Unraveling the Impact of Extracellular Vesicle-Depleted Serum on Endothelial Cell Characteristics over Time
Abstract
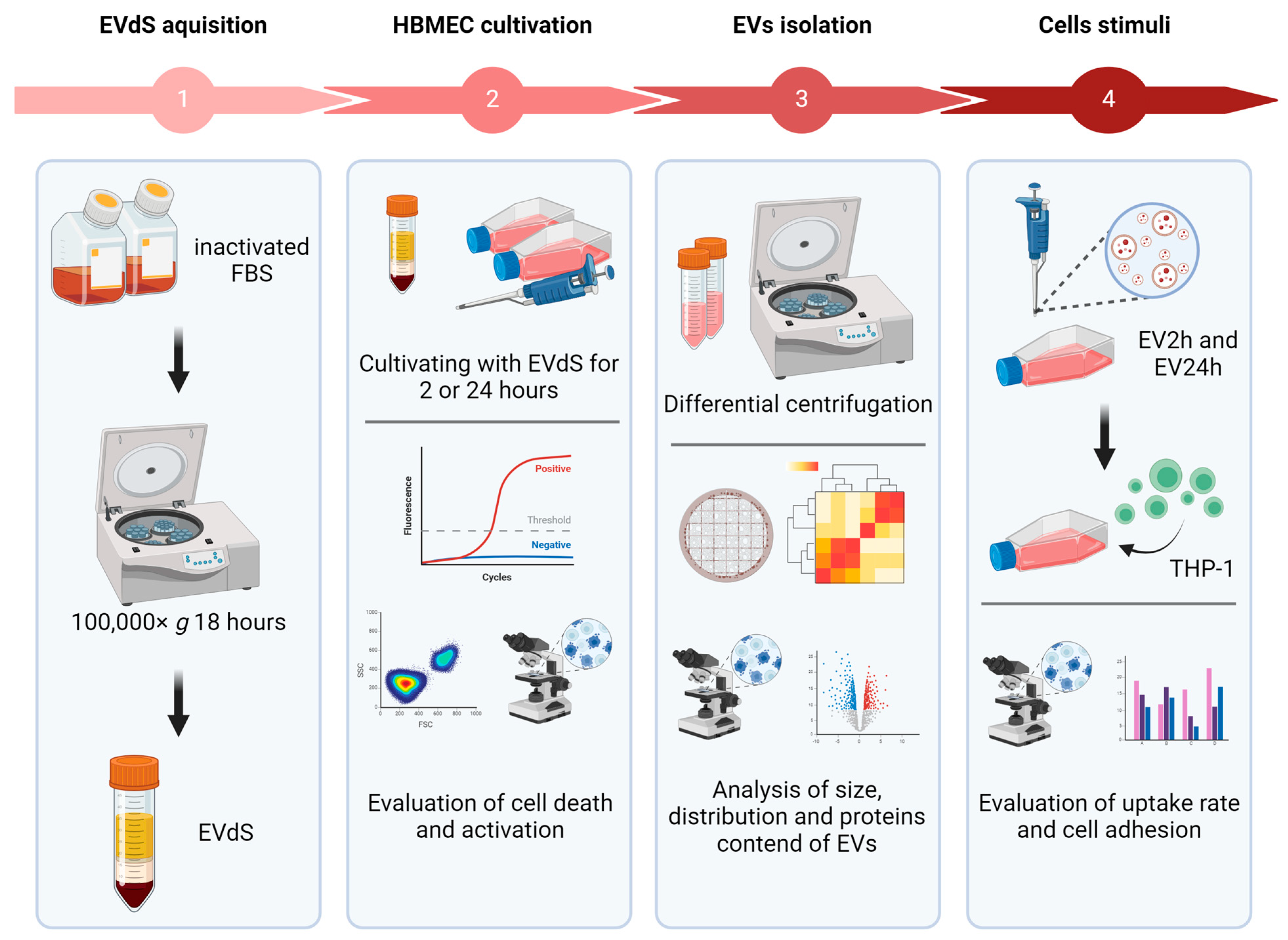
1. Introduction
2. Results
2.1. Endothelial Cells Grown in Media Depleted of EVs Exhibit an Increase in Early and Late Apoptosis
2.2. HBMEC Grown with EVdS Exhibit Increased Endothelial Activation
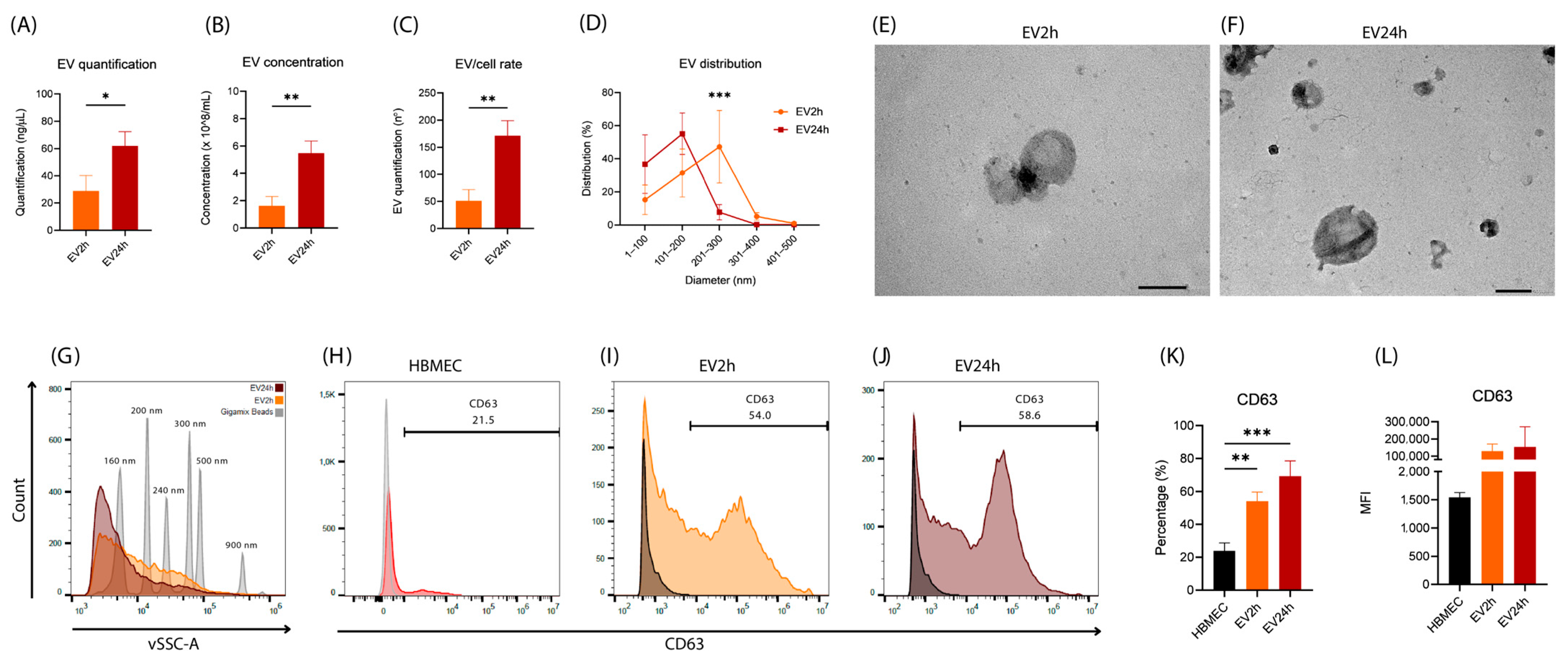
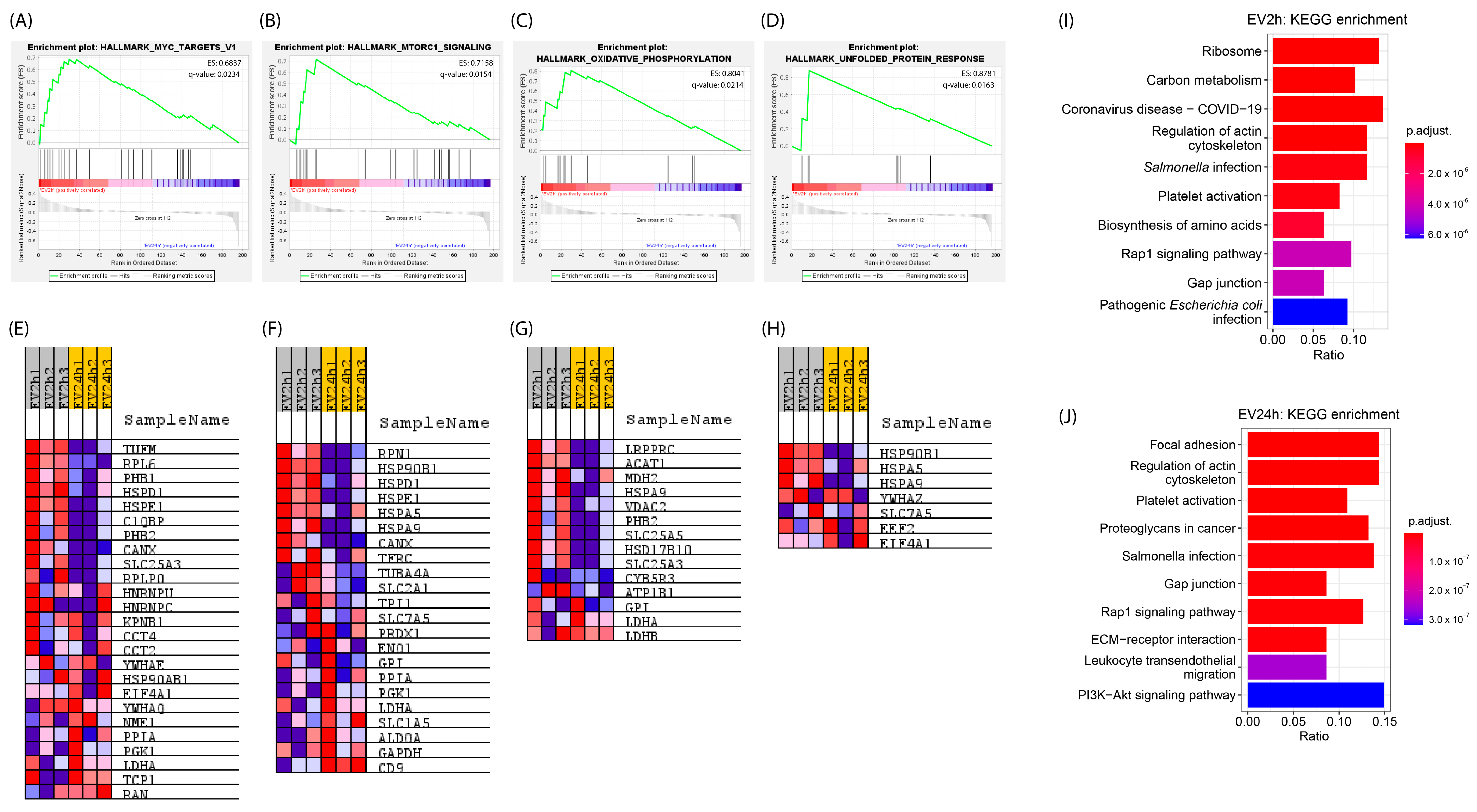
2.3. EVs Isolated from HBMEC Cultured with EVdS for 2 or 24 h Exhibited Distinct Physical Characteristics and Protein Contents
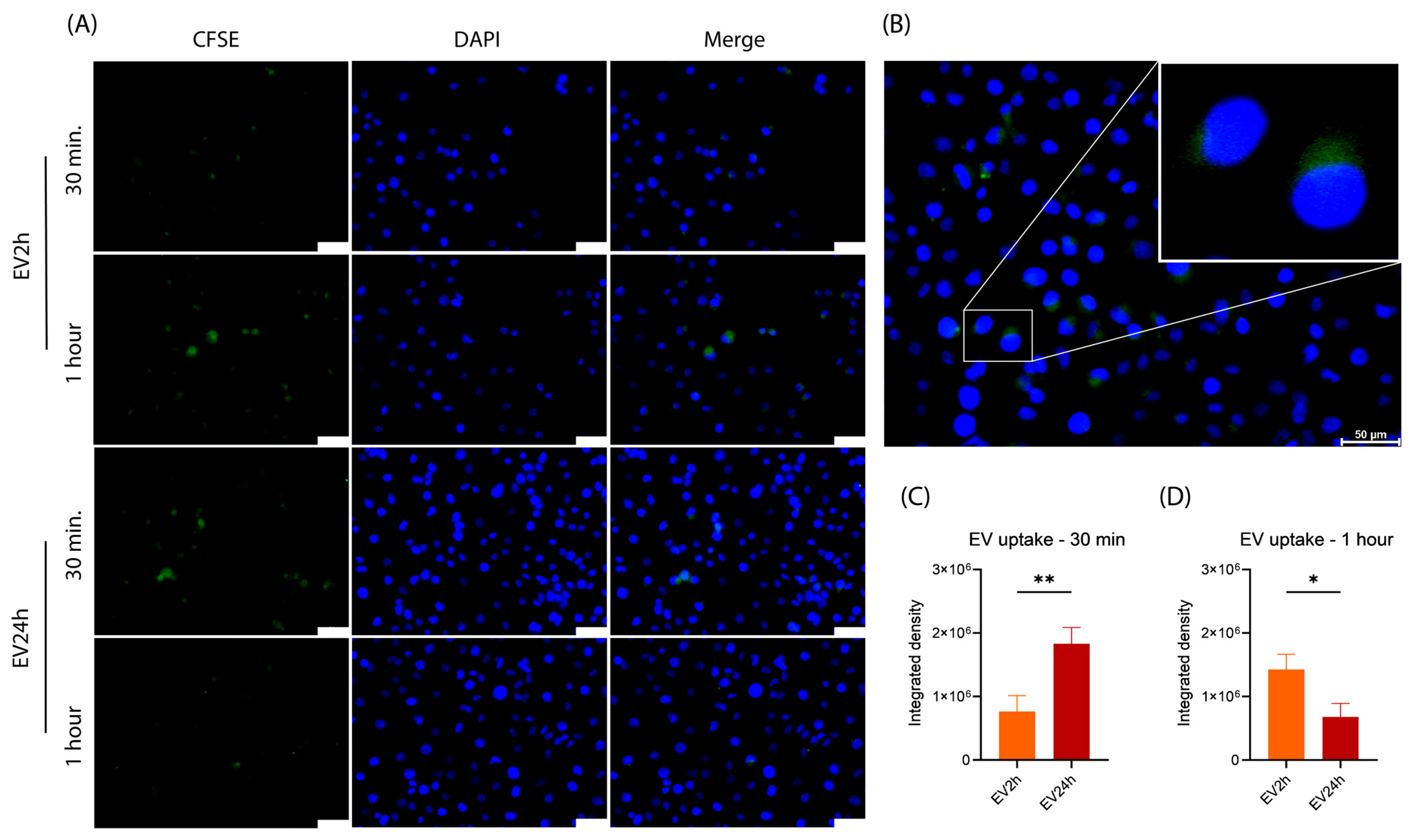
2.4. The Uptake of EV2h and EV24h by HBMEC Differs over Time
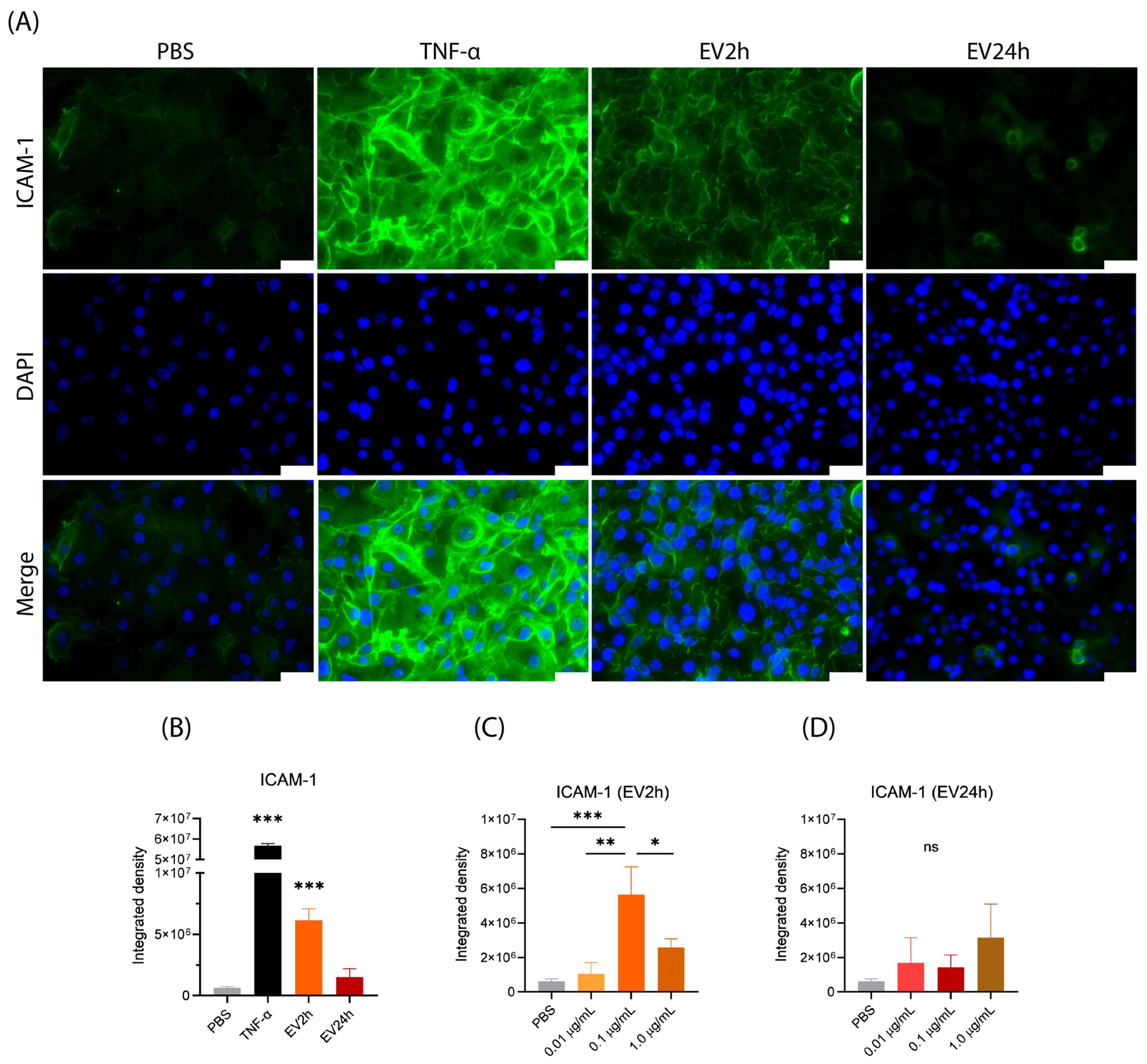
2.5. EV2h Enhances ICAM-1 Expression on HBMEC, Promoting THP-1 Adhesion
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. EV Isolation
4.3. Characterization of EVs
4.4. Evaluation of Endothelial Cell Activation and Death
4.5. Evaluation of the Biological Activity of EVs
4.6. Statistical Evaluation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hernanz, R.; Martínez-Revelles, S.; Palacios, R.; Martín, A.; Cachofeiro, V.; Aguado, A.; García-Redondo, L.; Barrús, M.T.; De Batista, P.R.; Briones, A.M.; et al. Toll-like receptor 4 contributes to vascular remodelling and endothelial dysfunction in angiotensin II-induced hypertension. Br. J. Pharmacol. 2015, 172, 3159–3176. [Google Scholar] [CrossRef] [PubMed]
- Nör, J.E.; Christensen, J.; Mooney, D.J.; Polverini, P.J. Vascular endothelial growth factor (VEGF)-mediated angiogenesis is associated with enhanced endothelial cell survival and induction of Bcl-2 expression. Am. J. Pathol. 1999, 154, 375–384. [Google Scholar] [CrossRef] [PubMed]
- Wilhelms, D.B.; Kirilov, M.; Mirrasekhian, E.; Eskilsson, A.; Kugelberg, U.O.; Klar, C.; Ridder, D.A.; Herschman, H.R.; Schwaninger, M.; Blomqvist, A.; et al. Deletion of Prostaglandin E2 Synthesizing Enzymes in Brain Endothelial Cells Attenuates Inflammatory Fever. J. Neurosci. 2014, 34, 11684–11690. [Google Scholar] [CrossRef] [PubMed]
- Michiels, C.; Arnould, T.; Knott, I.; Dieu, M.; Remacle, J. Stimulation of prostaglandin synthesis by human endothelial cells exposed to hypoxia. Am. J. Physiol. Cell Physiol. 1993, 264, C866–C874. [Google Scholar] [CrossRef] [PubMed]
- Lopes Pinheiro, M.A.; Kamermans, A.; Garcia-Vallejo, J.J.; Van Het Hof, B.; Wierts, L.; O’Toole, T.; Boeve, D.; Verstege, M.; Van Der Pol, S.M.A.; Van Kooyk, Y.; et al. Internalization and presentation of myelin antigens by the brain endothelium guides antigen-specific T cell migration. elife 2016, 5, 13149. [Google Scholar] [CrossRef]
- Cunningham, A.C.; Zhang, J.G.; Moy, J.V.; Ali, S.; Kirby, J.A. A comparison of the antigen-presenting capabilities of class II MHC-expressing human lung epithelial and endothelial cells. Immunology 1997, 91, 458–463. [Google Scholar] [CrossRef] [PubMed]
- Schoergenhofer, C.; Schwameis, M.; Gelbenegger, G.; Buchtele, N.; Thaler, B.; Mussbacher, M.; Schabbauer, G.; Wojta, J.; Jilma-Stohlawetz, P.; Jilma, B. Inhibition of Protease-Activated Receptor (PAR1) Reduces Activation of the Endothelium, Coagulation, Fibrinolysis and Inflammation during Human Endotoxemia. Thromb. Haemost. 2018, 118, 1176–1184. [Google Scholar] [CrossRef] [PubMed]
- Manz, X.D.; Albers, H.J.; Symersky, P.; Aman, J.; van der Meer, A.D.; Bogaard, H.J.; Szulcek, R. In vitro microfluidic disease model to study whole blood-endothelial interactions and blood clot dynamics in real-time. J. Vis. Exp. 2020, 2020, e61068. [Google Scholar] [CrossRef]
- Helms, H.C.; Abbott, N.J.; Burek, M.; Cecchelli, R.; Couraud, P.-O.; Deli, M.A.; Förster, C.; Galla, H.J.; Romero, I.A.; Shusta, E.V.; et al. In vitro models of the blood–brain barrier: An overview of commonly used brain endothelial cell culture models and guidelines for their use. J. Cereb. Blood Flow. Metab. 2016, 36, 862–890. [Google Scholar] [CrossRef]
- Shilo, M.; Sharon, A.; Baranes, K.; Motiei, M.; Lellouche, J.P.M.; Popovtzer, R. The effect of nanoparticle size on the probability to cross the blood-brain barrier: An in-vitro endothelial cell model. J. Nanobiotechnol. 2015, 13, 19. [Google Scholar] [CrossRef]
- De Assis, M.C.; Da Costa, A.O.; Barja-Fidalgo, T.C.; Plotkowski, M.C. Human endothelial cells are activated by interferon-γ plus tumour necrosis factor-α to kill intracellular Pseudomonas aeruginosa. Immunology 2000, 101, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Drevets, D.A.; Sawyer, R.T.; Potter, T.A.; Campbell, P.A. Listeria monocytogenes infects human endothelial cells by two distinct mechanisms. Infect. Immun. 1995, 63, 4268–4276. [Google Scholar] [CrossRef] [PubMed]
- Calvert, J.K.; Helbig, K.J.; Dimasi, D.; Cockshell, M.; Beard, M.R.; Pitson, S.M.; Bonder, C.S.; Carr, J.M. Dengue Virus Infection of Primary Endothelial Cells in Endothelial Cells Function and Is Restricted by Interferon-Stimulated Responses. J. Interf. Cytokine Res. 2015, 35, 654–665. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, N.F.; Wilson, G.K.; Murray, J.; Hu, K.; Lewis, A.; Reynolds, G.M.; Stamataki, Z.; Meredith, L.W.; Rowe, I.A.; Luo, G.; et al. Hepatitis C virus infects the endothelial cells of the blood-brain barrier. Gastroenterology 2012, 142, 634–643.e6. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Spellberg, B.; Phan, Q.T.; Fu, Y.; Fu, Y.; Lee, A.S.; Edwards, J.E.; Filler, S.G.; Ibrahim, A.S. The endothelial cell receptor GRP78 is required for mucormycosis pathogenesis in diabetic mice. J. Clin. Invest 2010, 120, 1914–1924. [Google Scholar] [CrossRef]
- Knight, B.C.; Brunton, C.L.; Modi, N.C.; Wallace, G.R.; Stanford, M.R. The effect of Toxoplasma gondii infection on expression of chemokines by rat retinal vascular endothelial cells. J. Neuroimmunol. 2005, 160, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Li, Y.; Huang, X.; Liu, K.; Wang, Q.D.; Chen, A.F.; Sun, K.; Lui, K.O.; Zhou, B. Seamless Genetic Recording of Transiently Activated Mesenchymal Gene Expression in Endothelial Cells During Cardiac Fibrosis. Circulation 2021, 144, 2004–2020. [Google Scholar] [CrossRef] [PubMed]
- White, L.E.; Cui, Y.; Shelak, C.M.F.; Lie, M.L.; Hassoun, H.T. Lung endothelial cell apoptosis during ischemic acute kidney injury. Shock 2012, 38, 320–327. [Google Scholar] [CrossRef]
- Liu, J.Y.; Yao, J.; Li, X.M.; Song, Y.C.; Wang, X.Q.; Li, Y.J.; Yan, B.; Jiang, Q. Pathogenic role of lncRNA-MALAT1 in endothelial cell dysfunction in diabetes mellitus. Cell Death Dis. 2014, 5, e1506. [Google Scholar] [CrossRef]
- Antonetti, D.A.; Barber, A.J.; Khin, S.; Lieth, E.; Tarbell, J.M.; Gardner, T.W.; the Penn State Retina Research Group. Vascular Endothelial Growth Factor Decreases Occludin in Retinal Endothelial Cells. Diabetes 1998, 47, 1953–1960. [Google Scholar] [CrossRef]
- Paone, S.; Baxter, A.A.; Hulett, M.D.; Poon, I.K.H. Endothelial cell apoptosis and the role of endothelial cell-derived extracellular vesicles in the progression of atherosclerosis. Cell. Mol. Life Sci. 2019, 76, 1093–1106. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, S.; Niida, S.; Azuma, E.; Yanagibashi, T.; Muramatsu, M.; Huang, T.T.; Sagara, H.; Higaki, S.; Ikutani, M.; Nagai, Y.; et al. Inflammation-induced endothelial cell-derived extracellular vesicles modulate the cellular status of pericytes. Sci. Rep. 2015, 5, 8505. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.W.; Lee, H.M.; Lee, T.H.; Kang, C.; Kleinman, H.K.; Gho, Y.S. Extracellular membrane vesicles from tumor cells promote angiogenesis via sphingomyelin. Cancer Res. 2002, 62, 6312–6317. [Google Scholar] [PubMed]
- Théry, C.; Regnault, A.; Garin, J.; Wolfers, J.; Zitvogel, L.; Ricciardi-Castagnoli, P.; Raposo, G.; Amigorena, S. Molecular characterization of dendritic cell-derived exosomes: Selective accumulation of the heat shock protein hsc73. J. Cell Biol. 1999, 147, 599–610. [Google Scholar] [CrossRef] [PubMed]
- Cocucci, E.; Meldolesi, J. Ectosomes and exosomes: Shedding the confusion between extracellular vesicles. Trends Cell Biol. 2015, 25, 364–372. [Google Scholar] [CrossRef] [PubMed]
- Bobrie, A.; Colombo, M.; Krumeich, S.; Raposo, G.; Théry, C. Diverse subpopulations of vesicles secreted by different intracellular mechanisms are present in exosome preparations obtained by differential ultracentrifugation. J. Extracell. Vesicles 2012, 1, 18397. [Google Scholar] [CrossRef] [PubMed]
- Bonsergent, E.; Grisard, E.; Buchrieser, J.; Schwartz, O.; Théry, C.; Lavieu, G. Quantitative characterization of extracellular vesicle uptake and content delivery within mammalian cells. Nat. Commun. 2021, 12, 1864. [Google Scholar] [CrossRef] [PubMed]
- Kowal, J.; Arras, G.; Colombo, M.; Jouve, M.; Morath, J.P.; Primdal-Bengtson, B.; Dingli, F.; Loew, D.; Tkach, M.; Théry, C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. USA 2016, 113, E968–E977. [Google Scholar] [CrossRef] [PubMed]
- Lázaro-Ibáñez, E.; Sanz-Garcia, A.; Visakorpi, T.; Escobedo-Lucea, C.; Siljander, P.; Ayuso-Sacido, Á.; Yliperttula, M. Different gDNA Content in the Subpopulations of Prostate Cancer Extracellular Vesicles: Apoptotic Bodies, Microvesicles, and Exosomes. Prostate 2014, 74, 1379–1390. [Google Scholar] [CrossRef]
- Rodrigues, M.L.; Nakayasu, E.S.; Oliveira, D.L.; Nimrichter, L.; Nosanchuk, J.D.; Almeida, I.C.; Casadevall, A. Extracellular Vesicles Produced by Cryptococcus neoformans Contain Protein Components Associated with Virulence. Eukariotic Cell 2008, 7, 58–67. [Google Scholar] [CrossRef]
- Berumen Sánchez, G.; Bunn, K.E.; Pua, H.H.; Rafat, M. Extracellular vesicles: Mediators of intercellular communication in tissue injury and disease. Cell Commun. Signal. 2021, 19, 104. [Google Scholar] [CrossRef] [PubMed]
- Pitt, J.M.; Kroemer, G.; Zitvogel, L. Extracellular vesicles: Masters of intercellular communication and potential clinical interventions. J. Clin. Invest 2016, 126, 1139–1143. [Google Scholar] [CrossRef] [PubMed]
- French, S.L.; Butov, K.R.; Allaeys, I.; Canas, J.; Morad, G.; Davenport, P.; Laroche, A.; Trubina, N.M.; Italiano, J.E.; Moses, M.A.; et al. Platelet-derived extracellular vesicles infiltrate and modify the bone marrow during inflammation. Blood Adv. 2020, 4, 3011–3023. [Google Scholar] [CrossRef] [PubMed]
- Bei, Y.; Xu, T.; Lv, D.; Yu, P.; Xu, J.; Che, L.; Das, A.; Tigges, J.; Toxavidis, V.; Ghiran, I.; et al. Exercise-induced circulating extracellular vesicles protect against cardiac ischemia–reperfusion injury. Basic Res. Cardiol. 2017, 112, 38. [Google Scholar] [CrossRef] [PubMed]
- Greening, D.W.; Xu, R.; Ji, H.; Tauro, B.J.; Simpson, R.J. A protocol for exosome isolation and characterization: Evaluation of ultracentrifugation, density-gradient separation, and immunoaffinity capture methods. In Methods in Molecular Biology; Humana Press: New York, NY, USA, 2015; Volume 1295, ISBN 9781493925506. [Google Scholar]
- Menck, K.; Bleckmann, A.; Schulz, M.; Ries, L.; Binder, C. Isolation and characterization of microvesicles from peripheral blood. J. Vis. Exp. 2017, 2017, e55057. [Google Scholar] [CrossRef]
- Jimenez, L.; Yu, H.; McKenzie, A.J.; Franklin, J.L.; Patton, J.G.; Liu, Q.; Weaver, A.M. Quantitative Proteomic Analysis of Small and Large Extracellular Vesicles (EVs) Reveals Enrichment of Adhesion Proteins in Small EVs. J. Proteome Res. 2019, 18, 947–959. [Google Scholar] [CrossRef]
- Lischnig, A.; Bergqvist, M.; Ochiya, T.; Lässer, C. Quantitative Proteomics Identifies Proteins Enriched in Large and Small Extracellular Vesicles. Mol. Cell. Proteomics 2022, 21, 100273. [Google Scholar] [CrossRef]
- Ciardiello, C.; Migliorino, R.; Leone, A.; Budillon, A. Large extracellular vesicles: Size matters in tumor progression. Cytokine Growth Factor. Rev. 2020, 51, 69–74. [Google Scholar] [CrossRef]
- Welsh, J.A.; Goberdhan, D.C.I.; O’Driscoll, L.; Buzas, E.I.; Blenkiron, C.; Bussolati, B.; Cai, H.; Di Vizio, D.; Driedonks, T.A.P.; Erdbrügger, U.; et al. Minimal information for studies of extracellular vesicles (MISEV2023): From basic to advanced approaches. J. Extracell. Vesicles 2024, 13, e12404. [Google Scholar] [CrossRef]
- Li, Y.; He, X.; Li, Q.; Lai, H.; Zhang, H.; Hu, Z.; Li, Y.; Huang, S. EV-origin: Enumerating the tissue-cellular origin of circulating extracellular vesicles using exLR profile. Comput. Struct. Biotechnol. J. 2020, 18, 2851–2859. [Google Scholar] [CrossRef]
- Aatonen, M.T.; öhman, T.; Nyman, T.A.; Laitinen, S.; Grönholm, M.; Siljander, P.R.M. Isolation and characterization of platelet-derived extracellular vesicles. J. Extracell. Vesicles 2014, 3, 24692. [Google Scholar] [CrossRef] [PubMed]
- Tessandier, N.; Melki, I.; Cloutier, N.; Allaeys, I.; Miszta, A.; Tan, S.; Milasan, A.; Michel, S.; Benmoussa, A.; Lévesque, T.; et al. Platelets Disseminate Extracellular Vesicles in Lymph in Rheumatoid Arthritis. Arterioscler. Thromb. Vasc. Biol. 2020, 40, 929–942. [Google Scholar] [CrossRef] [PubMed]
- Usman, W.M.; Pham, T.C.; Kwok, Y.Y.; Vu, L.T.; Ma, V.; Peng, B.; Chan, Y.S.; Wei, L.; Chin, S.M.; Azad, A.; et al. Efficient RNA drug delivery using red blood cell extracellular vesicles. Nat. Commun. 2018, 9, 2359. [Google Scholar] [CrossRef] [PubMed]
- Sampaio, N.G.; Emery, S.J.; Garnham, A.L.; Tan, Q.Y.; Sisquella, X.; Pimentel, M.A.; Jex, A.R.; Regev-Rudzki, N.; Schofield, L.; Eriksson, E.M. Extracellular vesicles from early stage Plasmodium falciparum-infected red blood cells contain PfEMP1 and induce transcriptional changes in human monocytes. Cell. Microbiol. 2018, 20, e12822. [Google Scholar] [CrossRef] [PubMed]
- Liao, T.L.; Chen, Y.M.; Tang, K.T.; Chen, P.K.; Liu, H.J.; Chen, D.Y. MicroRNA-223 inhibits neutrophil extracellular traps formation through regulating calcium influx and small extracellular vesicles transmission. Sci. Rep. 2021, 11, 15676. [Google Scholar] [CrossRef]
- Puhm, F.; Afonyushkin, T.; Resch, U.; Obermayer, G.; Rohde, M.; Penz, T.; Schuster, M.; Wagner, G.; Rendeiro, A.F.; Melki, I.; et al. Mitochondria are a subset of extracellular vesicles released by activated monocytes and induce type I IFN and TNF responses in endothelial cells. Circ. Res. 2019, 125, 43–52. [Google Scholar] [CrossRef]
- Njock, M.S.; Cheng, H.S.; Dang, L.T.; Nazari-Jahantigh, M.; Lau, A.C.; Boudreau, E.; Roufaiel, M.; Cybulsky, M.I.; Schober, A.; Fish, J.E. Endothelial cells suppress monocyte activation through secretion of extracellular vesicles containing antiinflammatory microRNAs. Blood 2015, 125, 3202–3212. [Google Scholar] [CrossRef]
- Khandagale, A.; Åberg, M.; Wikström, G.; Bergström Lind, S.; Shevchenko, G.; Björklund, E.; Siegbahn, A.; Christersson, C. Role of Extracellular Vesicles in Pulmonary Arterial Hypertension: Modulation of Pulmonary Endothelial Function and Angiogenesis. Arterioscler. Thromb. Vasc. Biol. 2020, 40, 2293–2309. [Google Scholar] [CrossRef] [PubMed]
- Palviainen, M.; Saraswat, M.; Varga, Z.; Kitka, D.; Neuvonen, M.; Puhka, M.; Joenväärä, S.; Renkonen, R.; Nieuwland, R.; Takatalo, M.; et al. Extracellular vesicles from human plasma and serum are carriers of extravesicular cargo—Implications for biomarker discovery. PLoS ONE 2020, 15, e0236439. [Google Scholar] [CrossRef]
- Liang, K.; Liu, F.; Fan, J.; Sun, D.; Liu, C.; Lyon, C.J.; Bernard, D.W.; Li, Y.; Yokoi, K.; Katz, M.H.; et al. Nanoplasmonic quantification of tumour-derived extracellular vesicles in plasma microsamples for diagnosis and treatment monitoring. Nat. Biomed. Eng. 2017, 1, 21. [Google Scholar] [CrossRef]
- Yoshioka, Y.; Konishi, Y.; Kosaka, N.; Katsuda, T.; Kato, T.; Ochiya, T. Comparative marker analysis of extracellular vesicles in different human cancer types. J. Extracell. Vesicles 2013, 2, 9. [Google Scholar] [CrossRef] [PubMed]
- Rakic, D.; Joksimovic Jovic, J.; Jakovljevic, V.; Zivkovic, V.; Nikolic, M.; Sretenovic, J.; Nikolic, M.; Jovic, N.; Bicanin Ilic, M.; Arsenijevic, P.; et al. High Fat Diet Exaggerate Metabolic and Reproductive PCOS Features by Promoting Oxidative Stress: An Improved EV Model in Rats. Medicina 2023, 59, 1104. [Google Scholar] [CrossRef] [PubMed]
- Monsel, A.; Zhu, Y.G.; Gudapati, V.; Lim, H.; Lee, J.W. Mesenchymal stem cell derived secretome and extracellular vesicles for acute lung injury and other inflammatory lung diseases. Expert Opin. Biol. Ther. 2016, 16, 859–871. [Google Scholar] [CrossRef] [PubMed]
- Sherman, C.D.; Lodha, S.; Sahoo, S. EV Cargo Sorting in Therapeutic Development for Cardiovascular Disease. Cells 2021, 10, 1500. [Google Scholar] [CrossRef] [PubMed]
- Kuczler, M.D.; Zieren, R.C.; Dong, L.; de Reijke, T.M.; Pienta, K.J.; Amend, S.R. Advancements in the identification of EV derived mRNA biomarkers for liquid biopsy of clear cell renal cell carcinomas. Urology 2022, 160, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Oyajobi, B.O.; Franchin, G.; Williams, P.J.; Pulkrabek, D.; Gupta, A.; Munoz, S.; Grubbs, B.; Zhao, M.; Chen, D.; Sherry, B.; et al. Dual effects of macrophage inflammatory protein-1α on osteolysis and tumor burden in the murine 5TGM1 model of myeloma bone disease. Blood 2003, 102, 311–319. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [PubMed]
- Lehrich, B.M.; Liang, Y.; Khosravi, P.; Federoff, H.J.; Fiandaca, M.S. Fetal bovine serum-derived extracellular vesicles persist within vesicle-depleted culture media. Int. J. Mol. Sci. 2018, 19, 3538. [Google Scholar] [CrossRef] [PubMed]
- Shelke, G.V.; Lässer, C.; Gho, Y.S.; Lötvall, J. Importance of exosome depletion protocols to eliminate functional and RNA-containing extracellular vesicles from fetal bovine serum. J. Extracell. Vesicles 2014, 3, 8. [Google Scholar] [CrossRef]
- Kornilov, R.; Puhka, M.; Mannerström, B.; Hiidenmaa, H.; Peltoniemi, H.; Siljander, P.; Seppänen-Kaijansinkko, R.; Kaur, S. Efficient ultrafiltration-based protocol to deplete extracellular vesicles from fetal bovine serum. J. Extracell. Vesicles 2018, 7, 1422674. [Google Scholar] [CrossRef]
- Auber, M.; Fröhlich, D.; Drechsel, O.; Karaulanov, E.; Krämer-Albers, E.M. Serum-free media supplements carry miRNAs that co-purify with extracellular vesicles. J. Extracell. Vesicles 2019, 8, 1656042. [Google Scholar] [CrossRef] [PubMed]
- Aswad, H.; Jalabert, A.; Rome, S. Depleting extracellular vesicles from fetal bovine serum alters proliferation and differentiation of skeletal muscle cells in vitro. BMC Biotechnol. 2016, 16, 32. [Google Scholar] [CrossRef]
- Liao, Z.; Muth, D.C.; Eitan, E.; Travers, M.; Learman, L.N.; Lehrmann, E.; Witwer, K.W. Serum extracellular vesicle depletion processes affect release and infectivity of HIV-1 in culture. Sci. Rep. 2017, 7, 2558. [Google Scholar] [CrossRef] [PubMed]
- Chaderjian, W.B.; Chin, E.T.; Harris, R.J.; Etcheverry, T.M. Effect of copper sulfate on performance of a serum-free CHO cell culture process and the level of free thiol in the recombinant antibody expressed. Biotechnol. Prog. 2005, 21, 550–553. [Google Scholar] [CrossRef]
- Zhang, L.; Shen, H.; Zhang, Y. Fed-batch culture of hybridoma cells in serum-free medium using an optimized feeding strategy. J. Chem. Technol. Biotechnol. 2004, 79, 171–181. [Google Scholar] [CrossRef]
- Deville, S.; Berckmans, P.; Van Hoof, R.; Lambrichts, I.; Salvati, A.; Nelissen, I. Comparison of extracellular vesicle isolation and storage methods using high-sensitivity flow cytometry. PLoS ONE 2021, 16, e0245835. [Google Scholar] [CrossRef] [PubMed]
- Picciotto, S.; Barone, M.E.; Fierli, D.; Aranyos, A.; Adamo, G.; Božič, D.; Romancino, D.P.; Stanly, C.; Parkes, R.; Morsbach, S.; et al. Isolation of extracellular vesicles from microalgae: Towards the production of sustainable and natural nanocarriers of bioactive compounds. Biomater. Sci. 2021, 9, 2917–2930. [Google Scholar] [CrossRef]
- Wowk, P.F.; Zardo, M.L.; Miot, H.T.; Goldenberg, S.; Carvalho, P.C.; Mörking, P.A. Proteomic profiling of extracellular vesicles secreted from Toxoplasma gondii. Proteomics 2017, 17, 15–16. [Google Scholar] [CrossRef]
- Caeiro, L.D.; Alba-Soto, C.D.; Rizzi, M.; Solana, M.E.; Rodriguez, G.; Chidichimo, A.M.; Rodriguez, M.E.; Sánchez, D.O.; Levy, G.V.; Tekiel, V. The protein family TcTASV-C is a novel Trypanosoma cruzi virulence factor secreted in extracellular vesicles by trypomastigotes and highly expressed in bloodstream forms. PLoS Negl. Trop. Dis. 2018, 12, e0006475. [Google Scholar] [CrossRef]
- Maia, M.M.; da Cruz, A.B.; Taniwaki, N.N.; Namiyama, G.M.; Gava, R.; Gomes, A.H.S.; Kanamura, C.T.; Barbo, M.L.P.; Pereira-Chioccola, V.L. Immunization with extracellular vesicles excreted by Toxoplasma gondii confers protection in murine infection, activating cellular and humoral responses. Int. J. Parasitol. 2021, 51, 559–569. [Google Scholar] [CrossRef]
- Robaye, B.; Mosselmans, R.; Fiers, W.; Dumont, J.E.; Galand, P. Tumor necrosis factor induces apoptosis (programmed cell death) in normal endothelial cells in vitro. Am. J. Pathol. 1991, 138, 447–453. [Google Scholar] [PubMed]
- Hosseinkhani, B.; Kuypers, S.; van den Akker, N.M.S.; Molin, D.G.M.; Michiels, L. Extracellular vesicles work as a functional inflammatory mediator between vascular endothelial cells and immune cells. Front. Immunol. 2018, 9, 1789. [Google Scholar] [CrossRef]
- Eitan, E.; Zhang, S.; Witwer, K.W.; Mattson, M.P. Extracellular vesicle-depleted fetal bovine and human sera have reduced capacity to support cell growth. J. Extracell. Vesicles 2015, 4, 26373. [Google Scholar] [CrossRef] [PubMed]
- Bi, X.; Song, J.; Gao, J.; Zhao, J.; Wang, M.; Scipione, C.A.; Koschinsky, M.L.; Wang, Z.V.; Xu, S.; Fu, G. Activation of liver X receptor attenuates lysophosphatidylcholine-induced IL-8 expression in endothelial cells via the NF-κB pathway and SUMOylation. J. Cell. Mol. Med. 2016, 20, 2249–2258. [Google Scholar] [CrossRef]
- Lehmann, U.; Schmitz, J.; Weissenbach, M.; Sobota, R.M.; Hörtner, M.; Friederichs, K.; Behrmann, I.; Tsiaris, W.; Sasaki, A.; Schneider-Mergener, J.; et al. SHP2 and SOCS3 contribute to Tyr-759-dependent attenuation of interleukin-6 signaling through gp130. J. Biol. Chem. 2003, 278, 661–671. [Google Scholar] [CrossRef] [PubMed]
- Dela Paz, N.G.; Walshe, T.E.; Leach, L.L.; Saint-geniez, M.; Amore, P.A.D. Role of shear-stress-induced VEGF expression in endothelial cell survival. J. Cell Sci. 2011, 125, 831–843. [Google Scholar] [CrossRef] [PubMed]
- Lacorre, D.; Baekkevold, E.S.; Garrido, I.; Brandtzaeg, P.; Haraldsen, G. Plasticity of endothelial cells: Rapid dedifferentiation of freshly isolated high endothelial venule endothelial cells outside the lymphoid tissue microenvironment. Blood 2004, 103, 4164–4172. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.J.; Lee, H.J.; Welday, A.C.; Song, E.; Cho, J.; Sharp, P.E.; Jung, M.W.; Blair, H.T. Stress-induced alterations in hippocampal plasticity, place cells, and spatial memory. Proc. Natl. Acad. Sci. USA 2007, 104, 18297–18302. [Google Scholar] [CrossRef] [PubMed]
- Shubin, A.D.; Sharipol, A.; Felong, T.J.; Weng, P.; Schutrum, B.E.; Joe, D.S.; Aure, M.H.; Benoit, D.S.W.; Ovitt, C.E. Stress or injury induces cellular plasticity in salivary gland acinar cells. Cell Tissue Res. 2020, 380, 487–497. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Guan, B.; Alzahrani, M.R.; Gao, Z.; Gao, L.; Bracey, S.; Wu, J.; Mbow, C.A.; Jobava, R.; Haataja, L.; et al. Adaptation to chronic ER stress enforces pancreatic β -cell plasticity. Nat. Commun. 2022, 13, 4621. [Google Scholar] [CrossRef]
- Hogg, N.; Browning, J.; Howard, T.; Winterford, C.; Fitzpatrick, D.; Gobé, G.; Browning, J.; Howard, T.; Winterford, C.; Fitzpatrick, D. Apoptosis in Vascular Endothelial Cells Caused by Serum Deprivation, Oxidative Stress and Transforming Growth Factor-β. J. Endothel. Cell Res. 1999, 7, 35–49. [Google Scholar] [CrossRef] [PubMed]
- Sheu, M.L.; Ho, F.M.; Yang, R.; Sen; Chao, K.F.; Lin, W.W. High Glucose Induces Human Endothelial Cell Apoptosis Through a Phosphoinositide 3-Kinase–Regulated Cyclooxygenase-2 Pathway. Arter. Thromb. Vasc. Biol. 2005, 25, 539–545. [Google Scholar] [CrossRef] [PubMed]
- Pollman, M.J.; Naumovski, L.; Gibbons, G.H. Vascular Cell Apoptosis Cell Type–Specific Modulation by Transforming Growth Factor-b1 in Endothelial Cells Versus Smooth Muscle Cells. Basic Sci. Rep. 1999, 99, 2019–2026. [Google Scholar] [CrossRef]
- Regal-McDonald, K.; Somarathna, M.; Lee, T.; Litovsky, S.H.; Barnes, J.; Peretik, J.M.; Traylor, J.G.; Orr, A.W.; Patel, R.P. Assessment of ICAM-1 N-glycoforms in mouse and human models of endothelial dysfunction. PLoS ONE 2020, 15, e0230358. [Google Scholar] [CrossRef]
- Kuravi, S.J.; Harrison, P.; Rainger, G.E.; Nash, G.B. Ability of Platelet-Derived Extracellular Vesicles to Promote Neutrophil-Endothelial Cell Interactions. Inflammation 2019, 42, 290–305. [Google Scholar] [CrossRef] [PubMed]
- Zhong, M.; Gadek, T.R.; Bui, M.; Shen, W.; Burnier, J.; Barr, K.J.; Hanan, E.J.; Oslob, J.D.; Yu, C.H.; Zhu, J.; et al. Discovery and Development of Potent LFA-1/ICAM-1 Antagonist SAR 1118 as an Ophthalmic Solution for Treating Dry Eye. Am. Chem. Soc. 2012, 3, 2003–2206. [Google Scholar] [CrossRef] [PubMed]
- Yacyshyn, B.R.; Beth, M.; Yacyshyn, B.; Jewell, L.; Tami, J.A.; Bennett, C.F.; Kisner, D.L.; Shanahan, W.R. A Placebo-Controlled Trial of ICAM-1 Antisense Oligonucleotide in the Treatment of Crohn’s Disease. Gastroenterology 1998, 114, 1133–1142. [Google Scholar] [CrossRef]
- Maurer, C.; Friess, H.; Kretschmann, B.; Wildi, S.; Müller, C.; Graber, L.H.; Schilling, M.; Büchler, M.W. Over-Expression of ICAM-1, VCAM-1 and ELAM-1 Might Influence Tumor Progression in Colorectal Cancer. Int. J. Cancer (Pred. Oncol.) 1998, 79, 76–81. [Google Scholar] [CrossRef]
- Guo, W.; Liu, S.; Cheng, Y.; Lu, L.; Shi, J.; Xu, G.; Li, N.; Cheng, K.; Wu, M.; Cheng, S.; et al. ICAM-1—Related Noncoding RNA in Cancer Stem Cells Maintains ICAM-1 Expression in Hepatocellular Carcinoma. Clin. Cancer Res. 2016, 22, 2041–2050. [Google Scholar] [CrossRef]
- Hess, D.C.; Howard, E.; Cheng, C.; Carroll, J.; Hill, D.W. Hypertonic mannitol loading of NF-κB transcription factor decoys in human brain microvascular endothelial cells blocks upregulation of ICAM-1. Stroke 2000, 31, 1179–1186. [Google Scholar] [CrossRef]
- Zhu, Y.P.; Shen, T.; Lin, Y.J.; Chen, B.D.; Ruan, Y.; Cao, Y.; Qiao, Y.; Man, Y.; Wang, S.; Li, J. Astragalus polysaccharides suppress ICAM-1 and VCAM-1 expression in TNF-α-treated human vascular endothelial cells by blocking NF-κB activation. Acta Pharmacol. Sin. 2013, 34, 1036–1042. [Google Scholar] [CrossRef] [PubMed]
- Herrera-Zelada, N.; Zúñiga-Cuevas, Ú.; Ramírez-Reyes, A.; Norambuena-Soto, I.; Venegas-Zamora, L.; Troncoso, M.F.; Hernández, A.; Sánchez, G.; Pedrozo, Z.; Lavandero, S.; et al. Endothelial activation impairs the function of small extracellular vesicles. Front. Pharmacol. 2023, 14, 1143888. [Google Scholar] [CrossRef] [PubMed]
- Cashikar, A.G.; Hanson, P.I. A cell-based assay for CD63-containing extracellular vesicles. PLoS ONE 2019, 14, e0220007. [Google Scholar] [CrossRef] [PubMed]
- Hagey, D.W.; Ojansivu, M.; Bostancioglu, B.R.; Saher, O.; Bost, J.P.; Gustafsson, M.O.; Gramignoli, R.; Svahn, M.; Gupta, D.; Stevens, M.M.; et al. The cellular response to extracellular vesicles is dependent on their cell source and dose. Sci. Adv. 2023, 9, eadh1168. [Google Scholar] [CrossRef] [PubMed]
- Dogrammatzis, C.; Deschamps, T. Biogenesis of Extracellular Vesicles during Herpes Simplex. J. Viromol. 2019, 93, 1–15. [Google Scholar]
- Stassen, F.R.M.; Eijck, P.H.; van Savelkoul, P.H.M.; Wouters, E.F.M.; Rohde, G.G.U.; Briedé, J.J.; Reynaert, N.L.; Kok, T.M.; De Benedikter, B.J. Cell Type- and Exposure-Specific Modulation of CD63/CD81- Positive and Tissue Factor-Positive Extracellular Vesicle Release in response to Respiratory Toxicants. Oxid. Med. Cell. Longev. 2019, 2019, 9. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Ji, D.; Yin, Y.; Wang, S.; Song, K.; Pan, Q.; Zhang, Q.; Yang, L. Proteomic profiling of serum small extracellular vesicles reveals immune signatures of children with pneumonia. Transl. Pediatr. 2022, 11, 891–908. [Google Scholar] [CrossRef] [PubMed]
- Blaser, M.C.; Buffolo, F.; Halu, A.; Turner, M.E.; Schlotter, F.; Higashi, H.; Pantano, L.; Clift, C.L.; Saddic, L.A.; Atkins, S.K.; et al. Multiomics of Tissue Extracellular Vesicles Identifies Unique Modulators of Atherosclerosis and Calcific Aortic Valve Stenosis. Circulation 2023, 148, 63402. [Google Scholar] [CrossRef] [PubMed]
- Palviainen, M.; Saari, H.; Kärkkäinen, O.; Pekkinen, J.; Auriola, S.; Yliperttula, M.; Puhka, M.; Hanhineva, K.; Siljander, P.R.M. Metabolic signature of extracellular vesicles depends on the cell culture conditions. J. Extracell. Vesicles 2019, 8, 1596669. [Google Scholar] [CrossRef]
- Serrano-Pertierra, E.; Oliveira-Rodríguez, M.; Rivas, M.; Oliva, P.; Villafani, J.; Navarro, A.; Blanco-López, M.C.; Cernuda-Morollón, E. Characterization of plasma-derived extracellular vesicles isolated by different methods: A comparison study. Bioengineering 2019, 6, 8. [Google Scholar] [CrossRef]
- Vujić, T.; Schvartz, D.; Furlani, I.L.; Meister, I.; González-Ruiz, V.; Rudaz, S.; Sanchez, J.C. Oxidative Stress and Extracellular Matrix Remodeling Are Signature Pathways of Extracellular Vesicles Released upon Morphine Exposure on Human Brain Microvascular Endothelial Cells. Cells 2022, 11, 3926. [Google Scholar] [CrossRef] [PubMed]
- András, I.E.; Sewell, B.B.; Toborek, M. HIV-1 and amyloid beta remodel proteome of brain endothelial extracellular vesicles. Int. J. Mol. Sci. 2020, 21, 2741. [Google Scholar] [CrossRef] [PubMed]
- Droste, M.; Tertel, T.; Jeruschke, S.; Dittrich, R.; Kontopoulou, E.; Walkenfort, B.; Börger, V.; Hoyer, P.F.; Büscher, A.K.; Thakur, B.K.; et al. Single extracellular vesicle analysis performed by imaging flow cytometry and nanoparticle tracking analysis evaluate the accuracy of urinary extracellular vesicle preparation techniques differently. Int. J. Mol. Sci. 2021, 22, 12436. [Google Scholar] [CrossRef] [PubMed]
- Cvjetkovic, A.; Lötvall, J.; Lässer, C. The influence of rotor type and centrifugation time on the yield and purity of extracellular vesicles. J. Extracell. Vesicles 2014, 3, 23111. [Google Scholar] [CrossRef] [PubMed]
- Tan, K.L.; Chia, W.C.; How, C.W.; Tor, Y.S.; Show, P.L.; Looi, Q.H.D.; Foo, J.B. Benchtop Isolation and Characterisation of Small Extracellular Vesicles from Human Mesenchymal Stem Cells. Mol. Biotechnol. 2021, 63, 780–791. [Google Scholar] [CrossRef]
- Takafuji, Y.; Tatsumi, K.; Ishida, M.; Kawao, N.; Okada, K.; Kaji, H. Extracellular vesicles secreted from mouse muscle cells suppress osteoclast formation: Roles of mitochondrial energy metabolism. Bone 2020, 134, 115298. [Google Scholar] [CrossRef] [PubMed]
- Morhayim, J.; Van De Peppel, J.; Demmers, J.A.A.; Kocer, G.; Nigg, A.L.; Van Driel, M.; Chiba, H.; Van Leeuwen, J.P. Proteomic signatures of extracellular vesicles secreted by nonmineralizing and mineralizing human osteoblasts and stimulation of tumor cell growth. FASEB J. 2015, 29, 274–285. [Google Scholar] [CrossRef] [PubMed]
- Feng, D.; Zhao, W.L.; Ye, Y.Y.; Bai, X.C.; Liu, R.Q.; Chang, L.F.; Zhou, Q.; Sui, S.F. Cellular internalization of exosomes occurs through phagocytosis. Traffic 2010, 11, 675–687. [Google Scholar] [CrossRef]
- Ekström, K.; Valadi, H.; Sjöstrand, M.; Malmhäll, C.; Bossios, A.; Eldh, M.; Lötvall, J. Characterization of mRNA and microRNA in human mast cell-derived exosomes and their transfer to other mast cells and blood CD34 progenitor cells. J. Extracell. Vesicles 2012, 1, 18389. [Google Scholar] [CrossRef]
- Pužar Dominkuš, P.; Stenovec, M.; Sitar, S.; Lasič, E.; Zorec, R.; Plemenitaš, A.; Žagar, E.; Kreft, M.; Lenassi, M. PKH26 labeling of extracellular vesicles: Characterization and cellular internalization of contaminating PKH26 nanoparticles. Biochim. Biophys. Acta-Biomembr. 2018, 1860, 1350–1361. [Google Scholar] [CrossRef]
- Dehghani, M.; Gulvin, S.M.; Flax, J.; Gaborski, T.R. Systematic Evaluation of PKH Labelling on Extracellular Vesicle Size by Nanoparticle Tracking Analysis. Sci. Rep. 2020, 10, 9533. [Google Scholar] [CrossRef]
- Stremersch, S.; Brans, T.; Braeckmans, K.; De Smedt, S.; Raemdonck, K. Nucleic acid loading and fluorescent labeling of isolated extracellular vesicles requires adequate purification. Int. J. Pharm. 2018, 548, 783–792. [Google Scholar] [CrossRef]
- Joubert, M.K.; Luo, Q.; Nashed-Samuel, Y.; Wypych, J.; Narhi, L.O. Classification and characterization of therapeutic antibody aggregates. J. Biol. Chem. 2011, 286, 25118–25133. [Google Scholar] [CrossRef] [PubMed]
- Toribio, V.; Morales, S.; López-martín, S.; Cardeñes, B.; Cabañas, C.; Yáñez-mó, M. Development of a quantitative method to measure EV uptake. Sci. Rep. 2019, 9, 10522. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, C.; Shah, N.; Ishii, M.; Annamalai, B.; Brandon, C.; Rodgers, J.; Nowling, T.; Rohrer, B. Mechanisms of extracellular vesicle uptake in stressed retinal pigment epithelial cell monolayers. Biochim. Biophys. Acta-Mol. Basis Dis. 2020, 1866, 165608. [Google Scholar] [CrossRef]
- Miyazawa, B.; Trivedi, A.; Togarrati, P.P.; Potter, D.; Baimukanova, G.; Vivona, L.; Lin, M.; Lopez, E.; Callcut, R.; Srivastava, A.K.; et al. Regulation of endothelial cell permeability by platelet-derived extracellular vesicles. J. Trauma. Acute Care Surg. 2019, 86, 931–942. [Google Scholar] [CrossRef]
- Bian, S.; Zhang, L.; Duan, L.; Wang, X.; Min, Y.; Yu, H. Extracellular vesicles derived from human bone marrow mesenchymal stem cells promote angiogenesis in a rat myocardial infarction model. J. Mol. Med. 2014, 92, 387–397. [Google Scholar] [CrossRef]
- Hosseinkhani, B.; van den Akker, N.M.S.; Molin, D.G.M.; Michiels, L. (Sub)populations of extracellular vesicles released by TNF-α –triggered human endothelial cells promote vascular inflammation and monocyte migration. J. Extracell. Vesicles 2020, 9, 1801153. [Google Scholar] [CrossRef] [PubMed]
- Kalani, A.; Kamat, P.K.; Chaturvedi, P.; Tyagi, S.C.; Tyagi, N. Curcumin-primed exosomes mitigate endothelial cell dysfunction during hyperhomocysteinemia. Life Sci. 2014, 107, 8. [Google Scholar] [CrossRef]
- Kawakami, A.; Aikawa, M.; Nitta, N.; Yoshida, M.; Libby, P.; Sacks, F.M. Apolipoprotein CIII-induced THP-1 cell adhesion to endothelial cells involves pertussis toxin-sensitive G protein- and protein kinase Cα-mediated nuclear factor-κB activation. Arterioscler. Thromb. Vasc. Biol. 2007, 27, 219–225. [Google Scholar] [CrossRef]
- Noguchi, N.; Hanyu, R.; Nonaka, A.; Okimoto, Y.; Kodama, T. Inhibition of THP-1 cell adhesion to endothelial cells by α-tocopherol and α-tocotrienol is dependent on intracellular concentration of the antioxidants. Free Radic. Biol. Med. 2003, 34, 1614–1620. [Google Scholar] [CrossRef] [PubMed]
- Grell, M.; Douni, E.; Wajant, H.; Löhden, M.; Clauss, M.; Maxeiner, B.; Georgopoulos, S.; Lesslauer, W.; Kollias, G.; Pfizenmaier, K.; et al. The transmembrane form of tumor necrosis factor is the prime activating ligand of the 80 kDa tumor necrosis factor receptor. Cell 1995, 83, 793–802. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Zhao, Z.; Yang, C.; Song, C. Transfer of microRNA-221 from mesenchymal stem cell-derived extracellular vesicles inhibits atherosclerotic plaque formation. Transl. Res. 2020, 226, 83–95. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.W.; Sung, H.C.; Lin, S.R.; Wu, C.W.; Lee, C.W.; Lee, I.T.; Yang, Y.F.; Yu, I.S.; Lin, S.W.; Chiang, M.H.; et al. Resveratrol attenuates ICAM-1 expression and monocyte adhesiveness to TNF-α-treated endothelial cells: Evidence for an anti-inflammatory cascade mediated by the MIR-221/222/AMPK/p38/NF-7kappa;B pathway. Sci. Rep. 2017, 7, srep44689. [Google Scholar] [CrossRef]
- Hu, G.; Gong, A.Y.; Liu, J.; Zhou, R.; Deng, C.; Chen, X.M. miR-221 suppresses ICAM-1 translation and regulates interferon-γ- induced ICAM-1 expression in human cholangiocytes. Am. J. Physiol. Gastrointest. Liver Physiol. 2010, 298, 542–551. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garcia, L.F.C.; Wowk, P.F.; Albrecht, L. Unraveling the Impact of Extracellular Vesicle-Depleted Serum on Endothelial Cell Characteristics over Time. Int. J. Mol. Sci. 2024, 25, 4761. https://doi.org/10.3390/ijms25094761
Garcia LFC, Wowk PF, Albrecht L. Unraveling the Impact of Extracellular Vesicle-Depleted Serum on Endothelial Cell Characteristics over Time. International Journal of Molecular Sciences. 2024; 25(9):4761. https://doi.org/10.3390/ijms25094761
Chicago/Turabian StyleGarcia, Luiz Fernando Cardoso, Pryscilla Fanini Wowk, and Letusa Albrecht. 2024. "Unraveling the Impact of Extracellular Vesicle-Depleted Serum on Endothelial Cell Characteristics over Time" International Journal of Molecular Sciences 25, no. 9: 4761. https://doi.org/10.3390/ijms25094761
APA StyleGarcia, L. F. C., Wowk, P. F., & Albrecht, L. (2024). Unraveling the Impact of Extracellular Vesicle-Depleted Serum on Endothelial Cell Characteristics over Time. International Journal of Molecular Sciences, 25(9), 4761. https://doi.org/10.3390/ijms25094761





