Biofilm Formation, Modulation, and Transcriptomic Regulation Under Stress Conditions in Halomicronema sp.
Abstract
1. Introduction
2. Results and Discussion
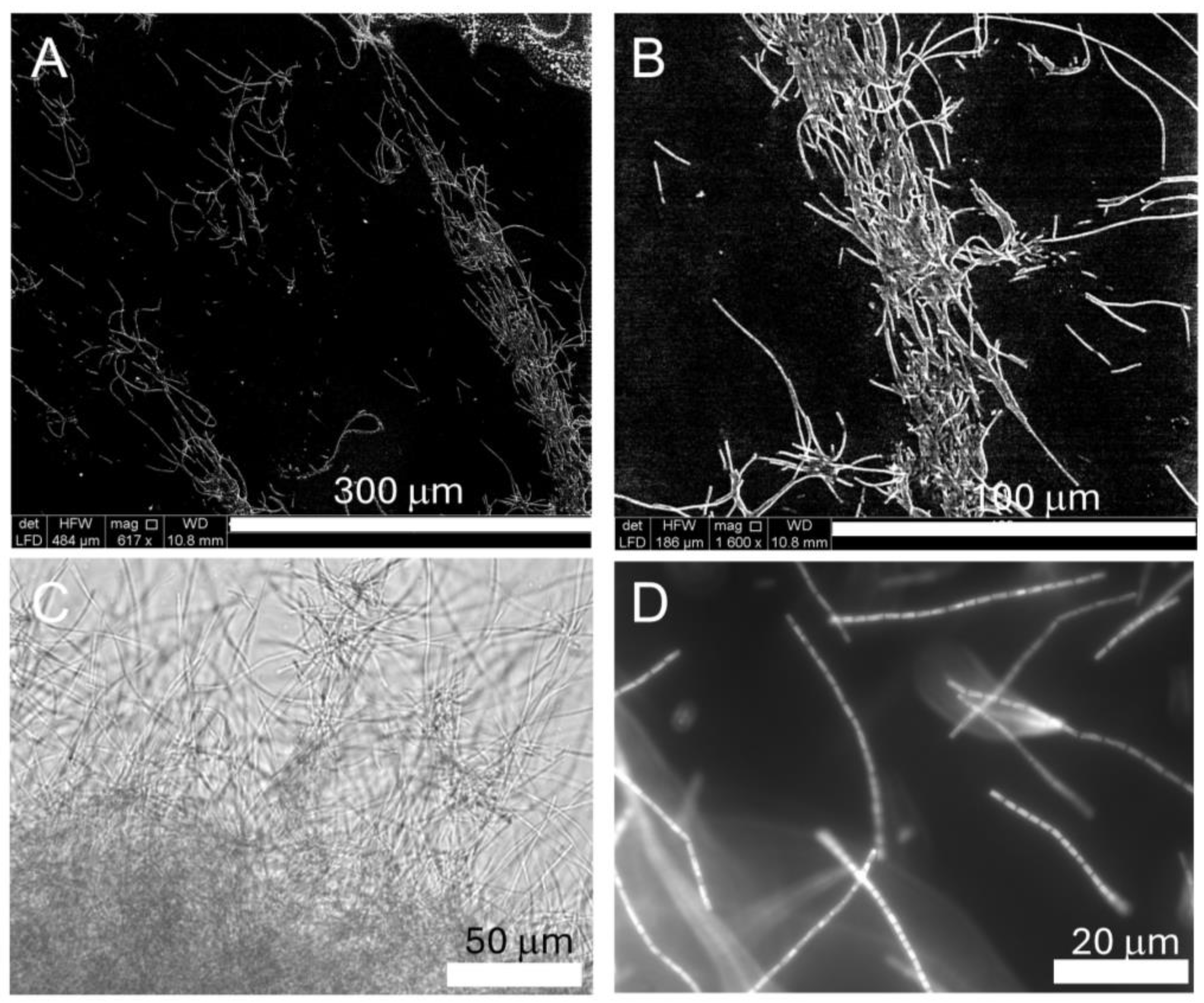
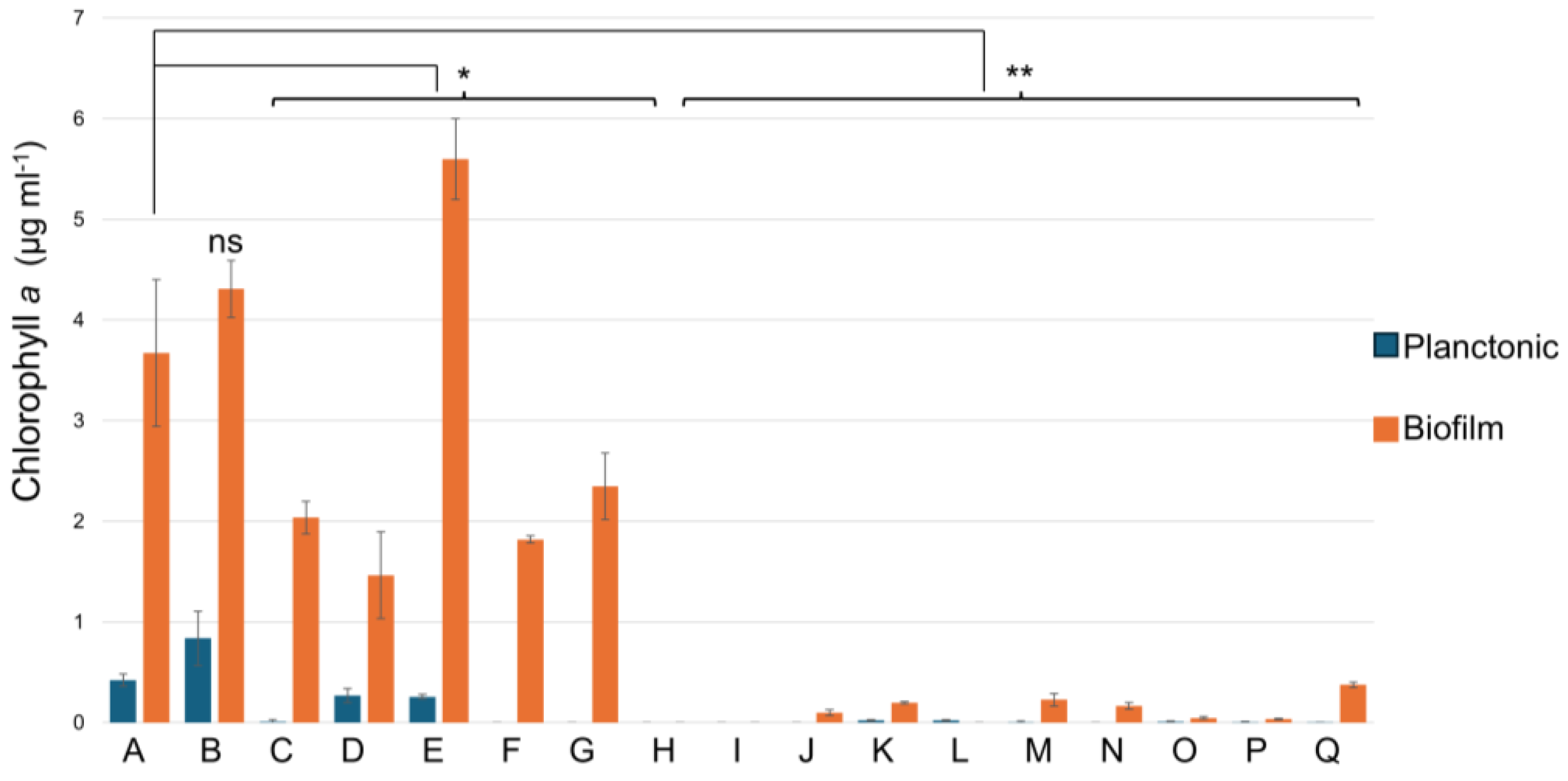
2.1. Growth of Halomicronema sp. and Biofilm Formation
2.2. Phenolic Content and Antioxidant Capacity
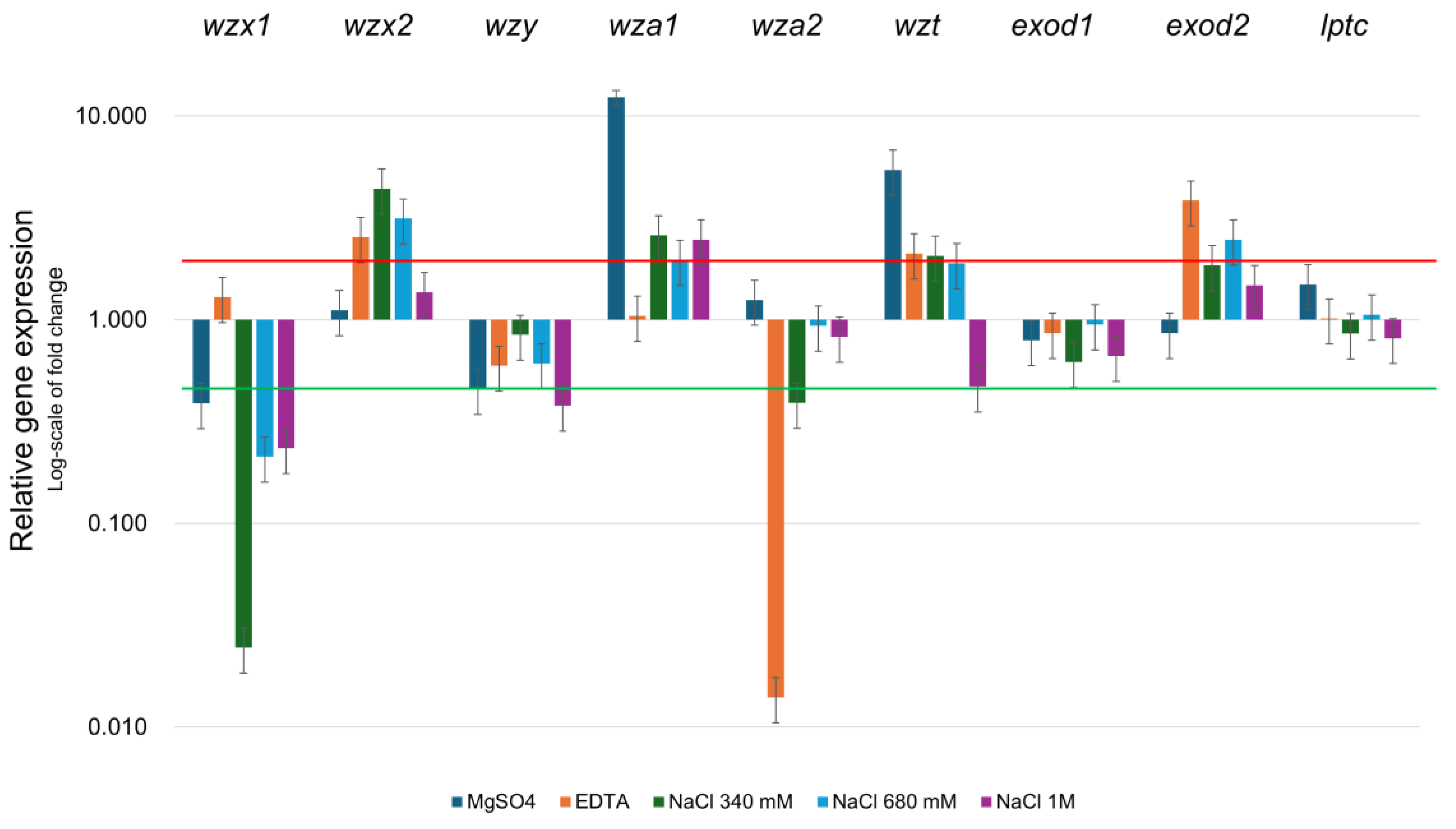
2.3. Expression Analysis of EPS-Related Genes
2.4. Transcriptomic Profile of Halomicronema sp.
2.5. Expression Analysis of Main Transcription Regulators
3. Materials and Methods
3.1. Growing Conditions
3.2. Microscopy
3.3. Total Phenolic Content, DPPH, and ABTS Assays
3.4. Gene Selection and Primer Design
3.5. Gene Expression Analysis by RTq-PCR
3.6. RNA Sequencing
3.7. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Martens, S. Handbook of Cyanobacterial Monitoring and Cyanotoxin Analysis. Adv. Oceanogr. Limnol. 2017, 8, 7221. [Google Scholar] [CrossRef]
- Durall, C.; Lindblad, P. Mechanisms of carbon fixation and engineering for increased carbon fixation in cyanobacteria. Algal Res. 2015, 11, 263–270. [Google Scholar] [CrossRef]
- Zakar, T.; Laczko-Dobos, H.; Toth, T.N.; Gombos, Z. Carotenoids assist in cyanobacterial photosystem II assembly and function. Front. Plant Sci. 2016, 7, 295. [Google Scholar] [CrossRef]
- Chamizo, S. Soil Inoculation with Cyanobacteria: Reviewing Its’ Potential for Agriculture Sustainability in Drylands. Agric. Res. Technol. Open Access J. 2018, 18, 556046. [Google Scholar] [CrossRef]
- Dahms, H.U.; Ying, X.; Pfeiffer, C. Antifouling potential of cyanobacteria: A mini-review. Biofouling 2006, 22, 317–327. [Google Scholar] [CrossRef]
- Chakdar, H.; Thapa, S.; Srivastava, A.; Shukla, P. Genomic and proteomic insights into the heavy metal bioremediation by cyanobacteria. J. Hazard. Mater. 2022, 424, 127609. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Xie, Y.; Sun, T.; Chen, L.; Zhang, W. Deciphering and engineering photosynthetic cyanobacteria for heavy metal bioremediation. Sci. Total Environ. 2021, 761, 144111. [Google Scholar] [CrossRef]
- Dang, H.; Lovell, C.R. Microbial Surface Colonization and Biofilm Development in Marine Environments. Microbiol. Mol. Biol. Rev. 2016, 80, 91–138. [Google Scholar] [CrossRef] [PubMed]
- Chittora, D.; Meena, M.; Barupal, T.; Swapnil, P. Cyanobacteria as a source of biofertilizers for sustainable agriculture. Biochem. Biophys. Rep. 2020, 22, 100737. [Google Scholar] [CrossRef] [PubMed]
- Cania, B.; Vestergaard, G.; Kublik, S.; Köhne, J.M.; Fischer, T.; Albert, A.; Winkler, B.; Schloter, M.; Schulz, S. Biological Soil Crusts from Different Soil Substrates Harbor Distinct Bacterial Groups with the Potential to Produce Exopolysaccharides and Lipopolysaccharides. Microb. Ecol. 2020, 79, 326–341. [Google Scholar] [CrossRef] [PubMed]
- Bolhuis, H.; Cretoiu, M.S.; Stal, L.J. Molecular ecology of microbial mats. FEMS Microbiol. Ecol. 2014, 90, 335–350. [Google Scholar] [PubMed]
- Flemming, H.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef] [PubMed]
- Flemming, H.C.; van Hullebusch, E.D.; Neu, T.R.; Nielsen, P.H.; Seviour, T.; Stoodley, P.; Wingender, J.; Wuertz, S. The biofilm matrix: Multitasking in a shared space. Nat. Rev. Microbiol. 2023, 21, 70–86. [Google Scholar] [CrossRef]
- Delattre, C.; Pierre, G.; Laroche, C.; Michaud, P. Production, extraction and characterization of microalgal and cyanobacterial exopolysaccharides. Biotechnol. Adv. 2016, 34, 1159–1179. [Google Scholar] [CrossRef]
- Laroche, C. Exopolysaccharides from Microalgae and Cyanobacteria: Diversity of Strains, Production Strategies, and Applications. Mar. Drugs 2022, 20, 336. [Google Scholar] [CrossRef] [PubMed]
- Kehr, J.C.; Dittmann, E. Biosynthesis and function of extracellular glycans in cyanobacteria. Life 2015, 5, 164–180. [Google Scholar] [CrossRef] [PubMed]
- Pereira, S.; Zille, A.; Micheletti, E.; Moradas-Ferreira, P.; De Philippis, R.; Tamagnini, P. Complexity of cyanobacterial exopolysaccharides: Composition, structures, inducing factors and putative genes involved in their biosynthesis and assembly. FEMS Microbiol. Rev. 2009, 33, 917–941. [Google Scholar] [CrossRef] [PubMed]
- Reeves, P.R.; Hobbs, M.; Valvano, M.A.; Skurnik, M.; Whitfield, C.; Coplin, D.; Kido, N.; Klena, J.; Maskell, D.; Raetz, C.R.H.; et al. Bacterial polysaccharide synthesis and gene nomenclature. Trends Microbiol. 1996, 4, 495–503. [Google Scholar] [CrossRef]
- Pereira, S.B.; Mota, R.; Vieira, C.P.; Vieira, J.; Tamagnini, P. Phylum-wide analysis of genes/proteins related to the last steps of assembly and export of extracellular polymeric substances (EPS) in cyanobacteria. Sci. Rep. 2015, 5, 14835. [Google Scholar] [CrossRef] [PubMed]
- Comte, S.; Guibaud, G.; Baudu, M. Biosorption properties of extracellular polymeric substances (EPS) resulting from activated sludge according to their type: Soluble or bound. Process Biochem. 2006, 41, 815–823. [Google Scholar] [CrossRef]
- Whitfield, C.; Stephen Trent, M. Biosynthesis and export of bacterial lipopolysaccharides. Annu. Rev. Biochem. 2014, 83, 99–128. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, D.C.; Cretoiu, M.S.; Stal, L.J.; Bolhuis, H. Seasonal development of a coastal microbial mat. Sci. Rep. 2019, 9, 9035. [Google Scholar] [CrossRef] [PubMed]
- Boden, J.S.; Grego, M.; Bolhuis, H.; Sánchez-Baracaldo, P. Draft genome sequences of three filamentous cyanobacteria isolated from brackish habitats. J. Genom. 2021, 9, 20–25. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Li, D.; Liu, Y. Salt tolerance of Microcoleus vaginatus Gom., a cyanobacterium isolated from desert algal crust, was enhanced by exogenous carbohydrates. J. Arid Environ. 2003, 55, 645–656. [Google Scholar] [CrossRef]
- Kharwar, S.; Mishra, A.K. Unraveling the complexities underlying sulfur deficiency and starvation in the cyanobacterium Anabaena sp. PCC 7120. Environ. Exp. Bot. 2020, 172, 103966. [Google Scholar] [CrossRef]
- Wu, R.X.; Zhang, Y.; Guo, Z.Q.; Zhao, B.; Guo, J.S. Role of Ca2+ and Mg2+ in changing biofilm structure and enhancing biofilm formation of P. stutzeri strain XL-2. Colloids Surf. B Biointerfaces 2022, 220, 112972. [Google Scholar] [CrossRef]
- Shen, Y.; Huang, P.C.; Huang, C.; Sun, P.; Monroy, G.L.; Wu, W.; Lin, J.; Espinosa-Marzal, R.M.; Boppart, S.A.; Liu, W.T.; et al. Effect of divalent ions and a polyphosphate on composition, structure, and stiffness of simulated drinking water biofilms. npj Biofilms Microbiomes 2018, 4, 15. [Google Scholar] [CrossRef] [PubMed]
- Finnegan, S.; Percival, S.L. EDTA: An Antimicrobial and Antibiofilm Agent for Use in Wound Care. Adv. Wound Care 2015, 4, 415–421. [Google Scholar] [CrossRef] [PubMed]
- Wickramasinghe, M.; Katyana, K.; Sewwandi, K.; Rathnayaka, I.; Magana-Arachchi, D.; Jayawardana, B.; Liyanage, R. Exploration of antioxidant activities, microstructural properties, and fatty acid composition of three cyanobacteria species. Biocatal. Agric. Biotechnol. 2024, 56, 103015. [Google Scholar] [CrossRef]
- Morone, J.; Lopes, G.; Preto, M.; Vasconcelos, V.; Martins, R. Exploitation of filamentous and picoplanktonic cyanobacteria for cosmetic applications: Potential to improve skin structure and preserve dermal matrix components. Mar. Drugs 2020, 18, 486. [Google Scholar] [CrossRef] [PubMed]
- Frazzini, S.; Scaglia, E.; Dell’anno, M.; Reggi, S.; Panseri, S.; Giromini, C.; Lanzoni, D.; Rossi, C.A.S.; Rossi, L. Antioxidant and Antimicrobial Activity of Algal and Cyanobacterial Extracts: An In Vitro Study. Antioxidants 2022, 11, 992. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.E.; Lee, J.H.; Park, S.M.; Kim, D.G. Symbiotic relationship between filamentous algae (Halomicronema sp.) and extracellular polymeric substance-producing algae (Chlamydomonas sp.) through biomimetic simulation of natural algal mats. Front. Microbiol. 2023, 14, 1176069. [Google Scholar] [CrossRef] [PubMed]
- Waditee-Sirisattha, R.; Kageyama, H. Halotolerance, stress mechanisms, and circadian clock of salt-tolerant cyanobacteria. Appl. Microbiol. Biotechnol. 2023, 107, 1129–1141. [Google Scholar] [CrossRef] [PubMed]
- Kageyama, H.; Waditee-Sirisattha, R. Halotolerance mechanisms in salt-tolerant cyanobacteria. In Advances in Applied Microbiology; Elsevier: Amsterdam, The Netherlands, 2023; Volume 124, pp. 55–117. ISBN 9780443192746. [Google Scholar]
- Hagemann, M. Molecular biology of cyanobacterial salt acclimation. FEMS Microbiol. Rev. 2011, 35, 87–123. [Google Scholar] [CrossRef] [PubMed]
- Marin, K.; Huckauf, J.; Fulda, S.; Hagemann, M. Salt-dependent expression of glucosylglycerol-phosphate synthase, involved in osmolyte synthesis in the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 2002, 184, 2870–2877. [Google Scholar] [CrossRef]
- Marin, K.; Suzuki, I.; Yamaguchi, K.; Ribbeck, K.; Yamamoto, H.; Kanesaki, Y.; Hagemann, M.; Murata, N. Identification of histidine kinases that act as sensors in the perception of salt stress in Synechocystis sp. PCC 6803. Proc. Natl. Acad. Sci. USA 2003, 100, 9061–9066. [Google Scholar] [CrossRef] [PubMed]
- Sinetova, M.A.; Los, D.A. Systemic analysis of stress transcriptomics of Synechocystis reveals common stress genes and their universal triggers. Mol. Biosyst. 2016, 12, 3254–3258. [Google Scholar] [CrossRef] [PubMed]
- Soumya, M.P.; Nampoothiri, K.M. An overview of functional genomics and relevance of glycosyltransferases in exopolysaccharide production by lactic acid bacteria. Int. J. Biol. Macromol. 2021, 184, 1014–1025. [Google Scholar] [CrossRef] [PubMed]
- Santos, M.; Pereira, S.B.; Flores, C.; Príncipe, C.; Couto, N.; Karunakaran, E.; Cravo, S.M.; Oliveira, P.; Tamagnini, P. Absence of KpsM (Slr0977) Impairs the Secretion of Extracellular Polymeric Substances (EPS) and Impacts Carbon Fluxes in Synechocystis sp. PCC 6803. mSphere 2021, 6, 10–128. [Google Scholar] [CrossRef]
- Keiler, K.C.; Ramadoss, N.S. Bifunctional transfer-messenger RNA. Biochimie 2011, 93, 1993–1997. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, Y.; Ueda, T. The role of SmpB protein in trans-translation. Proc. FEBS Lett. 2002, 514, 74–77. [Google Scholar] [CrossRef] [PubMed]
- Keiler, K.C.; Shapiro, L.; Williams, K.P. tmRNAs that encode proteolysis-inducing tags are found in all known bacterial genomes: A two-piece tmRNA functions in caulobacter. Proc. Natl. Acad. Sci. USA 2000, 97, 7778–7783. [Google Scholar] [CrossRef] [PubMed]
- Domínguez-Martín, M.A.; Sauer, P.V.; Kirst, H.; Sutter, M.; Bína, D.; Greber, B.J.; Nogales, E.; Polívka, T.; Kerfeld, C.A. Structures of a phycobilisome in light-harvesting and photoprotected states. Nature 2022, 609, 835–845. [Google Scholar] [CrossRef] [PubMed]
- Plohnke, N.; Seidel, T.; Kahmann, U.; Rögner, M.; Schneider, D.; Rexroth, S. The proteome and lipidome of Synechocystis sp. PCC 6803 cells grown under light-activated heterotrophic conditions. Mol. Cell. Proteom. 2015, 14, 572–584. [Google Scholar] [CrossRef]
- Huang, F.; Grauslys, A.; Huokko, T.; Caamaño Gutiérrez, E.; Jones, A.R.; Liu, L.N. Dynamic Changes in the Thylakoid Proteome of Cyanobacteria during Light-Regulated Thylakoid Membrane Development. Plants 2023, 12, 3967. [Google Scholar] [CrossRef]
- Tiwari, B. Phosphate metabolism in cyanobacteria: Fundamental prospective and applications. In Cyanobacteria: Metabolisms to Molecules; Elsevier: Amsterdam, The Netherlands, 2023; pp. 159–175. ISBN 9780443132315. [Google Scholar]
- Muro-Pastor, M.I.; Florencio, F.J. Regulation of ammonium assimilation in cyanobacteria. Plant Physiol. Biochem. 2003, 41, 595–603. [Google Scholar] [CrossRef]
- Qiu, G.W.; Koedooder, C.; Qiu, B.S.; Shaked, Y.; Keren, N. Iron transport in cyanobacteria—From molecules to communities. Trends Microbiol. 2022, 30, 229–240. [Google Scholar] [CrossRef]
- Rubin, M.; Berman-Frank, I.; Shaked, Y. Dust-and mineral-iron utilization by the marine dinitrogen-fixer Trichodesmium. Nat. Geosci. 2011, 4, 529–534. [Google Scholar] [CrossRef]
- Sullivan, D.M.; Bobay, B.G.; Kojetin, D.J.; Thompson, R.J.; Rance, M.; Strauch, M.A.; Cavanagh, J. Insights into the Nature of DNA Binding of AbrB-like Transcription Factors. Structure 2008, 16, 1702–1713. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wei, L.; Wu, Y.; Qiao, H.; Xu, W.; Zhang, Y.; Liu, X.; Wang, Q. YebC controls virulence by activating T3SS gene expression in the pathogen Edwardsiella piscicida. FEMS Microbiol. Lett. 2018, 365, fny137. [Google Scholar] [CrossRef]
- Colclough, A.L.; Scadden, J.; Blair, J.M.A. TetR-family transcription factors in Gram-negative bacteria: Conservation, variation and implications for efflux-mediated antimicrobial resistance. BMC Genom. 2019, 20, 731. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.; Li, C.; Xie, J. The underling mechanism of bacterial TetR/AcrR family transcriptional repressors. Cell. Signal. 2013, 25, 1608–1613. [Google Scholar] [CrossRef]
- Chen, J.; Xie, J. Role and regulation of bacterial LuxR-like regulators. J. Cell. Biochem. 2011, 112, 2694–2702. [Google Scholar] [CrossRef] [PubMed]
- Santos, C.L.; Correia-Neves, M.; Moradas-Ferreira, P.; Mendes, M.V. A Walk into the LuxR Regulators of Actinobacteria: Phylogenomic Distribution and Functional Diversity. PLoS ONE 2012, 7, 0046758. [Google Scholar] [CrossRef]
- Maddocks, S.E.; Oyston, P.C.F. Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology 2008, 154, 3609–3623. [Google Scholar] [CrossRef]
- Liu, G.F.; Wang, X.X.; Su, H.Z.; Lu, G.T. Progress on the GntR family transcription regulators in bacteria. Yi Chuan = Hered. 2021, 43, 66–73. [Google Scholar]
- Körner, H.; Sofia, H.J.; Zumft, W.G. Phylogeny of the bacterial superfamily of Crp-Fnr transcription regulators: Exploiting the metabolic spectrum by controlling alternative gene programs. FEMS Microbiol. Rev. 2003, 27, 559–592. [Google Scholar] [CrossRef] [PubMed]
- Soberón-Chávez, G.; Alcaraz, L.D.; Morales, E.; Ponce-Soto, G.Y.; Servín-González, L. The transcriptional regulators of the CRP family regulate different essential bacterial functions and can be inherited vertically and horizontally. Front. Microbiol. 2017, 8, 959. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Hu, Q.; Wei, R.; Li, R.; Zhao, D.; Ge, M.; Yao, Q.; Yu, X. The XRE family transcriptional regulator SrtR in streptococcus suis is involved in oxidant tolerance and virulence. Front. Cell. Infect. Microbiol. 2019, 9, 00452. [Google Scholar] [CrossRef]
- Dressaire, C.; Moreira, R.N.; Barahona, S.; de Matos, A.P.A.; Arraiano, C.M. BolA is a transcriptional switch that turns off motility and turns on biofilm development. mBio 2015, 6, e02352-14. [Google Scholar] [CrossRef]
- Zavrel, T.; Sinetova, M.; Cerven, J. Measurement of Chlorophyll a and Carotenoids Concentration in Cyanobacteria. Bio-Protocol 2015, 5, 1467. [Google Scholar] [CrossRef]
- Sendersky, E.; Simkovsky, R.; Golden, S.; Schwarz, R. Quantification of Chlorophyll as a Proxy for Biofilm Formation in the Cyanobacterium Synechococcus elongatus. Bio-Protocol 2017, 7, 2406. [Google Scholar] [CrossRef]
- Marmiroli, M.; Caldara, M.; Pantalone, S.; Malcevschi, A.; Maestri, E.; Marmiroli, N. Building a risk matrix for the safety assessment of wood derived biochars. Sci. Total Environ. 2022, 15, 156265. [Google Scholar] [CrossRef] [PubMed]
- Guerreiro, A.; Andrade, M.A.; Menezes, C.; Vilarinho, F.; Dias, E. Antioxidant and Cytoprotective Properties of Cyanobacteria: Potential for Biotechnological Applications. Toxins 2020, 12, 548. [Google Scholar] [CrossRef] [PubMed]
- Erkan, N.; Ayranci, G.; Ayranci, E. Antioxidant activities of rosemary (Rosmarinus officinalis L.) extract, blackseed (Nigella sativa L.) essential oil, carnosic acid, rosmarinic acid and sesamol. Food Chem. 2008, 110, 76–82. [Google Scholar] [CrossRef]
- Andrade, M.A.; Ribeiro-Santos, R.; Costa Bonito, M.C.; Saraiva, M.; Sanches-Silva, A. Characterization of rosemary and thyme extracts for incorporation into a whey protein based film. LWT 2018, 92, 497–508. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]

| ASSAY | Mean ± SD |
|---|---|
| Total Phenolic Compounds (mg GAE g−1 extract DW) | 0.7 ± 0.1 |
| DPPH Inhibition (PI%) | 63 ± 5 |
| ABTS assay (PI%) | 52 ± 4 |
| Transcription Factor Family | Role of the Transcription Factor Family | Protein Name | Gene_ID | Average Value * | Protein_ID |
|---|---|---|---|---|---|
| AbrB family transcriptional regulator | Essential for cell survival, spore formation, and biofilm development [51] | AbrB-06010 | JUK32_RS06010 | 5239 | WP_204137943.1 |
| YebC/PmpR family DNA-binding transcriptional regulator | Probably involved in regulation of quorum sensing, and virulence, regulates EPS and LPS production to form biofilm [52] | YebC-12275 | JUK32_RS12275 | 283 | WP_204139153.1 |
| TetR/AcrR family transcriptional regulator | Regulation of essential processes (metabolism virulence, resistance, and biofilm formation) [53,54] | TetR-01335 | JUK32_RS01335 | 768 | WP_204137057.1 |
| TetR-14575 | JUK32_RS14575 | 167 | WP_204139591.1 | ||
| TetR-16580 | JUK32_RS16580 | 1471 | WP_204139976.1 | ||
| TetR-19750 | JUK32_RS19750 | 517 | WP_204140582.1 | ||
| TetR-23190 | JUK32_RS23190 | 271 | WP_204141244.1 | ||
| TetR-11335 | JUK32_RS11335 | 702 | WP_239112565.1 | ||
| TetR-13695 | JUK32_RS13695 | 483 | WP_239112647.1 | ||
| LuxR C-terminal-related transcriptional regulator | Involved in quorum sensing, regulates virulence gene production, motility, plasmid transfer, and biofilm formation [55,56] | LuxR-02440 | JUK32_RS02440 | 445 | WP_204137261.1 |
| LuxR-04290 | JUK32_RS04290 | 935 | WP_204137618.1 | ||
| LuxR-05665 | JUK32_RS05665 | 653 | WP_239112365.1 | ||
| LysR family transcriptional regulator | Regulates virulence, metabolism, quorum sensing, and motility [57] | LysR-07280 | JUK32_RS07280 | 645 | WP_204138189.1 |
| LysR-08635 | JUK32_RS08635 | 1034 | WP_204138452.1 | ||
| LysR-09925 | JUK32_RS09925 | 266 | WP_204138702.1 | ||
| LysR-11580 | JUK32_RS11580 | 654 | WP_204139027.1 | ||
| LysR-12705 | JUK32_RS12705 | 613 | WP_204139229.1 | ||
| LysR-20905 | JUK32_RS20905 | 1630 | WP_204140807.1 | ||
| GntR family transcriptional regulator | Regulates motility, glucose metabolism, and virulence [58] | GntR-03040 | JUK32_RS03040 | 2277 | WP_239112277.1 |
| GntR-24680 | JUK32_RS24680 | 1551 | WP_239113044.1 | ||
| Crp/Fnr family transcriptional regulator | Regulates virulence, the enzymes that degrade aromatic rings, genes involved in nitrogen fixation and photosynthesis [59,60] | Crp-04910 | JUK32_RS04910 | 4875 | WP_204137735.1 |
| Crp-18565 | JUK32_RS18565 | 8743 | WP_204140370.1 | ||
| Crp-20005 | JUK32_RS20005 | 592 | WP_204140632.1 | ||
| Crp-24180 | JUK32_RS24180 | 3763 | WP_204141438.1 | ||
| XRE family transcriptional regulator | Unknown function, probably involved in the expression of virulence genes [61] | XRE-06870 | JUK32_RS06870 | 3544 | WP_204138108.1 |
| BolA family transcriptional regulator | Involved in stress response, cell growth, morphology and division, biofilm development, inhibit motility [62] | BolA-25875 | JUK32_RS25875 | 2291 | WP_204141766.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Caldara, M.; Bolhuis, H.; Marmiroli, M.; Marmiroli, N. Biofilm Formation, Modulation, and Transcriptomic Regulation Under Stress Conditions in Halomicronema sp. Int. J. Mol. Sci. 2025, 26, 673. https://doi.org/10.3390/ijms26020673
Caldara M, Bolhuis H, Marmiroli M, Marmiroli N. Biofilm Formation, Modulation, and Transcriptomic Regulation Under Stress Conditions in Halomicronema sp. International Journal of Molecular Sciences. 2025; 26(2):673. https://doi.org/10.3390/ijms26020673
Chicago/Turabian StyleCaldara, Marina, Henk Bolhuis, Marta Marmiroli, and Nelson Marmiroli. 2025. "Biofilm Formation, Modulation, and Transcriptomic Regulation Under Stress Conditions in Halomicronema sp." International Journal of Molecular Sciences 26, no. 2: 673. https://doi.org/10.3390/ijms26020673
APA StyleCaldara, M., Bolhuis, H., Marmiroli, M., & Marmiroli, N. (2025). Biofilm Formation, Modulation, and Transcriptomic Regulation Under Stress Conditions in Halomicronema sp. International Journal of Molecular Sciences, 26(2), 673. https://doi.org/10.3390/ijms26020673








