Secreted Aspartic Proteinases: Key Factors in Candida Infections and Host-Pathogen Interactions
Abstract
:1. Candida Pathogenic Yeasts—Their Clinical Relevance as Opportunistic Pathogens
2. Various Pathophysiological Functions of SAPs
2.1. Biofilm Formation
2.2. Tissue Invasion and Damage
2.2.1. Degradation of Host Barriers
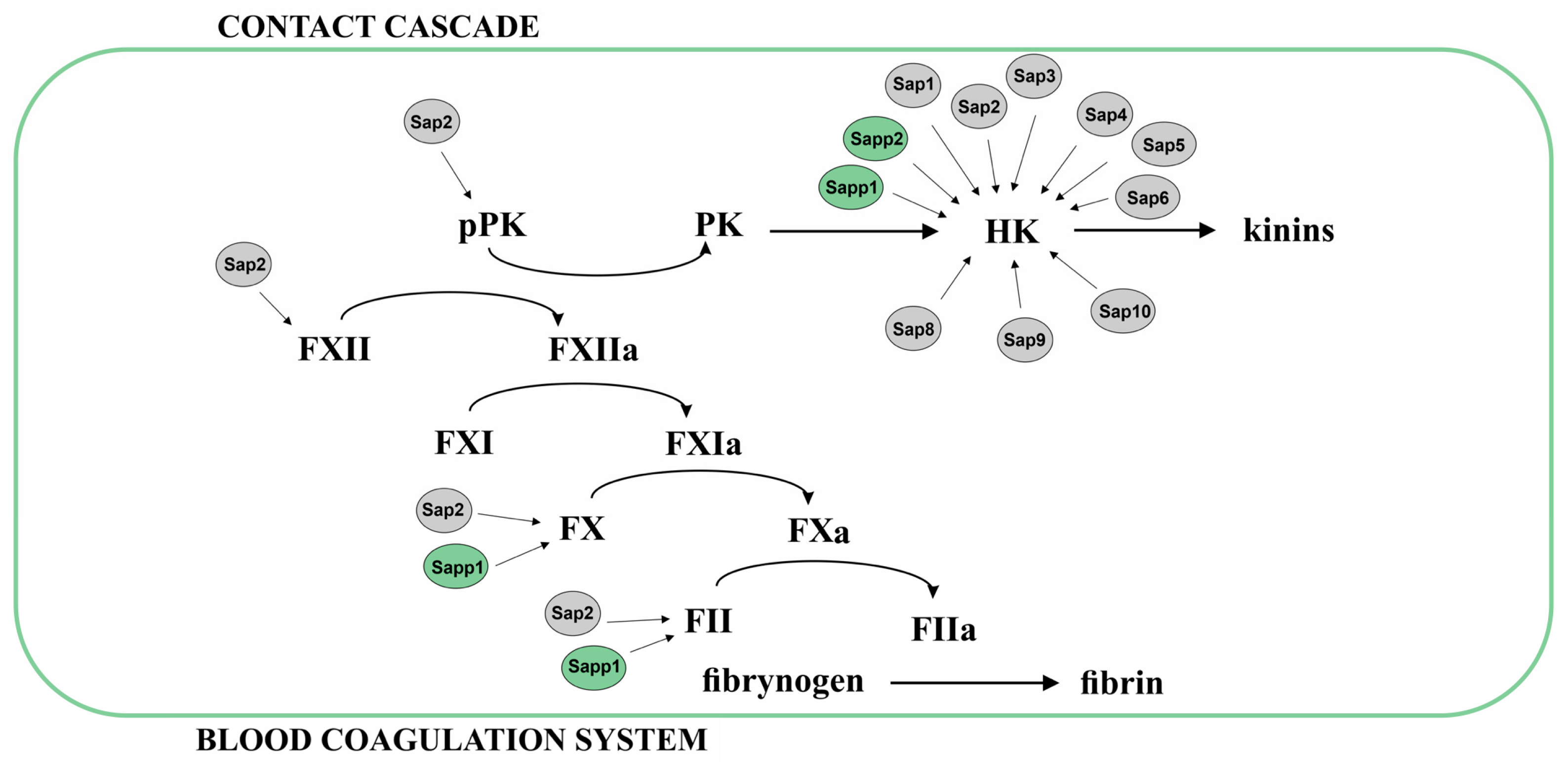
2.2.2. Proteolysis of Proteins from Coagulation Cascade, Contact System, and Inhibitors of Plasma Proteinases
2.3. Immune Evasion and Modulation
2.3.1. Interactions with Neutrophils
2.3.2. Interactions with Monocytes/Macrophages
2.3.3. Proteolysis of Complement, Antibodies, and Antimicrobial Peptides
2.4. Antifungal Resistance
2.5. Maintenance of Cellular Homeostasis
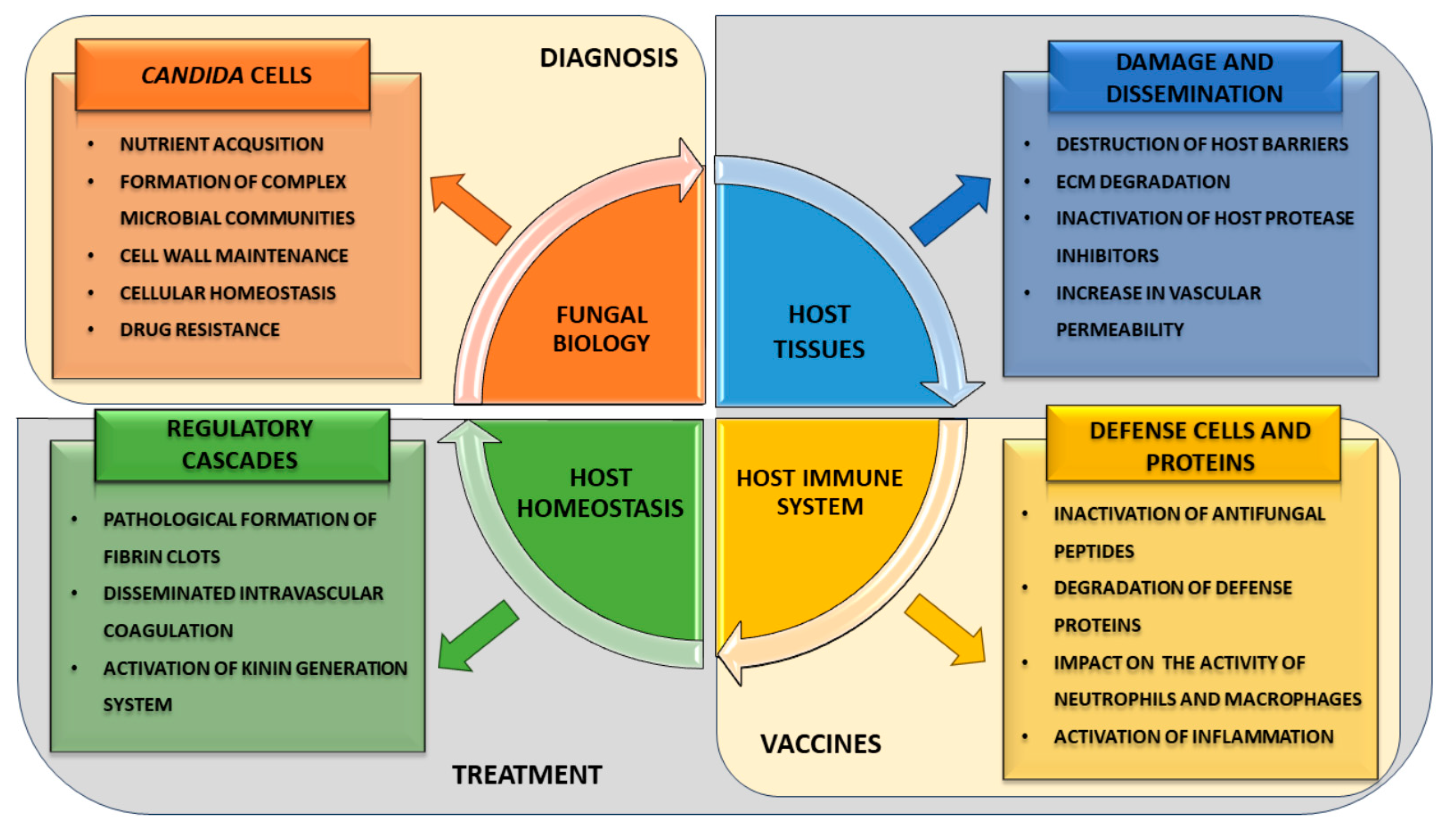
3. The Challenges and Opportunities for Developing Novel Strategies to Prevent and Treat Candida Infections
3.1. Diagnostic Potential of Saps
3.2. Saps as Components of Anti-Candida Vaccines
3.3. Protease Inhibitors as Prospective Agents Accompanying the Treatment of Candidiasis
4. Recommendations and Suggestions for Future Research Directions and Priorities on the Role of Saps in Candida Infections
Author Contributions
Funding
Conflicts of Interest
References
- Chen, C.; Huang, X. Candida albicans Commensalism and Human Diseases. In Mechanisms Underlying Host-Microbiome Interactions in Pathophysiology of Human Diseases; Sun, J., Dudeja, P.K., Eds.; Springer: Boston, MA, USA, 2018; pp. 247–278. ISBN 978-1-4939-7533-4. [Google Scholar]
- Kumamoto, C.A.; Gresnigt, M.S.; Hube, B. The Gut, the Bad and the Harmless: Candida albicans as a Commensal and Opportunistic Pathogen in the Intestine. Curr. Opin. Microbiol. 2020, 56, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Lopes, J.P.; Lionakis, M.S. Pathogenesis and Virulence of Candida albicans. Virulence 2022, 13, 89–121. [Google Scholar] [CrossRef] [PubMed]
- Proctor, D.M.; Drummond, R.A.; Lionakis, M.S.; Segre, J.A. One Population, Multiple Lifestyles: Commensalism and Pathogenesis in the Human Mycobiome. Cell Host Microbe 2023, 31, 539–553. [Google Scholar] [CrossRef] [PubMed]
- Logan, C.; Martin-Loeches, I.; Bicanic, T. Invasive Candidiasis in Critical Care: Challenges and Future Directions. Intensive Care Med. 2020, 46, 2001–2014. [Google Scholar] [CrossRef] [PubMed]
- McCarty, T.P.; White, C.M.; Pappas, P.G. Candidemia and Invasive Candidiasis. Infect. Dis. Clin. N. Am. 2021, 35, 389–413. [Google Scholar] [CrossRef] [PubMed]
- Soriano, A.; Honore, P.M.; Puerta-Alcalde, P.; Garcia-Vidal, C.; Pagotto, A.; Gonçalves-Bradley, D.C.; Verweij, P.E. Invasive Candidiasis: Current Clinical Challenges and Unmet Needs in Adult Populations. J. Antimicrob. Chemother. 2023, 78, 1569–1585. [Google Scholar] [CrossRef] [PubMed]
- Bongomin, F.; Gago, S.; Oladele, R.; Denning, D. Global and Multi-National Prevalence of Fungal Diseases—Estimate Precision. J. Fungi 2017, 3, 57. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, M.A.; Diekema, D.J. Epidemiology of Invasive Candidiasis: A Persistent Public Health Problem. Clin. Microbiol. Rev. 2007, 20, 133–163. [Google Scholar] [CrossRef] [PubMed]
- Arendrup, M.C. Candida and Candidaemia. Susceptibility and Epidemiology. Dan. Med. J. 2013, 60, B4698. [Google Scholar]
- Quindós, G.; Marcos-Arias, C.; San-Millán, R.; Mateo, E.; Eraso, E. The Continuous Changes in the Aetiology and Epidemiology of Invasive Candidiasis: From Familiar Candida albicans to Multiresistant Candida auris. Int. Microbiol. 2018, 21, 107–119. [Google Scholar] [CrossRef]
- Sharma, M.; Chakrabarti, A. Candidiasis and Other Emerging Yeasts. Curr. Fungal Infect. Rep. 2023, 17, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Polke, M.; Hube, B.; Jacobsen, I.D. Candida Survival Strategies. Adv. Appl. Microbiol. 2015, 91, 139–235. [Google Scholar] [CrossRef] [PubMed]
- Loaiza-Loeza, S.; Parra-Ortega, B.; Cancino-Díaz, J.C.; Illades-Aguiar, B.; Hernández-Rodríguez, C.H.; Villa-Tanaca, L. Differential Expression of Candida dubliniensis-Secreted Aspartyl Proteinase Genes (CdSAP1–4) under Different Physiological Conditions and during Infection of a Keratinocyte Culture. FEMS Immunol. Med. Microbiol. 2009, 56, 212–222. [Google Scholar] [CrossRef] [PubMed]
- Naglik, J.R.; Challacombe, S.J.; Hube, B. Candida albicans Secreted Aspartyl Proteinases in Virulence and Pathogenesis. Microbiol. Mol. Biol. Rev. 2003, 67, 400–428. [Google Scholar] [CrossRef] [PubMed]
- Monod, M.; Togni, G.; Hube, B.; Sanglard, D. Multiplicity of Genes Encoding Secreted Aspartic Proteinases in Candida Species. Mol. Microbiol. 1994, 13, 357–368. [Google Scholar] [CrossRef] [PubMed]
- Pichová, I.; Pavlícková, L.; Dostál, J.; Dolejsí, E.; Hrusková-Heidingsfeldová, O.; Weber, J.; Ruml, T.; Soucek, M. Secreted Aspartic Proteases of Candida albicans, Candida tropicalis, Candida parapsilosis and Candida lusitaniae. Inhibition with Peptidomimetic Inhibitors. Eur. J. Biochem. 2001, 268, 2669–2677. [Google Scholar] [CrossRef]
- Parra-Ortega, B.; Cruz-Torres, H.; Villa-Tanaca, L.; Hernández-Rodríguez, C. Phylogeny and Evolution of the Aspartyl Protease Family from Clinically Relevant Candida Species. Mem. Inst. Oswaldo Cruz 2009, 104, 505–512. [Google Scholar] [CrossRef]
- Singh, D.K.; Németh, T.; Papp, A.; Tóth, R.; Lukácsi, S.; Heidingsfeld, O.; Dostal, J.; Vágvölgyi, C.; Bajtay, Z.; Józsi, M.; et al. Functional Characterization of Secreted Aspartyl Proteases in Candida parapsilosis. mSphere 2019, 4, e00484-19. [Google Scholar] [CrossRef]
- Kwon-Chung, K.J.; Lehman, D.; Good, C.; Magee, P.T. Genetic Evidence for Role of Extracellular Proteinase in Virulence of Candida albicans. Infect. Immun. 1985, 49, 571–575. [Google Scholar] [CrossRef]
- Hube, B. Candida albicans Secreted Aspartyl Proteinases. Curr. Top. Med. Mycol. 1996, 7, 55–69. [Google Scholar]
- Fusek, M.; Smith, E.A.; Monod, M.; Foundling, S.I. Candida parapsilosis Expresses and Secretes Two Aspartic Proteinases. FEBS Lett. 1993, 327, 108–112. [Google Scholar] [CrossRef] [PubMed]
- Silva, S.; Henriques, M.; Oliveira, R.; Azeredo, J.; Malic, S.; Hooper, S.J.; Williams, D.W. Characterization of Candida parapsilosis Infection of an in Vitro Reconstituted Human Oral Epithelium. Eur. J. Oral Sci. 2009, 117, 669–675. [Google Scholar] [CrossRef] [PubMed]
- Zaugg, C.; Borg-von Zepelin, M.; Reichard, U.; Sanglard, D.; Monod, M. Secreted Aspartic Proteinase Family of Candida tropicalis. Infect. Immun. 2001, 69, 405–412. [Google Scholar] [CrossRef] [PubMed]
- Moran, G. Comparative Genomics Using Candida albicans DNA Microarrays Reveals Absence and Divergence of Virulence-Associated Genes in Candida dubliniensis. Microbiology 2004, 150, 3363–3382. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-S.; Lee, K.-T.; Bahn, Y.-S. Secreted Aspartyl Protease 3 Regulated by the Ras/cAMP/PKA Pathway Promotes the Virulence of Candida auris. Front. Cell. Infect. Microbiol. 2023, 13, 1257897. [Google Scholar] [CrossRef] [PubMed]
- Hube, B.; Sanglard, D.; Odds, F.C.; Hess, D.; Monod, M.; Schäfer, W.; Brown, A.J.; Gow, N.A. Disruption of Each of the Secreted Aspartyl Proteinase Genes SAP1, SAP2, and SAP3 of Candida albicans Attenuates Virulence. Infect. Immun. 1997, 65, 3529–3538. [Google Scholar] [CrossRef] [PubMed]
- Staib, P.; Lermann, U.; Blaβ-Warmuth, J.; Degel, B.; Würzner, R.; Monod, M.; Schirmeister, T.; Morschhäuser, J. Tetracycline-Inducible Expression of Individual Secreted Aspartic Proteases in Candida albicans Allows Isoenzyme-Specific Inhibitor Screening. Antimicrob. Agents Chemother. 2008, 52, 146–156. [Google Scholar] [CrossRef] [PubMed]
- Gil-Bona, A.; Monteoliva, L.; Gil, C. Global Proteomic Profiling of the Secretome of Candida albicans Ecm33 Cell Wall Mutant Reveals the Involvement of Ecm33 in Sap2 Secretion. J. Proteome Res. 2015, 14, 4270–4281. [Google Scholar] [CrossRef] [PubMed]
- Kaur, R.; Ma, B.; Cormack, B.P. A Family of Glycosylphosphatidylinositol-Linked Aspartyl Proteases Is Required for Virulence of Candida glabrata. Proc. Natl. Acad. Sci. USA 2007, 104, 7628–7633. [Google Scholar] [CrossRef]
- Kidd, S.E.; Abdolrasouli, A.; Hagen, F. Fungal Nomenclature: Managing Change Is the Name of the Game. Open Forum Infect. Dis. 2023, 10, ofac559. [Google Scholar] [CrossRef]
- Newport, G.; Agabian, N. KEX2 Influences Candida albicans Proteinase Secretion and Hyphal Formation. J. Biol. Chem. 1997, 272, 28954–28961. [Google Scholar] [CrossRef] [PubMed]
- Togni, G.; Sanglard, D.; Quadroni, M.; Foundling, S.I.; Monod, M. Acid Proteinase Secreted by Candida tropicalis: Functional Analysis of Preproregion Cleavages in C. tropicalis and Saccharomyces Cerevisiae. Microbiology 1996, 142, 493–503. [Google Scholar] [CrossRef]
- Dostál, J.; Dlouhá, H.; Maloň, P.; Pichová, I.; Hrušková-Heidingsfeldová, O. The Precursor of Secreted Aspartic Proteinase Sapp1p from Candida parapsilosis Can Be Activated Both Autocatalytically and by a Membrane-Bound Processing Proteinase. Biol. Chem. 2005, 386, 791–799. [Google Scholar] [CrossRef] [PubMed]
- Merkerová, M.; Dostál, J.; Hradilek, M.; Pichová, I.; Hrusková-Heidingsfeldová, O. Cloning and Characterization of Sapp2p, the Second Aspartic Proteinase Isoenzyme from Candida parapsilosis. FEMS Yeast Res. 2006, 6, 1018–1026. [Google Scholar] [CrossRef] [PubMed]
- Koelsch, G.; Tang, J.; Loy, J.A.; Monod, M.; Jackson, K.; Foundling, S.I.; Lin, X. Enzymic Characteristics of Secreted Aspartic Proteases of Candida albicans. Biochim. Biophys. Acta (BBA)-Protein Struct. Mol. Enzymol. 2000, 1480, 117–131. [Google Scholar] [CrossRef]
- Albrecht, A.; Felk, A.; Pichova, I.; Naglik, J.R.; Schaller, M.; de Groot, P.; Maccallum, D.; Odds, F.C.; Schäfer, W.; Klis, F.; et al. Glycosylphosphatidylinositol-Anchored Proteases of Candida albicans Target Proteins Necessary for Both Cellular Processes and Host-Pathogen Interactions. J. Biol. Chem. 2006, 281, 688–694. [Google Scholar] [CrossRef] [PubMed]
- Zarnowski, R.; Sanchez, H.; Covelli, A.S.; Dominguez, E.; Jaromin, A.; Bernhardt, J.; Mitchell, K.F.; Heiss, C.; Azadi, P.; Mitchell, A.; et al. Candida albicans Biofilm–Induced Vesicles Confer Drug Resistance through Matrix Biogenesis. PLoS Biol. 2018, 16, e2006872. [Google Scholar] [CrossRef]
- Kulig, K.; Karnas, E.; Woznicka, O.; Kuleta, P.; Zuba-Surma, E.; Pyza, E.; Osyczka, A.; Kozik, A.; Rapala-Kozik, M.; Karkowska-Kuleta, J. Insight into the Properties and Immunoregulatory Effect of Extracellular Vesicles Produced by Candida glabrata, Candida parapsilosis, and Candida tropicalis Biofilms. Front. Cell. Infect. Microbiol. 2022, 12, 879237. [Google Scholar] [CrossRef]
- Karkowska-Kuleta, J.; Kulig, K.; Karnas, E.; Zuba-Surma, E.; Woznicka, O.; Pyza, E.; Kuleta, P.; Osyczka, A.; Rapala-Kozik, M.; Kozik, A. Characteristics of Extracellular Vesicles Released by the Pathogenic Yeast-Like Fungi Candida glabrata, Candida parapsilosis and Candida tropicalis. Cells 2020, 9, E1722. [Google Scholar] [CrossRef]
- Karkowska-Kuleta, J.; Kulig, K.; Bras, G.; Stelmaszczyk, K.; Surowiec, M.; Kozik, A.; Karnas, E.; Barczyk-Woznicka, O.; Zuba-Surma, E.; Pyza, E.; et al. Candida albicans Biofilm-Derived Extracellular Vesicles Are Involved in the Tolerance to Caspofungin, Biofilm Detachment, and Fungal Proteolytic Activity. J. Fungi 2023, 9, 1078. [Google Scholar] [CrossRef]
- Martínez-López, R.; Hernáez, M.L.; Redondo, E.; Calvo, G.; Radau, S.; Pardo, M.; Gil, C.; Monteoliva, L. Candida albicans Hyphal Extracellular Vesicles Are Different from Yeast Ones, Carrying an Active Proteasome Complex and Showing a Different Role in Host Immune Response. Microbiol. Spectr. 2022, 10, e00698-22. [Google Scholar] [CrossRef]
- Fusek, M.; Smith, E.A.; Monod, M.; Dunn, B.M.; Foundling, S.I. Extracellular Aspartic Proteinases from Candida albicans, Candida tropicalis, and Candida parapsilosis Yeasts Differ Substantially in Their Specificities. Biochemistry 1994, 33, 9791–9799. [Google Scholar] [CrossRef] [PubMed]
- Aoki, W.; Kitahara, N.; Miura, N.; Morisaka, H.; Yamamoto, Y.; Kuroda, K.; Ueda, M. Comprehensive Characterization of Secreted Aspartic Proteases Encoded by a Virulence Gene Family in Candida albicans. J. Biochem. 2011, 150, 431–438. [Google Scholar] [CrossRef] [PubMed]
- Gogol, M.; Bochenska, O.; Zawrotniak, M.; Karkowska-Kuleta, J.; Zajac, D.; Rapala-Kozik, M. Roles of Candida albicans Aspartic Proteases in Host-Pathogen Interactions. In Pathophysiological Aspects of Proteases; Chakraborti, S., Dhalla, N.S., Eds.; Springer: Singapore, 2017; pp. 353–380. ISBN 978-981-10-6140-0. [Google Scholar]
- Schild, L.; Heyken, A.; de Groot, P.W.J.; Hiller, E.; Mock, M.; de Koster, C.; Horn, U.; Rupp, S.; Hube, B. Proteolytic Cleavage of Covalently Linked Cell Wall Proteins by Candida albicans Sap9 and Sap10. Eukaryot. Cell 2011, 10, 98–109. [Google Scholar] [CrossRef]
- Hrušková-Heidingsfeldová, O.; Dostál, J.; Majer, F.; Havlíková, J.; Hradilek, M.; Pichová, I. Two Aspartic Proteinases Secreted by the Pathogenic Yeast Candida Parapsilosis Differ in Expression Pattern and Catalytic Properties. Biol. Chem. 2009, 390, 259–268. [Google Scholar] [CrossRef]
- Lin, L.; Wang, M.; Zeng, J.; Mao, Y.; Qin, R.; Deng, J.; Ouyang, X.; Hou, X.; Sun, C.; Wang, Y.; et al. Sequence Variation of Candida albicans Sap2 Enhances Fungal Pathogenicity via Complement Evasion and Macrophage M2-Like Phenotype Induction. Adv. Sci. 2023, 10, 2206713. [Google Scholar] [CrossRef]
- Naglik, J.; Albrecht, A.; Bader, O.; Hube, B. Candida albicans Proteinases and Host/Pathogen Interactions. Cell. Microbiol. 2004, 6, 915–926. [Google Scholar] [CrossRef]
- Van Eck, N.J.; Waltman, L. Text Mining and Visualization Using VOSviewer. arXiv 2011, arXiv:1109.2058v1. [Google Scholar] [CrossRef]
- Ponde, N.O.; Lortal, L.; Ramage, G.; Naglik, J.R.; Richardson, J.P. Candida albicans Biofilms and Polymicrobial Interactions. Crit. Rev. Microbiol. 2021, 47, 91–111. [Google Scholar] [CrossRef]
- Pereira, R.; Santos Fontenelle, R.O.; Brito, E.H.S.; Morais, S.M. Biofilm of Candida albicans: Formation, Regulation and Resistance. J. Appl. Microbiol. 2021, 131, 11–22. [Google Scholar] [CrossRef]
- Winter, M.B.; Salcedo, E.C.; Lohse, M.B.; Hartooni, N.; Gulati, M.; Sanchez, H.; Takagi, J.; Hube, B.; Andes, D.R.; Johnson, A.D.; et al. Global Identification of Biofilm-Specific Proteolysis in Candida albicans. mBio 2016, 7, e01514-16. [Google Scholar] [CrossRef] [PubMed]
- Lohse, M.B.; Gulati, M.; Johnson, A.D.; Nobile, C.J. Development and Regulation of Single- and Multi-Species Candida albicans Biofilms. Nat. Rev. Microbiol. 2018, 16, 19–31. [Google Scholar] [CrossRef] [PubMed]
- Nobile, C.J.; Fox, E.P.; Nett, J.E.; Sorrells, T.R.; Mitrovich, Q.M.; Hernday, A.D.; Tuch, B.B.; Andes, D.R.; Johnson, A.D. A Recently Evolved Transcriptional Network Controls Biofilm Development in Candida albicans. Cell 2012, 148, 126–138. [Google Scholar] [CrossRef] [PubMed]
- Nailis, H.; Kucharíková, S.; Ricicová, M.; Van Dijck, P.; Deforce, D.; Nelis, H.; Coenye, T. Real-Time PCR Expression Profiling of Genes Encoding Potential Virulence Factors in Candida albicans Biofilms: Identification of Model-Dependent and -Independent Gene Expression. BMC Microbiol. 2010, 10, 114. [Google Scholar] [CrossRef] [PubMed]
- Joo, M.Y.; Shin, J.H.; Jang, H.-C.; Song, E.S.; Kee, S.J.; Shin, M.G.; Suh, S.P.; Ryang, D.W. Expression of SAP5 and SAP9 in Candida albicans Biofilms: Comparison of Bloodstream Isolates with Isolates from Other Sources. Med. Mycol. 2013, 51, 892–896. [Google Scholar] [CrossRef] [PubMed]
- Tumbarello, M.; Fiori, B.; Trecarichi, E.M.; Posteraro, P.; Losito, A.R.; De Luca, A.; Sanguinetti, M.; Fadda, G.; Cauda, R.; Posteraro, B. Risk Factors and Outcomes of Candidemia Caused by Biofilm-Forming Isolates in a Tertiary Care Hospital. PLoS ONE 2012, 7, e33705. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, R.; May, A.; Sherry, L.; Kean, R.; Williams, C.; Jones, B.L.; Burgess, K.V.; Heringa, J.; Abeln, S.; Brandt, B.W.; et al. Integrating Candida albicans Metabolism with Biofilm Heterogeneity by Transcriptome Mapping. Sci. Rep. 2016, 6, 35436. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Downs, D.; Ghosh, K.; Ghosh, A.K.; Staib, P.; Monod, M.; Tang, J. Candida albicans Secreted Aspartic Proteases 4-6 Induce Apoptosis of Epithelial Cells by a Novel Trojan Horse Mechanism. FASEB J. 2013, 27, 2132–2144. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Breindel, C.; Saraswat, D.; Cullen, P.J.; Edgerton, M. Candida albicans Sap6 Amyloid Regions Function in Cellular Aggregation and Zinc Binding, and Contribute to Zinc Acquisition. Sci. Rep. 2017, 7, 2908. [Google Scholar] [CrossRef]
- Fox, E.P.; Bui, C.K.; Nett, J.E.; Hartooni, N.; Mui, M.C.; Andes, D.R.; Nobile, C.J.; Johnson, A.D. An Expanded Regulatory Network Temporally Controls Candida albicans Biofilm Formation. Mol. Microbiol. 2015, 96, 1226–1239. [Google Scholar] [CrossRef]
- Li, F.; Svarovsky, M.J.; Karlsson, A.J.; Wagner, J.P.; Marchillo, K.; Oshel, P.; Andes, D.; Palecek, S.P. Eap1p, an Adhesin That Mediates Candida albicans Biofilm Formation In Vitro and In Vivo. Eukaryot. Cell 2007, 6, 931–939. [Google Scholar] [CrossRef]
- Nobbs, A.H.; Margaret Vickerman, M.; Jenkinson, H.F. Heterologous Expression of Candida albicans Cell Wall-Associated Adhesins in Saccharomyces cerevisiae Reveals Differential Specificities in Adherence and Biofilm Formation and in Binding Oral Streptococcus gordonii. Eukaryot. Cell 2010, 9, 1622–1634. [Google Scholar] [CrossRef] [PubMed]
- Fox, S.J.; Shelton, B.T.; Kruppa, M.D. Characterization of Genetic Determinants That Modulate Candida albicans Filamentation in the Presence of Bacteria. PLoS ONE 2013, 8, e71939. [Google Scholar] [CrossRef] [PubMed]
- Ramsook, C.B.; Tan, C.; Garcia, M.C.; Fung, R.; Soybelman, G.; Henry, R.; Litewka, A.; O’Meally, S.; Otoo, H.N.; Khalaf, R.A.; et al. Yeast Cell Adhesion Molecules Have Functional Amyloid-Forming Sequences. Eukaryot. Cell 2010, 9, 393–404. [Google Scholar] [CrossRef]
- Dutton, L.C.; Jenkinson, H.F.; Lamont, R.J.; Nobbs, A.H. Role of Candida albicans Secreted Aspartyl Protease Sap9 in Interkingdom Biofilm Formation. Pathog. Dis. 2016, 74, ftw005. [Google Scholar] [CrossRef] [PubMed]
- Silverman, R.J.; Nobbs, A.H.; Vickerman, M.M.; Barbour, M.E.; Jenkinson, H.F. Interaction of Candida albicans Cell Wall Als3 Protein with Streptococcus gordonii SspB Adhesin Promotes Development of Mixed-Species Communities. Infect. Immun. 2010, 78, 4644–4652. [Google Scholar] [CrossRef] [PubMed]
- Nobbs, A.H.; Jenkinson, H.F. Interkingdom Networking within the Oral Microbiome. Microbes Infect. 2015, 17, 484–492. [Google Scholar] [CrossRef]
- Uppuluri, P.; Chaturvedi, A.K.; Srinivasan, A.; Banerjee, M.; Ramasubramaniam, A.K.; Köhler, J.R.; Kadosh, D.; Lopez-Ribot, J.L. Dispersion as an Important Step in the Candida albicans Biofilm Developmental Cycle. PLoS Pathog. 2010, 6, e1000828. [Google Scholar] [CrossRef] [PubMed]
- Uppuluri, P.; Pierce, C.G.; Thomas, D.P.; Bubeck, S.S.; Saville, S.P.; Lopez-Ribot, J.L. The Transcriptional Regulator Nrg1p Controls Candida albicans Biofilm Formation and Dispersion. Eukaryot Cell 2010, 9, 1531–1537. [Google Scholar] [CrossRef]
- Uppuluri, P.; Acosta Zaldívar, M.; Anderson, M.Z.; Dunn, M.J.; Berman, J.; Lopez Ribot, J.L.; Köhler, J.R. Candida albicans Dispersed Cells Are Developmentally Distinct from Biofilm and Planktonic Cells. mBio 2018, 9, e01338-18. [Google Scholar] [CrossRef]
- Munro, C.A.; Hube, B. Anti-Fungal Therapy at the HAART of Viral Therapy. Trends Microbiol. 2002, 10, 173–177. [Google Scholar] [CrossRef] [PubMed]
- Cassone, A.; Cauda, R. HIV Proteinase Inhibitors: Do They Really Work against Candida in a Clinical Setting? Trends Microbiol. 2002, 10, 177–178. [Google Scholar] [CrossRef] [PubMed]
- Jalal, M.; Ansari, M.A.; Alzohairy, M.A.; Ali, S.G.; Khan, H.M.; Almatroudi, A.; Siddiqui, M.I. Anticandidal Activity of Biosynthesized Silver Nanoparticles: Effect on Growth, Cell Morphology, and Key Virulence Attributes of Candida Species. Int. J. Nanomed. 2019, 14, 4667–4679. [Google Scholar] [CrossRef] [PubMed]
- Lohse, M.B.; Gulati, M.; Craik, C.S.; Johnson, A.D.; Nobile, C.J. Combination of Antifungal Drugs and Protease Inhibitors Prevent Candida albicans Biofilm Formation and Disrupt Mature Biofilms. Front. Microbiol. 2020, 11, 1027. [Google Scholar] [CrossRef] [PubMed]
- Colina, A.R.; Aumont, F.; Deslauriers, N.; Belhumeur, P.; De Repentigny, L. Evidence for Degradation of Gastrointestinal Mucin by Candida albicans Secretory Aspartyl Proteinase. Infect. Immun. 1996, 64, 4514–4519. [Google Scholar] [CrossRef]
- De Repentigny, L.; Aumont, F.; Bernard, K.; Belhumeur, P. Characterization of Binding of Candida albicans to Small Intestinal Mucin and Its Role in Adherence to Mucosal Epithelial Cells. Infect. Immun. 2000, 68, 3172–3179. [Google Scholar] [CrossRef] [PubMed]
- Villar, C.C.; Kashleva, H.; Nobile, C.J.; Mitchell, A.P.; Dongari-Bagtzoglou, A. Mucosal Tissue Invasion by Candida albicans Is Associated with E-Cadherin Degradation, Mediated by Transcription Factor Rim101p and Protease Sap5p. Infect. Immun. 2007, 75, 2126–2135. [Google Scholar] [CrossRef] [PubMed]
- Tomlin, H.; Piccinini, A.M. A Complex Interplay between the Extracellular Matrix and the Innate Immune Response to Microbial Pathogens. Immunology 2018, 155, 186–201. [Google Scholar] [CrossRef] [PubMed]
- Naglik, J.R.; Rodgers, C.A.; Shirlaw, P.J.; Dobbie, J.L.; Fernandes-Naglik, L.L.; Greenspan, D.; Agabian, N.; Challacombe, S.J. Differential Expression of Candida albicans Secreted Aspartyl Proteinase and Phospholipase B Genes in Humans Correlates with Active Oral and Vaginal Infections. J. Infect. Dis. 2003, 188, 469–479. [Google Scholar] [CrossRef]
- Ray, T.L.; Payne, C.D. Comparative Production and Rapid Purification of Candida Acid Proteinase from Protein-Supplemented Cultures. Infect. Immun. 1990, 58, 508–514. [Google Scholar] [CrossRef]
- Morschhäuser, J.; Virkola, R.; Korhonen, T.K.; Hacker, J. Degradation of Human Subendothelial Extracellular Matrix by Proteinase-Secreting Candida albicans. FEMS Microbiol. Lett. 2006, 153, 349–355. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Liu, Y.; Zhang, H. Extracellular Matrix: An Important Regulator of Cell Functions and Skeletal Muscle Development. Cell Biosci. 2021, 11, 65. [Google Scholar] [CrossRef] [PubMed]
- Kaminishi, H.; Hamatake, H.; Cho, T.; Tamaki, T.; Suenaga, N.; Fujii, T.; Hagihara, Y.; Maeda, H. Activation of Blood Clotting Factors by Microbial Proteinases. FEMS Microbiol. Lett. 1994, 121, 327–332. [Google Scholar] [CrossRef] [PubMed]
- Rüchel, R. On the Renin-like Activity of Candida Proteinases and Activation of Blood Coagulation in vitro. Zentralbl Bakteriol. Mikrobiol. Hyg. A Med. Mikrobiol. Infekt. Parasitol. 1983, 255, 368–379. [Google Scholar] [CrossRef] [PubMed]
- Kaminishi, H.; Miyaguchi, H.; Tamaki, T.; Suenaga, N.; Hisamatsu, M.; Mihashi, I.; Matsumoto, H.; Maeda, H.; Hagihara, Y. Degradation of Humoral Host Defense by Candida albicans Proteinase. Infect. Immun. 1995, 63, 984–988. [Google Scholar] [CrossRef]
- Schmaier, A.H. The Contact Activation and Kallikrein/Kinin Systems: Pathophysiologic and Physiologic Activities. J. Thromb. Haemost. 2016, 14, 28–39. [Google Scholar] [CrossRef] [PubMed]
- Frick, I.-M.; Björck, L.; Herwald, H. The Dual Role of the Contact System in Bacterial Infectious Disease. Thromb. Haemost. 2007, 98, 497–502. [Google Scholar] [CrossRef] [PubMed]
- Kaminishi, H.; Tanaka, M.; Cho, T.; Maeda, H.; Hagihara, Y. Activation of the Plasma Kallikrein-Kinin System by Candida albicans Proteinase. Infect. Immun. 1990, 58, 2139–2143. [Google Scholar] [CrossRef] [PubMed]
- Rapala-Kozik, M.; Karkowska-Kuleta, J.; Ryzanowska, A.; Golda, A.; Barbasz, A.; Faussner, A.; Kozik, A. Degradation of Human Kininogens with the Release of Kinin Peptides by Extracellular Proteinases of Candida spp. Biol. Chem. 2010, 391, 823–830. [Google Scholar] [CrossRef]
- Bras, G.; Bochenska, O.; Rapala-Kozik, M.; Guevara-Lora, I.; Faussner, A.; Kozik, A. Extracellular Aspartic Protease SAP2 of Candida albicans Yeast Cleaves Human Kininogens and Releases Proinflammatory Peptides, Met-Lys-Bradykinin and Des-Arg(9)-Met-Lys-Bradykinin. Biol. Chem. 2012, 393, 829–839. [Google Scholar] [CrossRef]
- Bras, G.; Bochenska, O.; Rapala-Kozik, M.; Guevara-Lora, I.; Faussner, A.; Kamysz, W.; Kozik, A. Release of Biologically Active Kinin Peptides, Met-Lys-Bradykinin and Leu-Met-Lys-Bradykinin from Human Kininogens by Two Major Secreted Aspartic Proteases of Candida parapsilosis. Peptides 2013, 48, 114–123. [Google Scholar] [CrossRef] [PubMed]
- Kozik, A.; Gogol, M.; Bochenska, O.; Karkowska-Kuleta, J.; Wolak, N.; Kamysz, W.; Aoki, W.; Ueda, M.; Faussner, A.; Rapala-Kozik, M. Kinin Release from Human Kininogen by 10 Aspartic Proteases Produced by Pathogenic Yeast Candida albicans. BMC Microbiol. 2015, 15, 60. [Google Scholar] [CrossRef] [PubMed]
- Gogol, M.; Ostrowska, D.; Klaga, K.; Bochenska, O.; Wolak, N.; Aoki, W.; Ueda, M.; Kozik, A.; Rapala-Kozik, M. Inactivation of A1-Proteinase Inhibitor by Candida albicans Aspartic Proteases Favors the Epithelial and Endothelial Cell Colonization in the Presence of Neutrophil Extracellular Traps. Acta Biochim. Pol. 2016, 63, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Tsushima, H.; Mine, H.; Kawakami, Y.; Hyodoh, F.; Ueki, A. Candida albicans Aspartic Proteinase Cleaves and Inactivates Human Epidermal Cysteine Proteinase Inhibitor, Cystatin A. Microbiology 1994, 140, 167–171. [Google Scholar] [CrossRef] [PubMed]
- Müller-Esterl, W. Novel Functions of the Kininogens. Semin. Thromb. Hemost. 1987, 13, 115–126. [Google Scholar] [CrossRef] [PubMed]
- Lecaille, F.; Brömme, D.; Lalmanach, G. Biochemical Properties and Regulation of Cathepsin K Activity. Biochimie 2008, 90, 208–226. [Google Scholar] [CrossRef] [PubMed]
- Bradford, H.N.; Jameson, B.A.; Adam, A.A.; Wassell, R.P.; Colman, R.W. Contiguous Binding and Inhibitory Sites on Kininogens Required for the Inhibition of Platelet Calpain. J. Biol. Chem. 1993, 268, 26546–26551. [Google Scholar] [CrossRef] [PubMed]
- Borregaard, N. Neutrophils, from Marrow to Microbes. Immunity 2010, 33, 657–670. [Google Scholar] [CrossRef] [PubMed]
- Appelberg, R. Neutrophils and Intracellular Pathogens: Beyond Phagocytosis and Killing. Trends Microbiol. 2007, 15, 87–92. [Google Scholar] [CrossRef]
- Kannan, S. Role of Protease-Activated Receptors in Neutrophil Degranulation. Med. Hypotheses 2002, 59, 266–267. [Google Scholar] [CrossRef]
- Araźna, M.; Pruchniak, M.P.; Demkow, U. Reactive Oxygen Species, Granulocytes, and NETosis. Adv. Exp. Med. Biol. 2015, 836, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Branzk, N.; Lubojemska, A.; Hardison, S.E.; Wang, Q.; Gutierrez, M.G.; Brown, G.D.; Papayannopoulos, V. Neutrophils Sense Microbe Size and Selectively Release Neutrophil Extracellular Traps in Response to Large Pathogens. Nat. Immunol. 2014, 15, 1017–1025. [Google Scholar] [CrossRef] [PubMed]
- Zawrotniak, M.; Wojtalik, K.; Rapala-Kozik, M. Farnesol, a Quorum-Sensing Molecule of Candida albicans Triggers the Release of Neutrophil Extracellular Traps. Cells 2019, 8, 1611. [Google Scholar] [CrossRef] [PubMed]
- Zawrotniak, M.; Bochenska, O.; Karkowska-Kuleta, J.; Seweryn-Ozog, K.; Aoki, W.; Ueda, M.; Kozik, A.; Rapala-Kozik, M. Aspartic Proteases and Major Cell Wall Components in Candida albicans Trigger the Release of Neutrophil Extracellular Traps. Front. Cell. Infect. Microbiol. 2017, 7, 414. [Google Scholar] [CrossRef] [PubMed]
- Gabrielli, E.; Sabbatini, S.; Roselletti, E.; Kasper, L.; Perito, S.; Hube, B.; Cassone, A.; Vecchiarelli, A.; Pericolini, E. In Vivo Induction of Neutrophil Chemotaxis by Secretory Aspartyl Proteinases of Candida albicans. Virulence 2016, 5594, 819–825. [Google Scholar] [CrossRef]
- Hornbach, A.; Heyken, A.; Schild, L.; Hube, B.; Löffler, J.; Kurzai, O. The Glycosylphosphatidylinositol-Anchored Protease Sap9 Modulates the Interaction of Candida albicans with Human Neutrophils. Infect. Immun. 2009, 77, 5216–5224. [Google Scholar] [CrossRef] [PubMed]
- Naglik, J.R. Candida Immunity. New J. Sci. 2014, 2014, 390241. [Google Scholar] [CrossRef]
- Aoki, W.; Kitahara, N.; Miura, N.; Morisaka, H.; Yamamoto, Y.; Kuroda, K.; Ueda, M. Candida albicans Possesses Sap7 as a Pepstatin A-Insensitive Secreted Aspartic Protease. PLoS ONE 2012, 7, e32513. [Google Scholar] [CrossRef] [PubMed]
- Naglik, J.R.; Moyes, D.; Makwana, J.; Kanzaria, P.; Tsichlaki, E.; Weindl, G.G.; Tappuni, A.R.; Rodgers, C.A.; Woodman, A.J.; Challacombe, S.J.; et al. Quantitative Expression of the Candida albicans Secreted Aspartyl Proteinase Gene Family in Human Oral and Vaginal Candidiasis. Microbiology 2008, 154, 3266–3280. [Google Scholar] [CrossRef]
- Whitlock, B.B.; Gardai, S.; Fadok, V.; Bratton, D.; Henson, P.M. Differential Roles for Alpha(M)Beta(2) Integrin Clustering or Activation in the Control of Apoptosis via Regulation of Akt and ERK Survival Mechanisms. J. Cell Biol. 2000, 151, 1305–1320. [Google Scholar] [CrossRef]
- Behnen, M.; Leschczyk, C.; Moller, S.; Batel, T.; Klinger, M.; Solbach, W.; Laskay, T.; Möller, S.; Batel, T.; Klinger, M.; et al. Immobilized Immune Complexes Induce Neutrophil Extracellular Trap Release by Human Neutrophil Granulocytes via Fc RIIIB and Mac-1. J. Immunol. 2014, 193, 1954–1965. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Shang, A.; Guo, M.; Shen, L.; Han, Y.; Huang, X. The Advances in the Regulation of Immune Microenvironment by Candida albicans and Macrophage Cross-Talk. Front. Microbiol. 2022, 13, 1029966. [Google Scholar] [CrossRef] [PubMed]
- Heung, L.J. Monocytes and the Host Response to Fungal Pathogens. Front. Cell. Infect. Microbiol. 2020, 10, 34. [Google Scholar] [CrossRef] [PubMed]
- Pietrella, D.; Rachini, A.; Pandey, N.; Schild, L.; Netea, M.; Bistoni, F.; Hube, B.; Vecchiarelli, A. The Inflammatory Response Induced by Aspartic Proteases of Candida albicans Is Independent of Proteolytic Activity. Infect. Immun. 2010, 78, 4754–4762. [Google Scholar] [CrossRef] [PubMed]
- Pietrella, D.; Pandey, N.; Gabrielli, E.; Pericolini, E.; Perito, S.; Kasper, L.; Bistoni, F.; Cassone, A.; Hube, B.; Vecchiarelli, A. Secreted Aspartic Proteases of Candida albicans Activate the NLRP3 Inflammasome. Eur. J. Immunol. 2013, 43, 679–692. [Google Scholar] [CrossRef] [PubMed]
- Gross, O.; Poeck, H.; Bscheider, M.; Dostert, C.; Hannesschläger, N.; Endres, S.; Hartmann, G.; Tardivel, A.; Schweighoffer, E.; Tybulewicz, V.; et al. Syk Kinase Signalling Couples to the Nlrp3 Inflammasome for Anti-Fungal Host Defence. Nature 2009, 459, 433–436. [Google Scholar] [CrossRef]
- Hise, A.G.; Tomalka, J.; Ganesan, S.; Patel, K.; Hall, B.A.; Brown, G.D.; Fitzgerald, K.A. An Essential Role for the NLRP3 Inflammasome in Host Defense against the Human Fungal Pathogen Candida albicans. Cell Host Microbe 2009, 5, 487–497. [Google Scholar] [CrossRef]
- Gabrielli, E.; Pericolini, E.; Luciano, E.; Sabbatini, S.; Roselletti, E.; Perito, S.; Kasper, L.; Hube, B.; Vecchiarelli, A. Induction of Caspase-11 by Aspartyl Proteinases of Candida albicans and Implication in Promoting Inflammatory Response. Infect. Immun. 2015, 83, 1940–1948. [Google Scholar] [CrossRef]
- Borg-von Zepelin, M.; Beggah, S.; Boggian, K.; Sanglard, D.; Monod, M. The Expression of the Secreted Aspartyl Proteinases Sap4 to Sap6 from Candida albicans in Murine Macrophages. Mol. Microbiol. 1998, 28, 543–554. [Google Scholar] [CrossRef]
- Meiller, T.F.; Hube, B.; Schild, L.; Shirtliff, M.E.; Scheper, M.A.; Winkler, R.; Ton, A.; Jabra-Rizk, M.A. A Novel Immune Evasion Strategy of Candida albicans: Proteolytic Cleavage of a Salivary Antimicrobial Peptide. PLoS ONE 2009, 4, e5039. [Google Scholar] [CrossRef]
- Bochenska, O.; Rapala-Kozik, M.; Wolak, N.; Aoki, W.; Ueda, M.; Kozik, A. The Action of Ten Secreted Aspartic Proteases of Pathogenic Yeast Candida albicans on Major Human Salivary Antimicrobial Peptide, Histatin 5. Acta Biochim. Pol. 2016, 63, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Ikonomova, S.P.; Moghaddam-Taaheri, P.; Jabra-Rizk, M.A.; Wang, Y.; Karlsson, A.J. Engineering Improved Variants of the Antifungal Peptide Histatin 5 with Reduced Susceptibility to Candida albicans Secreted Aspartic Proteases and Enhanced Antimicrobial Potency. FEBS J. 2018, 285, 146–159. [Google Scholar] [CrossRef] [PubMed]
- Ikonomova, S.P.; Moghaddam-Taaheri, P.; Wang, Y.; Doolin, M.T.; Stroka, K.M.; Hube, B.; Karlsson, A.J. Effects of Histatin 5 Modifications on Antifungal Activity and Kinetics of Proteolysis. Protein Sci. 2020, 29, 480–493. [Google Scholar] [CrossRef] [PubMed]
- Moghaddam-Taaheri, P.; Leissa, J.A.; Eppler, H.B.; Jewell, C.M.; Karlsson, A.J. Histatin 5 Variant Reduces Candida albicans Biofilm Viability and Inhibits Biofilm Formation. Fungal Genet. Biol. 2021, 149, 103529. [Google Scholar] [CrossRef]
- Vandamme, D.; Landuyt, B.; Luyten, W.; Schoofs, L. A Comprehensive Summary of LL-37, the Factotum Human Cathelicidin Peptide. Cell. Immunol. 2012, 280, 22–35. [Google Scholar] [CrossRef] [PubMed]
- Rapala-Kozik, M.; Bochenska, O.; Zawrotniak, M.; Wolak, N.; Trebacz, G.; Gogol, M.; Ostrowska, D.; Aoki, W.; Ueda, M.; Kozik, A. Inactivation of the Antifungal and Immunomodulatory Properties of Human Cathelicidin LL-37 by Aspartic Proteases Produced by the Pathogenic Yeast Candida albicans. Infect. Immun. 2015, 83, 2518–2530. [Google Scholar] [CrossRef] [PubMed]
- Frick, I.-M.; Åkesson, P.; Herwald, H.; Mörgelin, M.; Malmsten, M.; Nägler, D.K.; Björck, L. The Contact System—A Novel Branch of Innate Immunity Generating Antibacterial Peptides. EMBO J. 2006, 25, 5569–5578. [Google Scholar] [CrossRef] [PubMed]
- Nordahl, E.A.; Rydengård, V.; Mörgelin, M.; Schmidtchen, A. Domain 5 of High Molecular Weight Kininogen Is Antibacterial. J. Biol. Chem. 2005, 280, 34832–34839. [Google Scholar] [CrossRef]
- Bochenska, O.; Rapala-Kozik, M.; Wolak, N.; Kamysz, W.; Grzywacz, D.; Aoki, W.; Ueda, M.; Kozik, A. Inactivation of Human Kininogen-Derived Antimicrobial Peptides by Secreted Aspartic Proteases Produced by the Pathogenic Yeast Candida albicans. Biol. Chem. 2015, 396, 1369–1375. [Google Scholar] [CrossRef]
- Rothstein, D.M.; Spacciapoli, P.; Tran, L.T.; Xu, T.; Roberts, F.D.; Dalla Serra, M.; Buxton, D.K.; Oppenheim, F.G.; Friden, P. Anticandida Activity Is Retained in P-113, a 12-Amino-Acid Fragment of Histatin 5. Antimicrob. Agents Chemother. 2001, 45, 1367–1373. [Google Scholar] [CrossRef]
- Cheng, K.-T.; Wu, C.-L.; Yip, B.-S.; Chih, Y.-H.; Peng, K.-L.; Hsu, S.-Y.; Yu, H.-Y.; Cheng, J.-W. The Interactions between the Antimicrobial Peptide P-113 and Living Candida albicans Cells Shed Light on Mechanisms of Antifungal Activity and Resistance. Int. J. Mol. Sci. 2020, 21, 2654. [Google Scholar] [CrossRef] [PubMed]
- Kalimuthu, S.; Pudipeddi, A.; Braś, G.; Tanner, J.A.; Rapala-Kozik, M.; Leung, Y.Y.; Neelakantan, P. A Heptadeca Amino Acid Peptide Subunit of Cathelicidin LL-37 Has Previously Unreported Antifungal Activity. APMIS 2023, 131, 584–600. [Google Scholar] [CrossRef] [PubMed]
- Germaine, G.R.; Tellefson, L.M. Effect of pH and Human Saliva on Protease Production by Candida albicans. Infect. Immun. 1981, 31, 323–326. [Google Scholar] [CrossRef]
- Rüchel, R. A Variety of Candida Proteinases and Their Possible Targets of Proteolytic Attack in the Host. Zentralbl Bakteriol. Mikrobiol. Hyg. A 1984, 257, 266–274. [Google Scholar] [CrossRef]
- Gropp, K.; Schild, L.; Schindler, S.; Hube, B.; Zipfel, P.F.; Skerka, C. The Yeast Candida albicans Evades Human Complement Attack by Secretion of Aspartic Proteases. Mol. Immunol. 2009, 47, 465–475. [Google Scholar] [CrossRef] [PubMed]
- Svoboda, E.; Schneider, A.E.; Sándor, N.; Lermann, U.; Staib, P.; Kremlitzka, M.; Bajtay, Z.; Barz, D.; Erdei, A.; Józsi, M. Secreted Aspartic Protease 2 of Candida albicans Inactivates Factor H and the Macrophage Factor H-Receptors CR3 (CD11b/CD18) and CR4 (CD11c/CD18). Immunol. Lett. 2015, 168, 13–21. [Google Scholar] [CrossRef]
- Valand, N.; Brunt, E.; Gazioglu, O.; Yesilkaya, H.; Mitchell, D.; Horley, N.; Arroo, R.; Kishore, U.; Wallis, R.; Venkatraman Girija, U. Inactivation of the Complement Lectin Pathway by Candida tropicalis Secreted Aspartyl Protease-1. Immunobiology 2022, 227, 152263. [Google Scholar] [CrossRef]
- Hirayasu, K.; Saito, F.; Suenaga, T.; Shida, K.; Arase, N.; Oikawa, K.; Yamaoka, T.; Murota, H.; Chibana, H.; Nakagawa, I.; et al. Microbially Cleaved Immunoglobulins Are Sensed by the Innate Immune Receptor LILRA2. Nat. Microbiol. 2016, 1, 16054. [Google Scholar] [CrossRef] [PubMed]
- Rüchel, R.; Tegeler, R.; Trost, M. A Comparison of Secretory Proteinases from Different Strains of Candida albicans. Med. Mycol. 1982, 20, 233–244. [Google Scholar] [CrossRef]
- Rüchel, R. Cleavage of Immunoglobulins by Pathogenic Yeasts of the Genus Candida. Microbiol. Sci. 1986, 3, 316–319. [Google Scholar]
- Wich, M.; Greim, S.; Ferreira-Gomes, M.; Krüger, T.; Kniemeyer, O.; Brakhage, A.A.; Jacobsen, I.D.; Hube, B.; Jungnickel, B. Functionality of the Human Antibody Response to Candida albicans. Virulence 2021, 12, 3137–3148. [Google Scholar] [CrossRef] [PubMed]
- Guevara-Lora, I.; Bras, G.; Karkowska-Kuleta, J.; González-González, M.; Ceballos, K.; Sidlo, W.; Rapala-Kozik, M. Plant-Derived Substances in the Fight Against Infections Caused by Candida Species. Int. J. Mol. Sci. 2020, 21, 6131. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Shukla, P.K. Amphotericin B Resistance Leads to Enhanced Proteinase and Phospholipase Activity and Reduced Germ Tube Formation in Candida albicans. Fungal Biol. 2010, 114, 189–197. [Google Scholar] [CrossRef] [PubMed]
- Feng, W.; Yang, J.; Pan, Y.; Xi, Z.; Qiao, Z.; Ma, Y. The Correlation of Virulence, Pathogenicity, and Itraconazole Resistance with SAP Activity in Candida albicans Strains. Can. J. Microbiol. 2016, 62, 173–178. [Google Scholar] [CrossRef] [PubMed]
- Kadry, A.A.; El-Ganiny, A.M.; El-Baz, A.M. Relationship between Sap Prevalence and Biofilm Formation among Resistant Clinical Isolates of Candida albicans. Afr. Health Sci. 2018, 18, 1166. [Google Scholar] [CrossRef] [PubMed]
- Gerges, M.A.; Fahmy, Y.A.; Hosny, T.; Gandor, N.H.; Mohammed, S.Y.; Mohamed, T.M.A.; Abdelmoteleb, N.E.M.; Esmaeel, N.E. Biofilm Formation and Aspartyl Proteinase Activity and Their Association with Azole Resistance Among Candida albicans Causing Vulvovaginal Candidiasis, Egypt. Infect. Drug Resist. 2023, 16, 5283–5293. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Wright, K.; Hurst, S.F.; Morrison, C.J. Enhanced Extracellular Production of Aspartyl Proteinase, a Virulence Factor, by Candida albicans Isolates Following Growth in Subinhibitory Concentrations of Fluconazole. Antimicrob. Agents Chemother. 2000, 44, 1200–1208. [Google Scholar] [CrossRef] [PubMed]
- Copping, V.M.S.; Barelle, C.J.; Hube, B.; Gow, N.A.R.; Brown, A.J.P.; Odds, F.C. Exposure of Candida albicans to Antifungal Agents Affects Expression of SAP2 and SAP9 Secreted Proteinase Genes. J. Antimicrob. Chemother. 2005, 55, 645–654. [Google Scholar] [CrossRef]
- Barelle, C.J.; Duncan, V.M.S.; Brown, A.J.P.; Gow, N.A.R.; Odds, F.C. Azole Antifungals Induce Up-Regulation of SAP4, SAP5 and SAP6 Secreted Proteinase Genes in Filamentous Candida albicans Cells In Vitro and In Vivo. J. Antimicrob. Chemother. 2008, 61, 315–322. [Google Scholar] [CrossRef]
- El-Houssaini, H.H.; Elnabawy, O.M.; Nasser, H.A.; Elkhatib, W.F. Correlation between Antifungal Resistance and Virulence Factors in Candida albicans Recovered from Vaginal Specimens. Microb. Pathog. 2019, 128, 13–19. [Google Scholar] [CrossRef]
- Figueiredo-Carvalho, M.H.G.; Ramos, L.D.S.; Barbedo, L.S.; Oliveira, J.C.A.D.; Santos, A.L.S.D.; Almeida-Paes, R.; Zancopé-Oliveira, R.M. Relationship between the Antifungal Susceptibility Profile and the Production of Virulence-Related Hydrolytic Enzymes in Brazilian Clinical Strains of Candida glabrata. Mediat. Inflamm. 2017, 2017, 8952878. [Google Scholar] [CrossRef] [PubMed]
- Askari, F.; Rasheed, M.; Kaur, R. The Yapsin Family of Aspartyl Proteases Regulate Glucose Homeostasis in Candida glabrata. J. Biol. Chem. 2022, 298, 101593. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, T.; Izumikawa, K.; Yamauchi, S.; Inamine, T.; Nagayoshi, Y.; Saijo, T.; Seki, M.; Kakeya, H.; Yamamoto, Y.; Yanagihara, K.; et al. The Glycosylphosphatidylinositol-Linked Aspartyl Protease Yps1 Is Transcriptionally Regulated by the Calcineurin-Crz1 and Slt2 MAPK Pathways in Candida glabrata: Transcriptional Regulation of YPS1 in C. Glabrata. FEMS Yeast Res. 2011, 11, 449–456. [Google Scholar] [CrossRef] [PubMed]
- Bairwa, G.; Rasheed, M.; Taigwal, R.; Sahoo, R.; Kaur, R. GPI (Glycosylphosphatidylinositol)-Linked Aspartyl Proteases Regulate Vacuole Homoeostasis in Candida glabrata. Biochem. J. 2014, 458, 323–334. [Google Scholar] [CrossRef] [PubMed]
- Bairwa, G.; Kaur, R. A Novel Role for a Glycosylphosphatidylinositol-anchored Aspartyl Protease, CgYps1, in the Regulation of pH Homeostasis in Candida glabrata. Mol. Microbiol. 2011, 79, 900–913. [Google Scholar] [CrossRef] [PubMed]
- Morrison, C.J.; Hurst, S.F.; Reiss, E. Competitive Binding Inhibition Enzyme-Linked Immunosorbent Assay That Uses the Secreted Aspartyl Proteinase of Candida albicans as an Antigenic Marker for Diagnosis of Disseminated Candidiasis. Clin. Diagn. Lab. Immunol. 2003, 10, 835–848. [Google Scholar] [CrossRef] [PubMed]
- Mavor, A.; Thewes, S.; Hube, B. Systemic Fungal Infections Caused by Candida Species: Epidemiology, Infection Process and Virulence Attributes. CDT 2005, 6, 863–874. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, Y.; Gao, X.; Zhi Gang, J.; Liu, J.; Dong, S. Detection of Candida albicans Sap2 in Cancer Patient Serum Samples by an Indirect Competitive Enzyme-Linked Immunosorbent Assay for the Diagnosis of Candidiasis. Indian J. Pathol. Microbiol. 2013, 56, 243. [Google Scholar] [CrossRef]
- De Bernardis, F.; Agatensi, L.; Ross, I.K.; Emerson, G.W.; Lorenzini, R.; Sullivan, P.A.; Cassone, A. Evidence for a Role for Secreted Aspartate Proteinase of Candida albicans in Vulvovaginal Candidiasis. J. Infect. Dis. 1990, 161, 1276–1283. [Google Scholar] [CrossRef]
- Louie, A.; Dixon, D.M.; el-Maghrabi, E.A.; Burnett, J.W.; Baltch, A.L.; Smith, R.P. Relationship between Candida albicans Epidermolytic Proteinase Activity and Virulence in Mice. J. Med. Vet. Mycol. 1994, 32, 59–64. [Google Scholar] [CrossRef]
- Schaller, M.; Schackert, C.; Korting, H.C.; Januschke, E.; Hube, B. Invasion of Candida albicans Correlates with Expression of Secreted Aspartic Proteinases during Experimental Infection of Human Epidermis. J. Investig. Dermatol. 2000, 114, 712–717. [Google Scholar] [CrossRef] [PubMed]
- Rüchel, R.; Böning-Stutzer, B.; Mari, A. A Synoptical Approach to the Diagnosis of Candidosis, Relying on Serological Antigen and Antibody Tests, on Culture, and on Evaluation of Clinical Data. Mycoses 1988, 31, 87–106. [Google Scholar] [CrossRef] [PubMed]
- Na, B.-K.; Song, C.-Y. Use of Monoclonal Antibody in Diagnosis of Candidiasis Caused by Candida albicans: Detection of Circulating Aspartyl Proteinase Antigen. Clin. Diagn. Lab. Immunol. 1999, 6, 924–929. [Google Scholar] [CrossRef] [PubMed]
- Aoki, W.; Kitahara, N.; Fujita, A.; Shibasaki, S.; Morisaka, H.; Kuroda, K.; Ueda, M. Detection of Candida albicans by Using a Designed Fluorescence-Quenched Peptide. J. Biosci. Bioeng. 2013, 116, 573–575. [Google Scholar] [CrossRef] [PubMed]
- Gheit, M.I.; Mohamed, T.M.; Abdelwahab, M.A. Evaluation of Polyclonal Antiserum Against Secretory Aspartyl Proteinase of Candida albicans as a Potential Serodiagnostic Tool for Invasive Candidiasis. Turk. J. Immunol. 2023, 11, 66–73. [Google Scholar] [CrossRef]
- Shukla, M.; Chandley, P.; Kaur, H.; Ghosh, A.K.; Rudramurthy, S.M.; Rohatgi, S. Expression and Purification along with Evaluation of Serological Response and Diagnostic Potential of Recombinant Sap2 Protein from C. parapsilosis for Use in Systemic Candidiasis. J. Fungi 2021, 7, 999. [Google Scholar] [CrossRef] [PubMed]
- Cassone, A.; Boccanera, M.; Adriani, D.; Santoni, G.; De Bernardis, F. Rats Clearing a Vaginal Infection by Candida albicans Acquire Specific, Antibody-Mediated Resistance to Vaginal Reinfection. Infect. Immun. 1995, 63, 2619–2624. [Google Scholar] [CrossRef] [PubMed]
- De Bernardis, F.; Boccanera, M.; Adriani, D.; Spreghini, E.; Santoni, G.; Cassone, A. Protective Role of Antimannan and Anti-Aspartyl Proteinase Antibodies in an Experimental Model of Candida albicans Vaginitis in Rats. Infect. Immun. 1997, 65, 3399–3405. [Google Scholar] [CrossRef] [PubMed]
- De Bernardis, F.; Boccanera, M.; Adriani, D.; Girolamo, A.; Cassone, A. Intravaginal and Intranasal Immunizations Are Equally Effective in Inducing Vaginal Antibodies and Conferring Protection against Vaginal Candidiasis. Infect. Immun. 2002, 70, 2725–2729. [Google Scholar] [CrossRef]
- Sandini, S.; La Valle, R.; Deaglio, S.; Malavasi, F.; Cassone, A.; De Bernardis, F. A Highly Immunogenic Recombinant and Truncated Protein of the Secreted Aspartic Proteases Family (rSap2t) of Candida albicans as a Mucosal Anticandidal Vaccine. FEMS Immunol. Med. Microbiol. 2011, 62, 215–224. [Google Scholar] [CrossRef]
- De Bernardis, F.; Amacker, M.; Arancia, S.; Sandini, S.; Gremion, C.; Zurbriggen, R.; Moser, C.; Cassone, A. A Virosomal Vaccine against Candidal Vaginitis: Immunogenicity, Efficacy and Safety Profile in Animal Models. Vaccine 2012, 30, 4490–4498. [Google Scholar] [CrossRef] [PubMed]
- Moser, C.; Amacker, M.; Zurbriggen, R. Influenza Virosomes as a Vaccine Adjuvant and Carrier System. Expert Rev. Vaccines 2011, 10, 437–446. [Google Scholar] [CrossRef] [PubMed]
- Mellid-Carballal, R.; Gutierrez-Gutierrez, S.; Rivas, C.; Garcia-Fuentes, M. Viral Protein-Based Nanoparticles (Part 2): Pharmaceutical Applications. Eur. J. Pharm. Sci. 2023, 189, 106558. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, N.; Singh, A.; Upadhyay, A.K.; Mannan, M.A. Design of a Multi-Epitope Vaccine against the Pathogenic Fungi Candida tropicalis Using an in Silico Approach. J. Genet. Eng. Biotechnol. 2022, 20, 140. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, N.; Magdaleno, J.S.L.; Ranjan, S.; Wani, A.K.; Grewal, R.K.; Oliva, R.; Shaikh, A.R.; Cavallo, L.; Chawla, M. Secreted Aspartyl Proteinases Targeted Multi-Epitope Vaccine Design for Candida dubliniensis Using Immunoinformatics. Vaccines 2023, 11, 364. [Google Scholar] [CrossRef]
- Shukla, M.; Rohatgi, S. Vaccination with Secreted Aspartyl Proteinase 2 Protein from Candida parapsilosis Can Enhance Survival of Mice during C. tropicalis -Mediated Systemic Candidiasis. Infect. Immun. 2020, 88, e00312-20. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Samaranayake, L.P.; Leung, W.K.; Sullivan, P.A. Inhibition of Growth and Secreted Aspartyl Proteinase Production in Candida albicans by Lysozyme. J. Med. Microbiol. 1999, 48, 721–730. [Google Scholar] [CrossRef]
- Wagener, J.; Schneider, J.J.; Baxmann, S.; Kalbacher, H.; Borelli, C.; Nuding, S.; Küchler, R.; Wehkamp, J.; Kaeser, M.D.; Mailänder-Sanchez, D.; et al. A Peptide Derived from the Highly Conserved Protein GAPDH Is Involved in Tissue Protection by Different Antifungal Strategies and Epithelial Immunomodulation. J. Investig. Dermatol. 2013, 133, 144–153. [Google Scholar] [CrossRef] [PubMed]
- Rüchel, R.; Ritter, B.; Schaffrinski, M. Modulation of Experimental Systemic Murine Candidosis by Intravenous Pepstatin. Zentralblatt Bakteriol. 1990, 273, 391–403. [Google Scholar] [CrossRef]
- Backman, D.; Danielson, U.H. Kinetic and Mechanistic Analysis of the Association and Dissociation of Inhibitors Interacting with Secreted Aspartic Acid Proteases 1 and 2 from Candida albicans. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2003, 1646, 184–195. [Google Scholar] [CrossRef]
- Cadicamo, C.D.; Mortier, J.; Wolber, G.; Hell, M.; Heinrich, I.E.; Michel, D.; Semlin, L.; Berger, U.; Korting, H.C.; Höltje, H.-D.; et al. Design, Synthesis, Inhibition Studies, and Molecular Modeling of Pepstatin Analogues Addressing Different Secreted Aspartic Proteinases of Candida albicans. Biochem. Pharmacol. 2013, 85, 881–887. [Google Scholar] [CrossRef] [PubMed]
- Dostál, J.; Brynda, J.; Hrušková-Heidingsfeldová, O.; Sieglová, I.; Pichová, I.; Řezáčová, P. The Crystal Structure of the Secreted Aspartic Protease 1 from Candida parapsilosis in Complex with Pepstatin A. J. Struct. Biol. 2009, 167, 145–152. [Google Scholar] [CrossRef]
- Dostál, J.; Brynda, J.; Vaňková, L.; Zia, S.R.; Pichová, I.; Heidingsfeld, O.; Lepšík, M. Structural Determinants for Subnanomolar Inhibition of the Secreted Aspartic Protease Sapp1p from Candida Parapsilosis. J. Enzym. Inhib. Med. Chem. 2021, 36, 914–921. [Google Scholar] [CrossRef]
- Gruber, A.; Berlit, J.; Speth, C.; Lass-Flörl, C.; Kofler, G.; Nagl, M.; Borg-von Zepelin, M.; Dierich, M.P.; Würzner, R. Dissimilar Attenuation of Candida albicans Virulence Properties by Human Immunodeficiency Virus Type 1 Protease Inhibitors. Immunobiology 1999, 201, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Gruber, A.; Speth, C.; Lukasser-Vogl, E.; Zangerle, R.; Borg-von Zepelin, M.; Dierich, M.P.; Würzner, R. Human Immunodeficiency Virus Type 1 Protease Inhibitor Attenuates Candida albicans Virulence Properties in Vitro. Immunopharmacology 1999, 41, 227–234. [Google Scholar] [CrossRef]
- Korting, H.C.; Schaller, M.; Eder, G.; Hamm, G.; Böhmer, U.; Hube, B. Effects of the Human Immunodeficiency Virus (HIV) Proteinase Inhibitors Saquinavir and Indinavir on In Vitro Activities of Secreted Aspartyl Proteinases of Candida albicans Isolates from HIV-Infected Patients. Antimicrob. Agents Chemother. 1999, 43, 2038–2042. [Google Scholar] [CrossRef]
- Cassone, A.; De Bernardis, F.; Torosantucci, A.; Tacconelli, E.; Tumbarello, M.; Cauda, R. In Vitro and In Vivo Anticandidal Activity of Human Immunodeficiency Virus Protease Inhibitors. J. Infect. Dis. 1999, 180, 448–453. [Google Scholar] [CrossRef] [PubMed]
- Borg-von Zepelin, M.; Meyer, I.; Thomssen, R.; Würzner, R.; Sanglard, D.; Telenti, A.; Monod, M. HIV-Protease Inhibitors Reduce Cell Adherence of Candida albicans Strains by Inhibition of Yeast Secreted Aspartic Proteases. J. Investig. Dermatol. 1999, 113, 747–751. [Google Scholar] [CrossRef] [PubMed]
- Bektić, J.; Lell, C.P.; Fuchs, A.; Stoiber, H.; Speth, C.; Lass-Flörl, C.; Borg-von Zepelin, M.; Dierich, M.P.; Würzner, R. HIV Protease Inhibitors Attenuate Adherence of Candida albicans to Epithelial Cells in Vitro. FEMS Immunol. Med. Microbiol. 2001, 31, 65–71. [Google Scholar] [CrossRef]
- Blanco, M.T.; Hurtado, C.; Pérez-giraldo, C.; Morán, F.J.; González-velasco, C.; Gómezgarcía, A.C. Effect of Ritonavir and Saquinavir on Candida albicans Growth Rate and In Vitro Activity of Aspartyl Proteinases. Med. Mycol. 2003, 41, 167–170. [Google Scholar] [CrossRef]
- Schaller, M.; Bein, M.; Korting, H.C.; Baur, S.; Hamm, G.; Monod, M.; Beinhauer, S.; Hube, B. The Secreted Aspartyl Proteinases Sap1 and Sap2 Cause Tissue Damage in an In Vitro Model of Vaginal Candidiasis Based on Reconstituted Human Vaginal Epithelium. Infect. Immun. 2003, 71, 3227–3234. [Google Scholar] [CrossRef]
- Santos, A.L.S.; Braga-Silva, L.A.; Gonçalves, D.S.; Ramos, L.S.; Oliveira, S.S.C.; Souza, L.O.P.; Oliveira, V.S.; Lins, R.D.; Pinto, M.R.; Muñoz, J.E.; et al. Repositioning Lopinavir, an HIV Protease Inhibitor, as a Promising Antifungal Drug: Lessons Learned from Candida albicans—In Silico, In Vitro and In Vivo Approaches. J. Fungi 2021, 7, 424. [Google Scholar] [CrossRef] [PubMed]
- Asencio, M.A.; Garduño, E.; Pérez-Giraldo, C.; Blanco, M.T.; Hurtado, C.; Gómez-García, A.C. Exposure to Therapeutic Concentrations of Ritonavir, but Not Saquinavir, Reduces Secreted Aspartyl Proteinase of Candida parapsilosis. Chemotherapy 2005, 51, 252–255. [Google Scholar] [CrossRef]
- Zhang, Z.; ElSohly, H.N.; Jacob, M.R.; Pasco, D.S.; Walker, L.A.; Clark, A.M. Natural Products Inhibiting Candida albicans Secreted Aspartic Proteases from Lycopodium Cernuum. J. Nat. Prod. 2002, 65, 979–985. [Google Scholar] [CrossRef]
- Höfling, J.F.; Mardegan, R.C.; Anibal, P.C.; Furletti, V.F.; Foglio, M.A. Evaluation of Antifungal Activity of Medicinal Plant Extracts Against Oral Candida albicans and Proteinases. Mycopathologia 2011, 172, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Zhang, N.; Zhang, S.; Zhang, L.; Liu, Q. Phloretin Inhibited the Pathogenicity and Virulence Factors against Candida albicans. Bioengineered 2021, 12, 2420–2431. [Google Scholar] [CrossRef] [PubMed]
- Christopeit, T.; Øverbø, K.; Danielson, U.; Nilsen, I. Efficient Screening of Marine Extracts for Protease Inhibitors by Combining FRET Based Activity Assays and Surface Plasmon Resonance Spectroscopy Based Binding Assays. Mar. Drugs 2013, 11, 4279–4293. [Google Scholar] [CrossRef]
- Gu, W.; Guo, D.; Zhang, L.; Xu, D.; Sun, S. The Synergistic Effect of Azoles and Fluoxetine against Resistant Candida albicans Strains Is Attributed to Attenuating Fungal Virulence. Antimicrob. Agents Chemother. 2016, 60, 6179–6188. [Google Scholar] [CrossRef]
- Falkensammer, B.; Pleyer, L.; Ressler, S.; Berg, A.; Borg-von Zepelin, M.; Nagl, M.; Lass-Flörl, C.; Speth, C.; Dierich, M.P.; Würzner, R. Basidiomycete Metabolites Attenuate Virulence Properties of Candida albicans in Vitro. Mycoses 2008, 51, 505–514. [Google Scholar] [CrossRef]
- Jebali, A.; Hajjar, F.H.E.; Hekmatimoghaddam, S.; Kazemi, B.; De La Fuente, J.M.; Rashidi, M. Triangular Gold Nanoparticles Conjugated with Peptide Ligands: A New Class of Inhibitor for Candida albicans Secreted Aspartyl Proteinase. Biochem. Pharmacol. 2014, 90, 349–355. [Google Scholar] [CrossRef]
- Goldman, R.C.; Frost, D.J.; Capobianco, J.O.; Kadam, S.; Rasmussen, R.R.; Abad-Zapatero, C. Antifungal Drug Targets: Candida Secreted Aspartyl Protease and Fungal Wall Beta-Glucan Synthesis. Infect. Agents Dis. 1995, 4, 228–247. [Google Scholar] [PubMed]
- Abad-Zapatero, C.; Goldman, R.; Muchmore, S.W.; Hutchins, C.; Stewart, K.; Navaza, J.; Payne, C.D.; Ray, T.L. Structure of a Secreted Aspartic Protease from C. albicans Complexed with a Potent Inhibitor: Implications for the Design of Antifungal Agents. Protein Sci. 1996, 5, 640–652. [Google Scholar] [CrossRef] [PubMed]
- Stewart, K.; Abad-Zapatero, C. Candida Proteases and Their Inhibition Prospects for Antifungal Therapy. Curr. Med. Chem. 2001, 8, 941–948. [Google Scholar] [CrossRef] [PubMed]
- Pranav Kumar, S.K.; Kulkarni, V.M. Insights into the Selective Inhibition of Candida albicans Secreted Aspartyl Protease: A Docking Analysis Study. Bioorganic Med. Chem. 2002, 10, 1153–1170. [Google Scholar] [CrossRef] [PubMed]
- Kathwate, G.H.; Karuppayil, S.M. Antifungal Properties of the Anti-Hypertensive Drug: Aliskiren. Arch. Oral Biol. 2013, 58, 1109–1115. [Google Scholar] [CrossRef]
- Gholam, G.M.; Firdausy, I.A. Molecular Docking Study of Natural Compounds from Red Betel (Piper Crocatum Ruiz & Pav) as Inhibitor of Secreted Aspartic Proteinase 5 (Sap 5) in Candida albicans. Sasambo J. Pharm. 2022, 3, 97–104. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bras, G.; Satala, D.; Juszczak, M.; Kulig, K.; Wronowska, E.; Bednarek, A.; Zawrotniak, M.; Rapala-Kozik, M.; Karkowska-Kuleta, J. Secreted Aspartic Proteinases: Key Factors in Candida Infections and Host-Pathogen Interactions. Int. J. Mol. Sci. 2024, 25, 4775. https://doi.org/10.3390/ijms25094775
Bras G, Satala D, Juszczak M, Kulig K, Wronowska E, Bednarek A, Zawrotniak M, Rapala-Kozik M, Karkowska-Kuleta J. Secreted Aspartic Proteinases: Key Factors in Candida Infections and Host-Pathogen Interactions. International Journal of Molecular Sciences. 2024; 25(9):4775. https://doi.org/10.3390/ijms25094775
Chicago/Turabian StyleBras, Grazyna, Dorota Satala, Magdalena Juszczak, Kamila Kulig, Ewelina Wronowska, Aneta Bednarek, Marcin Zawrotniak, Maria Rapala-Kozik, and Justyna Karkowska-Kuleta. 2024. "Secreted Aspartic Proteinases: Key Factors in Candida Infections and Host-Pathogen Interactions" International Journal of Molecular Sciences 25, no. 9: 4775. https://doi.org/10.3390/ijms25094775
APA StyleBras, G., Satala, D., Juszczak, M., Kulig, K., Wronowska, E., Bednarek, A., Zawrotniak, M., Rapala-Kozik, M., & Karkowska-Kuleta, J. (2024). Secreted Aspartic Proteinases: Key Factors in Candida Infections and Host-Pathogen Interactions. International Journal of Molecular Sciences, 25(9), 4775. https://doi.org/10.3390/ijms25094775




