Analysing the Cyanobacterial PipX Interaction Network Using NanoBiT Complementation in Synechococcus elongatus PCC7942
Abstract
1. Introduction
2. Results and Discussion
2.1. Reporter Constructs and Strains Used to Analyse PipX-PII and PipX-NtcA Interactions in S. elongatus
| Plasmid | Description, Relevant Characteristics | Reference or Source |
|---|---|---|
| pUAGC280 | Ptrc into NSI, ApR SmR | [53] |
| pUAGC1160 | (PpipX:pipX:FL:SmBiT) into NSI, ApR SmR | This work |
| pUAGC1161 | (PpipX:pipX:FL:SmBiT PglnB:glnB:FL:LgBiT) into NSI, ApR SmR | This work |
| pUAGC1162 | (PpipX:pipX16ta>gc:FL:SmBiT PglnB:glnB:FL:LgBiT) into NSI, ApR SmR | This work |
| pUAGC1163 | (PpipX:pipX:FL:SmBiT PntcA:ntcA:FL:LgBiT) into NSI, ApR SmR | This work |
| pUAGC1164 | (PpipX:pipX16ta>gc:FL:SmBiT PntcA:ntcA:FL:LgBiT) into NSI, ApR SmR | This work |
| PII-ST-FL-LgBiT | PterR:PII-StrepTag-FL-LgBiT | [19] |
| pUAGC126 | pipX replaced with cat, ApR CmR | [38] |
| pPM128 | CK2 (+) into glnB, KmR | [54] |
2.2. PII-LgBiT and PipX-SmBiT Retain Their Regulatory Features in S. elongatus
2.3. PipX-PII and PipX-NtcA Reporters Respond in Opposite Ways to a Drop in the Intracellular ATP Levels in S. elongatus
2.4. PipX-PII and PipX-NtcA Reporters Respond in Opposite Ways to the 2-OG Levels in S. elongatus
2.5. PipX Levels Decrease in the Absence of Combined Nitrogen in S. elongatus
2.6. The PipX Point Mutation Y6A Drastically Impairs PipX-PII and PipX-NtcA Complexes in S. elongatus
2.7. PII Plays a Positive Regulatory Role on PipX-NtcA Complexes during Their Initial Response to Nitrogen Deprivation
2.8. Additional Players May Affect PipX-PII and PipX-NtcA Complexes in S. elongatus
2.9. The NanoBit Approach in the Context of Cyanobacterial Interaction Networks
3. Materials and Methods
3.1. Plasmid Construction
3.2. Cyanobacterial Transformation and Strain Verification
3.3. Cyanobacterial Growth and Culture Conditions
3.4. Bioluminiscence Assays
3.5. Intracellular ATP Content Determination
3.6. Protein Extraction and Immunodetection
3.7. Computational Methods
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Blank, C.E.; Sánchez-Baracaldo, P. Timing of Morphological and Ecological Innovations in the Cyanobacteria—A Key to Understanding the Rise in Atmospheric Oxygen. Geobiology 2010, 8, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-W.; Noh, J.-H.; Choi, D.-H.; Yun, M.; Bhavya, P.S.; Kang, J.-J.; Lee, J.-H.; Kim, K.-W.; Jang, H.-K.; Lee, S.-H. Picocyanobacterial Contribution to the Total Primary Production in the Northwestern Pacific Ocean. Water 2021, 13, 1610. [Google Scholar] [CrossRef]
- Khan, S.; Fu, P. Biotechnological Perspectives on Algae: A Viable Option for next Generation Biofuels. Curr. Opin. Biotechnol. 2020, 62, 146–152. [Google Scholar] [CrossRef] [PubMed]
- Forchhammer, K.; Selim, K.A. Carbon/Nitrogen Homeostasis Control in Cyanobacteria. FEMS Microbiol. Rev. 2020, 44, 33–53. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.-C.; Zhou, C.-Z.; Burnap, R.L.; Peng, L. Carbon/Nitrogen Metabolic Balance: Lessons from Cyanobacteria. Trends Plant Sci. 2018, 23, 1116–1130. [Google Scholar] [CrossRef] [PubMed]
- Rubin, B.E.; Wetmore, K.M.; Price, M.N.; Diamond, S.; Shultzaberger, R.K.; Lowe, L.C.; Curtin, G.; Arkin, A.P.; Deutschbauer, A.; Golden, S.S. The Essential Gene Set of a Photosynthetic Organism. Proc. Natl. Acad. Sci. USA 2015, 112, E6634–E6643. [Google Scholar] [CrossRef] [PubMed]
- Welkie, D.G.; Rubin, B.E.; Chang, Y.-G.; Diamond, S.; Rifkin, S.A.; LiWang, A.; Golden, S.S. Genome-Wide Fitness Assessment during Diurnal Growth Reveals an Expanded Role of the Cyanobacterial Circadian Clock Protein KaiA. Proc. Natl. Acad. Sci. USA 2018, 115, E7174–E7183. [Google Scholar] [CrossRef] [PubMed]
- Burillo, S.; Luque, I.; Fuentes, I.; Contreras, A. Interactions between the Nitrogen Signal Transduction Protein PII and N -Acetyl Glutamate Kinase in Organisms That Perform Oxygenic Photosynthesis. J. Bacteriol. 2004, 186, 3346–3354. [Google Scholar] [CrossRef] [PubMed]
- Espinosa, J.; Forchhammer, K.; Burillo, S.; Contreras, A. Interaction Network in Cyanobacterial Nitrogen Regulation: PipX, a Protein That Interacts in a 2-oxoglutarate Dependent Manner with PII and NtcA. Mol. Microbiol. 2006, 61, 457–469. [Google Scholar] [CrossRef]
- Esteves-Ferreira, A.A.; Inaba, M.; Fort, A.; Araújo, W.L.; Sulpice, R. Nitrogen Metabolism in Cyanobacteria: Metabolic and Molecular Control, Growth Consequences and Biotechnological Applications. Crit. Rev. Microbiol. 2018, 44, 541–560. [Google Scholar] [CrossRef]
- Forchhammer, K.; Selim, K.A.; Huergo, L.F. New Views on PII Signaling: From Nitrogen Sensing to Global Metabolic Control. Trends Microbiol. 2022, 30, 722–735. [Google Scholar] [CrossRef] [PubMed]
- Herrero, A.; Muro-Pastor, A.M.; Flores, E. Nitrogen Control in Cyanobacteria. J. Bacteriol. 2001, 183, 411–425. [Google Scholar] [CrossRef]
- Lee, H.-M.; Flores, E.; Herrero, A.; Houmard, J.; Tandeau de Marsac, N. A Role for the Signal Transduction Protein PII in the Control of Nitrate/Nitrite Uptake in a Cyanobacterium. FEBS Lett. 1998, 427, 291–295. [Google Scholar] [CrossRef]
- Forchhammer, K.; Hedler, A. Phosphoprotein PII from Cyanobacteria. Eur. J. Biochem. 1997, 244, 869–875. [Google Scholar] [CrossRef]
- Huergo, L.F.; Dixon, R. The Emergence of 2-Oxoglutarate as a Master Regulator Metabolite. Microbiol. Mol. Biol. Rev. 2015, 79, 419–435. [Google Scholar] [CrossRef] [PubMed]
- Kamberov, E.S.; Atkinson, M.R.; Ninfa, A.J. The Escherichia coli PII Signal Transduction Protein Is Activated upon Binding 2-Ketoglutarate and ATP. J. Biol. Chem. 1995, 270, 17797–17807. [Google Scholar] [CrossRef] [PubMed]
- Heinrich, A.; Maheswaran, M.; Ruppert, U.; Forchhammer, K. The Synechococcus elongatus PII Signal Transduction Protein Controls Arginine Synthesis by Complex Formation with N-acetyl-L-glutamate Kinase. Mol. Microbiol. 2004, 52, 1303–1314. [Google Scholar] [CrossRef]
- Labella, J.I.; Cantos, R.; Salinas, P.; Espinosa, J.; Contreras, A. Distinctive Features of PipX, a Unique Signaling Protein of Cyanobacteria. Life 2020, 10, 79. [Google Scholar] [CrossRef]
- Rozbeh, R.; Forchhammer, K. Split NanoLuc Technology Allows Quantitation of Interactions between PII Protein and Its Receptors with Unprecedented Sensitivity and Reveals Transient Interactions. Sci. Rep. 2021, 11, 12535. [Google Scholar] [CrossRef]
- Espinosa, J.; Rodríguez-Mateos, F.; Salinas, P.; Lanza, V.F.; Dixon, R.; de la Cruz, F.; Contreras, A. PipX, the Coactivator of NtcA, Is a Global Regulator in Cyanobacteria. Proc. Natl. Acad. Sci. USA 2014, 111, 201404030–201404097. [Google Scholar] [CrossRef] [PubMed]
- Giner-Lamia, J.; Robles-Rengel, R.; Hernández-Prieto, M.A.; Muro-Pastor, M.I.; Florencio, F.J.; Futschik, M.E. Identification of the Direct Regulon of NtcA during Early Acclimation to Nitrogen Starvation in the Cyanobacterium synechocystis sp. PCC 6803. Nucleic Acids Res. 2017, 45, 11800–11820. [Google Scholar] [CrossRef] [PubMed]
- Camargo, S.; Valladares, A.; Forchhammer, K.; Herrero, A. Effects of PipX on NtcA-dependent Promoters and Characterization of the Cox3 Promoter Region in the Heterocyst-forming Cyanobacterium anabaena sp. PCC 7120. FEBS Lett. 2014, 588, 1787–1794. [Google Scholar] [CrossRef] [PubMed]
- Domínguez-Martín, M.A.; López-Lozano, A.; Clavería-Gimeno, R.; Velázquez-Campoy, A.; Seidel, G.; Burkovski, A.; Díez, J.; García-Fernández, J.M. Differential NtcA Responsiveness to 2-Oxoglutarate Underlies the Diversity of C/N Balance Regulation in Prochlorococcus. Front. Microbiol. 2018, 8, 2641. [Google Scholar] [CrossRef] [PubMed]
- Tanigawa, R.; Shirokane, M.; Maeda, S.; Omata, T.; Tanaka, K.; Takahashi, H. Transcriptional Activation of NtcA-Dependent Promoters of Synechococcus sp. PCC 7942 by 2-Oxoglutarate in Vitro. Proc. Natl. Acad. Sci. USA 2002, 99, 4251–4255. [Google Scholar] [CrossRef]
- Zhao, M.-X.; Jiang, Y.-L.; He, Y.-X.; Chen, Y.-F.; Teng, Y.-B.; Chen, Y.; Zhang, C.-C.; Zhou, C.-Z. Structural Basis for the Allosteric Control of the Global Transcription Factor NtcA by the Nitrogen Starvation Signal 2-Oxoglutarate. Proc. Natl. Acad. Sci. USA 2010, 107, 12487–12492. [Google Scholar] [CrossRef] [PubMed]
- Llácer, J.L.; Espinosa, J.; Castells, M.A.; Contreras, A.; Forchhammer, K.; Rubio, V. Structural Basis for the Regulation of NtcA-Dependent Transcription by Proteins PipX and PII. Proc. Natl. Acad. Sci. USA 2010, 107, 15397–15402. [Google Scholar] [CrossRef]
- Zhao, M.-X.; Jiang, Y.-L.; Xu, B.-Y.; Chen, Y.; Zhang, C.-C.; Zhou, C.-Z. Crystal Structure of the Cyanobacterial Signal Transduction Protein PII in Complex with PipX. J. Mol. Biol. 2010, 402, 552–559. [Google Scholar] [CrossRef] [PubMed]
- Espinosa, J.; Castells, M.A.; Laichoubi, K.B.; Contreras, A. Mutations at pipX Suppress Lethality of P. II -Deficient Mutants of Synechococcus elongatus PCC 7942. J. Bacteriol. 2009, 191, 4863–4869. [Google Scholar] [CrossRef] [PubMed]
- Espinosa, J.; Castells, M.A.; Laichoubi, K.B.; Forchhammer, K.; Contreras, A. Effects of Spontaneous Mutations in pipX Functions and Regulatory Complexes on the Cyanobacterium Synechococcus elongatus Strain PCC 7942. Microbiology 2010, 156, 1517–1526. [Google Scholar] [CrossRef][Green Version]
- Forcada-Nadal, A.; Llácer, J.L.; Contreras, A.; Marco-Marín, C.; Rubio, V. The PII-NAGK-PipX-NtcA Regulatory Axis of Cyanobacteria: A Tale of Changing Partners, Allosteric Effectors and Non-Covalent Interactions. Front. Mol. Biosci. 2018, 5, 91. [Google Scholar] [CrossRef]
- Laichoubi, K.B.; Espinosa, J.; Castells, M.A.; Contreras, A. Mutational Analysis of the Cyanobacterial Nitrogen Regulator PipX. PLoS ONE 2012, 7, e35845. [Google Scholar] [CrossRef] [PubMed]
- Forcada-Nadal, A.; Palomino-Schätzlein, M.; Neira, J.L.; Pineda-Lucena, A.; Rubio, V. The PipX Protein, When Not Bound to Its Targets, Has Its Signaling C-Terminal Helix in a Flexed Conformation. Biochemistry 2017, 56, 3211–3224. [Google Scholar] [CrossRef] [PubMed]
- Espinosa, J.; Labella, J.I.; Cantos, R.; Contreras, A. Energy Drives the Dynamic Localization of Cyanobacterial Nitrogen Regulators during Diurnal Cycles. Environ. Microbiol. 2018, 20, 1240–1252. [Google Scholar] [CrossRef]
- Zeth, K.; Fokina, O.; Forchhammer, K. Structural Basis and Target-Specific Modulation of ADP Sensing by the Synechococcus elongatus PII Signaling Protein. J. Biol. Chem. 2014, 289, 8960–8972. [Google Scholar] [CrossRef] [PubMed]
- Cantos, R.; Labella, J.I.; Espinosa, J.; Contreras, A. The Nitrogen Regulator PipX Acts in cis to Prevent Operon Polarity. Environ. Microbiol. Rep. 2019, 11, 495–507. [Google Scholar] [CrossRef] [PubMed]
- Jerez, C.; Salinas, P.; Llop, A.; Cantos, R.; Espinosa, J.; Labella, J.I.; Contreras, A. Regulatory Connections Between the Cyanobacterial Factor PipX and the Ribosome Assembly GTPase EngA. Front. Microbiol. 2021, 12, 781760. [Google Scholar] [CrossRef] [PubMed]
- Labella, J.I.; Obrebska, A.; Espinosa, J.; Salinas, P.; Forcada-Nadal, A.; Tremiño, L.; Rubio, V.; Contreras, A. Expanding the Cyanobacterial Nitrogen Regulatory Network: The GntR-Like Regulator PlmA Interacts with the PII-PipX Complex. Front. Microbiol. 2016, 7, 1677. [Google Scholar] [CrossRef] [PubMed]
- Labella, J.I.; Cantos, R.; Espinosa, J.; Forcada-Nadal, A.; Rubio, V.; Contreras, A. PipY, a Member of the Conserved COG0325 Family of PLP-Binding Proteins, Expands the Cyanobacterial Nitrogen Regulatory Network. Front. Microbiol. 2017, 8, 1244. [Google Scholar] [CrossRef] [PubMed]
- Labella, J.I.; Llop, A.; Contreras, A. The Default Cyanobacterial Linked Genome: An Interactive Platform Based on Cyanobacterial Linkage Networks to Assist Functional Genomics. FEBS Lett. 2020, 594, 1661–1674. [Google Scholar] [CrossRef]
- Riediger, M.; Spät, P.; Bilger, R.; Voigt, K.; Maček, B.; Hess, W.R. Analysis of a Photosynthetic Cyanobacterium Rich in Internal Membrane Systems via Gradient Profiling by Sequencing (Grad-Seq). Plant Cell 2021, 33, 248–269. [Google Scholar] [CrossRef]
- Llop, A.; Bibak, S.; Cantos, R.; Salinas, P.; Contreras, A. The Ribosome Assembly GTPase EngA Is Involved in Redox Signaling in Cyanobacteria. Front. Microbiol. 2023, 14, 1242616. [Google Scholar] [CrossRef] [PubMed]
- Laichoubi, K.B.; Beez, S.; Espinosa, J.; Forchhammer, K.; Contreras, A. The Nitrogen Interaction Network in Synechococcus WH5701, a Cyanobacterium with Two PipX and Two PII-like Proteins. Microbiology 2011, 157, 1220–1228. [Google Scholar] [CrossRef] [PubMed]
- Dixon, A.S.; Schwinn, M.K.; Hall, M.P.; Zimmerman, K.; Otto, P.; Lubben, T.H.; Butler, B.L.; Binkowski, B.F.; Machleidt, T.; Kirkland, T.A.; et al. NanoLuc Complementation Reporter Optimized for Accurate Measurement of Protein Interactions in Cells. ACS Chem. Biol. 2016, 11, 400–408. [Google Scholar] [CrossRef] [PubMed]
- Kashima, D.; Kageoka, M.; Kimura, Y.; Horikawa, M.; Miura, M.; Nakakido, M.; Tsumoto, K.; Nagamune, T.; Kawahara, M. A Novel Cell-Based Intracellular Protein–Protein Interaction Detection Platform (SOLIS) for Multimodality Screening. ACS Synth. Biol. 2021, 10, 990–999. [Google Scholar] [CrossRef] [PubMed]
- Pipchuk, A.; Yang, X. Using Biosensors to Study Protein–Protein Interaction in the Hippo Pathway. Front. Cell Dev. Biol. 2021, 9, 660137. [Google Scholar] [CrossRef] [PubMed]
- Sicking, M.; Jung, M.; Lang, S. Lights, Camera, Interaction: Studying Protein–Protein Interactions of the ER Protein Translocase in Living Cells. Int. J. Mol. Sci. 2021, 22, 10358. [Google Scholar] [CrossRef]
- Bardelang, P.; Murray, E.J.; Blower, I.; Zandomeneghi, S.; Goode, A.; Hussain, R.; Kumari, D.; Siligardi, G.; Inoue, K.; Luckett, J.; et al. Conformational Analysis and Interaction of the Staphylococcus aureus Transmembrane Peptidase AgrB with Its AgrD Propeptide Substrate. Front. Chem. 2023, 11, 1113885. [Google Scholar] [CrossRef] [PubMed]
- Oliveira Paiva, A.M.; Friggen, A.H.; Qin, L.; Douwes, R.; Dame, R.T.; Smits, W.K. The Bacterial Chromatin Protein HupA Can Remodel DNA and Associates with the Nucleoid in Clostridium difficile. J. Mol. Biol. 2019, 431, 653–672. [Google Scholar] [CrossRef]
- Rozbeh, R.; Forchhammer, K. In Vivo Detection of Metabolic Fluctuations in Real Time Using the NanoBiT Technology Based on PII Signalling Protein Interactions. Int. J. Mol. Sci. 2024, 25, 3409. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Takatani, N.; Aichi, M.; Maeda, S.; Omata, T. Evaluation of the Effects of PII Deficiency and the Toxicity of PipX on Growth Characteristics of the PII-Less Mutant of the Cyanobacterium Synechococcus elongatus. Plant Cell Physiol. 2013, 54, 1504–1514. [Google Scholar] [CrossRef]
- Espinosa, J.; Forchhammer, K.; Contreras, A. Role of the Synechococcus PCC 7942 Nitrogen Regulator Protein PipX in NtcA-Controlled Processes. Microbiology 2007, 153, 711–718. [Google Scholar] [CrossRef]
- Guerreiro, A.C.L.; Benevento, M.; Lehmann, R.; van Breukelen, B.; Post, H.; Giansanti, P.; Maarten Altelaar, A.F.; Axmann, I.M.; Heck, A.J.R. Daily Rhythms in the Cyanobacterium Synechococcus elongatus Probed by High-Resolution Mass Spectrometry–Based Proteomics Reveals a Small Defined Set of Cyclic Proteins. Mol. Cell. Proteom. 2014, 13, 2042–2055. [Google Scholar] [CrossRef] [PubMed]
- Moronta-Barrios, F.; Espinosa, J.; Contreras, A. Negative Control of Cell Size in the Cyanobacterium Synechococcus elongatus PCC 7942 by the Essential Response Regulator RpaB. FEBS Lett. 2013, 587, 504–509. [Google Scholar] [CrossRef]
- Forchhammer, K.; Tandeau de Marsac, N. Functional Analysis of the Phosphoprotein PII (GlnB Gene Product) in the Cyanobacterium Synechococcus sp. Strain PCC 7942. J. Bacteriol. 1995, 177, 2033–2040. [Google Scholar] [CrossRef] [PubMed]
- Bullock, W.O.; Fernandez, J.M.; Short, J.M. XL1-Blue—A High-Efficiency Plasmid Transforming recA Escherichia coli Strain with β-Galactosidase Selection. Biotechniques 1987, 5, 376–379. [Google Scholar]
- Llop, A.; Tremiño, L.; Cantos, R.; Contreras, A. The Signal Transduction Protein PII Controls the Levels of the Cyanobacterial Protein PipX. Microorganisms 2023, 11, 2379. [Google Scholar] [CrossRef]
- Takano, S.; Tomita, J.; Sonoike, K.; Iwasaki, H. The Initiation of Nocturnal Dormancy in Synechococcus as an Active Process. BMC Biol. 2015, 13, 36. [Google Scholar] [CrossRef] [PubMed]
- Laurent, S.; Chen, H.; Bédu, S.; Ziarelli, F.; Peng, L.; Zhang, C.-C. Nonmetabolizable Analogue of 2-Oxoglutarate Elicits Heterocyst Differentiation under Repressive Conditions in Anabaena sp. PCC 7120. Proc. Natl. Acad. Sci. USA 2005, 102, 9907–9912. [Google Scholar] [CrossRef]
- Muro-Pastor, M.I.; Reyes, J.C.; Florencio, F.J. Cyanobacteria Perceive Nitrogen Status by Sensing Intracellular 2-Oxoglutarate Levels. J. Biol. Chem. 2001, 276, 38320–38328. [Google Scholar] [CrossRef]
- Salinas, P.; Bibak, S.; Cantos, R.; Tremiño, L.; Jerez, C.; Mata, T.; Contreras, A. Studies on the PII-PipX-NtcA Regulatory Axis of Cyanobacteria Provide Novel Insights into the Advantages and Limitations of Two-Hybrid Systems for Protein Interactions. Int. J. Mol. Sci. 2024. [Google Scholar] [CrossRef]
- Huang, H.; Jedynak, B.M.; Bader, J.S. Where Have All the Interactions Gone? Estimating the Coverage of Two-Hybrid Protein Interaction Maps. PLoS Comput. Biol. 2007, 3, e214. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-C.; Rajagopala, S.V.; Stellberger, T.; Uetz, P. Exhaustive Benchmarking of the Yeast Two-Hybrid System. Nat. Methods 2010, 7, 667–668. [Google Scholar] [CrossRef]
- Battesti, A.; Bouveret, E. The Bacterial Two-Hybrid System Based on Adenylate Cyclase Reconstitution in Escherichia Coli. Methods 2012, 58, 325–334. [Google Scholar] [CrossRef]
- Lin, J.-S.; Lai, E.-M. Protein–Protein Interactions: Yeast Two Hybrid. In Bacterial Secretion Systems. Methods in Molecular Biology; Journet, L., Cascales, E., Eds.; Humana: New York, NY, USA, 2024; Volume 2715, pp. 235–246. [Google Scholar] [CrossRef]
- Clontech. MatchmakerTM GAL4 Two-Hybrid System 3 & Libraries User Manual; Clontech Laboratories, Inc.: Mountain View, CA, USA, 2007. [Google Scholar]
- Karimova, G.; Gauliard, E.; Davi, M.; Ouellette, S.P.; Ladant, D. Protein–Protein Interaction: Bacterial Two Hybrid. In Bacterial Secretion Systems. Methods in Molecular Biology; Journet, L., Cascales, E., Eds.; Humana: New York, NY, USA, 2024; Volume 2715, pp. 207–224. [Google Scholar] [CrossRef]
- Mehla, J.; Caufield, J.H.; Sakhawalkar, N.; Uetz, P. A Comparison of Two-Hybrid Approaches for Detecting Protein–Protein Interactions. Methods Enzymol. 2017, 586, 333–358. [Google Scholar] [CrossRef] [PubMed]
- Gibson, D.G.; Young, L.; Chuang, R.-Y.; Venter, J.C.; Hutchison, C.A.; Smith, H.O. Enzymatic Assembly of DNA Molecules up to Several Hundred Kilobases. Nat. Methods 2009, 6, 343–345. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Naismith, J.H. An Efficient One-Step Site-Directed Deletion, Insertion, Single and Multiple-Site Plasmid Mutagenesis Protocol. BMC Biotechnol. 2008, 8, 91. [Google Scholar] [CrossRef]
- Taton, A.; Erikson, C.; Yang, Y.; Rubin, B.E.; Rifkin, S.A.; Golden, J.W.; Golden, S.S. The Circadian Clock and Darkness Control Natural Competence in Cyanobacteria. Nat. Commun. 2020, 11, 1688. [Google Scholar] [CrossRef] [PubMed]
- Rippka, R.; Deruelles, J.; Waterbury, J.B.; Herdman, M.; Stanier, R.Y. Generic Assignments, Strain Histories and Properties of Pure Cultures of Cyanobacteria. Microbiology 1979, 111, 1–61. [Google Scholar] [CrossRef]
- Doello, S.; Burkhardt, M.; Forchhammer, K. The Essential Role of Sodium Bioenergetics and ATP Homeostasis in the Developmental Transitions of a Cyanobacterium. Curr. Biol. 2021, 31, 1606–1615. [Google Scholar] [CrossRef]
- RStudio. RStudio: Integrated Development for R. 2020. Available online: http://www.rstudio.com/ (accessed on 1 March 2024).

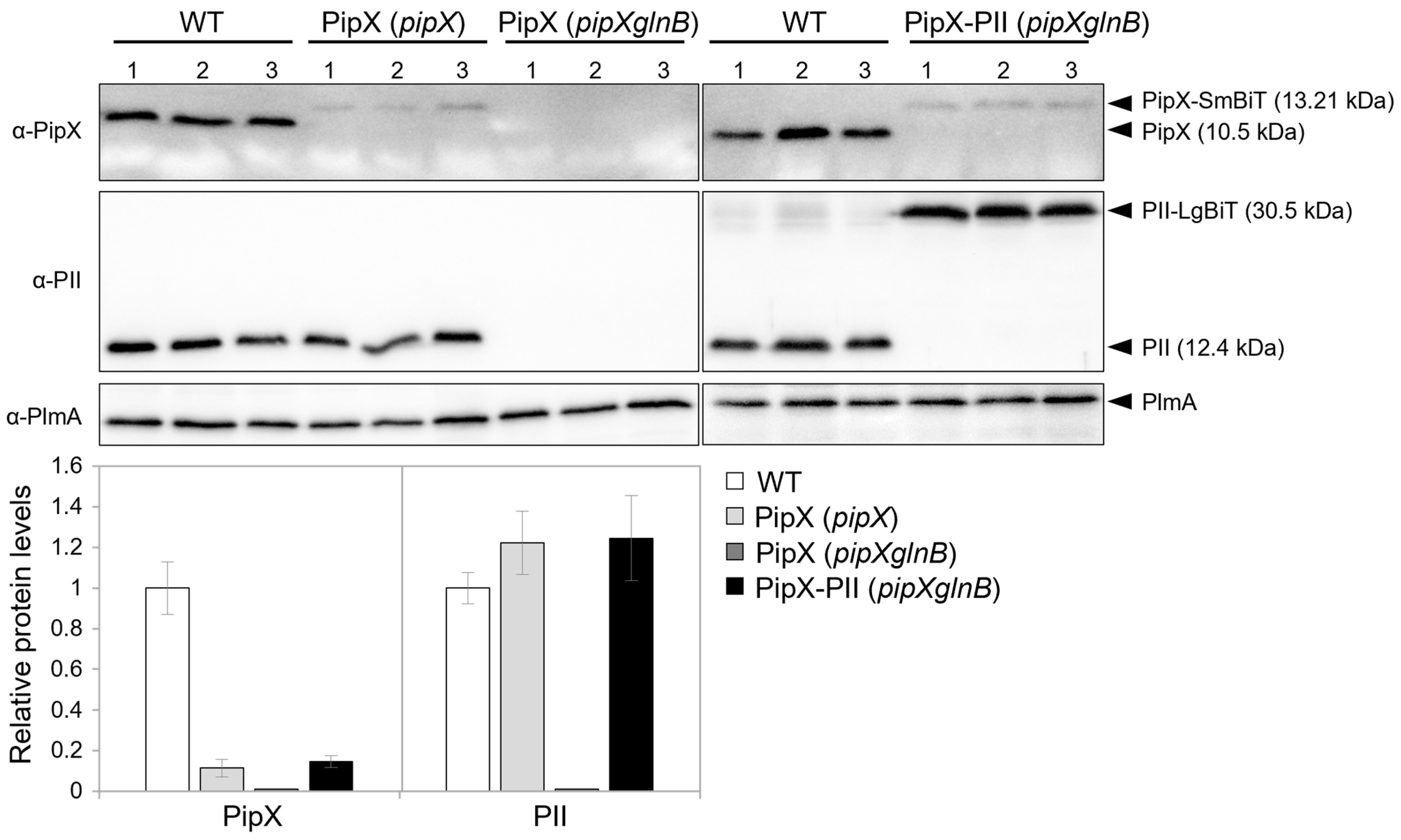
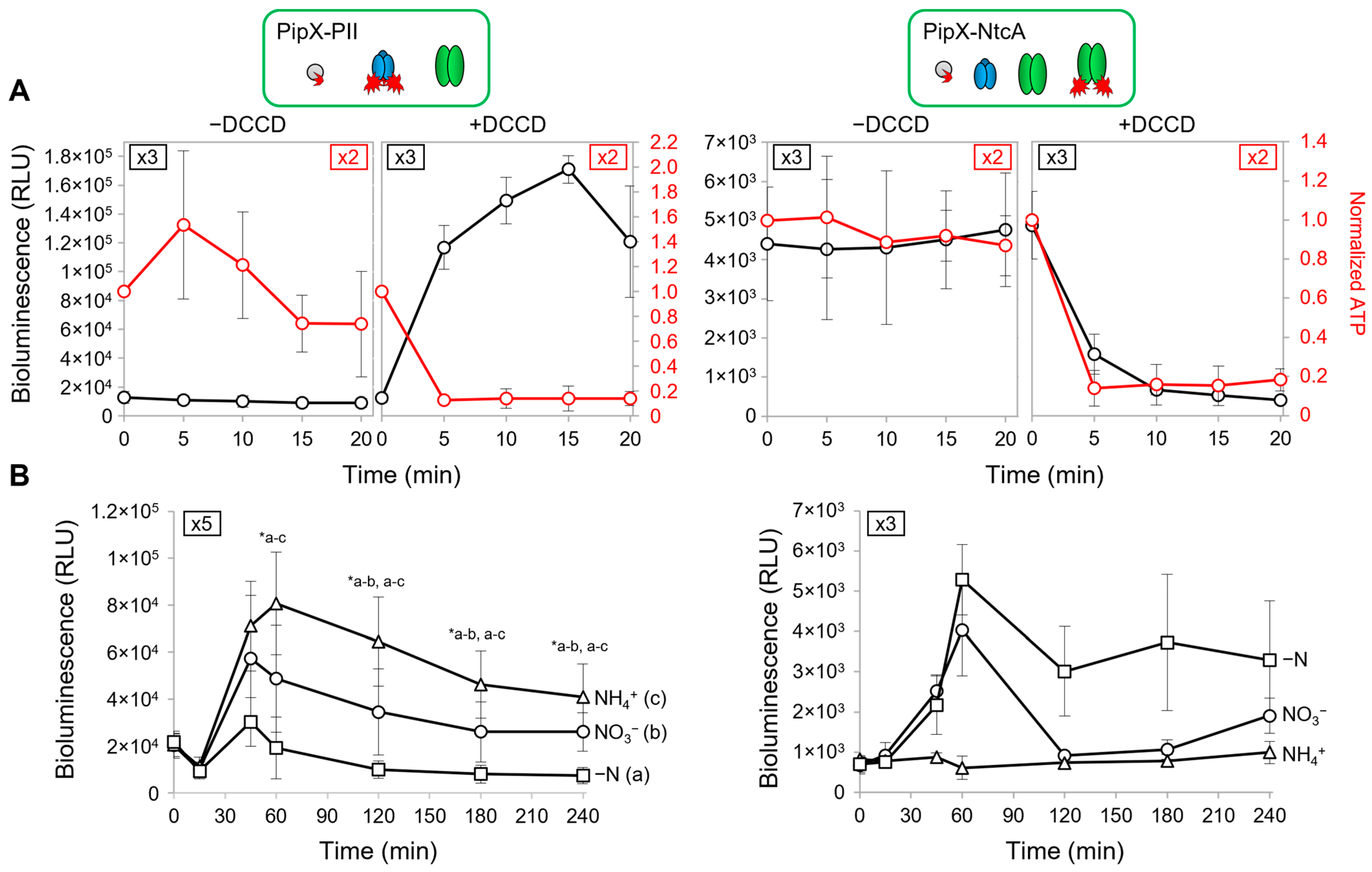
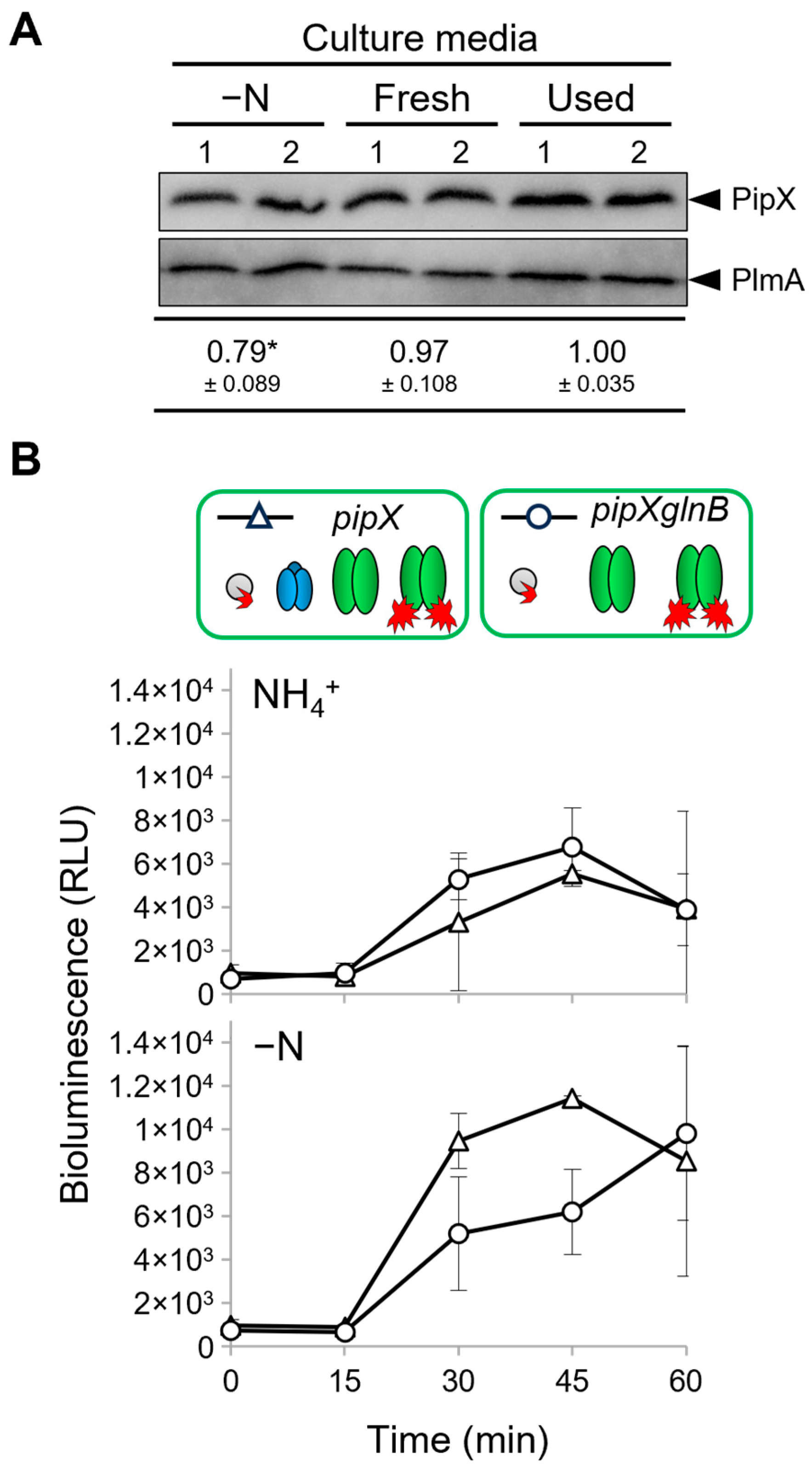
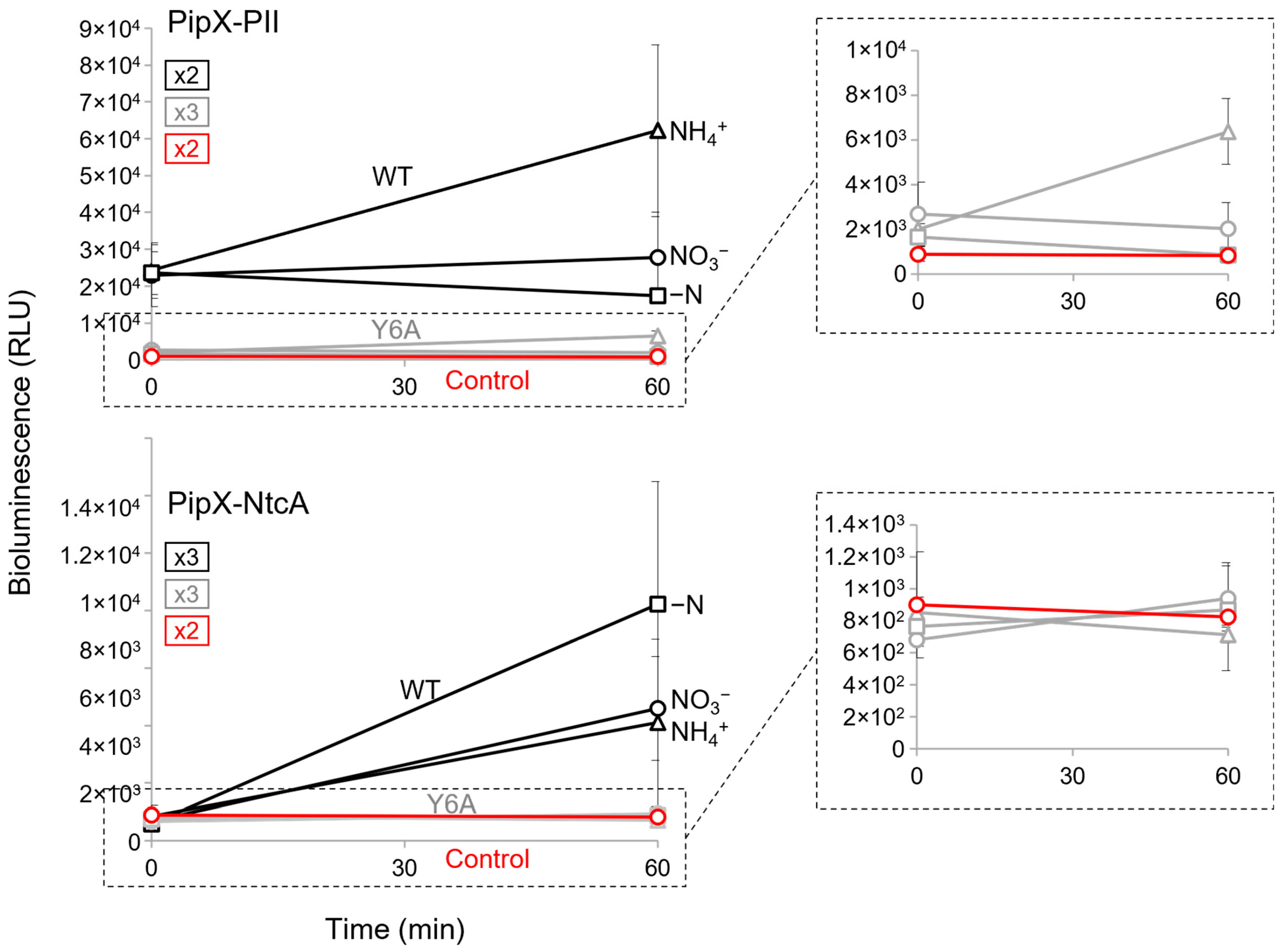
| Strain | Genotype, Relevant Characteristics | Reference or Source |
|---|---|---|
| E. coli XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F ́ proAB lacIqZ∆M15 Tn10 (TetR)] | [55] |
| E. coli TOP10 | F− mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 nupG recA1 araD139 Δ(ara-leu)7697 galE15 galK16 rpsL (StrR) endA1 λ− | Invitrogen |
| WT | Wild-type S. elongatus PCC7942 | Pasteur Culture Collection |
| pipX | ΔpipX::cat, CmR | [38] |
| pipXglnB | ΔpipX::cat glnB::CK.2, CmR KmR | [33] |
| pipX 1SPipX-SmBiT | ΔpipX::cat NSI::(PpipX:pipX:FL:SmBiT), SmR CmR | This work |
| pipXglnB 1SPipX-SmBiT | ΔpipX::cat glnB::CK2 NSI::(PpipX:pipX:FL:SmBiT), SmR KmR CmR | This work |
| pipXglnB 1SPipXSmBiT-PIILgBiT | ΔpipX::cat glnB::CK2 NSI::(PpipX:pipX:FL:SmBiT PglnB:glnB:FL:LgBiT), SmR CmR KmR | This work |
| pipXglnB 1SPipXY6ASmBiT-PIILgBiT | ΔpipX::cat glnB::CK2 NSI::(PpipX:pipX16ta>gc:FL:SmBiT PglnB:glnB:FL:LgBiT), SmR CmR KmR | This work |
| pipX 1SPipXSmBiT-NtcALgBiT | ΔpipX::cat NSI::(PpipX:pipX:FL:SmBiT PntcA:ntcA:FL:LgBiT), SmR CmR | This work |
| pipX 1SPipXY6ASmBiT-NtcALgBiT | ΔpipX::cat NSI::(PpipX:pipX16ta>gc:FL:SmBiT PntcA:ntcA:FL:LgBiT), SmR CmR | This work |
| pipXglnB 1SPipXSmBiT-NtcALgBiT | ΔpipX::cat glnB::CK2 NSI::(PpipX:pipX:FL:SmBiT PntcA:ntcA:FL:LgBiT), SmR CmR KmR | This work |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jerez, C.; Llop, A.; Salinas, P.; Bibak, S.; Forchhammer, K.; Contreras, A. Analysing the Cyanobacterial PipX Interaction Network Using NanoBiT Complementation in Synechococcus elongatus PCC7942. Int. J. Mol. Sci. 2024, 25, 4702. https://doi.org/10.3390/ijms25094702
Jerez C, Llop A, Salinas P, Bibak S, Forchhammer K, Contreras A. Analysing the Cyanobacterial PipX Interaction Network Using NanoBiT Complementation in Synechococcus elongatus PCC7942. International Journal of Molecular Sciences. 2024; 25(9):4702. https://doi.org/10.3390/ijms25094702
Chicago/Turabian StyleJerez, Carmen, Antonio Llop, Paloma Salinas, Sirine Bibak, Karl Forchhammer, and Asunción Contreras. 2024. "Analysing the Cyanobacterial PipX Interaction Network Using NanoBiT Complementation in Synechococcus elongatus PCC7942" International Journal of Molecular Sciences 25, no. 9: 4702. https://doi.org/10.3390/ijms25094702
APA StyleJerez, C., Llop, A., Salinas, P., Bibak, S., Forchhammer, K., & Contreras, A. (2024). Analysing the Cyanobacterial PipX Interaction Network Using NanoBiT Complementation in Synechococcus elongatus PCC7942. International Journal of Molecular Sciences, 25(9), 4702. https://doi.org/10.3390/ijms25094702







