The Use of Dansyl Chloride to Probe Protein Structure and Dynamics
Abstract
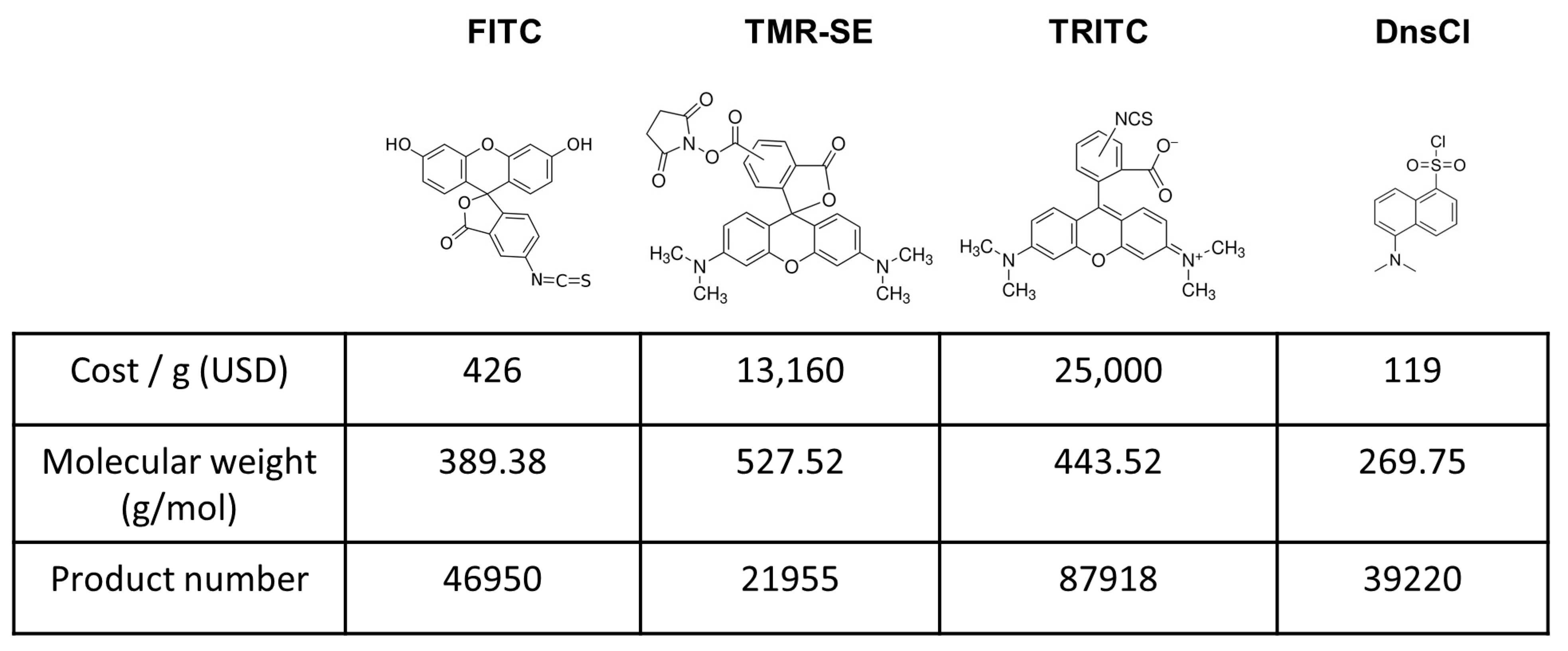
1. Introduction
2. Results
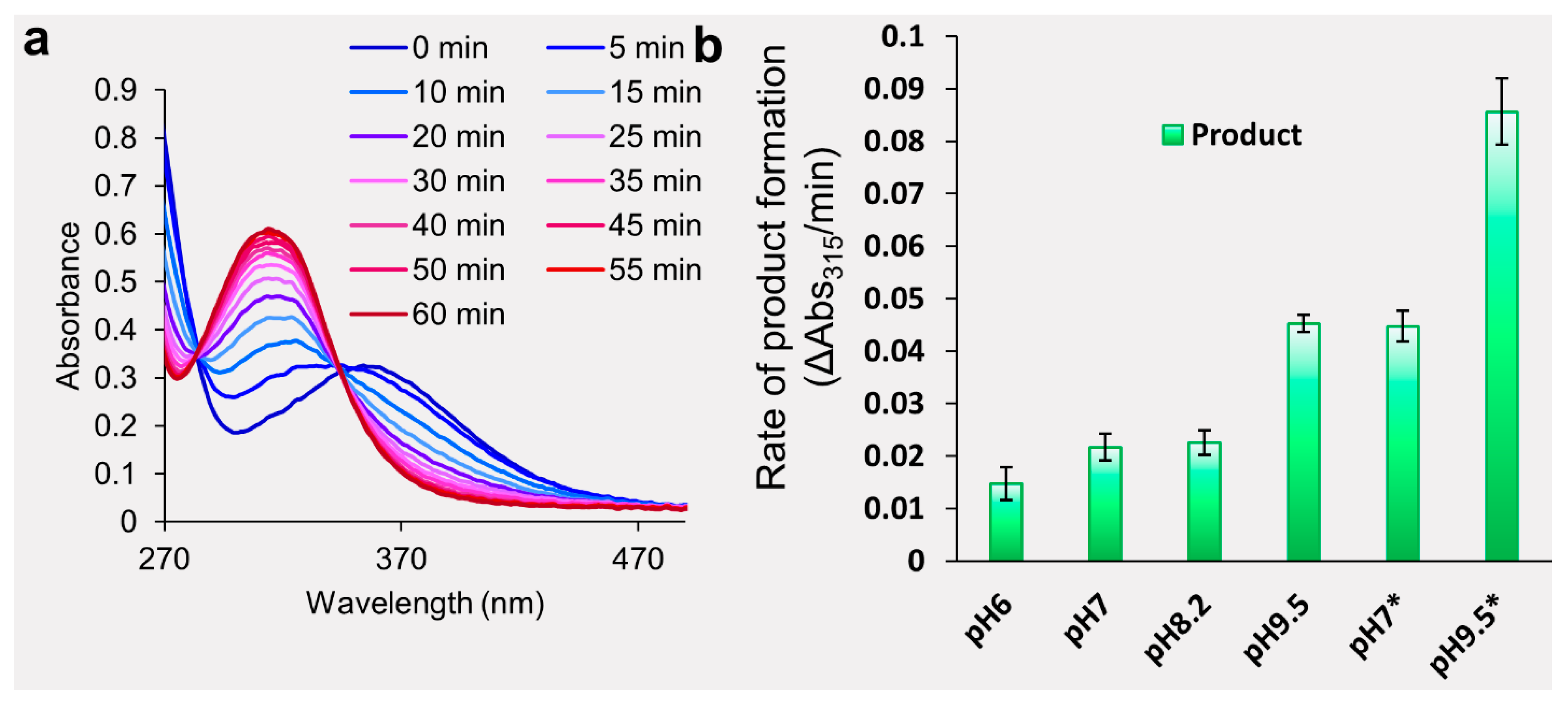
2.1. Evaluation of Dansyl Chloride Stability in Aqueous Solutions
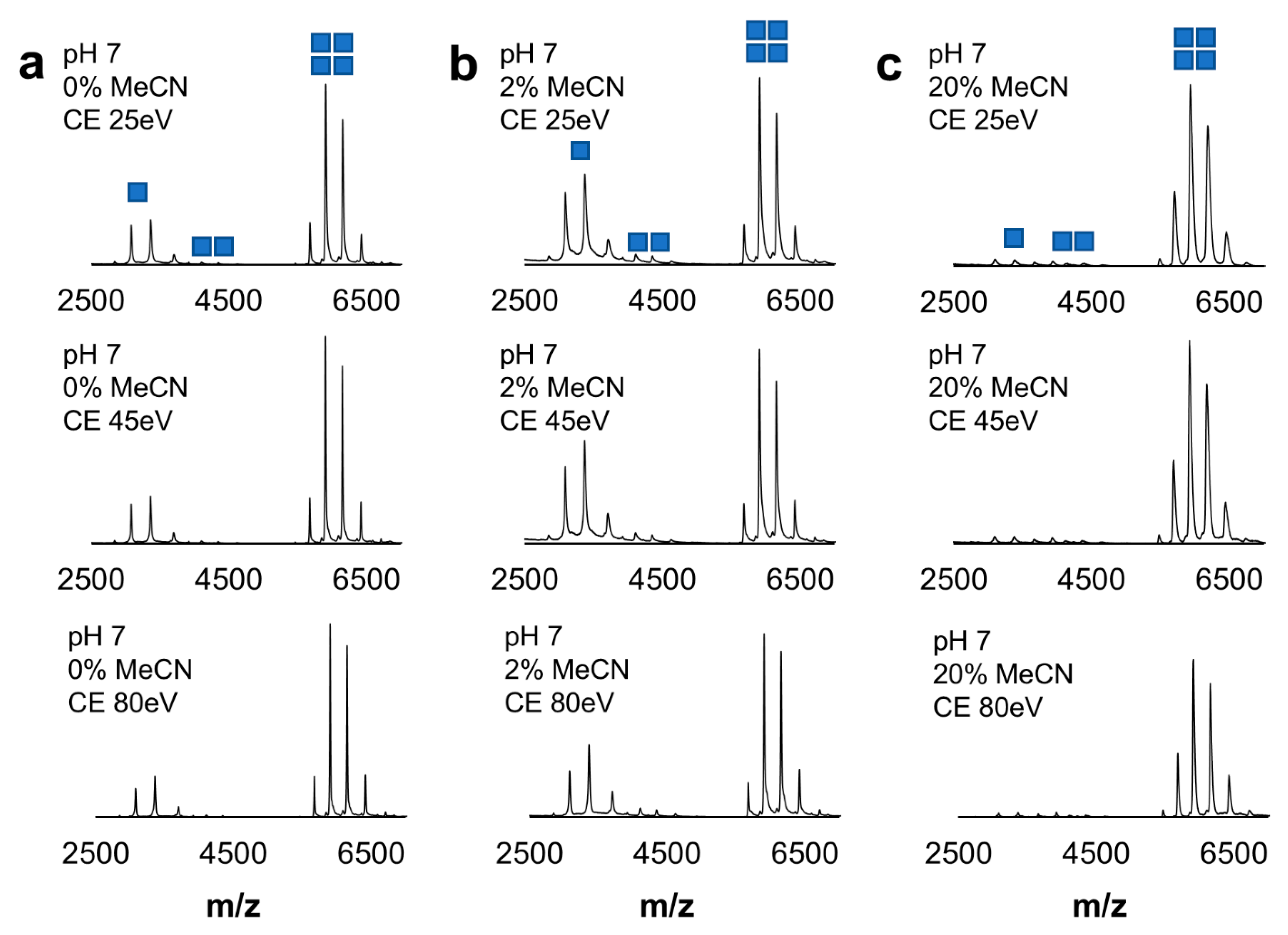
2.2. Characterization of Model Protein Systems Stability in Labeling Conditions
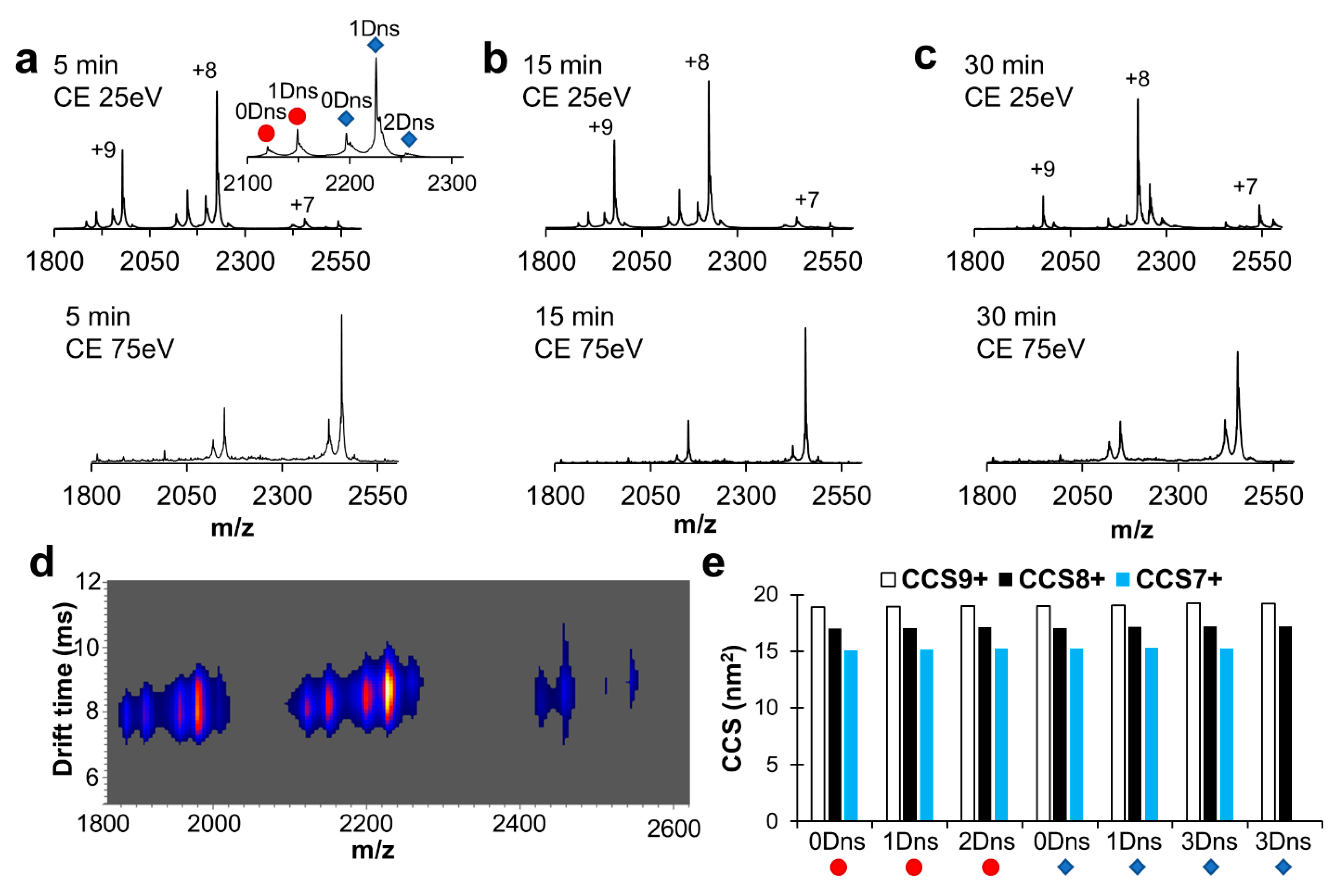
2.3. Robustness of Protein Dansylation Reaction
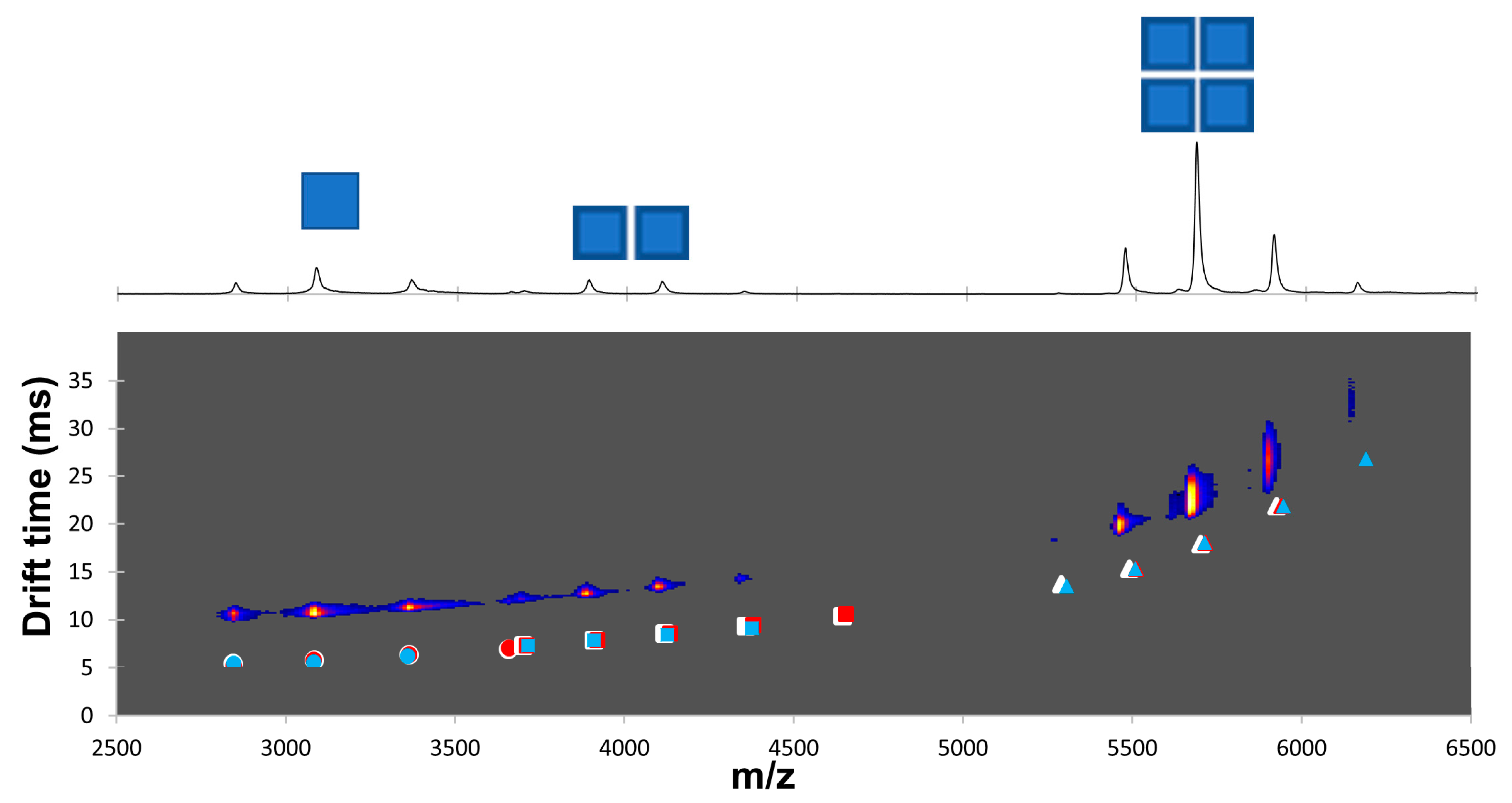
2.4. Dansylation-Induced Structural Changes
3. Discussion
4. Materials and Methods
4.1. Chemical Stability of the Dansyl Chloride Probe
4.2. Protein Analysis with Native Mass Spectrometry and Ion Mobility Mass Spectrometry
4.3. Evaluation of Labeling Conditions
4.4. Stability of Model Proteins After Label Incorporation
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Morris, R.; Black, K.A.; Stollar, E.J. Uncovering Protein Function: From Classification to Complexes. Essays Biochem. 2022, 66, 255–285. [Google Scholar] [CrossRef]
- Alberts, B.; Johnson, A.; Lewis, J.; Raff, M.; Roberts, K.; Walter, P. Analyzing Protein Structure and Function; National Library of Medicine: Bethesda, MD, USA, 2002. [Google Scholar]
- Mendoza, V.L.; Vachet, R.W. Probing Protein Structure by Amino Acid-Specific Covalent Labeling and Mass Spectrometry. Mass. Spectrom. Rev. 2009, 28, 785. [Google Scholar] [CrossRef]
- Limpikirati, P.; Liu, T.; Vachet, R.W. Covalent Labeling-Mass Spectrometry with Non-Specific Reagents for Studying Protein Structure and Interactions. Methods 2018, 144, 79–93. [Google Scholar] [CrossRef] [PubMed]
- Dill, K.A.; Ozkan, S.B.; Shell, M.S.; Weikl, T.R. The Protein Folding Problem. Annu. Rev. Biophys. 2008, 37, 289–316. [Google Scholar] [CrossRef] [PubMed]
- Tantipanjaporn, A.; Wong, M.K. Development and Recent Advances in Lysine and N-Terminal Bioconjugation for Peptides and Proteins. Molecules 2023, 28, 1083. [Google Scholar] [CrossRef]
- Przybylski, M.; Glocker, M.O.; Nestel, U.; Schnaible, V.; Blüggel, M.; Diederichs, K.; Weckesser, J.; Schad, M.; Schmid, A.; Welte, W.; et al. X-Ray Crystallographic and Mass Spectrometric Structure Determination and Functional Characterization of Succinylated Porin from Rhodobacter Capsulatus: Implications for Ion Selectivity and Single-Channel Conductance. Protein Sci. 1996, 5, 1477–1489. [Google Scholar] [CrossRef] [PubMed]
- Legrand, D.; Mazurier, J.; Maes, P.; Rochard, E.; Montreuil, J.; Spik, G. Inhibition of the Specific Binding of Human Lactotransferrin to Human Peripheral-Blood Phytohaemagglutinin-Stimulated Lymphocytes by Fluorescein Labelling and Location of the Binding Site. Biochem. J. 1991, 276, 733–738. [Google Scholar] [CrossRef] [PubMed]
- Jacobsen, M.T.; Fairhead, M.; Fogelstrand, P.; Howarth, M. Amine Landscaping to Maximize Protein-Dye Fluorescence and Ultrastable Protein-Ligand Interaction. Cell Chem. Biol. 2017, 24, 1040–1047.e4. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Medintz, I.L.; Goldman, E.R.; Lassman, M.E.; Mauro, J.M. A Fluorescence Resonance Energy Transfer Sensor Based on Maltose Binding Protein. Bioconjugate Chem. 2003, 14, 909–918. [Google Scholar] [CrossRef]
- Shrestha, D.; Jenei, A.; Nagy, P.; Vereb, G.; Szöllősi, J. Understanding FRET as a Research Tool for Cellular Studies. Int. J. Mol. Sci. 2015, 16, 6718–6756. [Google Scholar] [CrossRef]
- Kim, D.; Cho, S.W.; Jun, Y.W.; Ahn, K.H. Fluorescent Labeling of Lysine Residues in Protein Using 8-Thiomethyl-BODIPY. Bull. Korean Chem. Soc. 2017, 38, 995–996. [Google Scholar] [CrossRef]
- Sánchez-Rico, C.; Voith von Voithenberg, L.; Warner, L.; Lamb, D.C.; Sattler, M. Effects of Fluorophore Attachment on Protein Conformation and Dynamics Studied by SpFRET and NMR Spectroscopy. Chem. A Eur. J. 2017, 23, 14267–14277. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.K.; Tang, J.; Taylor, D.; Bothner, B.; Johnson, J.E. Small Compounds Targeted to Subunit Interfaces Arrest Maturation in a Nonenveloped, Icosahedral Animal Virus. J. Virol. 2004, 78, 7208–7216. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Seiler, N. Use of the Dansyl Reaction in Biochemical Analysis. In Methods of Biochemical Analysis; Glick, D., Ed.; Interscience Publishers: Palo Alto, CA, USA, 1970; Volume 18, pp. 259–338. [Google Scholar]
- Gros, C.; Labouesse, B. Study of the Dansylation Reaction of Amino Acids, Peptides and Proteins. Eur. J. Biochem. 1969, 7, 463–470. [Google Scholar] [CrossRef]
- Cheney, A.M.; Costello, S.M.; Pinkham, N.V.; Waldum, A.; Broadaway, S.C.; Cotrina-Vidal, M.; Mergy, M.; Tripet, B.; Kominsky, D.J.; Grifka-Walk, H.M.; et al. Gut Microbiome Dysbiosis Drives Metabolic Dysfunction in Familial Dysautonomia. Nat. Commun. 2023, 14, 218. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Alowaifeer, A.; Kerner, P.; Balasubramanian, N.; Patterson, A.; Christian, W.; Tarver, A.; Dore, J.E.; Hatzenpichler, R.; Bothner, B.; et al. Aerobic Bacterial Methane Synthesis. Proc. Natl. Acad. Sci. USA 2021, 118, e2019229118. [Google Scholar] [CrossRef] [PubMed]
- Harris, E.L.V. Identification of N-Terminal Amino Acids by High—Performance Liquid Chromatography. In Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 1988; Volume 3, pp. 49–56. [Google Scholar] [CrossRef]
- Takeuchi, T. HPLC of Amino Acids as Dansyl and Dabsyl Derivatives. J. Chromatogr. Libr. 2005, 70, 229–241. [Google Scholar] [CrossRef]
- Walker, J.M. The Dansyl-Edman Method for Peptide Sequencing. In Experiments in Molecular Biology; Humana Press: Totowa, NJ, USA, 1984; Volume 1, pp. 213–220. [Google Scholar] [CrossRef]
- Smith, B.J.; Walker, J.M. The Dansyl Method for Identifying N-Terminal Amino Acids. In Protein Sequencing Protocols; Humana Press: Totowa, NJ, USA, 2003; pp. 189–196. [Google Scholar] [CrossRef]
- Schut, G.J.; Mohamed-Raseek, N.; Tokmina-Lukaszewska, M.; Mulder, D.W.; Nguyen, D.M.N.; Lipscomb, G.L.; Hoben, J.P.; Patterson, A.; Lubner, C.E.; King, P.W.; et al. The Catalytic Mechanism of Electron-Bifurcating Electron Transfer Flavoproteins (ETFs) Involves an Intermediary Complex with NAD+. J. Biol. Chem. 2019, 294, 3271–3283. [Google Scholar] [CrossRef]
- Brown, K.A.; Guo, Z.; Tokmina-Lukaszewska, M.; Scott, L.W.; Lubner, C.E.; Smolinski, S.; Mulder, D.W.; Bothner, B.; King, P.W. The Oxygen Reduction Reaction Catalyzed by Synechocystis Sp. PCC 6803 Flavodiiron Proteins. Sustain. Energy Fuels 2019, 3, 3191–3200. [Google Scholar] [CrossRef]
- Kosoy, A.; Möller, C.; Perdomo, D.; Bubis, J. Chemical Modification of Transducin with Dansyl Chloride Hinders Its Binding to Light-Activated Rhodopsin. J. Biochem. Mol. Biol. 2004, 37, 260–267. [Google Scholar] [CrossRef] [PubMed]
- Patrascu, B.; Mocanu, S.; Coman, A.; Madalan, A.M.; Popescu, C.; Paun, A.; Matache, M.; Ionita, P. Synthesis of Fluorescent Dansyl Derivatives of Methoxyamine and Diphenylhydrazine as Free Radical Precursors. Int. J. Mol. Sci. 2020, 21, 3559. [Google Scholar] [CrossRef]
- Roy-Lachapelle, A.; Solliec, M.; Sauvé, S. Determination of BMAA and Three Alkaloid Cyanotoxins in Lake Water Using Dansyl Chloride Derivatization and High-Resolution Mass Spectrometry. Anal. Bioanal. Chem. 2015, 407, 5487–5501. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, Y.J.; Chien, K.Y.; Chang, C.M.; Hung, C.Y.; Li, L.; Chiang, W.F.; Lee, C.C.; Chang, C.H.; Chang, Y.H.; Yu, J.S.; et al. Development of a Method for Dansylation of Metabolites Using Organic Solvent—Compatible Buffer Systems for Amine/Phenol Submetabolome Analysis. Anal. Chim. Acta 2022, 1189, 339218. [Google Scholar] [CrossRef] [PubMed]
- Mantoanelli, J.O.F.; Gonçalves, L.M.; Pereira, E.A. Dansyl Chloride as a Derivatizing Agent for the Analysis of Biogenic Amines by CZE-UV. Chromatographia 2020, 83, 767–778. [Google Scholar] [CrossRef]
- Cal, P.M.S.D.; Vicente, J.B.; Pires, E.; Coelho, A.V.; Veiros, L.F.; Cordeiro, C.; Gois, P.M.P. Iminoboronates: A New Strategy for Reversible Protein Modification. J. Am. Chem. Soc. 2012, 134, 10299–10305. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Li, L. Dansylhydrazine Isotope Labeling LC-MS for Comprehensive Carboxylic Acid Submetabolome Profiling. Anal. Chem. 2018, 90, 13514–13522. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.I. Fluorimetric Studies of Actin Labeled with Dansyl Aziridine. Arch. Biochem. Biophys. 1978, 185, 285–299. [Google Scholar] [CrossRef]
- Luo, M.L.; Jackson, R.N.; Denny, S.R.; Tokmina-Lukaszewska, M.; Maksimchuk, K.R.; Lin, W.; Bothner, B.; Wiedenheft, B.; Beisel, C.L. The CRISPR RNA-Guided Surveillance Complex in Escherichia Coli Accommodates Extended RNA Spacers. Nucleic Acids Res. 2016, 44, 7385–7394. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bush, M.F.; Hall, Z.; Giles, K.; Hoyes, J.; Robinson, C.V.; Ruotolo, B.T. Collision Cross Sections of Proteins and Their Complexes: A Calibration Framework and Database for Gas-Phase Structural Biology. Anal. Chem. 2010, 82, 9557–9565. [Google Scholar] [CrossRef] [PubMed]
- Bush Lab. Collision Cross Section Database. Available online: https://depts.washington.edu/bushlab/ccsdatabase/ (accessed on 3 May 2020).


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Larson, J.; Tokmina-Lukaszewska, M.; Malone, J.; Hasenoehrl, E.J.; Kelly, W.; Fang, X.; White, A.; Patterson, A.; Bothner, B. The Use of Dansyl Chloride to Probe Protein Structure and Dynamics. Int. J. Mol. Sci. 2025, 26, 456. https://doi.org/10.3390/ijms26020456
Larson J, Tokmina-Lukaszewska M, Malone J, Hasenoehrl EJ, Kelly W, Fang X, White A, Patterson A, Bothner B. The Use of Dansyl Chloride to Probe Protein Structure and Dynamics. International Journal of Molecular Sciences. 2025; 26(2):456. https://doi.org/10.3390/ijms26020456
Chicago/Turabian StyleLarson, James, Monika Tokmina-Lukaszewska, Jadyn Malone, Ethan J. Hasenoehrl, Will Kelly, Xuelan Fang, Aidan White, Angela Patterson, and Brian Bothner. 2025. "The Use of Dansyl Chloride to Probe Protein Structure and Dynamics" International Journal of Molecular Sciences 26, no. 2: 456. https://doi.org/10.3390/ijms26020456
APA StyleLarson, J., Tokmina-Lukaszewska, M., Malone, J., Hasenoehrl, E. J., Kelly, W., Fang, X., White, A., Patterson, A., & Bothner, B. (2025). The Use of Dansyl Chloride to Probe Protein Structure and Dynamics. International Journal of Molecular Sciences, 26(2), 456. https://doi.org/10.3390/ijms26020456






