for systematic review. PLoS ONE 2022, 17, e0279039. [Google Scholar] [CrossRef] [PubMed]
- Ouzzani, M.; Hammady, H.; Fedorowicz, Z.; Elmagarmid, A. Rayyan-a web and mobile app for systematic reviews. Syst. Rev. 2016, 5, 210. [Google Scholar] [CrossRef] [PubMed]
- Taldaev, A.; Terekhov, R.; Nikitin, I.; Zhevlakova, A.; Selivanova, I. Insights into the Pharmacological Effects of Flavonoids: The Systematic Review of Computer Modeling. Int. J. Mol. Sci. 2022, 23, 6023. [Google Scholar] [CrossRef]
- Birari, R.; Gupta, S.; Mohan, C.; Bhutani, K. Antiobesity and lipid lowering effects of Glycyrrhiza chalcones: Experimental and computational studies. Phytomedicine 2011, 18, 795–801. [Google Scholar] [CrossRef] [PubMed]
- Coronado-Cáceres, L.; Rabadán-Chávez, G.; Mojica, L.; Hernández-Ledesma, B.; Quevedo-Corona, L.; Cervantes, E.L. Cocoa (Theobroma cacao L.) Seed Proteins’ Anti-Obesity Potential through Lipase Inhibition Using In Silico, In Vitro and In Vivo Models. Foods 2020, 9, 1359. [Google Scholar] [CrossRef]
- El-Korany, S.; Helmy, O.; El-Halawany, A.; Ragab, Y.; Zedan, H. Kojic acid repurposing as a pancreatic lipase inhibitor and the optimization of its production from a local Aspergillus oryzae soil isolate. BMC Biotechnol. 2020, 20, 52. [Google Scholar] [CrossRef] [PubMed]
- Yakaiah, V.; Dakshinamoorthi, A.; Ty, S. Novel Aspects in Inhibiting Pancreatic Lipase with Potential New Compound from Nutmeg in Connection with Obesity—In Vitro, In Silico, In Vivo and Ex Vivo Studies. Maedica 2021, 16, 445–452. [Google Scholar] [CrossRef]
- Elekofehinti, O.; Lawal, A.; Ejelonu, O.; Molehin, O.; Famusiwa, C. Involvement of fat mass and obesity gene (FTO) in the anti-obesity action of Annona muricata Annonaceae: In silico and in vivo studies. J. Diabetes Metab. Disord. 2020, 19, 197–204. [Google Scholar] [CrossRef]
- Yaccoubi, F.; El-Naggar, M.; Abdelrazek, F.M.; Gomha, S.M.; Farghaly, M.S.; Abolibda, T.Z.; Ali, L.A.; Abdelmonsef, A.H. Pyrido-pyrimido-thiadiazinones: Green synthesis, molecular docking studies and biological investigation as obesity inhibitors. J. Taibah Univ. Sci. 2022, 16, 1275–1286. [Google Scholar] [CrossRef]
- Fajriaty, I.; Ih, H.; Fidrianny, I.; Kurniati, N.F.; Reynaldi, M.A.; Adnyana, I.K.; Rommy, R.; Kurniawan, F.; Tjahjono, D.H. In Vivo Pharmacodynamics of Calophyllum soulattri as Antiobesity with In Silico Molecular Docking and ADME/Pharmacokinetic Prediction Studies. Pharmaceuticals 2023, 16, 191. [Google Scholar] [CrossRef]
- Khan, A.S.; Hichami, A.; Murtaza, B.; Louillat-Habermeyer, M.-L.; Ramseyer, C.; Azadi, M.; Yesylevskyy, S.; Mangin, F.; Lirussi, F.; Leemput, J.; et al. Novel Fat Taste Receptor Agonists Curtail Progressive Weight Gain in Obese Male Mice. Cell Mol. Gastroenterol. Hepatol. 2023, 15, 633–663. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Seo, H.J.; Lee, S.-H.; Jung, M.E.; Park, J.-H.; Park, H.-J.; Yoo, J.; Yun, H.; Na, J.; Kang, S.Y.; et al. Biarylpyrazolyl Oxadiazole as Potent, Selective, Orally Bioavailable Cannabinoid-1 Receptor Antagonists for the Treatment of Obesity. J. Med. Chem. 2008, 51, 7216–7233. [Google Scholar] [CrossRef]
- Ghareb, N.; El-Sayed, N.; Abdelhameed, R.; Yamada, K.; Elgawish, M. Toward a treatment of diabesity: Rational design, synthesis and biological evaluation of benzene-sulfonamide derivatives as a new class of PTP-1B inhibitors. Bioorganic Chem. 2019, 86, 322–338. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Hu, M.; Tang, M.; Gao, C.; Wang, H.; Man, S.; Lu, F. Oligosaccharide and short-chain fatty acid: A double-edged sword in obese mice by regulating food intake and fat synthesis. Food Res. Int. 2022, 159, 111619. [Google Scholar] [CrossRef] [PubMed]
- Kang, N.; Oh, S.; Kim, S.-Y.; Ahn, H.; Son, M.; Heo, S.-J.; Byun, K.; Jeon, Y.-J. Anti-obesity effects of Ishophloroglucin A from the brown seaweed Ishige okamurae (Yendo) via regulation of leptin signal in ob/ob mice. Algal Res. 2022, 61, 102533. [Google Scholar] [CrossRef]
- Giordano, D.; Biancaniello, C.; Argenio, M.; Facchiano, A. Drug Design by Pharmacophore and Virtual Screening Approach. Pharmaceuticals 2022, 15, 646. [Google Scholar] [CrossRef]
- Sun, D.; Gao, W.; Hu, H.; Zhou, S. Why 90% of clinical drug development fails and how to improve it? Acta Pharm. Sin. B 2022, 12, 3049–3062. [Google Scholar] [CrossRef] [PubMed]
- Somda, D.; Kpordze, S.W.; Jerpkorir, M.; Mahora, M.C.; Ndungu, J.W.; Kamau, S.W.; Arthur, V.; Elbasyouni, A. The Role of Bioinformatics in Drug Discovery: A Comprehensive Overview. Drug Metabolism and Pharmacokinetics. In Drug Metabolism and Pharmacokinetics; IntechOpen: Rijeka, Croatia, 2023. [Google Scholar] [CrossRef]
- Suleiman, J.; Mohamed, M.; Bakar, A. A systematic review on different models of inducing obesity in animals: Advantages and limitations. J. Adv. Vet. Anim. Res. 2020, 7, 103. [Google Scholar] [CrossRef] [PubMed]
- Preguiça, I.; Alves, A.; Nunes, S.; Fernandes, R.; Gomes, P.; Viana, S.D.; Reis, F. Diet-induced rodent models of obesity-related metabolic disorders—A guide to a translational perspective. Obes. Rev. 2020, 21, e13081. [Google Scholar] [CrossRef] [PubMed]
- Dias, M.d.M.e.; dos Reis, S.A.; da Conceição, L.L.; Sediyama, C.M.N.d.O.; Pereira, S.S.; de Oliveira, L.L.; Peluzio, M.D.C.G.; Martinez, J.A.; Milagro, F.I. Diet-induced obesity in animal models: Points to consider and influence on metabolic markers. Diabetol. Metab. Syndr. 2021, 13, 32. [Google Scholar] [CrossRef]
- Madden, J.; Enoch, S.; Paini, A.; Cronin, M. A Review of In Silico Tools as Alternatives to Animal Testing: Principles, Resources and Applications. Altern. Lab. Anim. 2020, 48, 146–172. [Google Scholar] [CrossRef] [PubMed]
- Greco, M.; De Santo, M.; Comandè, A.; Belsito, E.L.; Andò, S.; Liguori, A.; Leggio, A. Leptin-Activity Modulators and Their Potential Pharmaceutical Applications. Biomolecules 2021, 11, 1045. [Google Scholar] [CrossRef] [PubMed]
- Andreoli, M.; Donato, J.; Cakir, I.; Perello, M. Leptin resensitisation: A reversion of leptin-resistant states. J. Endocrinol. 2019, 241, 81–96. [Google Scholar] [CrossRef] [PubMed]
- Daval, M.; Foufelle, F.; Ferré, P. Functions of AMP-activated protein kinase in adipose tissue. J. Physiol. 2006, 574, 55. [Google Scholar] [CrossRef] [PubMed]
- Minokoshi, Y.; Kim, Y.-B.; Peroni, O.D.; Fryer, L.G.D.; Müller, C.; Carling, D.; Kahn, B.B. Leptin stimulates fatty-acid oxidation by activating AMP-activated protein kinase. Nature 2002, 415, 339–343. [Google Scholar] [CrossRef]
- Fediuc, S.; Gaidhu, M.; Ceddia, R. Regulation of AMP-activated protein kinase and acetyl-CoA carboxylase phosphorylation by palmitate in skeletal muscle cells. J. Lipid Res. 2006, 47, 412–420. [Google Scholar] [CrossRef] [PubMed]
- Tarasiuk, O.; Miceli, M.; Di Domizio, A.; Nicolini, G. AMPK and Diseases: State of the Art Regulation by AMPK-Targeting Molecules. Biology 2022, 11, 1041. [Google Scholar] [CrossRef]
- Minokoshi, Y.; Alquier, T.; Furukawa, N.; Kim, Y.-B.; Lee, A.; Xue, B.; Mu, J.; Foufelle, F.; Ferré, P.; Birnbaum, M.J.; et al. AMP-kinase regulates food intake by responding to hormonal and nutrient signals in the hypothalamus. Nature 2004, 428, 569–574. [Google Scholar] [CrossRef]
- Vohra, M.; Benchoula, K.; Serpell, C.; Hwa, W. AgRP/NPY and POMC neurons in the arcuate nucleus and their potential role in treatment of obesity. Eur. J. Pharmacol. 2022, 915, 174611. [Google Scholar] [CrossRef]
- Yu, Y.; Wu, Y.; Szabo, A.; Wu, Z.; Wang, H.; Li, D.; Huang, X.-F. Teasaponin Reduces Inflammation and Central Leptin Resistance in Diet-Induced Obese Male Mice. Endocrinology 2013, 154, 3130–3140. [Google Scholar] [CrossRef]
- Lund, I.; Hansen, J.; Andersen, H.; Møller, N.; Billestrup, N. Mechanism of protein tyrosine phosphatase 1B-mediated inhibition of leptin signalling. J. Mol. Endocrinol. 2005, 34, 339–351. [Google Scholar] [CrossRef] [PubMed]
- Campos-Almazán, M.I.; Hernández-Campos, A.; Castillo, R.; Sierra-Campos, E.; Valdez-Solana, M.; Avitia-Domínguez, C.; Téllez-Valencia, A. Computational Methods in Cooperation with Experimental Approaches to Design Protein Tyrosine Phosphatase 1B Inhibitors in Type 2 Diabetes Drug Design: A Review of the Achievements of This Century. Pharmaceuticals 2022, 15, 866. [Google Scholar] [CrossRef] [PubMed]
- You-Ten, K.E.; Muise, E.S.; Itié, A.; Michaliszyn, E.; Wagner, J.; Jothy, S.; Lapp, W.S.; Tremblay, M.L. Impaired Bone Marrow Microenvironment and Immune Function in T Cell Protein Tyrosine Phosphatase–deficient Mice. J. Exp. Med. 1997, 186, 683–693. [Google Scholar] [CrossRef] [PubMed]
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.-G.; et al. N6-Methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011, 7, 885–887. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Roundtree, I.; He, C. Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 2017, 18, 31. [Google Scholar] [CrossRef]
- Yang, Z.; Yu, G.; Zhu, X.; Peng, T.; Lv, Y. Critical roles of FTO-mediated mRNA m6A demethylation in regulating adipogenesis and lipid metabolism: Implications in lipid metabolic disorders. Genes Dis. 2022, 9, 51–61. [Google Scholar] [CrossRef]
- Church, C.; Moir, L.; McMurray, F.; Girard, C.; Banks, G.T.; Teboul, L.; Wells, S.; Brüning, J.C.; Nolan, P.M.; Ashcroft, F.M.; et al. Overexpression of Fto leads to increased food intake and results in obesity. Nat. Genet. 2010, 42, 1086–1092. [Google Scholar] [CrossRef]
- Wu, R.; Chen, Y.; Liu, Y.; Zhuang, L.; Chen, W.; Zeng, B.; Liao, X.; Guo, G.; Wang, Y.; Wang, X. m6A methylation promotes white-to-beige fat transition by facilitating Hif1a translation. EMBO Rep. 2021, 22, e52348. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.; Xiao, W.; Ju, D.; Sun, B.; Hou, N.; Liu, Q.; Wang, Y.; Zhao, H.; Gao, C.; Zhang, S.; et al. Identification of entacapone as a chemical inhibitor of FTO mediating metabolic regulation through FOXO1. Sci. Transl. Med. 2019, 11, eaau7116. [Google Scholar] [CrossRef]
- El-Yassimi, A.; Hichami, A.; Besnard, P.; Khan, N. Linoleic Acid Induces Calcium Signaling, Src Kinase Phosphorylation, and Neurotransmitter Release in Mouse CD36-positive Gustatory Cells. J. Biol. Chem. 2008, 283, 12949–12959. [Google Scholar] [CrossRef]
- Proserpio, C.; Laureati, M.; Bertoli, S.; Battezzati, A.; Pagliarini, E. Determinants of Obesity in Italian Adults: The Role of Taste Sensitivity, Food Liking, and Food Neophobia. Chem. Senses 2015, 41, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Huang, T. Recent development in acetyl-CoA carboxylase inhibitors and their potential as novel drugs. Future Med. Chem. 2020, 12, 533–561. [Google Scholar] [CrossRef] [PubMed]
- Abu-Elheiga, L.; Almarza-Ortega, D.; Baldini, A.; Wakil, S. Human Acetyl-CoA Carboxylase 2. J. Biol. Chem. 1997, 272, 10669–10677. [Google Scholar] [CrossRef] [PubMed]
- Wakil, S.; Abu-Elheiga, L. Fatty acid metabolism: Target for metabolic syndrome. J. Lipid Res. 2009, 50, S138–S143. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan, S.; Yates, A.; Porter, R. The Peripheral Cannabinoid Receptor Type 1 (CB1) as a Molecular Target for Modulating Body Weight in Man. Molecules 2021, 26, 6178. [Google Scholar] [CrossRef]
- Lange, J.; Kruse, C. Keynote review: Medicinal chemistry strategies to CB1 cannabinoid receptor antagonists. Drug Discov. Today 2005, 10, 693–702. [Google Scholar] [CrossRef] [PubMed]
- Cinar, R.; Iyer, M.; Kunos, G. The therapeutic potential of second and third generation CB1R antagonists. Pharmacol. Ther. 2020, 208, 107477. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Godlewski, G.; Jourdan, T.; Liu, Z.; Cinar, R.; Xiong, K.; Kunos, G. Cannabinoid-1 Receptor Antagonism Improves Glycemic Control and Increases Energy Expenditure Through Sirtuin-1/Mechanistic Target of Rapamycin Complex 2 and 5′Adenosine Monophosphate–Activated Protein Kinase Signaling. Hepatology 2019, 69, 1535–1548. [Google Scholar] [CrossRef]
- Argueta, D.A.; Perez, P.A.; Makriyannis, A.; Di Patrizio, N. Cannabinoid CB1 receptors inhibit gut-brain satiation signaling in diet-induced obesity. Front. Physiol. 2019, 10, 704. [Google Scholar] [CrossRef]
- Senin, L.L.; Al-Massadi, O.; Folgueira, C.; Castelao, C.; Pardo, M.; Barja-Fernandez, S.; Roca-Rivada, A.; Amil, M.; Crujeiras, A.B.; Garcia-Caballero, T.; et al. The Gastric CB1 Receptor Modulates Ghrelin Production through the mTOR Pathway to Regulate Food Intake. PLoS ONE 2013, 8, e80339. [Google Scholar] [CrossRef]
- Tam, J.; Szanda, G.; Drori, A.; Liu, Z.; Cinar, R.; Kashiwaya, Y.; Reitman, M.L.; Kunos, G. Peripheral cannabinoid-1 receptor blockade restores hypothalamic leptin signaling. Mol. Metab. 2017, 6, 1113–1125. [Google Scholar] [CrossRef] [PubMed]
- European Medicines Agency. Public Statement on Zimulti (Rimonabant: Withdrawal of the Marketing Authorisation in the European Union; European Medicines Agency: London, UK, 2009.
- Christensen, R.; Kristensen, P.; Bartels, E.; Bliddal, H.; Astrup, A. Efficacy and safety of the weight-loss drug rimonabant: A meta-analysis of randomised trials. Lancet 2007, 370, 1706–1713. [Google Scholar] [CrossRef] [PubMed]
- Quarta, C.; Cota, D. Anti-obesity therapy with peripheral CB1 blockers: From promise to safe practice. Int. J. Obes. 2020, 44, 2179–2193. [Google Scholar] [CrossRef] [PubMed]

| Abbreviation | Descriptor | Elements of the Question |
|---|---|---|
| P: | Problem | Therapeutic targets used in the treatment of obesity. |
| E: | Exposure | Obesity. |
| Co: | Context | In silico studies |
| Database | Search Equation |
|---|---|
| PubMed Selected filter: Title/Abstract | (((therapeutic target[Title/Abstract] OR target[Title/Abstract] OR treatment[Title/Abstract]) AND (Obesity[Title/Abstract] OR Obese[Title/Abstract])) AND (in silico[Title/Abstract] OR computer simulation[Title/Abstract])) AND (molecular dynamic simulation[Title/Abstract] OR molecular dynamic[Title/Abstract] OR molecular docking simulation[Title/Abstract] OR molecular docking[Title/Abstract]) |
| Science Direct Selected filter: Title, abstract or author-specified keywords | (obesity OR obese) AND (in silico OR computer simulation OR molecular dynamic simulation OR molecular dynamic OR molecular docking simulation OR molecular docking) |
| Scopus Selected filter: Article title, abstract, keywords | TITLE-ABS-KEY (“therapeutic target” OR target OR treatment) AND TITLE-ABS-KEY (obesity OR obese) AND TITLE-ABS-KEY (“in silico” OR “computer simulation”) AND TITLE-ABS-KEY (“molecular dynamic simulation” OR “molecular dynamic” OR “molecular docking simulation” OR “molecular docking”) |
| Embase Selected filter: Title, abstract or author keywords | (obesity:ti,ab,kw OR obese*:ti,ab,kw) AND (‘in silico’:ti,ab,kw OR ‘computer simulation*’:ti,ab,kw) AND (‘molecular dynamic* simulation*’:ti,ab,kw OR ‘molecular dynamic*’:ti,ab,kw OR ‘molecular docking* simulation*’:ti,ab,kw OR ‘molecular docking*’:ti,ab,kw) |
| Web of Science | ((ALL=(Obesity OR Obese*)) AND ALL=(in silico OR computer simulation*)) AND ALL=(molecular dynamic* simulation* OR molecular dynamic* OR molecular docking simulation* OR molecular docking*) |
| Virtual Health Library Selected filter: Title, abstract, subject | (therapeutic* target* OR target* OR treatment*) AND (obesity OR obese*) AND (in silico OR computer simulation*) AND (molecular dynamic* simulation* OR molecular dynamic* OR molecular docking* simulation* OR molecular docking*) |
| Study/ Year/References | Study In Silico | In Vivo Reassessment | Possible Molecular Mechanism of Application | ||
|---|---|---|---|---|---|
| Methodology | Main Residues in the Interaction Interface of the Most Promising Agent/Main Results of Docking or Molecular Dynamics | Lineages/Diet | Treatment/Effects | ||
| Human Pancreatic Lipase (HPL) | |||||
| Birari et al., 2011 [32] | Docking Software FlexX | Gly76, Asp79, Thr115, His151, Ser152, Phe215, His263, Arg256 Docking score (Kcal/mol) −23.7 and −21.9 | Male Sprague Dawley Rats HF diet | Isolated agent ↓ Weight gain and Serum TG and CT | Inhibitor |
| Coronado-Cáceres et al., 2020 [33] | Docking AutoDockTools vina version 4.5 | Asn88, Asn92, Lys239, Arg265, Tyr267, Thr271, Ser333, Asp 331 Affinity (Kcal/mol) −6.5 | Male Wistar rats HF diet | Cocoa Seed Proteins ↑ Total fecal lipids and fecal TG ↓ Fat absorption rate Ø Body weight and fecal CT | |
| El-Korany et al., 2020 [34] | Docking Molecular Operating Environment 2015.10 version | Lys80, Asn84, Trp252 Docking score (Kcal/mol) −3.7 | Male Sprague Dawley Rats HF diet | Methanolic fraction ↓ Body weight, food consumption, serum TG and CT, and liver weight | |
| Yakaiah et al., 2021 [35] | Docking Autodock4 | Phe77 E Leu 153 Binding energy (Kcal/mol): −6.2 | Male and female albino rats CD diet | Ethanol extract ↑ Total fecal lipids ↓ Volume and size of adipocytes | |
| Fat Mass and Obesity-Associated Protein (FTO) | |||||
| Elekofehinti et al., 2020 [36] | Docking Autodock vina | Arg96, Tyr108, Leu109, Val228, His231, Asp233 Binding energy (Kcal/mol) −9.2 | Male Wistar rats HF diet | Aqueous extract ↓ FTO mRNA expression ↑ STAT3 mRNA expression | Inhibitor |
| Yaccoubi et al., 2022 [37] | Docking PyRx | Lys216, Leu91 and Asn101 Affinity (Kcal/mol): −11.6 and −10.6 | Male Wistar rats HF diet | Isolated agent ↓ Hepatic steatosis and volume of adipocytes | |
| Fajriaty et al., 2023 [38] | Docking/molecular dynamics Autodock version 4.2 and Amber16 | Tyr108, Glu 234, Ser 229 Docking—Binding energy (Kcal/mol): −9.74 Molecular dynamics—Free energy (Kcal/mol): −34.21 | Rats HF diet | Ethanol extract ↓ Weight gain, kidney fat, anal fat, and CT | |
| CD36 Receptor | |||||
| Khan et al., 2023 [39] | Docking Quick Vina version 2 | Val61, Asn250, Leu251, Lys252, Phe266, Ala267, Ser268, Pro269, Val270, Glu271, Asn275, Asp295, Lys369, Leu371, Asn383, Thr385, Thr387 e Glu418 Docking score (kcal/mol): CD36: −8.4 | Male C57BL/6J mice HF diet | Isolated agent ↓ Daily fat-rich food intake, body weight gain, body fat mass, liver weight, and mRNA expression of CD36, Leptin, SREBP1c, SCD1, FAS, and PPAR-γ, LPS, LDL, and IL-6 blood concentrations ↑ Pancreatic-bile secretion, blood concentrations (GLP-1, CCK, PYY and HDL) Ø Lean body mass, serum TG and size of adipocytes | Agonist |
| Type 1 Cannabinoid Receptor (CB1) | |||||
| Lee et al., 2008 [40] | Docking Surflex-dock | Thr197, Asp366, Lys192, Ser383 Docking score: Uninformed | Male C57BL/6J mice WD diet | Isolated agent ↓ Body weight | Selective antagonist |
| Protein Tyrosine Phosphatase 1B (PTP1B) | |||||
| Ghareb et al., 2019 [41] | Docking Glide version 10.1 | Arg24, Arg254, Gln262, Arg47, Asp48, Ala217, Ala27, Ser28, Asp29, Phe52, Cys32, Met258 Docking score (KJ/mol): −5.40 and −4.64 | Male Wistar rats HF diet | Isolated agent: ↓ Body weight, Homa-IR Index, serum TG, serum CT, and blood glucose and insulin | Inhibitor |
| Acetyl-Coa Carboxylase (ACC) | |||||
| Chen et al., 2022 [42] | Docking AutoDock vina version 1.1.2 | Uninformed Affinity (Kcal/mol): Result presented in the form of a graph. | Male C57BL/6J mice HF diet | Isolated agent ↑ Food consumption, liver ACCase, and serum levels of ACBP, GLP-1, and PYY ↓ Body weight and hepatic Malonyl-CoA | Inhibitor |
| Leptin Receptor (LEPR) | |||||
| Kang et al., 2022 [43] | Docking CDOCKER | Leu471, Tyr472, Leu505 e Leu506, Leu530, His467, Ser469, Ser470, Arg615 Binding energy (Kcal/mol)—264.3 Leu2, Leu3, Cys6, Lys29, Lys31, Tyr35, e His 37 Binding energy (Kcal/mol)—776.573 | Male C57BL/6J-ob/ob mice Control Diet | Isolated agent ↓ Body weight, volume of adipocytes in white adipose tissue, liver lipid droplets, and serum TG and CT ↑ mRNA expression of 4EBP and FoxO1, and phosphorylation of JAK2, STAT3, STAT5, ERK1/2, AKT, and mTOR in the cytoplasm of the hypothalamus. | Agonist |
| Target | Experimental Structure | Theoretical Structure Obtained by Modeling | ||||
|---|---|---|---|---|---|---|
| PDB ID | Resolution | Method | Year | Source/Sequence Code | Template Structures | |
| Acetyl-CoA carboxylase (ACC) | 4ASI | 2.80 Å | X-ray diffraction | 2012 | NA | NA |
| Fat mass and obesity-associated protein (FTO) | 3LFM | 2.50 Å | X-ray diffraction | 2010 | NA | NA |
| Protein tyrosine phosphatase 1B (PTP1B) | 2F70 | 2.12 Å | X-ray diffraction | 2005 | NA | NA |
| Pancreatic lipase (PL) | 1LPB | 2.46 Å | X-ray diffraction | 1994 | NA | NA |
| Leptin receptor (LEPR) CRH2 subdomain Leptin receptor (LEPR) IGD subdomain | 3V6O | 1.95 Å | X-ray diffraction | 2011 | NA | NA |
| 1I1R | 2.40 Å | X-ray diffraction | 2001 | NA | NA | |
| CD36 Extracellular domain CD36 Transmembrane helices | 5LGD | 2.07 Å | X-ray diffraction | 2016 | NA | NA |
| NA | NA | NA | NA | UniProt ID: P16671 | NA | |
| Type 1 cannabinoid receptor (CB1) | NA | NA | NA | NA | Uniprot ID: P21554 | PDB ID: 1F88 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Medeiros, W.F.; Gomes, A.F.T.; Aguiar, A.J.F.C.; de Queiroz, J.L.C.; Bezerra, I.W.L.; da Silva-Maia, J.K.; Piuvezam, G.; Morais, A.H.d.A. Anti-Obesity Therapeutic Targets Studied In Silico and In Vivo: A Systematic Review. Int. J. Mol. Sci. 2024, 25, 4699. https://doi.org/10.3390/ijms25094699
de Medeiros WF, Gomes AFT, Aguiar AJFC, de Queiroz JLC, Bezerra IWL, da Silva-Maia JK, Piuvezam G, Morais AHdA. Anti-Obesity Therapeutic Targets Studied In Silico and In Vivo: A Systematic Review. International Journal of Molecular Sciences. 2024; 25(9):4699. https://doi.org/10.3390/ijms25094699
Chicago/Turabian Stylede Medeiros, Wendjilla F., Ana Francisca T. Gomes, Ana Júlia F. C. Aguiar, Jaluza Luana C. de Queiroz, Ingrid Wilza L. Bezerra, Juliana Kelly da Silva-Maia, Grasiela Piuvezam, and Ana Heloneida de A. Morais. 2024. "Anti-Obesity Therapeutic Targets Studied In Silico and In Vivo: A Systematic Review" International Journal of Molecular Sciences 25, no. 9: 4699. https://doi.org/10.3390/ijms25094699
APA Stylede Medeiros, W. F., Gomes, A. F. T., Aguiar, A. J. F. C., de Queiroz, J. L. C., Bezerra, I. W. L., da Silva-Maia, J. K., Piuvezam, G., & Morais, A. H. d. A. (2024). Anti-Obesity Therapeutic Targets Studied In Silico and In Vivo: A Systematic Review. International Journal of Molecular Sciences, 25(9), 4699. https://doi.org/10.3390/ijms25094699





