Expression and Localization Profiles of Tight Junction Proteins in Immune Cells Depend on Their Activation Status
Abstract
1. Introduction
2. Results
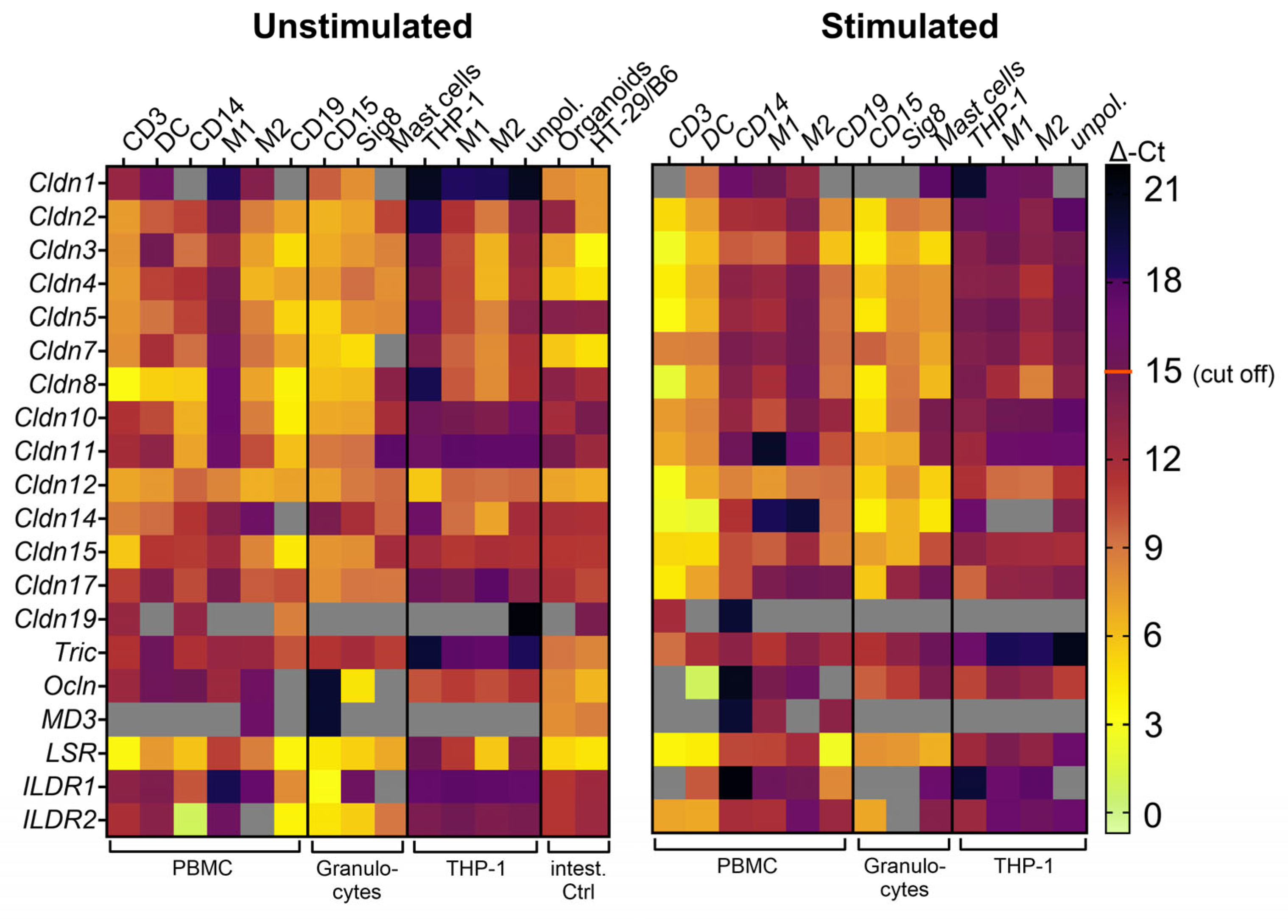
2.1. Primary Peripheral Blood Immune Cells Possess Distinct mRNA Expression Profiles of TJ Proteins That Change upon Stimulation
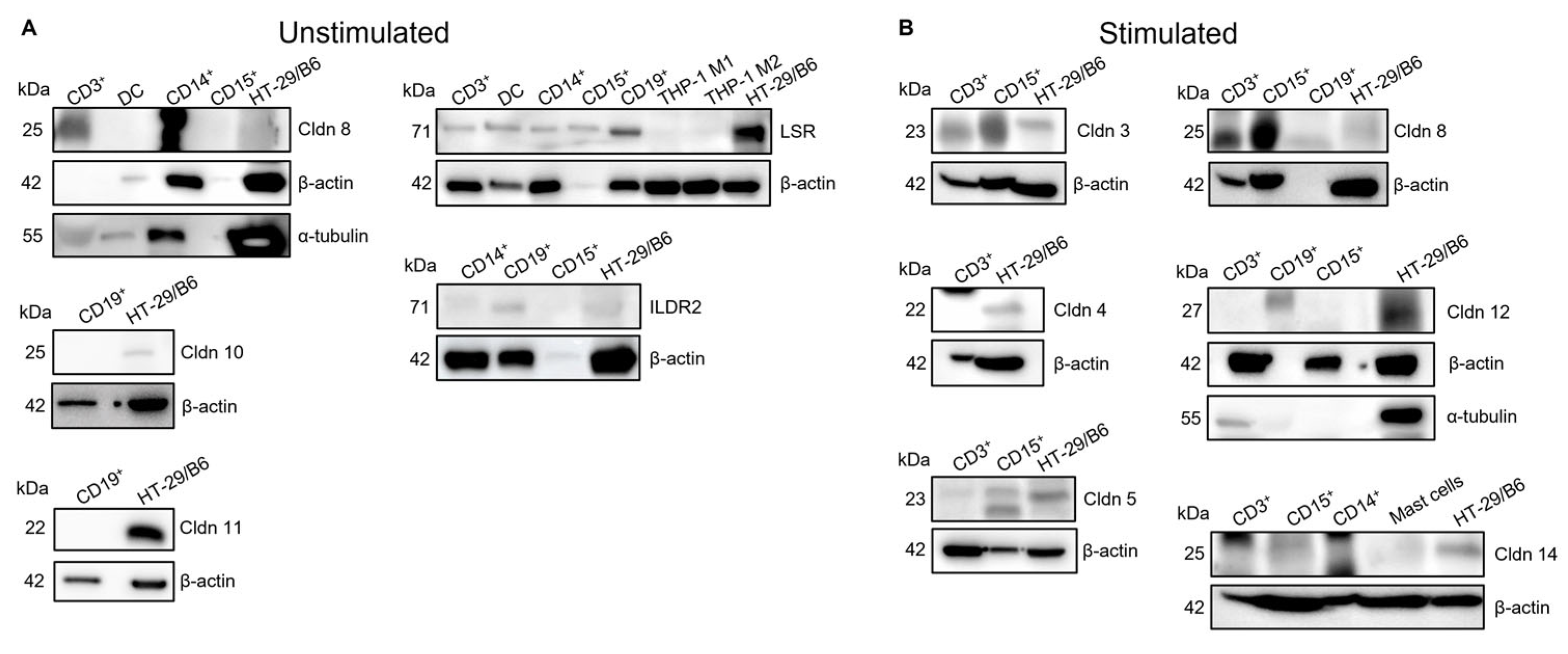
2.2. Expression of Some TJ Proteins Can Be Confirmed also at Protein Level
2.3. Claudin-Binding Domain of Clostridium Perfringens Enterotoxin May Bind Only to Claudins of Stimulated Immune Cells
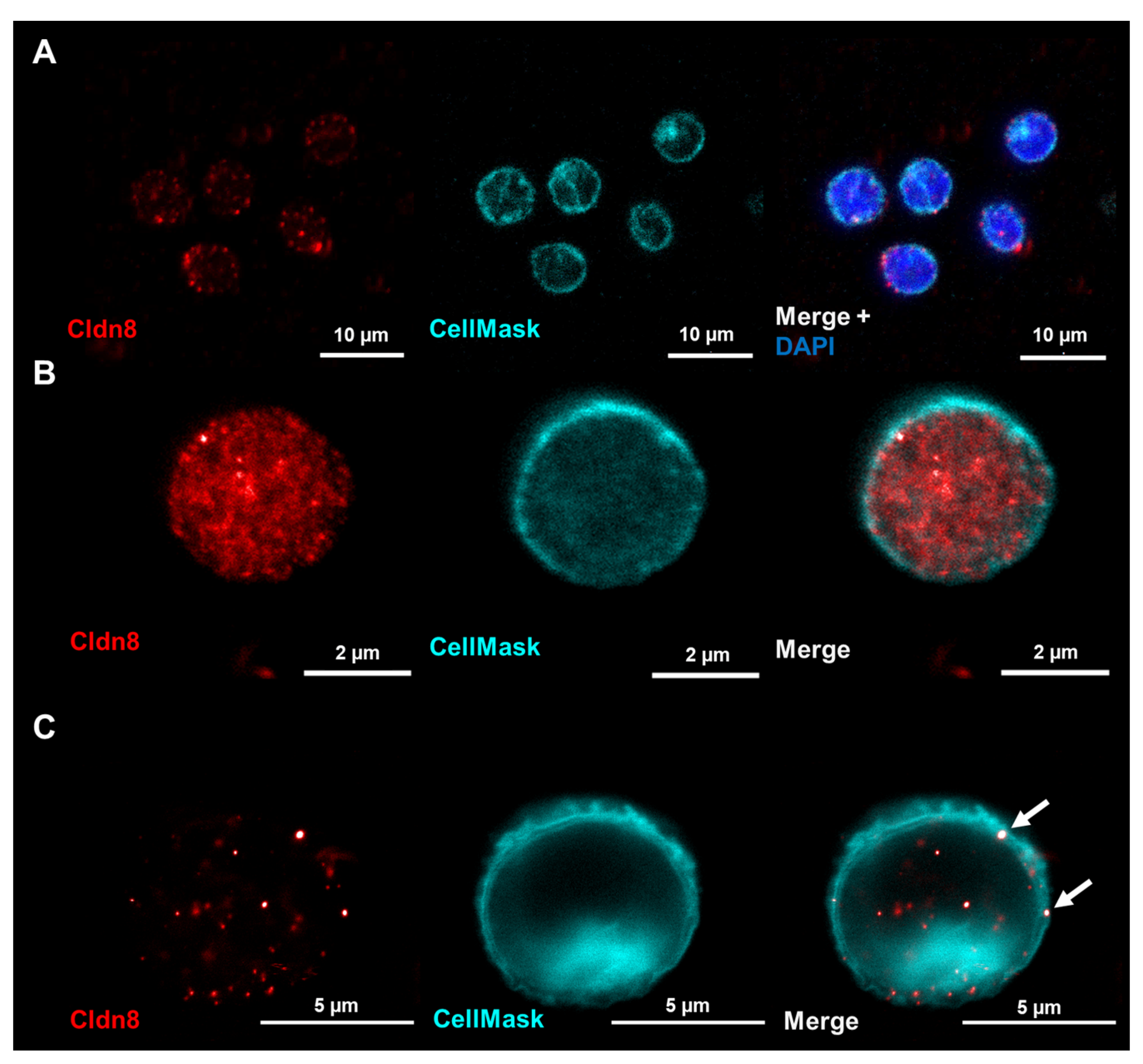
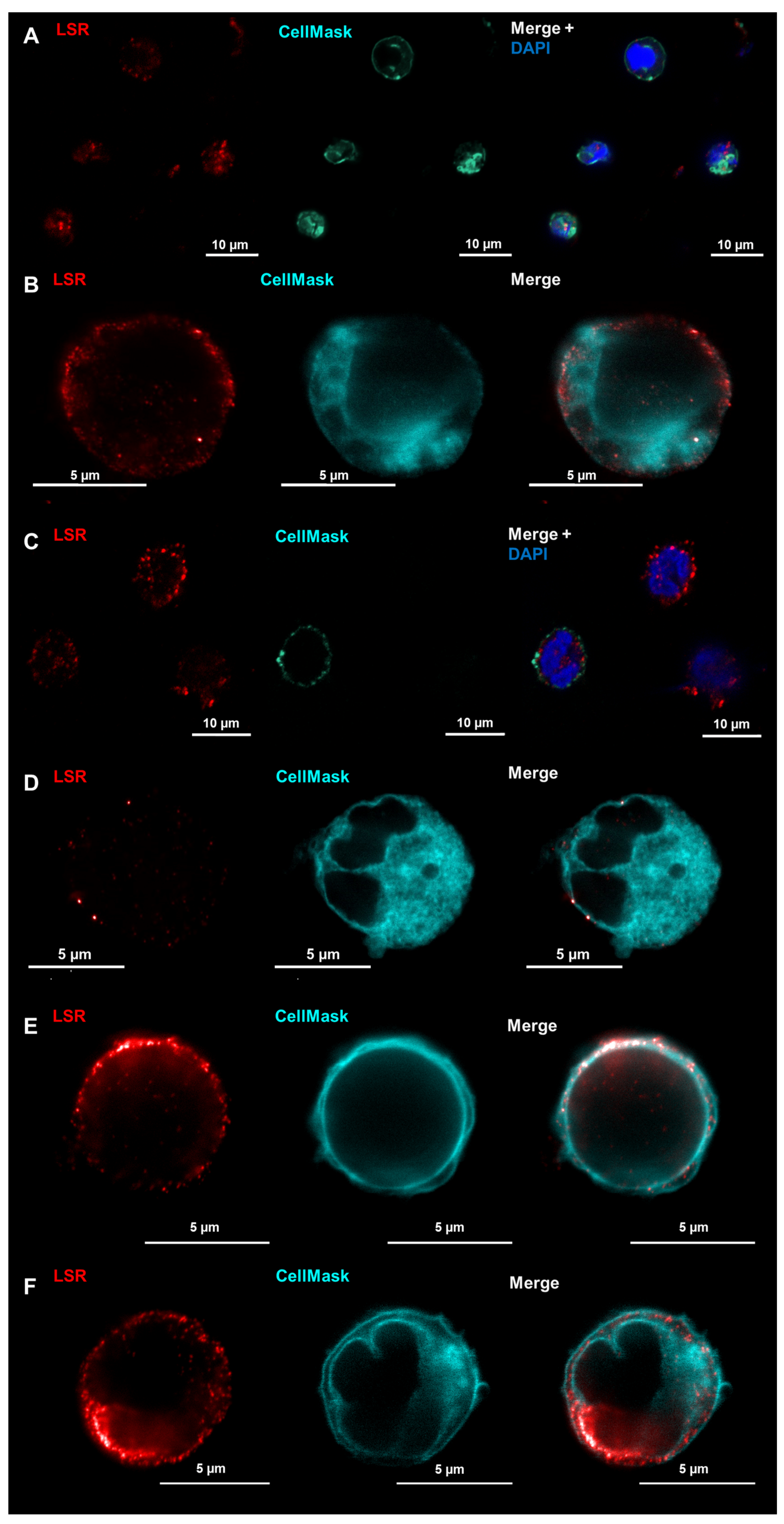
2.4. LSR Is Located in Membranes of Unstimulated Immune Cells While Claudin-8 Only Appears in Membranes after Immune Cell Activation
3. Discussion
- Immune cells possess specific expression patterns for TJ proteins at the mRNA level that change upon activation, but only a few are expressed at the protein level
- Claudin-8 is localized within immune cell membranes only upon activation
- LSR may have general functions for immune cells, being present in the membranes in unstimulated and stimulated cells
4. Materials and Methods
4.1. Immune Cell Isolation
4.2. Cell Culture
4.2.1. T Cells
4.2.2. THP-1
4.2.3. Monocytes
4.2.4. B Cells
4.2.5. Dendritic Cells
4.2.6. Immune Cell Activation
4.3. Fluorescence-Activated Cell Sorting (FACS)
4.4. RNA Isolation and Real-Time qPCR
4.5. Protein Isolation and Western Blotting
4.6. Immunofluorescence Staining
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Baranov, M.; Ter Beest, M.; Reinieren-Beeren, I.; Cambi, A.; Figdor, C.G.; van den Bogaart, G. Podosomes of dendritic cells facilitate antigen sampling. J. Cell Sci. 2014, 127, 1052–1064. [Google Scholar] [CrossRef] [PubMed]
- Murdoch, J.R.; Lloyd, C.M. Chronic inflammation and asthma. Mutat. Res. Fundam. Mol. Mech. Mutagen. 2010, 690, 24–39. [Google Scholar] [CrossRef] [PubMed]
- Ramos, G.P.; Papadakis, K.A. Mechanisms of Disease: Inflammatory Bowel Diseases. Mayo Clin. Proc. 2019, 94, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Engelhardt, B.; Wolburg, H. Mini review: Transendothelial migration of leukocytes: Through the front door or around the side of the house? Eur. J. Immunol. 2004, 34, 2955–2963. [Google Scholar] [CrossRef] [PubMed]
- Burns, A.R.; Walker, D.C.; Brown, E.S.; Thurmon, L.T.; Bowden, R.A.; Keese, C.R.; Simon, S.I.; Entman, M.L.; Smith, C.W. Neutrophil transendothelial migration is independent of tight junctions and occurs preferentially at tricellular corners. J. Immunol. 1997, 159, 2893–2903. [Google Scholar] [CrossRef] [PubMed]
- Castro Dias, M.; Odriozola Quesada, A.; Soldati, S.; Bösch, F.; Gruber, I.; Hildbrand, T.; Sönmez, D.; Khire, T.; Witz, G.; McGrath, J.L.; et al. Brain endothelial tricellular junctions as novel sites for T cell diapedesis across the blood-brain barrier. J. Cell Sci. 2021, 134, jcs253880. [Google Scholar] [CrossRef] [PubMed]
- Mineta, K.; Yamamoto, Y.; Yamazaki, Y.; Tanaka, H.; Tada, Y.; Saito, K.; Tamura, A.; Igarashi, M.; Endo, T.; Takeuchi, K.; et al. Predicted expansion of the claudin multigene family. FEBS Lett. 2011, 585, 606–612. [Google Scholar] [CrossRef] [PubMed]
- Günzel, D.; Fromm, M. Claudins and other tight junction proteins. Compr. Physiol. 2012, 2, 1819–1852. [Google Scholar] [CrossRef] [PubMed]
- Krug, S.M.; Günzel, D.; Conrad, M.P.; Lee, I.F.; Amasheh, S.; Fromm, M.; Yu, A.S. Charge-selective claudin channels. Ann. N. Y. Acad. Sci. 2012, 1257, 20–28. [Google Scholar] [CrossRef] [PubMed]
- Staehelin, L.A. Further observations on the fine structure of freeze-cleaved tight junctions. J. Cell Sci. 1973, 13, 763–786. [Google Scholar] [CrossRef] [PubMed]
- Krug, S.M.; Amasheh, S.; Richter, J.F.; Milatz, S.; Günzel, D.; Westphal, J.K.; Huber, O.; Schulzke, J.D.; Fromm, M. Tricellulin Fprms a Barrier to Macromolecules in Tricellular Tight Junctions without Affecting Ion Permeability. Mol. Biol. Cell 2009, 20, 4524–4530. [Google Scholar]
- Krug, S.M. Contribution of the tricellular tight junction to paracellular permeability in leaky and tight epithelia. Ann. N. Y. Acad. Sci. 2017, 1397, 219–230. [Google Scholar] [CrossRef] [PubMed]
- Ayala-Torres, C.; Krug, S.M.; Schulzke, J.D.; Rosenthal, R.; Fromm, M. Tricellulin Effect on Paracellular Water Transport. Int. J. Mol. Sci. 2019, 20, 5700. [Google Scholar] [CrossRef] [PubMed]
- Ayala-Torres, C.; Krug, S.M.; Rosenthal, R.; Fromm, M. Angulin-1 (LSR) Affects Paracellular Water Transport, However Only in Tight Epithelial Cells. Int. J. Mol. Sci. 2021, 22, 7827. [Google Scholar] [CrossRef] [PubMed]
- Sung, S.S.; Fu, S.M.; Rose, C.E., Jr.; Gaskin, F.; Ju, S.T.; Beaty, S.R. A major lung CD103 (alphaE)-beta7 integrin-positive epithelial dendritic cell population expressing Langerin and tight junction proteins. J. Immunol. 2006, 176, 2161–2172. [Google Scholar] [CrossRef] [PubMed]
- Ogasawara, N.; Kojima, T.; Go, M.; Fuchimoto, J.; Kamekura, R.; Koizumi, J.; Ohkuni, T.; Masaki, T.; Murata, M.; Tanaka, S.; et al. Induction of JAM-A during differentiation of human THP-1 dendritic cells. Biochem. Biophys. Res. Commun. 2009, 389, 543–549. [Google Scholar] [CrossRef] [PubMed]
- Mariano, C.; Silva, S.L.; Pereira, P.; Fernandes, A.; Brites, D.; Brito, M.A. Evidence of tricellulin expression by immune cells, particularly microglia. Biochem. Biophys. Res. Commun. 2011, 409, 799–802. [Google Scholar] [CrossRef] [PubMed]
- Bauer, H.; Stelzhammer, W.; Fuchs, R.; Weiger, T.M.; Danninger, C.; Probst, G.; Krizbai, I.A. Astrocytes and Neurons Express the Tight Junction-Specific Protein Occludin in Vitro. Exp. Cell Res. 1999, 250, 434–438. [Google Scholar] [CrossRef] [PubMed]
- Blank, F.; Wehrli, M.; Lehmann, A.; Baum, O.; Gehr, P.; von Garnier, C.; Rothen-Rutishauser, B.M. Macrophages and dendritic cells express tight junction proteins and exchange particles in an in vitro model of the human airway wall. Immunobiology 2011, 216, 86–95. [Google Scholar] [CrossRef] [PubMed]
- Dustin, M.L.; Springer, T. Lymphocytes Function-associated Antigen-1 (LFA-1) Interaction with Intercellular Adhesion Molecule-1 (ICAM-1) is One of At Least Three Mechanisms for Lymphocyte Adhesion to Cultured Enduthelial Cells. J. Cell Biol. 1988, 107, 321–331. [Google Scholar] [CrossRef] [PubMed]
- Ostermann, G.; Weber, K.S.C.; Zernecke, A.; Schröder, A.; Weber, C. JAM-I is a ligand of the β2 integrin LFA-I involved in transendothelial migration of leukocytes. Nat. Immunol. 2002, 3, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Kubo, A.; Nagao, K.; Yokouchi, M.; Sasaki, H.; Amagai, M. External antigen uptake by Langerhans cells with reorganization of epidermal tight junction barriers. J. Exp. Med. 2009, 206, 2937–2946. [Google Scholar] [CrossRef] [PubMed]
- Protze, J.; Eichner, M.; Piontek, A.; Dinter, S.; Rossa, J.; Blecharz, K.G.; Vajkoczy, P.; Piontek, J.; Krause, G. Directed structural modification of Clostridium perfringens enterotoxin to enhance binding to claudin-5. Cell. Mol. Life Sci. 2015, 72, 1417–1432. [Google Scholar] [CrossRef] [PubMed]
- Beier, L.S.; Rossa, J.; Woodhouse, S.; Bergmann, S.; Kramer, H.B.; Protze, J.; Eichner, M.; Piontek, A.; Vidal-y-Sy, S.; Brandner, J.M.; et al. Use of modified clostridium perfringens enterotoxin fragments for claudin targeting in liver and skin cells. Int. J. Mol. Sci. 2019, 20, 4774. [Google Scholar] [CrossRef] [PubMed]
- Beier, L.S.; Waldow, A.; Khomeijani Farahani, S.; Mannweiler, R.; Vidal-Y-Sy, S.; Brandner, J.M.; Piontek, J.; Günzel, D. Claudin targeting as an effective tool for directed barrier modulation of the viable epidermis. Ann. New York Acad. Sci. 2022, 1517, 251–265. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, A.; Saito, Y.; Kondoh, M.; Matsushita, K.; Krug, S.M.; Suzuki, H.; Tsujino, H.; Li, X.; Aoyama, H.; Matsuhisa, K.; et al. Creation and biochemical analysis of a broad-specific claudin binder. Biomaterials 2012, 33, 3464–3474. [Google Scholar] [CrossRef] [PubMed]
- Rescigno, M.; Urbano, M.; Valzasina, B.; Francolini, M.; Rotta, G.; Bonasio, R.; Granucci, F.; Kraehenbuhl, J.-P.; Ricciardi-Castagnoli, P. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat. Immunol. 2001, 2, 361–367. [Google Scholar] [CrossRef] [PubMed]
- Garneau, N.L.; Wilusz, J.; Wilusz, C.J. The highways and byways of mRNA decay. Nat. Rev. Mol. Cell Biol. 2007, 8, 113–126. [Google Scholar] [CrossRef] [PubMed]
- Thomson, D.W.; Dinger, M.E. Endogenous microRNA sponges: Evidence and controversy. Nat. Rev. Genet. 2016, 17, 272–283. [Google Scholar] [CrossRef]
- Anderson, P.; Kedersha, N. RNA granules: Post-transcriptional and epigenetic modulators of gene expression. Nat. Rev. Mol. Cell Biol. 2009, 10, 430–436. [Google Scholar] [CrossRef] [PubMed]
- Darido, C.; Buchert, M.; Pannequin, J.; Bastide, P.; Zalzali, H.; Mantamadiotis, T.; Bourgaux, J.-F.o.; Garambois, V.R.; Jay, P.; Blache, P.; et al. Defective Claudin-7 Regulation by Tcf-4 and Sox-9 Disrupts the Polarity and Increases the Tumorigenicity of Colorectal Cancer Cells. Cancer Res. 2008, 68, 4258–4268. [Google Scholar] [CrossRef] [PubMed]
- Hagen, S.J. Non-canonical functions of claudin proteins: Beyond the regulation of cell-cell adhesions. Tissue Barriers 2017, 5, e1327839. [Google Scholar] [CrossRef]
- Weber, C.R.; Nalle, S.C.; Tretiakova, M.; Rubin, D.T.; Turner, J.R. Claudin-1 and claudin-2 expression is elevated in inflammatory bowel disease and may contribute to early neoplastic transformation. Lab. Investig. 2008, 88, 1110–1120. [Google Scholar] [CrossRef] [PubMed]
- Dhawan, P.; Singh, A.B.; Deane, N.G.; No, Y.; Shiou, S.R.; Schmidt, C.; Neff, J.; Washington, M.K.; Beauchamp, R.D. Claudin-1 regulates cellular transformation and metastatic behavior in colon cancer. J. Clin. Investig. 2005, 115, 1765–1776. [Google Scholar] [CrossRef] [PubMed]
- Ikari, A.; Watanabe, R.; Sato, T.; Taga, S.; Shimobaba, S.; Yamaguchi, M.; Yamazaki, Y.; Endo, S.; Matsunaga, T.; Sugatani, J. Nuclear distribution of claudin-2 increases cell proliferation in human lung adenocarcinoma cells. Biochim. Et Biophys. Acta (BBA) Mol. Cell Res. 2014, 1843, 2079–2088. [Google Scholar] [CrossRef] [PubMed]
- Todd, M.C.; Petty, H.M.; King, J.M.; Piana Marshall, B.N.; Sheller, R.A.; Cuevas, M.E. Overexpression and delocalization of claudin-3 protein in MCF-7 and MDA-MB-415 breast cancer cell lines. Oncol. Lett. 2015, 10, 156–162. [Google Scholar] [CrossRef]
- Cuevas, M.E.; Gaska, J.M.; Gist, A.C.; King, J.M.; Sheller, R.A.; Todd, M.C. Estrogen-dependent expression and subcellular localization of the tight junction protein claudin-4 in HEC-1A endometrial cancer cells. Int. J. Oncol. 2015, 47, 650–656. [Google Scholar] [CrossRef] [PubMed]
- Geissmann, F.; Manz, M.G.; Jung, S.; Sieweke, M.H.; Merad, M.; Ley, K. Development of monocytes, macrophages, and dendritic cells. Science 2010, 327, 656–661. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Jeremiah Bell, J.; Bhandoola, A. T-cell lineage determination. Immunol. Rev. 2010, 238, 12–22. [Google Scholar] [CrossRef] [PubMed]
- Kumar, B.V.; Connors, T.J.; Farber, D.L. Human T Cell Development, Localization, and Function throughout Life. Immunity 2018, 48, 202–213. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Liu, Y.J. Development of dendritic-cell lineages. Immunity 2007, 26, 741–750. [Google Scholar] [CrossRef] [PubMed]
- Masuda, S.; Oda, Y.; Sasaki, H.; Ikenouchi, J.; Higashi, T.; Akashi, M.; Nishi, E.; Furuse, M. LSR defines cell corners for tricellular tight junction formation in epithelial cells. J. Cell Sci. 2011, 124, 548–555. [Google Scholar] [CrossRef] [PubMed]
- Funauchi, M.; Serada, S.; Hiramatsu, K.; Funajima, E.; Kanda, M.; Nagase, Y.; Nakagawa, S.; Ohkawara, T.; Fujimoto, M.; Suzuki, Y.; et al. Tumor cell-expressed lipolysis-stimulated lipoprotein receptor negatively regulates T-cell function. Int. J. Cancer 2024, 154, 425–433. [Google Scholar] [CrossRef] [PubMed]
- Akuthota, P.; Capron, K.; Weller, P.F. Eosinophil purification from peripheral blood. Methods Mol. Biol. 2014, 1178, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Waldow, A.; Beier, L.S.; Arndt, J.; Schallenberg, S.; Vollbrecht, C.; Bischoff, P.; Farrera-Sal, M.; Loch, F.N.; Bojarski, C.; Schumann, M.; et al. cCPE Fusion Proteins as Molecular Probes to Detect Claudins and Tight Junction Dysregulation in Gastrointestinal Cell Lines, Tissue Explants and Patient-Derived Organoids. Pharmaceutics 2023, 15, 1980. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]

| Primary antibodies and fluorescence labels | ||
| Antibody | Concentration | Supplier |
| Anti-huβ-actin | WB 1:1000 | Sigma-Aldrich Chemie GmbH, Traufkirchen, Germany, CAT: A544 |
| Anti-huCD1c PB | FACS 1:200 | BioLegend, San Diego, CA, USA, CAT: 331507 |
| Anti-huCD3 FITC | FACS 1:200 | BioLegend, San Diego, CA, USA, CAT: 317306 |
| Anti-huCD3 PE | FACS 1:200 | BioLegend, San Diego, CA, USA, CAT: 300307 |
| Anti-huCD14 AF700 | FACS 1:1600 | BioLegend, San Diego, CA, USA, CAT: 367113 |
| Anti-huCD11c PE | FACS 1:100 | BioLegend, San Diego, CA, USA, CAT: 301605 |
| Anti-huCD19 PB | FACS 1:50 | BioLegend, San Diego, CA, USA, CAT: 363035 |
| Anti-huCD19 PerCP | FACS 1:200 | BioLegend, San Diego, CA, USA, CAT: 363013 |
| Anti-huCD15 PE | FACS 1:50 | BioLegend, San Diego, CA, USA, CAT: 301905 |
| Anti-huCD123 APC | FACS 1:50 | BioLegend, San Diego, CA, USA, CAT: 306011 |
| Anti-huCD303 PE | FACS 1:200 | BioLegend, San Diego, CA, USA, CAT: 354203 |
| Anti-huClaudin-3 | WB 1:1000 | Invitrogen, Thermo Fisher Scientific GmbH, Dreireich, Germany, CAT: 325600 |
| Anti-huClaudin-5 | WB 1:1000 | Invitrogen, Thermo Fisher Scientific GmbH, Dreireich, Germany, CAT: MA5-32614 |
| Anti-huClaudin-8 | IF 1:200 WB 1:1000 | Invitrogen, Thermo Fisher Scientific GmbH, Dreireich, Germany, CAT: 710222 |
| Anti-huClaudin-11 | WB 1:1000 | BiCell Scientific, Maryland Heights, MO, USA, CAT: 00211 |
| Anti-huClaudin-12 | WB 1:1000 | IBL Interantional GmbH, Hamburg, Germany, CAT: JP18801 |
| Anti-huClaudin-14 | WB 1:1000 | Invitrogen, Thermo Fisher Scientific GmbH, Dreireich, Germany, CAT: 36-4200 |
| Anti-huILDR2 | WB 1:1000 | Sigma-Aldrich Chemie GmbH, Traufkirchen, Germany, CAT: HPA012815 |
| Anti-huLSR | IF 1:200 WB 1:1000 | Atlas antibodies AB, Bromma, Sweden, CAT: HPA007270 |
| Anti-huSiglec8 FITC | WB 1:1600 | Miltenyi Biotec, Bergisch Gladbach, Deutschland, CAT: 130-098-716 |
| Secondary antibodies and fluorescence labels | ||
| Antibody | Concentration | Supplier |
| 4′,6-diamidino-2-phenylindole (DAPI) | IF 1:500 | Roche Diagnostics GmbH, Mannheim, Germany, Cat. No.: 10236276001 |
| Anti-mouse | WB 1:10,000 | Invitrogen, Thermo Fisher Scientific GmbH, Dreireich, Germany, CAT: 31430 |
| Anti-rabbit | WB 1:10,000 | Invitrogen, Thermo Fisher Scientific GmbH, Dreireich, Germany, CAT: 31460 |
| Anti-rabbit AF647 | IF 1:250 | Invitrogen, Thermo Fisher Scientific GmbH, Dreireich, Germany, CAT: A-21245 |
| Anti-rabbit STARred | IF 1:250 | Abberior GmbH, Göttingen, Germany, CAT: STRED-1002-500UG |
| CellMask™ orange | IF 1:5000 | Thermo Fisher, Waltham, MA, USA, Cat. No.: C37608 |
| YFP-cCPE-SSS | FACS 10 ng/mL | Kind gift of PD Dr. Jörg Piontek, Charité—Universitätsmedizin Berlin, Clinical Physiology/Nutritional Medicine |
| YFP-cCPE-YALA | FACS 10 ng/mL | Kind gift of PD Dr. Jörg Piontek, Charité—Universitätsmedizin Berlin, Clinical Physiology/Nutritional Medicine |
| Zombie Aqua™ Fixable Viability Kit | FACS 1:1000 | BioLegend, San Diego, CA, USA, Cat. No.: 423101 |
| RT-qPCR Probes | ||
| Probe | Dye | Assay ID |
| huClaudin-1 | FAM | Hs00912957_m1 |
| huClaudin-2 | FAM | Hs00252666_s1 |
| huClaudin-3 | FAM | Hs00265816_s1 |
| huClaudin-4 | FAM | Hs00976831_s1 |
| huClaudin-5 | FAM | Hs00533949_s1 |
| huClaudin-7 | FAM | Hs00600772_s1 |
| huClaudin-8 | FAM | Hs00273282_s1 |
| huClaudin-10 | FAM | Hs00199599_m1 |
| huClaudin-11 | FAM | Hs00912957_m1 |
| huClaudin-12 | FAM | Hs00273258_s1 |
| huClaudin-14 | FAM | Hs00377953_m1 |
| huClaudin-15 | FAM | Hs00204982_m1 |
| huClaudin-17 | FAM | Hs00273276_s1 |
| huClaudin-19 | FAM | Hs00961709_m1 |
| huGAPDH | VIC | 4310884E |
| huILDR1 | FAM | Hs01111437_m1 |
| huILDR2 | FAM | Hs01025498_m1 |
| huLSR | FAM | Hs01076319_g1 |
| huOccludin | FAM | Hs01049883_m1 |
| huMarvelD3 | FAM | Hs00369354_m1 |
| huTricellulin | FAM | Hs00930631_m1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Voges, L.; Weiß, F.; Branco, A.-T.; Fromm, M.; Krug, S.M. Expression and Localization Profiles of Tight Junction Proteins in Immune Cells Depend on Their Activation Status. Int. J. Mol. Sci. 2024, 25, 4861. https://doi.org/10.3390/ijms25094861
Voges L, Weiß F, Branco A-T, Fromm M, Krug SM. Expression and Localization Profiles of Tight Junction Proteins in Immune Cells Depend on Their Activation Status. International Journal of Molecular Sciences. 2024; 25(9):4861. https://doi.org/10.3390/ijms25094861
Chicago/Turabian StyleVoges, Lena, Franziska Weiß, Ana-Teresa Branco, Michael Fromm, and Susanne M. Krug. 2024. "Expression and Localization Profiles of Tight Junction Proteins in Immune Cells Depend on Their Activation Status" International Journal of Molecular Sciences 25, no. 9: 4861. https://doi.org/10.3390/ijms25094861
APA StyleVoges, L., Weiß, F., Branco, A.-T., Fromm, M., & Krug, S. M. (2024). Expression and Localization Profiles of Tight Junction Proteins in Immune Cells Depend on Their Activation Status. International Journal of Molecular Sciences, 25(9), 4861. https://doi.org/10.3390/ijms25094861








