Genome-Wide Identification, Phylogenetic and Expression Analysis of Expansin Gene Family in Medicago sativa L.
Abstract
1. Introduction
2. Results
2.1. Phylogenetic Analysis of the Expansin Family Members in Alfalfa
2.2. The Features and Distributions of MsEXPs
2.3. Gene Structure of the MsEXPs and Conserved Motifs of the Putative Proteins
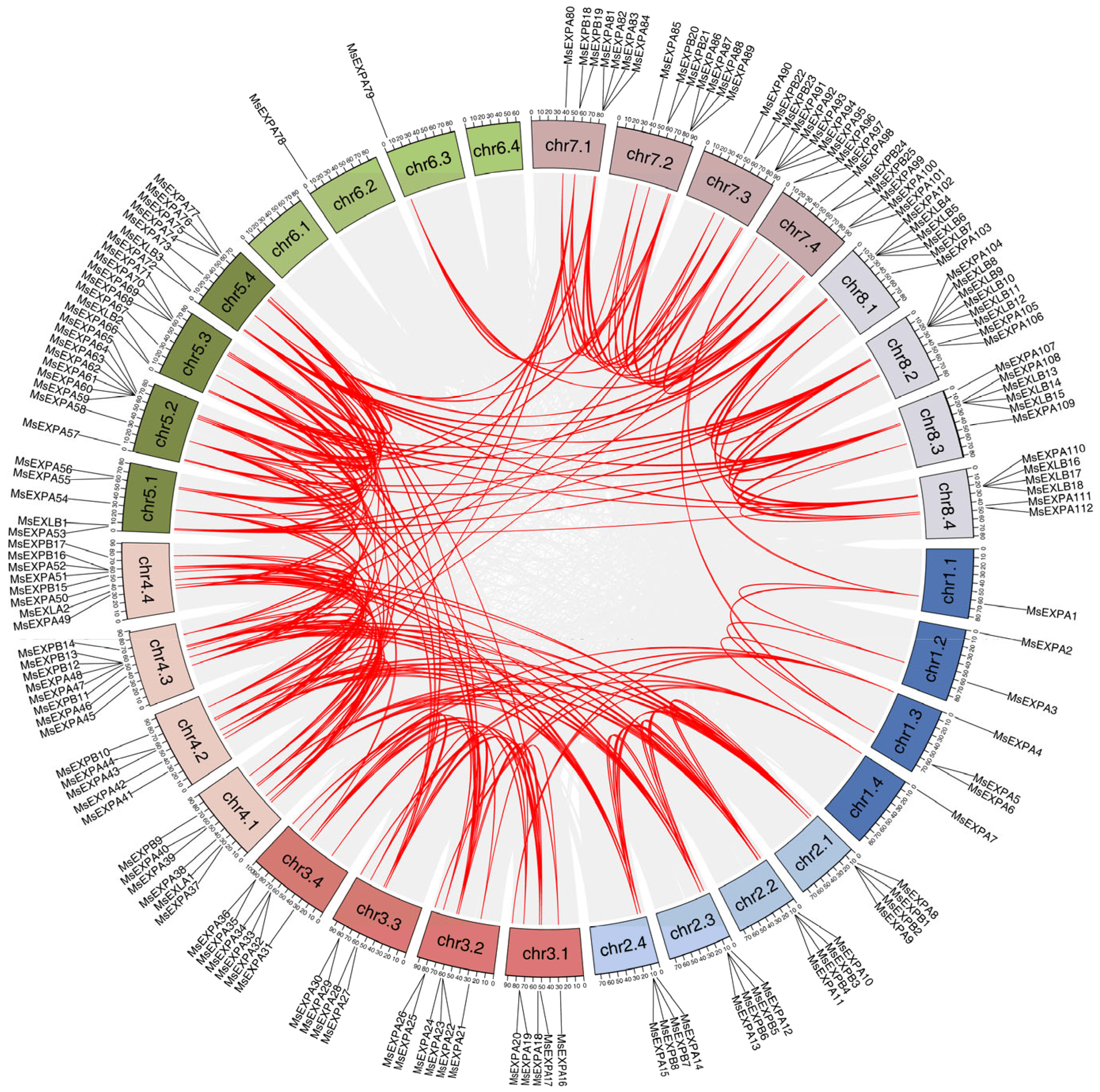
2.4. Gene Duplication and Synteny Analysis of MsEXPs
2.5. The Cis-Acting Elements Related to Abiotic Stresses Were Enriched in the Putative Promoters of MsEXPs
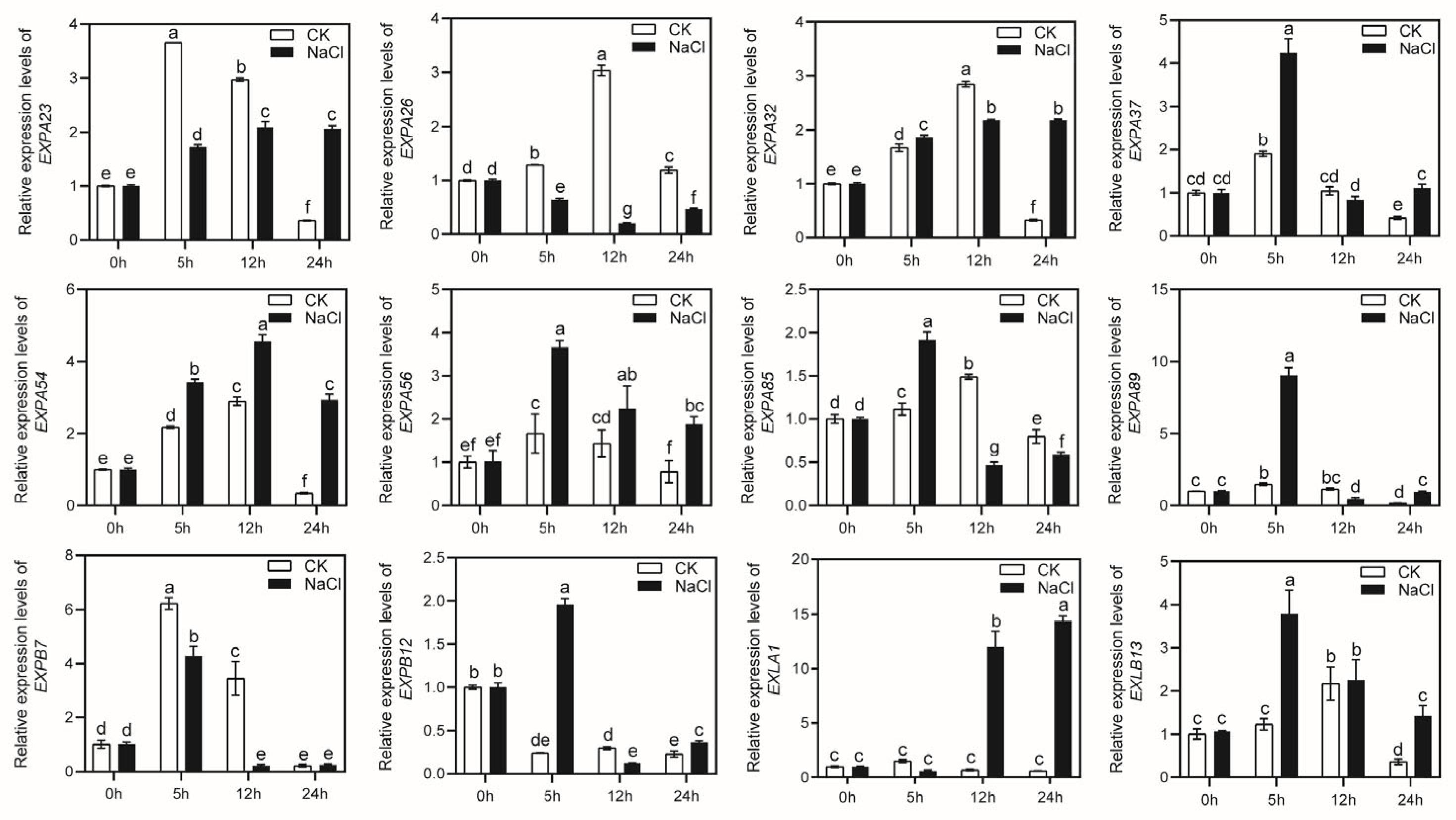
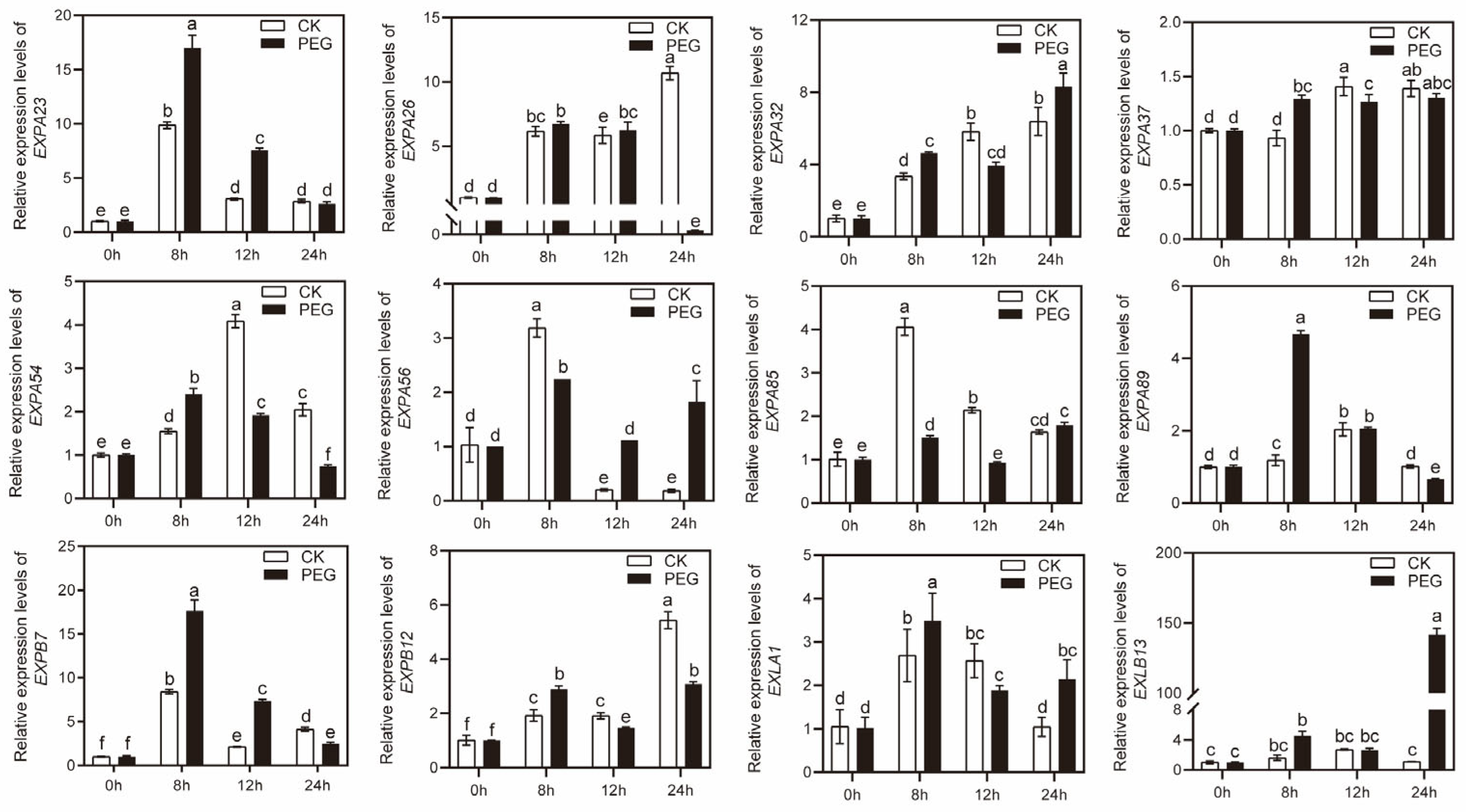
2.6. The Expression of MsEXPs under Drought and Salt Stress
3. Discussion
4. Materials and Methods
4.1. Identification of MsEXPs
4.2. Phylogenetic Analysis and Comparison of MsEXPs
4.3. Chromosomal Location and Structural Characterization Analysis of MsEXPs
4.4. Analysis of Cis-Acting Elements of MsEXPs
4.5. Gene Duplication Pattern and Collinearity Analysis of MsEXPs
4.6. Plant Material Growth Conditions and Stress Treatment
4.7. RNA Extraction and Real-Time Quantitative PCR (qRT-PCR)
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cosgrove, D.J. Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 2005, 6, 850–861. [Google Scholar] [CrossRef]
- Zhang, B.; Gao, Y.; Zhang, L.; Zhou, Y. The plant cell wall: Biosynthesis, construction, and functions. J. Integr. Plant Biol. 2021, 63, 251–272. [Google Scholar] [CrossRef]
- Zhu, J.K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef]
- Chebli, Y.; Bidhendi, A.J.; Kapoor, K.; Geitmann, A. Cytoskeletal regulation of primary plant cell wall assembly. Curr. Biol. 2021, 31, R681–R695. [Google Scholar] [CrossRef]
- Carpita, N.C.; Gibeaut, D.M. Structural models of primary cell walls in flowering plants: Consistency of molecular structure with the physical properties of the walls during growth. Plant J. 1993, 3, 1–30. [Google Scholar] [CrossRef]
- Vorwerk, S.; Somerville, S.; Somerville, C. The role of plant cell wall polysaccharide composition in disease resistance. Trends Plant Sci. 2004, 9, 203–209. [Google Scholar] [CrossRef]
- Wolf, S.; Mravec, J.; Greiner, S.; Mouille, G.; Höfte, H. Plant cell wall homeostasis is mediated by brassinosteroid feedback signaling. Curr. Biol. 2012, 22, 1732–1737. [Google Scholar] [CrossRef]
- Du, F.; Jiao, Y. Mechanical control of plant morphogenesis: Concepts and progress. Curr. Opin. Plant Biol. 2020, 57, 16–23. [Google Scholar] [CrossRef]
- Sun, W.; Yu, H.; Liu, M.; Ma, Z.; Chen, H. Evolutionary research on the expansin protein family during the plant transition to land provides new insights into the development of Tartary buckwheat fruit. BMC Genom. 2021, 22, 252. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Characterization of long-term extension of isolated cell walls from growing cucumber hypocotyls. Planta 1989, 177, 121–130. [Google Scholar] [CrossRef]
- McCann, M.C.; Wells, B.; Roberts, K. Complexity in the spatial localization and length distribution of plant cell-wall matrix polysaccharides. J. Microsc. 1992, 166, 123–136. [Google Scholar] [CrossRef]
- McQueen-Mason, S.; Durachko, D.M.; Cosgrove, D.J. Two endogenous proteins that induce cell wall extension in plants. Plant Cell 1992, 4, 1425–1433. [Google Scholar]
- Kende, H.; Bradford, K.; Brummell, D.; Cho, H.T.; Cosgrove, D.; Fleming, A.; Gehring, C.; Lee, Y.; McQueen-Mason, S.; Rose, J.; et al. Nomenclature for members of the expansin superfamily of genes and proteins. Plant Mol. Biol. 2004, 55, 311–314. [Google Scholar] [CrossRef]
- Cosgrove, D.J.; Bedinger, P.; Durachko, D.M. Group I allergens of grass pollen as cell wall-loosening agents. Proc. Natl. Acad. Sci. USA 1997, 94, 6559–6564. [Google Scholar] [CrossRef]
- Han, Z.; Liu, Y.; Deng, X.; Liu, D.; Liu, Y.; Hu, Y.; Yan, Y. Genome-wide identification and expression analysis of expansin gene family in common wheat (Triticum aestivum L.). BMC Genom. 2019, 20, 101. [Google Scholar] [CrossRef]
- Li, K.; Ma, B.; Shen, J.; Zhao, S.; Ma, X.; Wang, Z.; Fan, Y.; Tang, Q.; Wei, D. The evolution of the expansin gene family in Brassica species. Plant Physiol. Bioch. 2021, 167, 630–638. [Google Scholar] [CrossRef]
- Lee, Y.; Choi, D.; Kende, H. Expansins: Ever-expanding numbers and functions. Curr. Opin. Plant Biol. 2001, 4, 527–532. [Google Scholar] [CrossRef]
- Yennawar, N.H.; Li, L.C.; Dudzinski, D.M.; Tabuchi, A.; Cosgrove, D.J. Crystal structure and activities of EXPB1 (Zea m 1), a beta-expansin and group-1 pollen allergen from maize. Proc. Natl. Acad. Sci. USA 2006, 103, 14664–14671. [Google Scholar] [CrossRef]
- Boron, A.K.; Van Loock, B.; Suslov, D.; Markakis, M.N.; Verbelen, J.P.; Vissenberg, K. Over-expression of AtEXLA2 alters etiolated arabidopsis hypocotyl growth. Ann. Bot. 2015, 115, 67–80. [Google Scholar] [CrossRef]
- Guimaraes, L.A.; Mota, A.P.Z.; Araujo, A.C.G.; de Alencar Figueiredo, L.F.; Pereira, B.M.; de Passos Saraiva, M.A.; Silva, R.B.; Danchin, E.G.J.; Guimaraes, P.M.; Brasileiro, A.C.M. Genome-wide analysis of expansin superfamily in wild Arachis discloses a stress-responsive expansin-like B gene. Plant Mol. Biol. 2017, 94, 79–96. [Google Scholar] [CrossRef]
- Schipper, O.; Schaefer, D.; Reski, R.; Flemin, A. Expansins in the bryophyte Physcomitrella patens. Plant Mol. Biol. 2002, 50, 789–802. [Google Scholar] [CrossRef]
- Carey, R.E.; Cosgrove, D.J. Portrait of the expansin superfamily in Physcomitrella patens: Comparisons with angiosperm expansins. Ann. Bot. 2007, 99, 1131–1141. [Google Scholar] [CrossRef]
- Carey, R.E.; Hepler, N.K.; Cosgrove, D.J. Selaginella moellendorffii has a reduced and highly conserved expansin superfamily with genes more closely related to angiosperms than to bryophytes. BMC Plant Biol. 2013, 13, 4. [Google Scholar] [CrossRef]
- Guo, F.; Guo, J.; El-Kassaby, Y.A.; Wang, G. Genome-wide identification of expansin gene family and their response under hormone exposure in Ginkgo biloba L. Int. J. Mol. Sci. 2023, 24, 5901. [Google Scholar] [CrossRef]
- Sampedro, J.; Carey, R.E.; Cosgrove, D.J. Genome histories clarify evolution of the expansin superfamily: New insights from the poplar genome and pine ESTs. J. Plant Res. 2006, 119, 11–21. [Google Scholar] [CrossRef]
- Tan, J.; Wang, M.; Shi, Z.; Miao, X. OsEXPA10 mediates the balance between growth and resistance to biotic stress in rice. Plant Cell Rep. 2018, 37, 993–1002. [Google Scholar] [CrossRef]
- Santiago, T.R.; Pereira, V.M.; de Souza, W.R.; Steindorff, A.S.; Cunha, B.; Gaspar, M.; Fávaro, L.C.L.; Formighieri, E.F.; Kobayashi, A.K.; Molinari, H.B. Genome-wide identification, characterization and expression profile analysis of expansins gene family in sugarcane (Saccharum spp.). PLoS ONE 2018, 13, e0191081. [Google Scholar]
- Feng, X.; Li, C.; He, F.; Xu, Y.; Li, L.; Wang, X.; Chen, Q.; Li, F. Genome-wide identification of expansin genes in wild soybean (Glycine soja) and functional characterization of Expansin B1 (GsEXPB1) in soybean hair root. Int. J. Mol. Sci. 2022, 23, 5407. [Google Scholar] [CrossRef]
- Zhu, Y.; Wu, N.; Song, W.; Yin, G.; Qin, Y.; Yan, Y.; Hu, Y. Soybean (Glycine max) expansin gene superfamily origins: Segmental and tandem duplication events followed by divergent selection among subfamilies. BMC Plant Biol. 2014, 14, 93. [Google Scholar] [CrossRef]
- Magadum, S.; Banerjee, U.; Murugan, P.; Gangapur, D.; Ravikesavan, R. Gene duplication as a major force in evolution. J. Genet. 2013, 92, 155–161. [Google Scholar] [CrossRef]
- Choi, D.; Lee, Y.; Cho, H.T.; Kende, H. Regulation of expansin gene expression affects growth and development in transgenic rice plants. Plant Cell 2003, 15, 1386–1398. [Google Scholar] [CrossRef]
- Cho, H.T.; Cosgrove, D.J. Regulation of root hair initiation and expansin gene expression in Arabidopsis. Plant Cell 2002, 14, 3237–3253. [Google Scholar] [CrossRef]
- Lee, D.K.; Ahn, J.H.; Song, S.K.; Choi, Y.D.; Lee, J.S. Expression of an expansin gene is correlated with root elongation in soybean. Plant Physiol. 2003, 131, 985–997. [Google Scholar] [CrossRef]
- Pien, S.; Wyrzykowska, J.; McQueen-Mason, S.; Smart, C.; Fleming, A. Local expression of expansin induces the entire process of leaf development and modifies leaf shape. Proc. Natl. Acad. Sci. USA 2001, 98, 11812–11817. [Google Scholar] [CrossRef]
- Yan, A.; Wu, M.; Yan, L.; Hu, R.; Ali, I.; Gan, Y. AtEXP2 is involved in seed germination and abiotic stress response in Arabidopsis. PLoS ONE 2014, 9, e85208. [Google Scholar] [CrossRef]
- Pezzotti, M.; Feron, R.; Mariani, C. Pollination modulates expression of the PPAL gene, a pistil-specific beta-expansin. Plant Mol. Biol. 2002, 49, 187–197. [Google Scholar] [CrossRef]
- Li, X.; Zhao, J.; Walk, T.C.; Liao, H. Characterization of soybean β-expansin genes and their expression responses to symbiosis, nutrient deficiency, and hormone treatment. Appl. Microbiol. Biotechnol. 2014, 98, 2805–2817. [Google Scholar] [CrossRef]
- Li, X.; Zhao, J.; Tan, Z.; Zeng, R.; Liao, H. GmEXPB2, a cell wall β-Expansin, affects soybean nodulation through modifying root architecture and promoting nodule formation and development. Plant Physiol. 2015, 169, 2640–2653. [Google Scholar] [CrossRef]
- Han, Y.; Li, A.; Li, F.; Zhao, M.; Wang, W. Characterization of a wheat (Triticum aestivum L.) expansin gene, TaEXPB23, involved in the abiotic stress response and phytohormone regulation. Plant Physiol. Bioch. 2012, 54, 49–58. [Google Scholar] [CrossRef]
- Muthusamy, M.; Kim, J.Y.; Yoon, E.K.; Kim, J.A.; Lee, S.I. BrEXLB1, a Brassica rapa Expansin-Like B1 Gene is associated with root development, drought stress response, and seed germination. Genes 2020, 11, 404. [Google Scholar] [CrossRef]
- Dai, F.; Zhang, C.; Jiang, X.; Kang, M.; Yin, X.; Lü, P.; Zhang, X.; Zheng, Y.; Gao, J. RhNAC2 and RhEXPA4 are involved in the regulation of dehydration tolerance during the expansion of rose petals. Plant Physiol. 2012, 160, 2064–2082. [Google Scholar] [CrossRef]
- Marowa, P.; Ding, A.; Xu, Z.; Kong, Y. Overexpression of NtEXPA11 modulates plant growth and development and enhances stress tolerance in tobacco. Plant Physiol. Bioch. 2020, 151, 477–485. [Google Scholar] [CrossRef]
- Han, Y.; Chen, Y.; Yin, S.; Zhang, M.; Wang, W. Over-expression of TaEXPB23, a wheat expansin gene, improves oxidative stress tolerance in transgenic tobacco plants. J. Plant Physiol. 2015, 173, 62–71. [Google Scholar] [CrossRef]
- Li, F.; Xing, S.; Guo, Q.; Zhao, M.; Zhang, J.; Gao, Q.; Wang, G.; Wang, W. Drought tolerance through over-expression of the expansin gene TaEXPB23 in transgenic tobacco. J. Plant Physiol. 2011, 168, 960–966. [Google Scholar] [CrossRef]
- Harb, A.; Krishnan, A.; Ambavaram, M.M.; Pereira, A. Molecular and physiological analysis of drought stress in Arabidopsis reveals early responses leading to acclimation in plant growth. Plant Physiol. 2010, 154, 1254–1271. [Google Scholar] [CrossRef]
- Wolf, S. Plant cell wall signalling and receptor-like kinases. Biochem. J. 2017, 474, 471–492. [Google Scholar] [CrossRef]
- Bashline, L.; Lei, L.; Li, S.; Gu, Y. Cell wall, cytoskeleton, and cell expansion in higher plants. Mol. Plant 2014, 7, 586–600. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, G.; An, J.; Li, Q.; Chen, Y.; Zhao, X.; Wu, J.; Wang, Y.; Hao, Q.; Wang, W.; et al. Expansin gene TaEXPA2 positively regulates drought tolerance in transgenic wheat (Triticum aestivum L.). Plant Sci. 2020, 298, 110596. [Google Scholar] [CrossRef]
- He, X.; Zeng, J.; Cao, F.; Ahmed, I.M.; Zhang, G.; Vincze, E.; Wu, F. HvEXPB7, a novel β-expansin gene revealed by the root hair transcriptome of Tibetan wild barley, improves root hair growth under drought stress. J. Exp. Bot. 2015, 66, 7405–7419. [Google Scholar] [CrossRef]
- Sun, W.; Yao, M.; Wang, Z.; Chen, Y.; Zhan, J.; Yan, J.; Jiang, S.; Jian, S.; Chen, H.; Bu, T.; et al. Involvement of auxin-mediated CqEXPA50 contributes to salt tolerance in Quinoa (Chenopodium quinoa) by interaction with auxin pathway genes. Int. J. Mol. Sci. 2022, 23, 8480. [Google Scholar] [CrossRef]
- Geilfus, C.M.; Ober, D.; Eichacker, L.A.; Mühling, K.H.; Zörb, C. Down-regulation of ZmEXPB6 (Zea mays β-expansin 6) protein is correlated with salt-mediated growth reduction in the leaves of Z. mays L. J. Biol. Chem. 2015, 290, 11235–11245. [Google Scholar] [CrossRef]
- Peng, L.N.; Xu, Y.Q.; Wang, X.; Feng, X.; Zhao, Q.Q.; Feng, S.S.; Zhao, Z.Y.; Hu, B.Z.; Li, F.L. Overexpression of paralogues of the wheat expansin gene TaEXPA8 improves low-temperature tolerance in Arabidopsis. Plant Biol. 2019, 21, 1119–1131. [Google Scholar] [CrossRef]
- Feng, X.; Xu, Y.; Peng, L.; Yu, X.; Zhao, Q.; Feng, S.; Zhao, Z.; Li, F.; Hu, B. TaEXPB7-B, a β-expansin gene involved in low-temperature stress and abscisic acid responses, promotes growth and cold resistance in Arabidopsis thaliana. J. Plant Physiol. 2019, 240, 153004. [Google Scholar] [CrossRef]
- Chen, H.; Zeng, Y.; Yang, Y.; Huang, L.; Tang, B.; Zhang, H.; Hao, F.; Liu, W.; Li, Y.; Liu, Y.; et al. Allele-aware chromosome-level genome assembly and efficient transgene-free genome editing for the autotetraploid cultivated alfalfa. Nat. Commun. 2020, 11, 2494. [Google Scholar] [CrossRef]
- Sampedro, J.; Cosgrove, D.J. The expansin superfamily. Genome Biol. 2005, 6, 242. [Google Scholar] [CrossRef]
- Wang, Y.; Li, J.; Paterson, A.H. MCScanX-transposed: Detecting transposed gene duplications based on multiple colinearity scans. Bioinformatics 2013, 29, 1458–1460. [Google Scholar] [CrossRef]
- Abbasi, A.; Malekpour, M.; Sobhanverdi, S. The Arabidopsis expansin gene (AtEXPA18) is capable to ameliorate drought stress tolerance in transgenic tobacco plants. Mol. Biol. Rep. 2021, 48, 5913–5922. [Google Scholar] [CrossRef]
- Castillo, R.M.; Mizuguchi, K.; Dhanaraj, V.; Albert, A.; Blundell, T.L.; Murzin, A.G. A six-stranded double-psi beta barrel is shared by several protein superfamilies. Structure 1999, 7, 227–236. [Google Scholar] [CrossRef]
- Jiang, S.Y.; Jasmin, P.X.; Ting, Y.Y.; Ramachandran, S. Genome-wide identification and molecular characterization of Ole_e_I, Allerg_1 and Allerg_2 domain-containing pollen-allergen-like genes in Oryza sativa. DNA Res. 2005, 12, 167–179. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Plant expansins: Diversity and interactions with plant cell walls. Curr. Opin. Plant Biol. 2015, 25, 162–172. [Google Scholar] [CrossRef]
- Liu, W.; Lyu, T.; Xu, L.; Hu, Z.; Xiong, X.; Liu, T.; Cao, J. Complex molecular evolution and expression of Expansin gene families in three basic diploid species of Brassica. Int. J. Mol. Sci. 2020, 21, 3424. [Google Scholar] [CrossRef]
- Wu, W.; Zhu, S.; Chen, Q.; Lin, Y.; Tian, J.; Liang, C. Cell wall proteins play critical roles in plant adaptation to phosphorus deficiency. Int. J. Mol. Sci. 2019, 20, 5259. [Google Scholar] [CrossRef]
- Narváez-Barragán, D.A.; Tovar-Herrera, O.E.; Segovia, L.; Serrano, M.; Martinez-Anaya, C. Expansin-related proteins: Biology, microbe-plant interactions and associated plant-defense responses. Microbiology 2020, 166, 1007–1018. [Google Scholar] [CrossRef]
- Ling, L.; An, Y.; Wang, D.; Tang, L.; Du, B.; Shu, Y.; Bai, Y.; Guo, C. Proteomic analysis reveals responsive mechanisms for saline-alkali stress in alfalfa. Plant Physiol. Bioch. 2022, 170, 146–159. [Google Scholar] [CrossRef]
- Bacete, L.; Mélida, H.; Miedes, E.; Molina, A. Plant cell wall-mediated immunity: Cell wall changes trigger disease resistance responses. Plant J. 2018, 93, 614–636. [Google Scholar] [CrossRef]
- Abe, H.; Urao, T.; Ito, T.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 2003, 15, 63–78. [Google Scholar] [CrossRef]
- Wang, X.; Niu, Y.; Zheng, Y. Multiple functions of MYB transcription factors in abiotic stress responses. Int. J. Mol. Sci. 2021, 22, 6125. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Duvaud, S.; Gabella, C.; Lisacek, F.; Stockinger, H.; Ioannidis, V.; Durinx, C. Expasy, the Swiss Bioinformatics Resource Portal, as designed by its users. Nucleic Acids Res. 2021, 49, W216–W227. [Google Scholar] [CrossRef]
- Chou, K.C.; Shen, H.B. Cell-PLoc: A package of Web servers for predicting subcellular localization of proteins in various organisms. Nat. Protoc. 2008, 3, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Holub, E.B. The arms race is ancient history in Arabidopsis, the wildflower. Nat. Rev. Genet. 2001, 2, 516–527. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Zhang, Y.; Cui, J.; Wang, X.; Li, M.; Zhang, L.; Kang, J. Genome-Wide Identification, Phylogenetic and Expression Analysis of Expansin Gene Family in Medicago sativa L. Int. J. Mol. Sci. 2024, 25, 4700. https://doi.org/10.3390/ijms25094700
Li Y, Zhang Y, Cui J, Wang X, Li M, Zhang L, Kang J. Genome-Wide Identification, Phylogenetic and Expression Analysis of Expansin Gene Family in Medicago sativa L. International Journal of Molecular Sciences. 2024; 25(9):4700. https://doi.org/10.3390/ijms25094700
Chicago/Turabian StyleLi, Yajing, Yangyang Zhang, Jing Cui, Xue Wang, Mingna Li, Lili Zhang, and Junmei Kang. 2024. "Genome-Wide Identification, Phylogenetic and Expression Analysis of Expansin Gene Family in Medicago sativa L." International Journal of Molecular Sciences 25, no. 9: 4700. https://doi.org/10.3390/ijms25094700
APA StyleLi, Y., Zhang, Y., Cui, J., Wang, X., Li, M., Zhang, L., & Kang, J. (2024). Genome-Wide Identification, Phylogenetic and Expression Analysis of Expansin Gene Family in Medicago sativa L. International Journal of Molecular Sciences, 25(9), 4700. https://doi.org/10.3390/ijms25094700




