Retinoic Acid-Mediated Control of Energy Metabolism Is Essential for Lung Branching Morphogenesis
Abstract
1. Introduction
2. Results
2.1. Retinoic Acid Signaling Downregulation Decreases Lung Branching and Induces a Cystic-like Phenotype
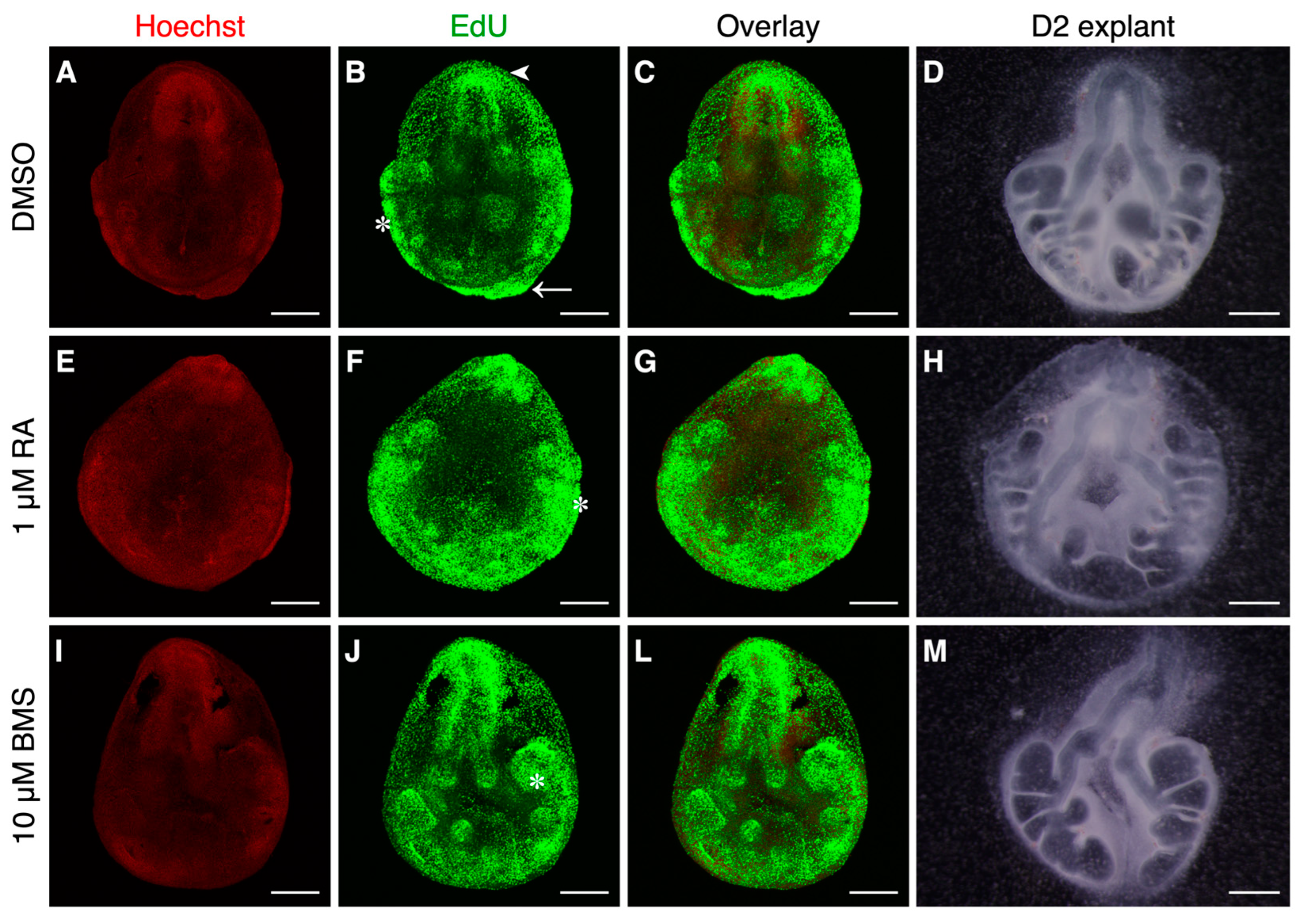
2.2. High Proliferation Is Associated with Active Branching Regions
2.3. Retinoic Acid Signaling Stimulation Requires Less Glucose Consumption
2.4. Retinoic Acid Signaling Controls Pyruvate Metabolism
2.5. Retinoic Acid Signaling Modulates Lactate Dehydrogenase Expression
2.6. Retinoic Acid Signaling Downregulation Increases Mitochondrial Function
2.7. Retinoic Acid Signaling Controls Fatty Acid Metabolism through AMPK
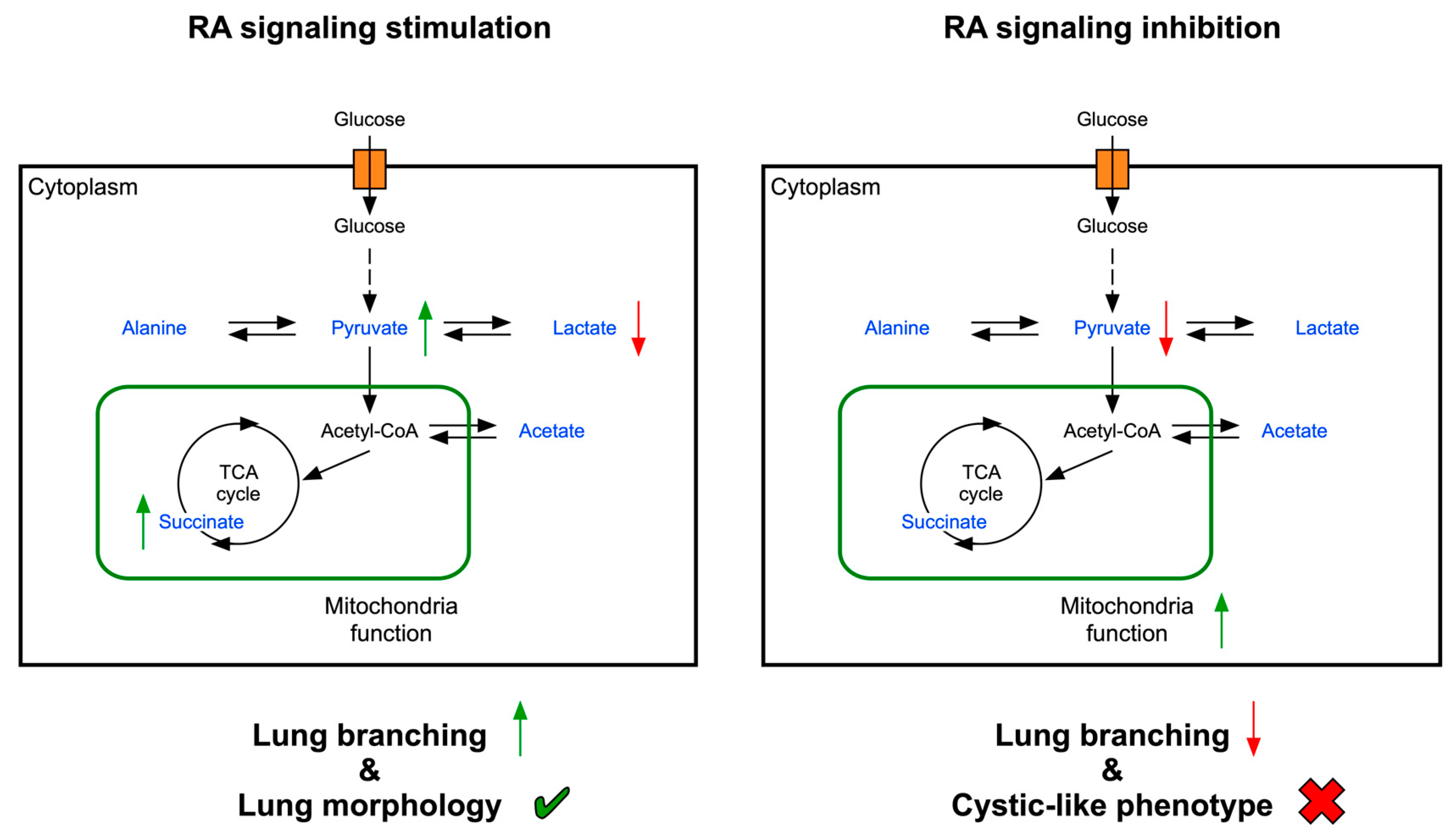
3. Discussion
4. Materials and Methods
4.1. Ethical Statement
4.2. Tissue Collection
4.3. Ex Vivo Lung Explant Culture
4.4. RNA Probes
4.5. Whole-Mount In Situ Hybridization
4.6. 1H-NMR Spectroscopy
4.7. Quantitative PCR
4.8. Western Blot
4.9. Proliferation Assay and Confocal Microscopy
4.10. Seahorse Analysis
4.11. mtDNA Copy Number
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Caldeira, I.; Fernandes-Silva, H.; Machado-Costa, D.; Correia-Pinto, J.; Moura, R.S. Developmental Pathways Underlying Lung Development and Congenital Lung Disorders. Cells 2021, 10, 2987. [Google Scholar] [CrossRef] [PubMed]
- Schittny, J.C. Development of the lung. Cell Tissue Res. 2017, 367, 427–444. [Google Scholar] [CrossRef] [PubMed]
- Harris, K.S.; Zhang, Z.; McManus, M.T.; Harfe, B.D.; Sun, X. Dicer function is essential for lung epithelium morphogenesis. Proc. Natl. Acad. Sci. USA 2006, 103, 2208–2213. [Google Scholar] [CrossRef] [PubMed]
- Gonzaga, S.; Henriques-Coelho, T.; Davey, M.; Zoltick, P.W.; Leite-Moreira, A.F.; Correia-Pinto, J.; Flake, A.W. Cystic adenomatoid malformations are induced by localized FGF10 overexpression in fetal rat lung. Am. J. Respir. Cell Mol. Biol. 2008, 39, 346–355. [Google Scholar] [CrossRef] [PubMed]
- Maina, J.N. A systematic study of the development of the airway (bronchial) system of the avian lung from days 3 to 26 of embryogenesis: A transmission electron microscopic study on the domestic fowl, Gallus gallus variant domesticus. Tissue Cell 2003, 35, 375–391. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.Y.; Varner, V.D.; Nelson, C.M. Apical constriction initiates new bud formation during monopodial branching of the embryonic chicken lung. Development 2013, 140, 3146–3155. [Google Scholar] [CrossRef] [PubMed]
- Maina, J.N. Comparative molecular developmental aspects of the mammalian- and the avian lungs, and the insectan tracheal system by branching morphogenesis: Recent advances and future directions. Front. Zool. 2012, 9, 16. [Google Scholar] [CrossRef]
- Metzger, R.J.; Klein, O.D.; Martin, G.R.; Krasnow, M.A. The branching programme of mouse lung development. Nature 2008, 453, 745–750. [Google Scholar] [CrossRef] [PubMed]
- Moura, R.S.; Correia-Pinto, J. Molecular aspects of avian lung development. In The Biology of the Avian Respiratory System; Maina, J.N., Ed.; Springer: Cham, Switzerland, 2017; pp. 129–146. [Google Scholar]
- Moura, R.S.; Silva-Goncalves, C.; Vaz-Cunha, P.; Correia-Pinto, J. Expression analysis of Shh signaling members in early stages of chick lung development. Histochem. Cell Biol. 2016, 146, 457–466. [Google Scholar] [CrossRef]
- Moura, R.S.; Coutinho-Borges, J.P.; Pacheco, A.P.; Damota, P.O.; Correia-Pinto, J. FGF signaling pathway in the developing chick lung: Expression and inhibition studies. PLoS ONE 2011, 6, e17660. [Google Scholar] [CrossRef]
- Moura, R.S.; Carvalho-Correia, E.; daMota, P.; Correia-Pinto, J. Canonical Wnt signaling activity in early stages of chick lung development. PLoS ONE 2014, 9, e112388. [Google Scholar] [CrossRef] [PubMed]
- Fernandes-Silva, H.; Vaz-Cunha, P.; Barbosa, V.B.; Silva-Goncalves, C.; Correia-Pinto, J.; Moura, R.S. Retinoic acid regulates avian lung branching through a molecular network. Cell. Mol. Life Sci. 2017, 74, 4599–4619. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, T.J.; Duester, G. Mechanisms of retinoic acid signalling and its roles in organ and limb development. Nat. Rev. Mol. Cell Biol. 2015, 16, 110–123. [Google Scholar] [CrossRef] [PubMed]
- Morrisey, E.E.; Hogan, B.L. Preparing for the first breath: Genetic and cellular mechanisms in lung development. Dev. Cell 2010, 18, 8–23. [Google Scholar] [CrossRef] [PubMed]
- Marquez, H.A.; Cardoso, W.V. Vitamin A-retinoid signaling in pulmonary development and disease. Mol. Cell. Pediatr. 2016, 3, 28. [Google Scholar] [CrossRef] [PubMed]
- Ghyselinck, N.B.; Duester, G. Retinoic acid signaling pathways. Development 2019, 146, dev167502. [Google Scholar] [CrossRef] [PubMed]
- Fernandes-Silva, H.; Araujo-Silva, H.; Correia-Pinto, J.; Moura, R.S. Retinoic Acid: A Key Regulator of Lung Development. Biomolecules 2020, 10, 152. [Google Scholar] [CrossRef] [PubMed]
- Marquez, H.A.; Chen, F. Retinoic Acid Signaling and Development of the Respiratory System. Subcell. Biochem. 2020, 95, 151–174. [Google Scholar] [CrossRef] [PubMed]
- Fernandes-Silva, H.; Alves, M.G.; Araujo-Silva, H.; Silva, A.M.; Correia-Pinto, J.; Oliveira, P.F.; Moura, R.S. Lung branching morphogenesis is accompanied by temporal metabolic changes towards a glycolytic preference. Cell Biosci. 2021, 11, 134. [Google Scholar] [CrossRef]
- Yeager, H., Jr.; Massaro, D. Glucose metabolism and glycoprotein synthesis by lung slices. J. Appl. Physiol. 1972, 32, 477–482. [Google Scholar] [CrossRef]
- Fisher, A.B. Normal and pathologic biochemistry of the lung. Environ. Health Perspect. 1976, 16, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Fisher, A.B. Intermediary metabolism of the lung. Environ. Health Perspect. 1984, 55, 149–158. [Google Scholar] [CrossRef] [PubMed]
- Krejci, A.; Tennessen, J.M. Metabolism in time and space—Exploring the frontier of developmental biology. Development 2017, 144, 3193–3198. [Google Scholar] [CrossRef] [PubMed]
- Miyazawa, H.; Aulehla, A. Revisiting the role of metabolism during development. Development 2018, 145, dev131110. [Google Scholar] [CrossRef] [PubMed]
- Teleman, A.A. Metabolism meets development at Wiston House. Development 2016, 143, 3045–3049. [Google Scholar] [CrossRef]
- Cable, J.; Pourquie, O.; Wellen, K.E.; Finley, L.W.S.; Aulehla, A.; Gould, A.P.; Teleman, A.; Tu, W.B.; Garrett, W.S.; Miguel-Aliaga, I.; et al. Metabolic decisions in development and disease—A Keystone Symposia report. Ann. N. Y. Acad. Sci. 2021, 1506, 55–73. [Google Scholar] [CrossRef] [PubMed]
- Slaninova, V.; Krafcikova, M.; Perez-Gomez, R.; Steffal, P.; Trantirek, L.; Bray, S.J.; Krejci, A. Notch stimulates growth by direct regulation of genes involved in the control of glycolysis and the tricarboxylic acid cycle. Open Biol. 2016, 6, 150155. [Google Scholar] [CrossRef] [PubMed]
- Nellas, I.; Iyer, K.V.; Iglesias-Artola, J.M.; Pippel, M.; Nadler, A.; Eaton, S.; Dye, N.A. Hedgehog signaling can enhance glycolytic ATP production in the Drosophila wing disc. EMBO Rep. 2022, 23, e54025. [Google Scholar] [CrossRef]
- Kuwabara, S.; Yamaki, M.; Yu, H.; Itoh, M. Notch signaling regulates the expression of glycolysis-related genes in a context-dependent manner during embryonic development. Biochem. Biophys. Res. Commun. 2018, 503, 803–808. [Google Scholar] [CrossRef]
- Oginuma, M.; Moncuquet, P.; Xiong, F.; Karoly, E.; Chal, J.; Guevorkian, K.; Pourquie, O. A Gradient of Glycolytic Activity Coordinates FGF and Wnt Signaling during Elongation of the Body Axis in Amniote Embryos. Dev. Cell 2017, 40, 342–353. [Google Scholar] [CrossRef]
- Pereira-Terra, P.; Moura, R.S.; Nogueira-Silva, C.; Correia-Pinto, J. Neuroendocrine factors regulate retinoic acid receptors in normal and hypoplastic lung development. J. Physiol. 2015, 593, 3301–3311. [Google Scholar] [CrossRef] [PubMed]
- Schuger, L.; Varani, J.; Mitra, R., Jr.; Gilbride, K. Retinoic acid stimulates mouse lung development by a mechanism involving epithelial-mesenchymal interaction and regulation of epidermal growth factor receptors. Dev. Biol. 1993, 159, 462–473. [Google Scholar] [CrossRef] [PubMed]
- Germain, P.; Gaudon, C.; Pogenberg, V.; Sanglier, S.; Van Dorsselaer, A.; Royer, C.A.; Lazar, M.A.; Bourguet, W.; Gronemeyer, H. Differential action on coregulator interaction defines inverse retinoid agonists and neutral antagonists. Chem. Biol. 2009, 16, 479–489. [Google Scholar] [CrossRef] [PubMed]
- Takayama, M.; Miyatake, K.; Nishida, E. Identification and characterization of retinoic acid-responsive genes in mouse kidney development. Genes Cells 2014, 19, 637–649. [Google Scholar] [CrossRef] [PubMed]
- Nadendla, E.; Teyssier, C.; Delfosse, V.; Vivat, V.; Krishnasamy, G.; Gronemeyer, H.; Bourguet, W.; Germain, P. An Unexpected Mode of Binding Defines BMS948 as a Full Retinoic Acid Receptor beta (RARbeta, NR1B2) Selective Agonist. PLoS ONE 2015, 10, e0123195. [Google Scholar] [CrossRef] [PubMed]
- Elgamal, R.M.; Bell, G.I.; Krause, S.C.T.; Hess, D.A. BMS 493 Modulates Retinoic Acid-Induced Differentiation During Expansion of Human Hematopoietic Progenitor Cells for Islet Regeneration. Stem Cells Dev. 2018, 27, 1062–1075. [Google Scholar] [CrossRef] [PubMed]
- Mendelsohn, C.; Lohnes, D.; Decimo, D.; Lufkin, T.; LeMeur, M.; Chambon, P.; Mark, M. Function of the retinoic acid receptors (RARs) during development (II). Multiple abnormalities at various stages of organogenesis in RAR double mutants. Development 1994, 120, 2749–2771. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D.; Cox, M. Lehninger Principles of Biochemistry, 5th ed.; Worth Publishers: New York, NY, USA, 2008. [Google Scholar]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef] [PubMed]
- Murphy, M.P.; O’Neill, L.A.J. Krebs Cycle Reimagined: The Emerging Roles of Succinate and Itaconate as Signal Transducers. Cell 2018, 174, 780–784. [Google Scholar] [CrossRef]
- Guo, Y.; Cho, S.W.; Saxena, D.; Li, X. Multifaceted Actions of Succinate as a Signaling Transmitter Vary with Its Cellular Locations. Endocrinol. Metab. 2020, 35, 36–43. [Google Scholar] [CrossRef]
- Ventura-Clapier, R.; Garnier, A.; Veksler, V. Transcriptional control of mitochondrial biogenesis: The central role of PGC-1alpha. Cardiovasc. Res. 2008, 79, 208–217. [Google Scholar] [CrossRef] [PubMed]
- Gureev, A.P.; Shaforostova, E.A.; Popov, V.N. Regulation of Mitochondrial Biogenesis as a Way for Active Longevity: Interaction Between the Nrf2 and PGC-1alpha Signaling Pathways. Front. Genet. 2019, 10, 435. [Google Scholar] [CrossRef] [PubMed]
- Amengual, J.; Ribot, J.; Bonet, M.L.; Palou, A. Retinoic acid treatment enhances lipid oxidation and inhibits lipid biosynthesis capacities in the liver of mice. Cell. Physiol. Biochem. 2010, 25, 657–666. [Google Scholar] [CrossRef] [PubMed]
- Amengual, J.; Ribot, J.; Bonet, M.L.; Palou, A. Retinoic acid treatment increases lipid oxidation capacity in skeletal muscle of mice. Obesity 2008, 16, 585–591. [Google Scholar] [CrossRef] [PubMed]
- Amengual, J.; Garcia-Carrizo, F.J.; Arreguin, A.; Musinovic, H.; Granados, N.; Palou, A.; Bonet, M.L.; Ribot, J. Retinoic Acid Increases Fatty Acid Oxidation and Irisin Expression in Skeletal Muscle Cells and Impacts Irisin In Vivo. Cell. Physiol. Biochem. 2018, 46, 187–202. [Google Scholar] [CrossRef] [PubMed]
- Garcia, D.; Shaw, R.J. AMPK: Mechanisms of Cellular Energy Sensing and Restoration of Metabolic Balance. Mol. Cell 2017, 66, 789–800. [Google Scholar] [CrossRef] [PubMed]
- Herzig, S.; Shaw, R.J. AMPK: Guardian of metabolism and mitochondrial homeostasis. Nat. Rev. Mol. Cell Biol. 2018, 19, 121–135. [Google Scholar] [CrossRef] [PubMed]
- Malpel, S.; Mendelsohn, C.; Cardoso, W.V. Regulation of retinoic acid signaling during lung morphogenesis. Development 2000, 127, 3057–3067. [Google Scholar] [CrossRef] [PubMed]
- Bellusci, S.; Grindley, J.; Emoto, H.; Itoh, N.; Hogan, B.L. Fibroblast growth factor 10 (FGF10) and branching morphogenesis in the embryonic mouse lung. Development 1997, 124, 4867–4878. [Google Scholar] [CrossRef]
- Carraro, G.; del Moral, P.M.; Warburton, D. Mouse embryonic lung culture, a system to evaluate the molecular mechanisms of branching. J. Vis. Exp. 2010, 40, e2035. [Google Scholar] [CrossRef]
- Yeganeh, B.; Bilodeau, C.; Post, M. Explant Culture for Studying Lung Development. Methods Mol. Biol. 2018, 1752, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Deng, B.; Jiang, X.; Cai, M.; Liu, N.; Zhang, S.; Tan, Y.; Huang, G.; Jin, W.; Liu, B.; et al. All-Trans-Retinoic Acid Suppresses Neointimal Hyperplasia and Inhibits Vascular Smooth Muscle Cell Proliferation and Migration via Activation of AMPK Signaling Pathway. Front. Pharmacol. 2019, 10, 485. [Google Scholar] [CrossRef] [PubMed]
- Ishijima, N.; Kanki, K.; Shimizu, H.; Shiota, G. Activation of AMP-activated protein kinase by retinoic acid sensitizes hepatocellular carcinoma cells to apoptosis induced by sorafenib. Cancer Sci. 2015, 106, 567–575. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.M.; Lee, J.O.; Jung, J.H.; Kim, J.H.; Park, S.H.; Park, J.M.; Kim, E.K.; Suh, P.G.; Kim, H.S. Retinoic acid leads to cytoskeletal rearrangement through AMPK-Rac1 and stimulates glucose uptake through AMPK-p38 MAPK in skeletal muscle cells. J. Biol. Chem. 2008, 283, 33969–33974. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xu, S.; Mihaylova, M.M.; Zheng, B.; Hou, X.; Jiang, B.; Park, O.; Luo, Z.; Lefai, E.; Shyy, J.Y.; et al. AMPK phosphorylates and inhibits SREBP activity to attenuate hepatic steatosis and atherosclerosis in diet-induced insulin-resistant mice. Cell Metab. 2011, 13, 376–388. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Zhang, M.; Wang, Y.; Kadambi, P.; Dave, V.; Lu, L.J.; Whitsett, J.A. A systems approach to mapping transcriptional networks controlling surfactant homeostasis. BMC Genom. 2010, 11, 451. [Google Scholar] [CrossRef] [PubMed]
- Michaille, J.J.; Kanzler, B.; Blanchet, S.; Garnier, J.M.; Dhouailly, D. Characterization of cDNAs encoding two chick retinoic acid receptor alpha isoforms and distribution of retinoic acid receptor alpha, beta and gamma transcripts during chick skin development. Int. J. Dev. Biol. 1995, 39, 587–596. [Google Scholar] [PubMed]
- Moura, R.S. Retinoic Acid as a Modulator of Proximal-Distal Patterning and Branching Morphogenesis of the Avian Lung. In Retinoid and Rexinoid Signaling; Springer: New York, NY, USA, 2019; pp. 209–224. [Google Scholar] [CrossRef]
- Alves, M.G.; Oliveira, P.F.; Martins, F.O.; Oliveira, P.J.; Carvalho, R.A. Gender-dependent metabolic remodeling during heart preservation in cardioplegic celsior and histidine buffer solution. J. Cardiovasc. Pharmacol. 2012, 60, 227–233. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Kling, D.E.; Lorenzo, H.K.; Trbovich, A.M.; Kinane, T.B.; Donahoe, P.K.; Schnitzer, J.J. MEK-1/2 inhibition reduces branching morphogenesis and causes mesenchymal cell apoptosis in fetal rat lungs. Am. J. Physiol. Lung Cell. Mol. Physiol. 2002, 282, L370–L378. [Google Scholar] [CrossRef]
- Martins, A.D.; Monteiro, M.P.; Silva, B.M.; Barros, A.; Sousa, M.; Carvalho, R.A.; Oliveira, P.F.; Alves, M.G. Metabolic dynamics of human Sertoli cells are differentially modulated by physiological and pharmacological concentrations of GLP-1. Toxicol. Appl. Pharmacol. 2019, 362, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Jin, L.; Long, K.; Tang, Q.; Ma, J.; Wang, X.; Zhu, L.; Jiang, A.; Tang, G.; Jiang, Y.; et al. Analysis of mitochondrial DNA sequence and copy number variation across five high-altitude species and their low-altitude relatives. Mitochondrial DNA B Resour. 2018, 3, 847–851. [Google Scholar] [CrossRef] [PubMed]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernandes-Silva, H.; Alves, M.G.; Garcez, M.R.; Correia-Pinto, J.; Oliveira, P.F.; Homem, C.C.F.; Moura, R.S. Retinoic Acid-Mediated Control of Energy Metabolism Is Essential for Lung Branching Morphogenesis. Int. J. Mol. Sci. 2024, 25, 5054. https://doi.org/10.3390/ijms25095054
Fernandes-Silva H, Alves MG, Garcez MR, Correia-Pinto J, Oliveira PF, Homem CCF, Moura RS. Retinoic Acid-Mediated Control of Energy Metabolism Is Essential for Lung Branching Morphogenesis. International Journal of Molecular Sciences. 2024; 25(9):5054. https://doi.org/10.3390/ijms25095054
Chicago/Turabian StyleFernandes-Silva, Hugo, Marco G. Alves, Marcia R. Garcez, Jorge Correia-Pinto, Pedro F. Oliveira, Catarina C. F. Homem, and Rute S. Moura. 2024. "Retinoic Acid-Mediated Control of Energy Metabolism Is Essential for Lung Branching Morphogenesis" International Journal of Molecular Sciences 25, no. 9: 5054. https://doi.org/10.3390/ijms25095054
APA StyleFernandes-Silva, H., Alves, M. G., Garcez, M. R., Correia-Pinto, J., Oliveira, P. F., Homem, C. C. F., & Moura, R. S. (2024). Retinoic Acid-Mediated Control of Energy Metabolism Is Essential for Lung Branching Morphogenesis. International Journal of Molecular Sciences, 25(9), 5054. https://doi.org/10.3390/ijms25095054






