The AP2/ERF Transcription Factor ERF56 Negatively Regulating Nitrate-Dependent Plant Growth in Arabidopsis
Abstract
1. Introduction
2. Results
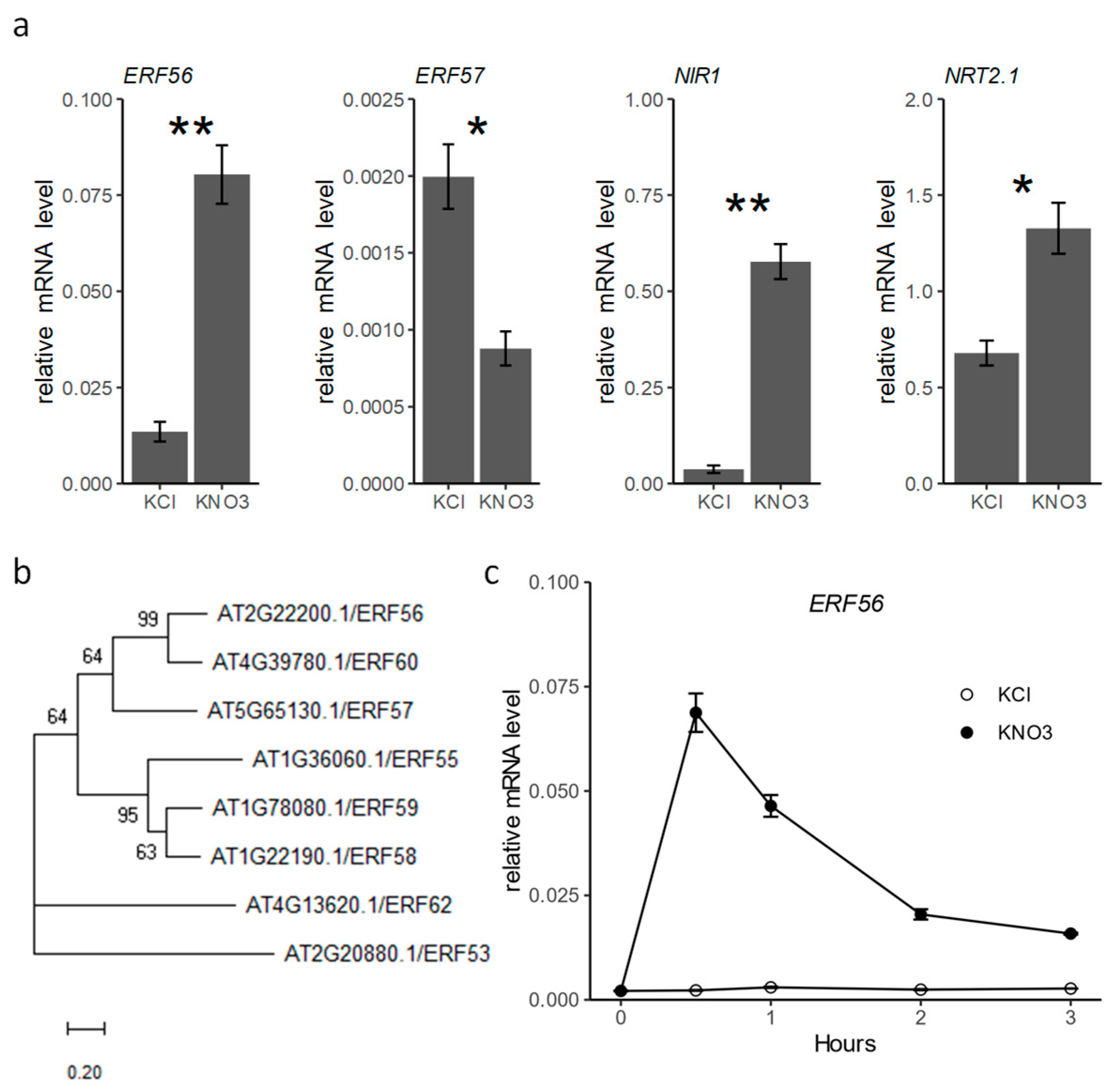
2.1. Transcription of ERF56 Responds Quickly to Nitrate Provision
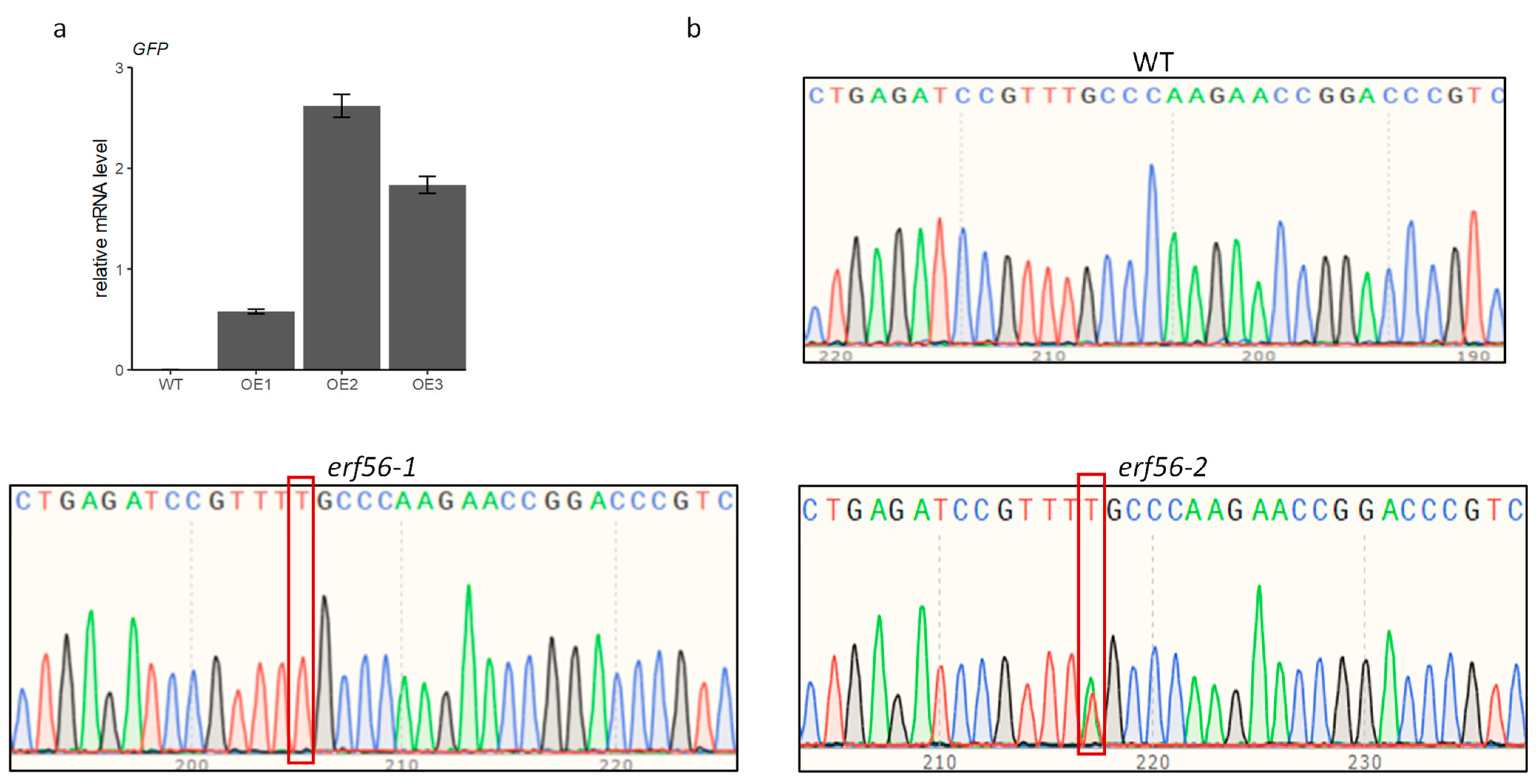
2.2. ERF56 Represses Nitrate-Dependent Plant Growth
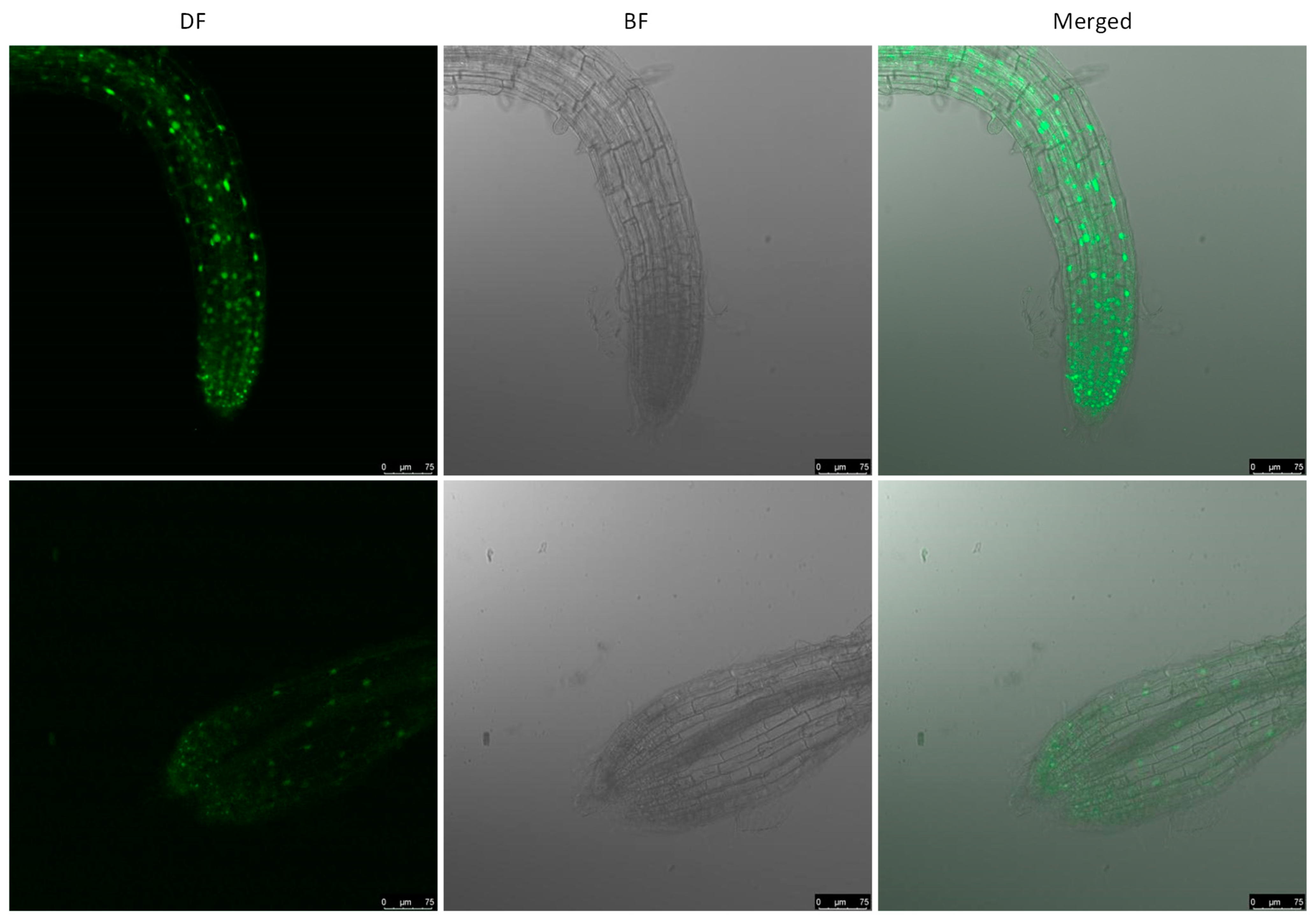
2.3. ERF56 Is a Nucleus-Localized Protein and Is Mainly Expressed in the Maturation Region of Roots and Anthers
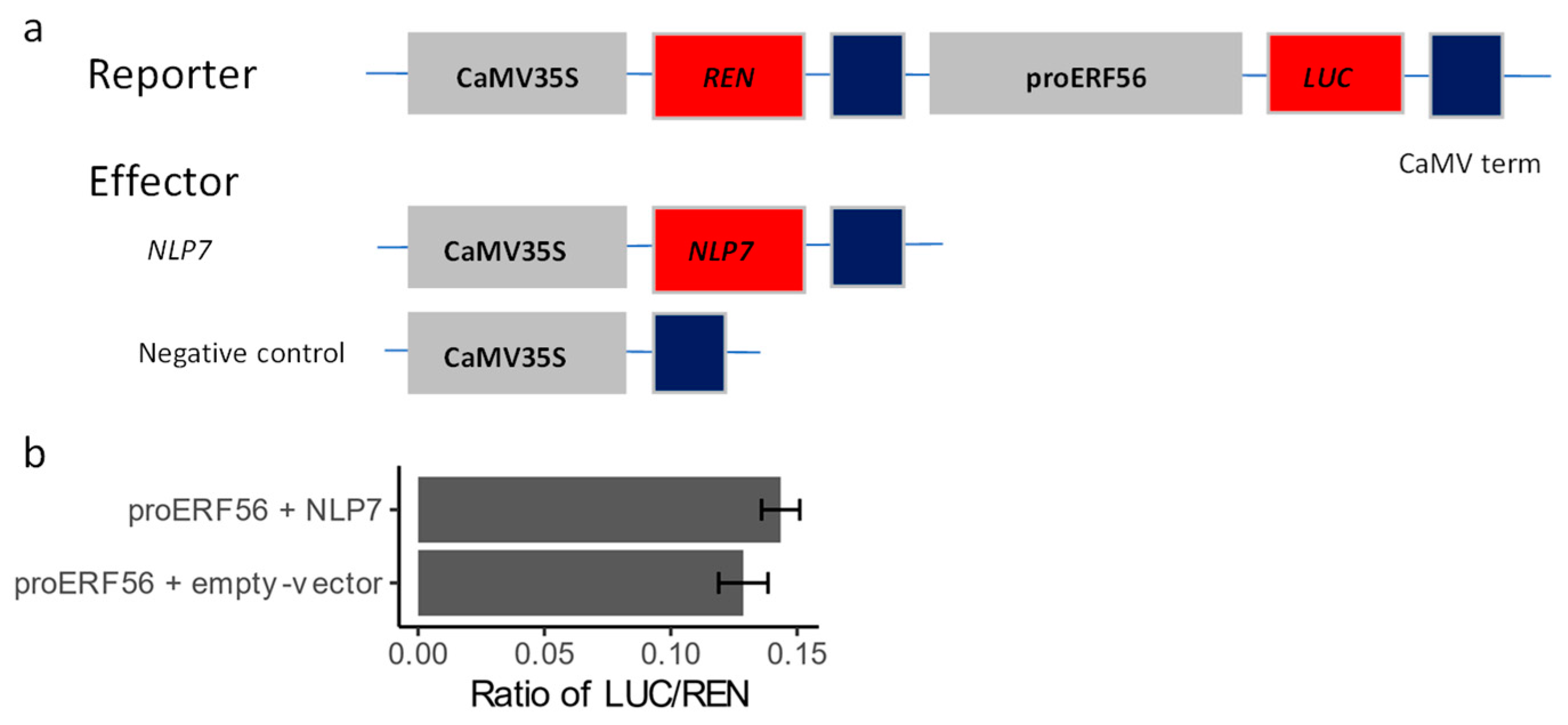
2.4. NLP7 Might Not Directly Target the Promoter of ERF56
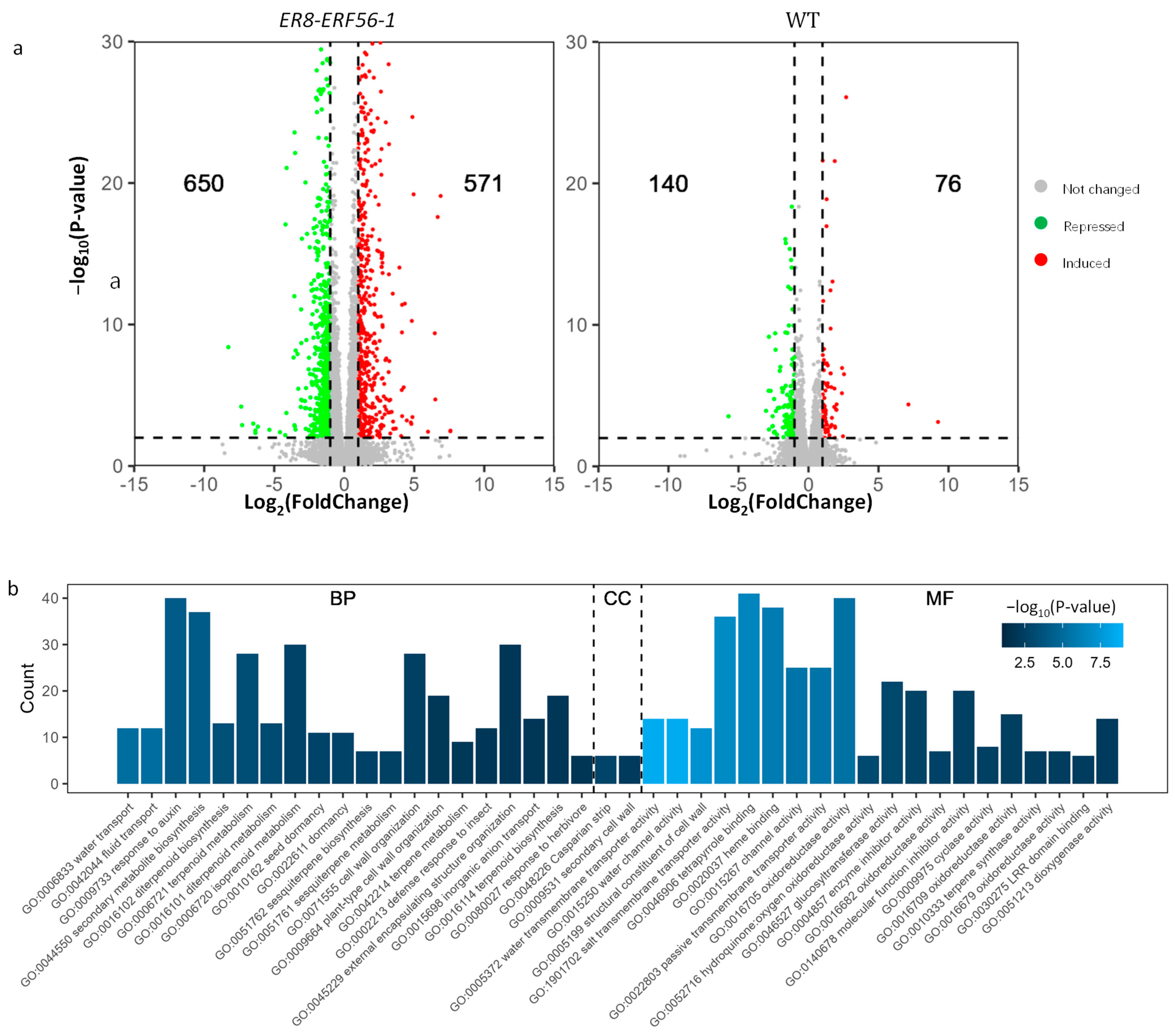
2.5. Genes Targeted by ERF56
2.6. Candidate Cis-Regulatory Elements Targeted by ERF56
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Genotyping of Erf56 Mutants
4.3. Growth Condition and Treatments
4.4. Root Growth Measurements
4.5. In Silico Analysis
4.6. Phylogenetic Analysis
4.7. RNA Extraction, Reverse Transcription, and Quantitative PCR
4.8. Transcriptome Analysis
4.9. Histochemical GUS Staining and Confocal Microscopy Analysis
4.10. Dual-Luciferase Assays
4.11. Cis-Regulatory Element Discovery
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Raun, W.R.; Johnson, G.V. Improving Nitrogen Use Efficiency for Cereal Production. Agron. J. 1999, 91, 357–363. [Google Scholar] [CrossRef]
- Dechorgnat, J.; Nguyen, C.T.; Armengaud, P.; Jossier, M.; Diatloff, E.; Filleur, S.; Daniel-Vedele, F. From the Soil to the Seeds: The Long Journey of Nitrate in Plants. J. Exp. Bot. 2011, 62, 1349–1359. [Google Scholar] [CrossRef] [PubMed]
- Tsay, Y.F.; Schroeder, J.I.; Feldmann, K.A.; Crawford, N.M. The Herbicide Sensitivity Gene CHL1 of Arabidopsis Encodes a Nitrate-Inducible Nitrate Transporter. Cell 1993, 72, 705–713. [Google Scholar] [CrossRef]
- Ho, C.-H.; Lin, S.-H.; Hu, H.-C.; Tsay, Y.-F. CHL1 Functions as a Nitrate Sensor in Plants. Cell 2009, 138, 1184–1194. [Google Scholar] [CrossRef]
- Riveras, E.; Alvarez, J.M.; Vidal, E.A.; Oses, C.; Vega, A.; Gutiérrez, R.A. The Calcium Ion Is a Second Messenger in the Nitrate Signaling Pathway of Arabidopsis. Plant Physiol. 2015, 169, 1397–1404. [Google Scholar] [CrossRef]
- Liu, K.-H.; Niu, Y.; Konishi, M.; Wu, Y.; Du, H.; Sun Chung, H.; Li, L.; Boudsocq, M.; McCormack, M.; Maekawa, S.; et al. Discovery of Nitrate-CPK-NLP Signalling in Central Nutrient-Growth Networks. Nature 2017, 545, 311–316. [Google Scholar] [CrossRef] [PubMed]
- Konishi, M.; Yanagisawa, S. Arabidopsis NIN-like Transcription Factors Have a Central Role in Nitrate Signalling. Nat. Commun. 2013, 4, 1617. [Google Scholar] [CrossRef] [PubMed]
- Marchive, C.; Roudier, F.; Castaings, L.; Bréhaut, V.; Blondet, E.; Colot, V.; Meyer, C.; Krapp, A. Nuclear Retention of the Transcription Factor NLP7 Orchestrates the Early Response to Nitrate in Plants. Nat. Commun. 2013, 4, 1713. [Google Scholar] [CrossRef]
- Konishi, M.; Okitsu, T.; Yanagisawa, S. Nitrate-Responsive NIN-like Protein Transcription Factors Perform Unique and Redundant Roles in Arabidopsis. J. Exp. Bot. 2021, 72, 5735–5750. [Google Scholar] [CrossRef]
- Castaings, L.; Camargo, A.; Pocholle, D.; Gaudon, V.; Texier, Y.; Boutet-Mercey, S.; Taconnat, L.; Renou, J.-P.; Daniel-Vedele, F.; Fernandez, E.; et al. The Nodule Inception-like Protein 7 Modulates Nitrate Sensing and Metabolism in Arabidopsis. Plant J. Cell Mol. Biol. 2009, 57, 426–435. [Google Scholar] [CrossRef]
- Liu, K.-H.; Liu, M.; Lin, Z.; Wang, Z.-F.; Chen, B.; Liu, C.; Guo, A.; Konishi, M.; Yanagisawa, S.; Wagner, G.; et al. NIN-like Protein 7 Transcription Factor Is a Plant Nitrate Sensor. Science 2022, 377, 1419–1425. [Google Scholar] [CrossRef] [PubMed]
- Varala, K.; Marshall-Colón, A.; Cirrone, J.; Brooks, M.D.; Pasquino, A.V.; Léran, S.; Mittal, S.; Rock, T.M.; Edwards, M.B.; Kim, G.J.; et al. Temporal Transcriptional Logic of Dynamic Regulatory Networks Underlying Nitrogen Signaling and Use in Plants. Proc. Natl. Acad. Sci. USA 2018, 115, 6494–6499. [Google Scholar] [CrossRef]
- Zhang, H.; Forde, B.G. An Arabidopsis MADS Box Gene That Controls Nutrient-Induced Changes in Root Architecture. Science 1998, 279, 407–409. [Google Scholar] [CrossRef]
- Guan, P.; Wang, R.; Nacry, P.; Breton, G.; Kay, S.A.; Pruneda-Paz, J.L.; Davani, A.; Crawford, N.M. Nitrate Foraging by Arabidopsis Roots Is Mediated by the Transcription Factor TCP20 through the Systemic Signaling Pathway. Proc. Natl. Acad. Sci. USA 2014, 111, 15267–15272. [Google Scholar] [CrossRef]
- Alvarez, J.M.; Riveras, E.; Vidal, E.A.; Gras, D.E.; Contreras-López, O.; Tamayo, K.P.; Aceituno, F.; Gómez, I.; Ruffel, S.; Lejay, L.; et al. Systems Approach Identifies TGA1 and TGA4 Transcription Factors as Important Regulatory Components of the Nitrate Response of Arabidopsis Thaliana Roots. Plant J. Cell Mol. Biol. 2014, 80, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Nakano, T.; Suzuki, K.; Fujimura, T.; Shinshi, H. Genome-Wide Analysis of the ERF Gene Family in Arabidopsis and Rice. Plant Physiol. 2006, 140, 411–432. [Google Scholar] [CrossRef] [PubMed]
- Brooks, M.D.; Cirrone, J.; Pasquino, A.V.; Alvarez, J.M.; Swift, J.; Mittal, S.; Juang, C.-L.; Varala, K.; Gutiérrez, R.A.; Krouk, G.; et al. Network Walking Charts Transcriptional Dynamics of Nitrogen Signaling by Integrating Validated and Predicted Genome-Wide Interactions. Nat. Commun. 2019, 10, 1569. [Google Scholar] [CrossRef] [PubMed]
- Scheible, W.-R.; Morcuende, R.; Czechowski, T.; Fritz, C.; Osuna, D.; Palacios-Rojas, N.; Schindelasch, D.; Thimm, O.; Udvardi, M.K.; Stitt, M. Genome-Wide Reprogramming of Primary and Secondary Metabolism, Protein Synthesis, Cellular Growth Processes, and the Regulatory Infrastructure of Arabidopsis in Response to Nitrogen. Plant Physiol. 2004, 136, 2483–2499. [Google Scholar] [CrossRef]
- Wang, R.; Okamoto, M.; Xing, X.; Crawford, N.M. Microarray Analysis of the Nitrate Response in Arabidopsis Roots and Shoots Reveals over 1,000 Rapidly Responding Genes and New Linkages to Glucose, Trehalose-6-Phosphate, Iron, and Sulfate Metabolism. Plant Physiol. 2003, 132, 556–567. [Google Scholar] [CrossRef] [PubMed]
- Sahu, S.S.; Loaiza, C.D.; Kaundal, R. Plant-mSubP: A Computational Framework for the Prediction of Single- and Multi-Target Protein Subcellular Localization Using Integrated Machine-Learning Approaches. AoB Plants 2020, 12, plz068. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Sheerin, D.J.; von Roepenack-Lahaye, E.; Stahl, M.; Hiltbrunner, A. The Phytochrome Interacting Proteins ERF55 and ERF58 Repress Light-Induced Seed Germination in Arabidopsis Thaliana. Nat. Commun. 2022, 13, 1656. [Google Scholar] [CrossRef]
- Hsu, P.-K.; Tsay, Y.-F. Two Phloem Nitrate Transporters, NRT1.11 and NRT1.12, Are Important for Redistributing Xylem-Borne Nitrate to Enhance Plant Growth. Plant Physiol. 2013, 163, 844–856. [Google Scholar] [CrossRef] [PubMed]
- Kiba, T.; Feria-Bourrellier, A.-B.; Lafouge, F.; Lezhneva, L.; Boutet-Mercey, S.; Orsel, M.; Bréhaut, V.; Miller, A.; Daniel-Vedele, F.; Sakakibara, H.; et al. The Arabidopsis Nitrate Transporter NRT2.4 Plays a Double Role in Roots and Shoots of Nitrogen-Starved Plants. Plant Cell 2012, 24, 245–258. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.-C.; Lin, C.-S.; Hsu, P.-K.; Lin, S.-H.; Tsay, Y.-F. The Arabidopsis Nitrate Transporter NRT1.7, Expressed in Phloem, Is Responsible for Source-to-Sink Remobilization of Nitrate. Plant Cell 2009, 21, 2750–2761. [Google Scholar] [CrossRef] [PubMed]
- Chopin, F.; Orsel, M.; Dorbe, M.-F.; Chardon, F.; Truong, H.-N.; Miller, A.J.; Krapp, A.; Daniel-Vedele, F. The Arabidopsis ATNRT2.7 Nitrate Transporter Controls Nitrate Content in Seeds. Plant Cell 2007, 19, 1590–1602. [Google Scholar] [CrossRef] [PubMed]
- Loqué, D.; Yuan, L.; Kojima, S.; Gojon, A.; Wirth, J.; Gazzarrini, S.; Ishiyama, K.; Takahashi, H.; von Wirén, N. Additive Contribution of AMT1;1 and AMT1;3 to High-Affinity Ammonium Uptake across the Plasma Membrane of Nitrogen-Deficient Arabidopsis Roots. Plant J. Cell Mol. Biol. 2006, 48, 522–534. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; Loqué, D.; Kojima, S.; Rauch, S.; Ishiyama, K.; Inoue, E.; Takahashi, H.; von Wirén, N. The Organization of High-Affinity Ammonium Uptake in Arabidopsis Roots Depends on the Spatial Arrangement and Biochemical Properties of AMT1-Type Transporters. Plant Cell 2007, 19, 2636–2652. [Google Scholar] [CrossRef]
- Ayadi, A.; David, P.; Arrighi, J.-F.; Chiarenza, S.; Thibaud, M.-C.; Nussaume, L.; Marin, E. Reducing the Genetic Redundancy of Arabidopsis PHOSPHATE TRANSPORTER1 Transporters to Study Phosphate Uptake and Signaling. Plant Physiol. 2015, 167, 1511–1526. [Google Scholar] [CrossRef] [PubMed]
- Chien, P.-S.; Chao, Y.-T.; Chou, C.-H.; Hsu, Y.-Y.; Chiang, S.-F.; Tung, C.-W.; Chiou, T.-J. Phosphate Transporter PHT1;1 Is a Key Determinant of Phosphorus Acquisition in Arabidopsis Natural Accessions. Plant Physiol. 2022, 190, 682–697. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Wallace, I.S.; Takano, J.; Roberts, D.M.; Fujiwara, T. NIP6; 1 Is a Boric Acid Channel for Preferential Transport of Boron to Growing Shoot Tissues in Arabidopsis. Plant Cell 2008, 20, 2860–2875. [Google Scholar] [CrossRef] [PubMed]
- Luan, M.; Zhao, F.; Han, X.; Sun, G.; Yang, Y.; Liu, J.; Shi, J.; Fu, A.; Lan, W.; Luan, S. Vacuolar Phosphate Transporters Contribute to Systemic Phosphate Homeostasis Vital for Reproductive Development in Arabidopsis. Plant Physiol. 2019, 179, 640–655. [Google Scholar] [CrossRef] [PubMed]
- Stanton, C.; Sanders, D.; Krämer, U.; Podar, D. Zinc in Plants: Integrating Homeostasis and Biofortification. Mol. Plant 2022, 15, 65–85. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wang, Y. Harnessing Hormone Gibberellin Knowledge for Plant Height Regulation. Plant Cell Rep. 2022, 41, 1945–1953. [Google Scholar] [CrossRef] [PubMed]
- Sampedro, J.; Cosgrove, D.J. The Expansin Superfamily. Genome Biol. 2005, 6, 242. [Google Scholar] [CrossRef][Green Version]
- Cosgrove, D.J. Growth of the Plant Cell Wall. Nat. Rev. Mol. Cell Biol. 2005, 6, 850–861. [Google Scholar] [CrossRef] [PubMed]
- Coculo, D.; Lionetti, V. The Plant Invertase/Pectin Methylesterase Inhibitor Superfamily. Front. Plant Sci. 2022, 13, 863892. [Google Scholar] [CrossRef] [PubMed]
- Minic, Z.; Jouanin, L. Plant Glycoside Hydrolases Involved in Cell Wall Polysaccharide Degradation. Plant Physiol. Biochem. PPB 2006, 44, 435–449. [Google Scholar] [CrossRef]
- Pelloux, J.; Rustérucci, C.; Mellerowicz, E.J. New Insights into Pectin Methylesterase Structure and Function. Trends Plant Sci. 2007, 12, 267–277. [Google Scholar] [CrossRef]
- Castilleux, R.; Plancot, B.; Vicré, M.; Nguema-Ona, E.; Driouich, A. Extensin, an Underestimated Key Component of Cell Wall Defence? Ann. Bot. 2021, 127, 709–713. [Google Scholar] [CrossRef]
- Bailey, T.L.; Elkan, C. Fitting a Mixture Model by Expectation Maximization to Discover Motifs in Biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994, 2, 28–36. [Google Scholar] [PubMed]
- Cortes, T.; Schubert, O.T.; Rose, G.; Arnvig, K.B.; Comas, I.; Aebersold, R.; Young, D.B. Genome-Wide Mapping of Transcriptional Start Sites Defines an Extensive Leaderless Transcriptome in Mycobacterium Tuberculosis. Cell Rep. 2013, 5, 1121–1131. [Google Scholar] [CrossRef] [PubMed]
- Danino, Y.M.; Even, D.; Ideses, D.; Juven-Gershon, T. The Core Promoter: At the Heart of Gene Expression. Biochim. Biophys. Acta 2015, 1849, 1116–1131. [Google Scholar] [CrossRef]
- Zhang, H.; Yu, F.; Xie, P.; Sun, S.; Qiao, X.; Tang, S.; Chen, C.; Yang, S.; Mei, C.; Yang, D.; et al. A Gγ Protein Regulates Alkaline Sensitivity in Crops. Science 2023, 379, eade8416. [Google Scholar] [CrossRef]
- Smirnoff, N.; Arnaud, D. Hydrogen Peroxide Metabolism and Functions in Plants. New Phytol. 2019, 221, 1197–1214. [Google Scholar] [CrossRef] [PubMed]
- Waszczak, C.; Carmody, M.; Kangasjärvi, J. Reactive Oxygen Species in Plant Signaling. Annu. Rev. Plant Biol. 2018, 69, 209–236. [Google Scholar] [CrossRef] [PubMed]
- Binenbaum, J.; Weinstain, R.; Shani, E. Gibberellin Localization and Transport in Plants. Trends Plant Sci. 2018, 23, 410–421. [Google Scholar] [CrossRef]
- Wang, Z.-P.; Xing, H.-L.; Dong, L.; Zhang, H.-Y.; Han, C.-Y.; Wang, X.-C.; Chen, Q.-J. Egg Cell-Specific Promoter-Controlled CRISPR/Cas9 Efficiently Generates Homozygous Mutants for Multiple Target Genes in Arabidopsis in a Single Generation. Genome Biol. 2015, 16, 144. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral Dip: A Simplified Method for Agrobacterium-Mediated Transformation of Arabidopsis Thaliana. Plant J. Cell Mol. Biol. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Hartig, S.M. Basic Image Analysis and Manipulation in ImageJ. Curr. Protoc. Mol. Biol. 2013, 102, 14–15. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods San Diego Calif. 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinforma. Oxf. Engl. 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A Fast Spliced Aligner with Low Memory Requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie Enables Improved Reconstruction of a Transcriptome from RNA-Seq Reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A Universal Enrichment Tool for Interpreting Omics Data. Innov. Camb. Mass 2021, 2, 100141. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yao, G.; Mu, C.; Yan, Z.; Ma, S.; Liu, X.; Sun, Y.; Hou, J.; Liu, Q.; Cao, B.; Shan, J.; et al. The AP2/ERF Transcription Factor ERF56 Negatively Regulating Nitrate-Dependent Plant Growth in Arabidopsis. Int. J. Mol. Sci. 2025, 26, 613. https://doi.org/10.3390/ijms26020613
Yao G, Mu C, Yan Z, Ma S, Liu X, Sun Y, Hou J, Liu Q, Cao B, Shan J, et al. The AP2/ERF Transcription Factor ERF56 Negatively Regulating Nitrate-Dependent Plant Growth in Arabidopsis. International Journal of Molecular Sciences. 2025; 26(2):613. https://doi.org/10.3390/ijms26020613
Chicago/Turabian StyleYao, Guoqi, Chunhua Mu, Zhenwei Yan, Shijun Ma, Xia Liu, Yue Sun, Jing Hou, Qiantong Liu, Bing Cao, Juan Shan, and et al. 2025. "The AP2/ERF Transcription Factor ERF56 Negatively Regulating Nitrate-Dependent Plant Growth in Arabidopsis" International Journal of Molecular Sciences 26, no. 2: 613. https://doi.org/10.3390/ijms26020613
APA StyleYao, G., Mu, C., Yan, Z., Ma, S., Liu, X., Sun, Y., Hou, J., Liu, Q., Cao, B., Shan, J., & Leng, B. (2025). The AP2/ERF Transcription Factor ERF56 Negatively Regulating Nitrate-Dependent Plant Growth in Arabidopsis. International Journal of Molecular Sciences, 26(2), 613. https://doi.org/10.3390/ijms26020613




