Decoding Codon Bias: The Role of tRNA Modifications in Tissue-Specific Translation
Abstract
:1. Introduction
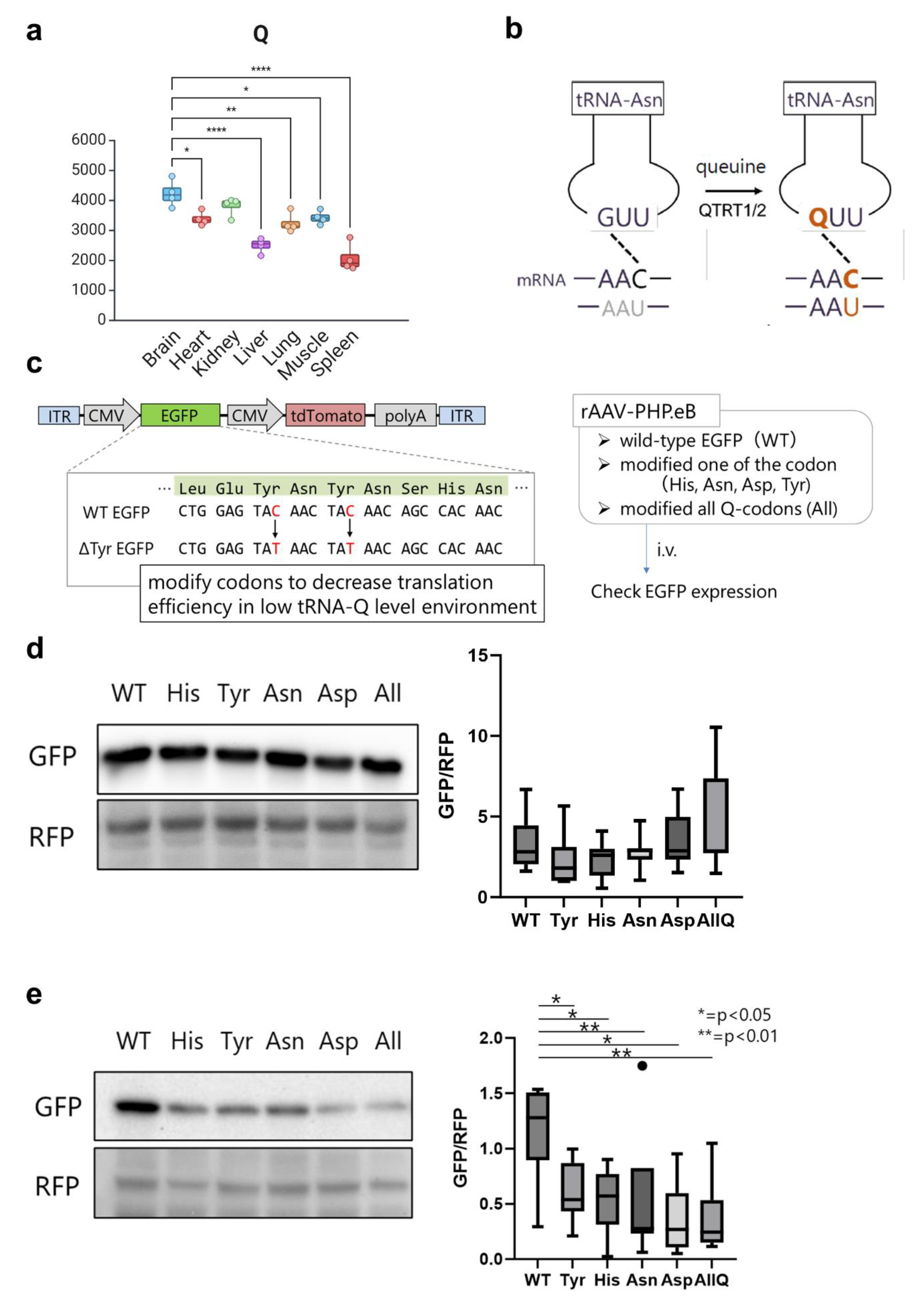
2. Results
- ➢
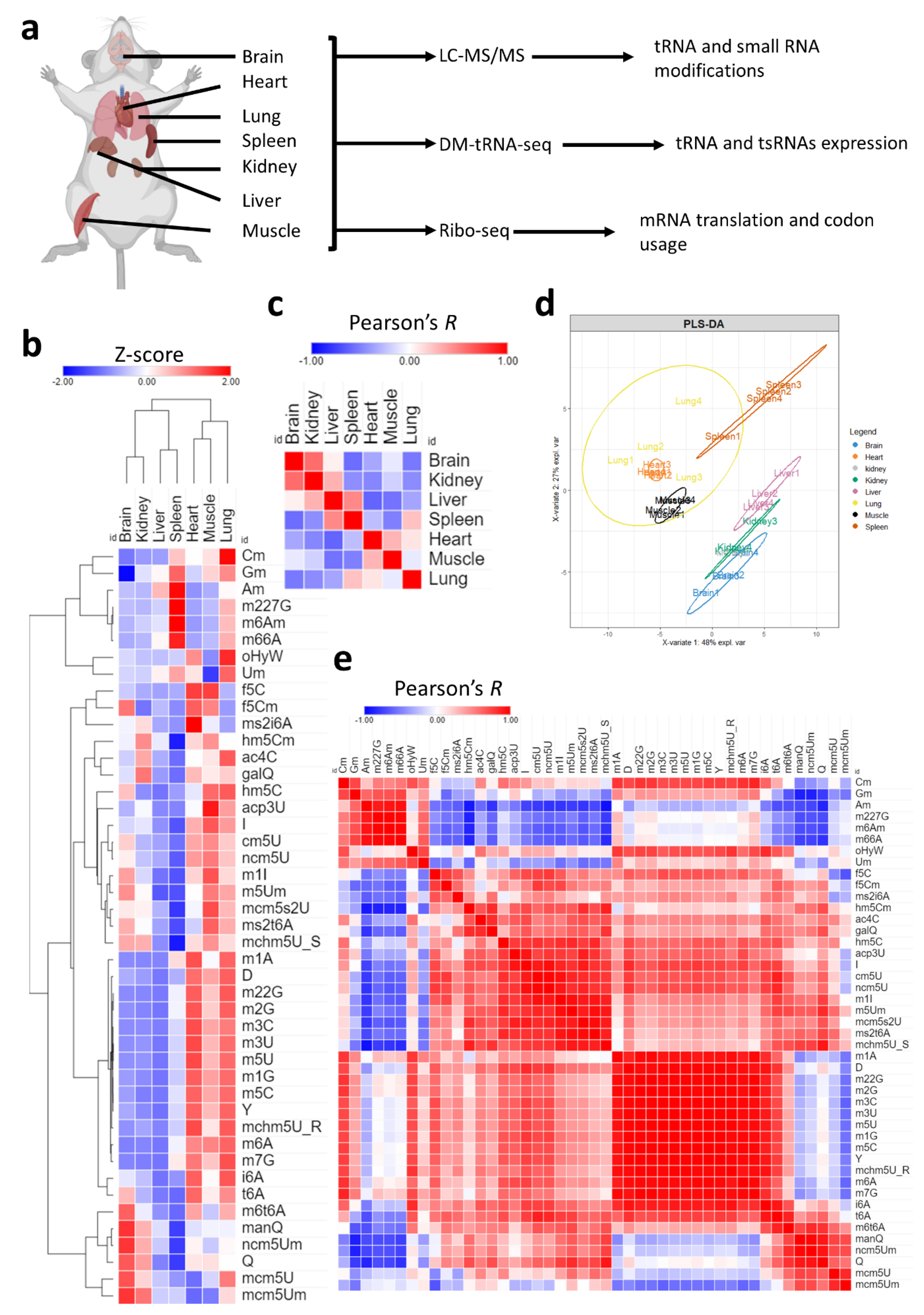
- tRNA and non-coding RNA modifications show tissue-level enrichment patterns
- ➢
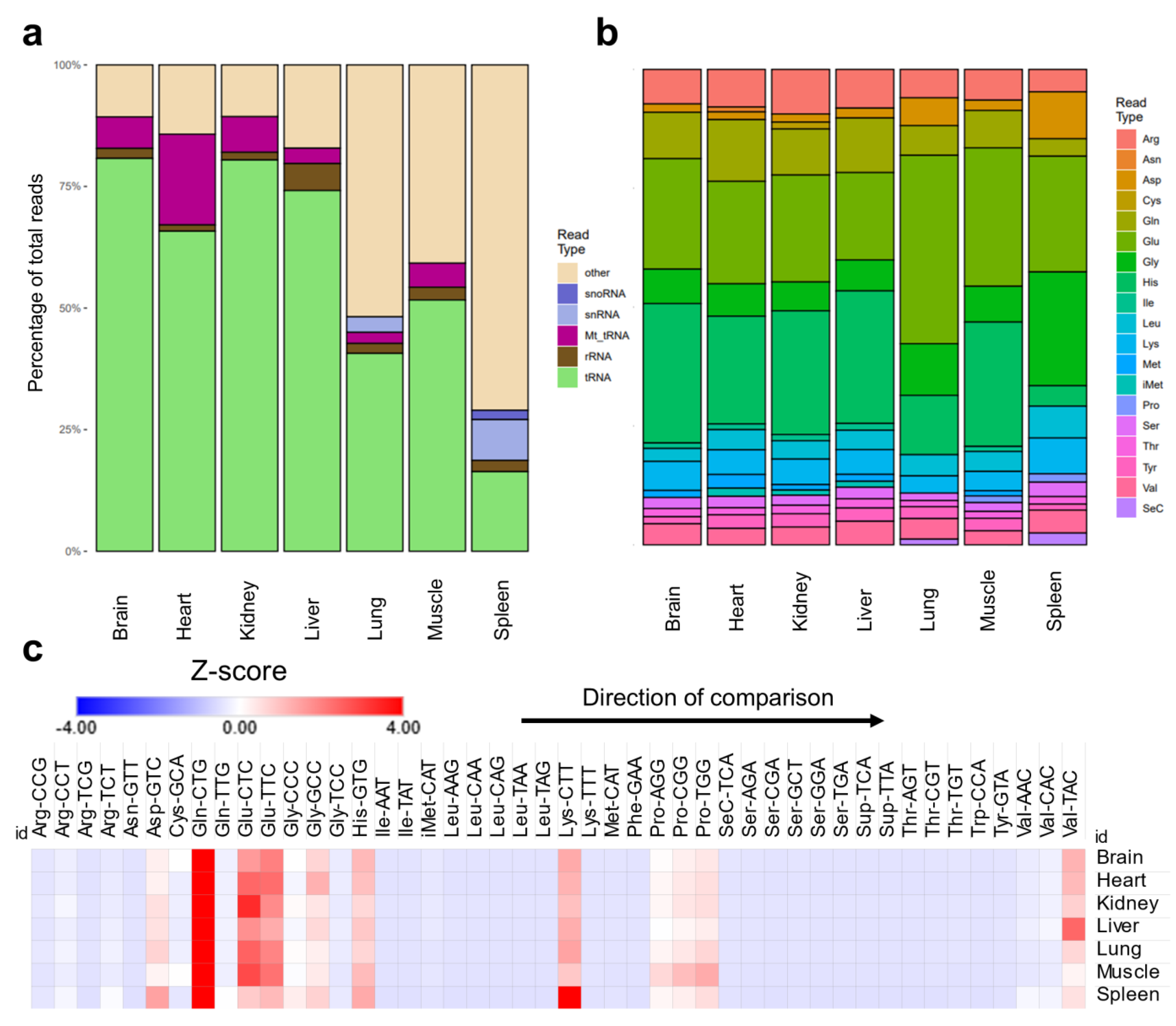
- tRNA isoacceptors are stably expressed equally across tissues.
- ➢
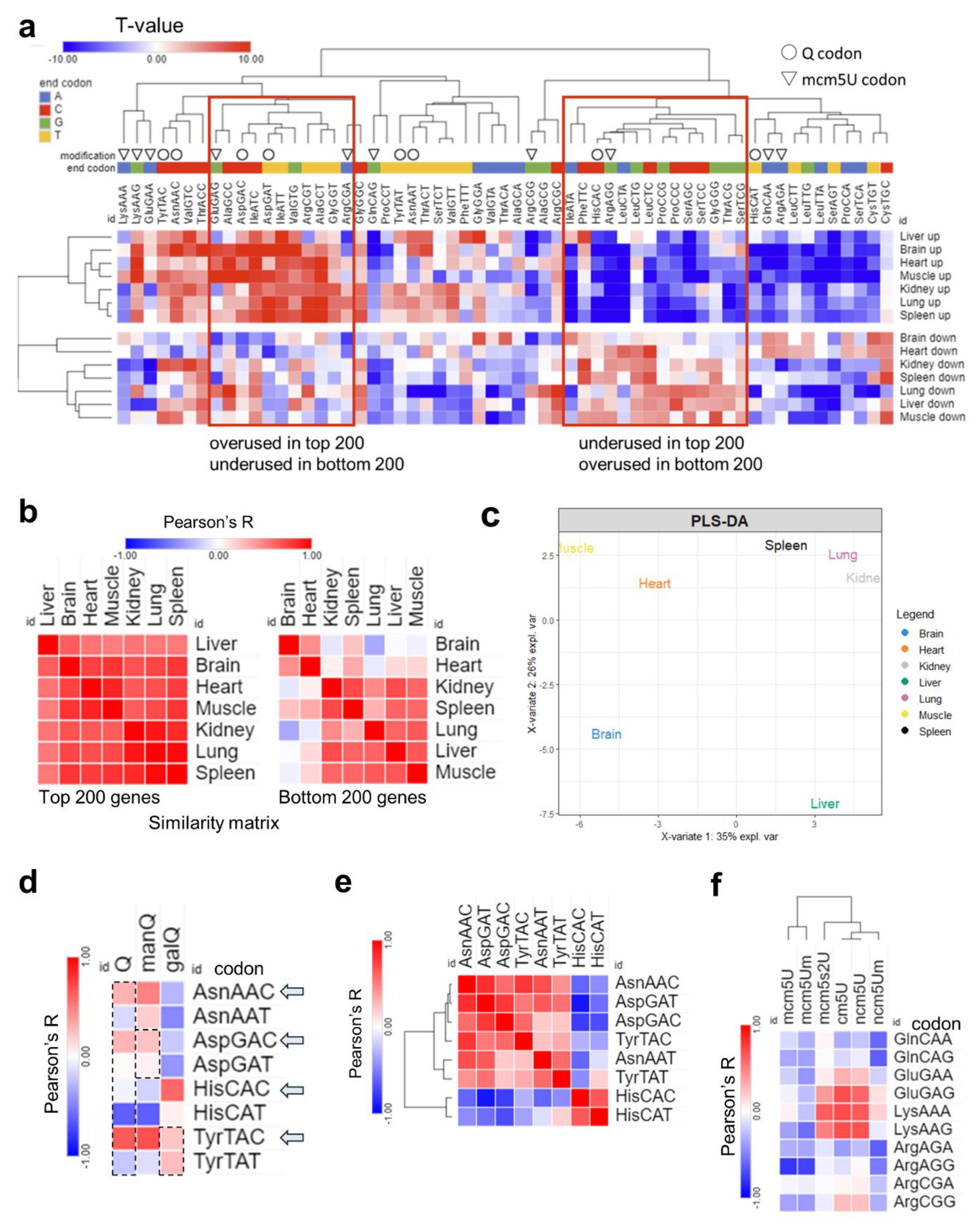
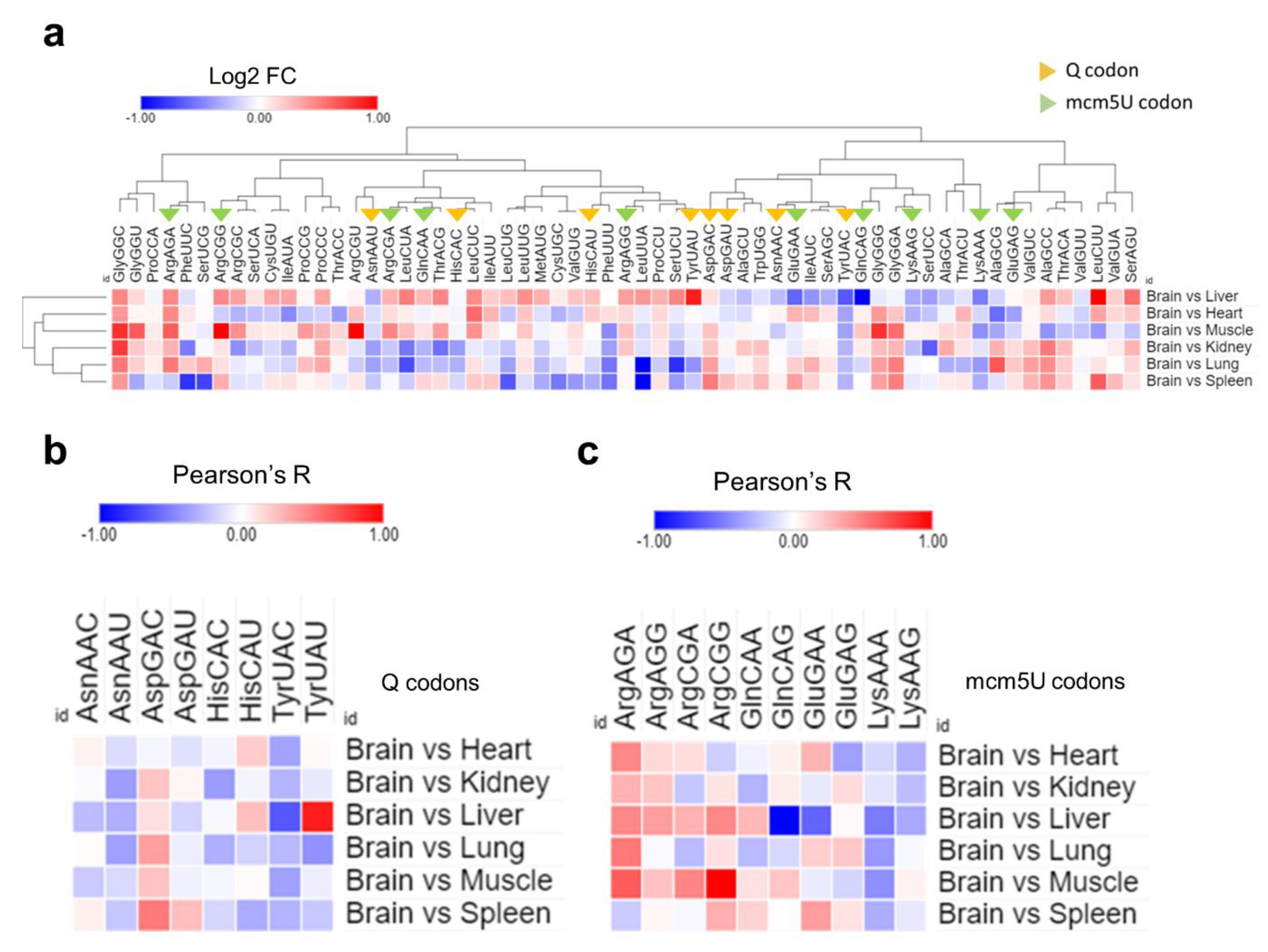
- Unique translational patterns observed in various tissues.
- ➢
- Tissue codon usage and optimality are governed by tRNA modifications levels.
- ➢
- tRNA modifications inform codon optimization algorithms for gene therapy.
3. Discussion
4. Methods
4.1. Animal and Sample Collection and RNA Isolation
4.2. Small RNA Quality Control
4.3. Quantitative Analysis of tRNA Modifications by Mass Spectrometry
4.4. Small RNA and tRNA Sequencing
4.5. Small RNA Sequencing Data Analysis
4.6. Ribosome Profiling
4.7. Ribosome Profiling Data Analysis
4.8. Codon Analysis
4.9. Generation of Adeno-Associated Viral (AAV) Vector Plasmids
- CTAGGAAGAGTACCATTGACGACATTGATTATTGACTAGTTATT
- AATCAATGTCTAGAGGCTCGAGCTCTT
- CGAGCCTCTAGACATTGATTATTGACTAGTTATT
- GGTGGCTTTAGGATCCGA
- TCGATATCAAGCTTATCGATAATCA
- GTCAATGGTACTCTTCCTAG
4.10. Viral Vector Production
4.11. In Vivo Virus Transduction
4.12. Western Blot
4.13. Data Visualization and Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Mao, Y.; Dong, L.; Liu, X.-M.; Guo, J.; Ma, H.; Shen, B.; Qian, S.-B. m6A in mRNA coding regions promotes translation via the RNA helicase-containing YTHDC2. Nat. Commun. 2019, 10, 5332. [Google Scholar] [CrossRef] [PubMed]
- Mauger, D.M.; Cabral, B.J.; Presnyak, V.; Su, S.V.; Reid, D.W.; Goodman, B.; Link, K.; Khatwani, N.; Reynders, J.; Moore, M.J.; et al. mRNA structure regulates protein expression through changes in functional half-life. Proc. Natl. Acad. Sci. USA 2019, 116, 24075–24083. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Hartmann, J.D.; Watzinger, P.; Klepper, A.; Peifer, C.; Kotter, P.; Lafontaine, D.L.J.; Entian, K.D. A single N(1)-methyladenosine on the large ribosomal subunit rRNA impacts locally its structure and the translation of key metabolic enzymes. Sci. Rep. 2018, 8, 11904. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T. The expanding world of tRNA modifications and their disease relevance. Nat. Rev. Mol. Cell Biol. 2021, 22, 375–392. [Google Scholar] [CrossRef]
- Nedialkova, D.D.; Leidel, S.A. Optimization of Codon Translation Rates via tRNA Modifications Maintains Proteome Integrity. Cell 2015, 161, 1606–1618. [Google Scholar] [CrossRef]
- Torrent, M.; Chalancon, G.; de Groot, N.S.; Wuster, A.; Madan Babu, M. Cells alter their tRNA abundance to selectively regulate protein synthesis during stress conditions. Sci. Signal. 2018, 11, eaat6409. [Google Scholar] [CrossRef]
- Goodarzi, H.; Nguyen, H.C.B.; Zhang, S.; Dill, B.D.; Molina, H.; Tavazoie, S.F. Modulated Expression of Specific tRNAs Drives Gene Expression and Cancer Progression. Cell 2016, 165, 1416–1427. [Google Scholar] [CrossRef]
- Liu, X.; Mei, W.; Padmanaban, V.; Alwaseem, H.; Molina, H.; Passarelli, M.C.; Tavora, B.; Tavazoie, S.F. A pro-metastatic tRNA fragment drives Nucleolin oligomerization and stabilization of its bound metabolic mRNAs. Mol. Cell 2022, 82, 2604–2617.e8. [Google Scholar] [CrossRef]
- Rosselló-Tortella, M.; Llinàs-Arias, P.; Sakaguchi, Y.; Miyauchi, K.; Davalos, V.; Setien, F.; Calleja-Cervantes, M.E.; Piñeyro, D.; Martínez-Gómez, J.; Guil, S.; et al. Epigenetic loss of the transfer RNA-modifying enzyme TYW2 induces ribosome frameshifts in colon cancer. Proc. Natl. Acad. Sci. USA 2020, 117, 20785–20793. [Google Scholar] [CrossRef]
- Zuko, A.; Mallik, M.; Thompson, R.; Spaulding, E.L.; Wienand, A.R.; Been, M.; Tadenev, A.L.D.; van Bakel, N.; Sijlmans, C.; Santos, L.A.; et al. tRNA overexpression rescues peripheral neuropathy caused by mutations in tRNA synthetase. Science 2021, 373, 1161–1166. [Google Scholar] [CrossRef]
- Dedon, P.C.; Begley, T.J. Dysfunctional tRNA reprogramming and codon-biased translation in cancer. Trends Mol. Med. 2022, 28, 964–978. [Google Scholar] [CrossRef] [PubMed]
- Matsumura, Y.; Wei, F.Y.; Sakai, J. Epitranscriptomics in metabolic disease. Nat. Metab. 2023, 5, 370–384. [Google Scholar] [CrossRef] [PubMed]
- Chan, P.P.; Lowe, T.M. GtRNAdb 2.0: An expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 2016, 44, D184–D189. [Google Scholar] [CrossRef] [PubMed]
- Crick, F.H. Codon—Anticodon pairing: The wobble hypothesis. J. Mol. Biol. 1966, 19, 548–555. [Google Scholar] [CrossRef] [PubMed]
- Huber, S.M.; Begley, U.; Sarkar, A.; Gasperi, W.; Davis, E.T.; Surampudi, V.; Lee, M.; Melendez, J.A.; Dedon, P.C.; Begley, T.J. Arsenite toxicity is regulated by queuine availability and oxidation-induced reprogramming of the human tRNA epitranscriptome. Proc. Natl. Acad. Sci. USA 2022, 119, e2123529119. [Google Scholar] [CrossRef]
- Rashad, S. Queuosine tRNA Modification: Connecting the Microbiome to the Translatome. Bioessays 2024, 2024, e2400213. [Google Scholar] [CrossRef]
- Murphy, F.V.; Ramakrishnan, V. Structure of a purine-purine wobble base pair in the decoding center of the ribosome. Nat. Struct. Mol. Biol. 2004, 11, 1251–1252. [Google Scholar] [CrossRef]
- Giguère, S.; Wang, X.; Huber, S.; Xu, L.; Warner, J.; Weldon, S.R.; Hu, J.; Phan, Q.A.; Tumang, K.; Prum, T.; et al. Antibody production relies on the tRNA inosine wobble modification to meet biased codon demand. Science 2024, 383, 205–211. [Google Scholar] [CrossRef]
- Cirzi, C.; Dyckow, J.; Legrand, C.; Schott, J.; Guo, W.; Perez Hernandez, D.; Hisaoka, M.; Parlato, R.; Pitzer, C.; van der Hoeven, F.; et al. Queuosine-tRNA promotes sex-dependent learning and memory formation by maintaining codon-biased translation elongation speed. EMBO J. 2023, 42, e112507. [Google Scholar] [CrossRef]
- Tuorto, F.; Legrand, C.; Cirzi, C.; Federico, G.; Liebers, R.; Muller, M.; Ehrenhofer-Murray, A.E.; Dittmar, G.; Grone, H.J.; Lyko, F. Queuosine-modified tRNAs confer nutritional control of protein translation. EMBO J. 2018, 37, e99777. [Google Scholar] [CrossRef]
- Zhao, X.; Ma, D.; Ishiguro, K.; Saito, H.; Akichika, S.; Matsuzawa, I.; Mito, M.; Irie, T.; Ishibashi, K.; Wakabayashi, K.; et al. Glycosylated queuosines in tRNAs optimize translational rate and post-embryonic growth. Cell 2023, 186, 5517–5535.e24. [Google Scholar] [CrossRef] [PubMed]
- Endres, L.; Begley, U.; Clark, R.; Gu, C.; Dziergowska, A.; Małkiewicz, A.; Melendez, J.A.; Dedon, P.C.; Begley, T.J. Alkbh8 Regulates Selenocysteine-Protein Expression to Protect against Reactive Oxygen Species Damage. PLoS ONE 2015, 10, e0131335. [Google Scholar] [CrossRef] [PubMed]
- Rashad, S.; Han, X.; Sato, K.; Mishima, E.; Abe, T.; Tominaga, T.; Niizuma, K. The stress specific impact of ALKBH1 on tRNA cleavage and tiRNA generation. RNA Biol. 2020, 17, 1092–1103. [Google Scholar] [CrossRef] [PubMed]
- Rashad, S.; Tominaga, T.; Niizuma, K. The cell and stress-specific canonical and noncanonical tRNA cleavage. J. Cell. Physiol. 2021, 236, 3710–3724. [Google Scholar] [CrossRef] [PubMed]
- Rashad, S.; Al-Mesitef, S.; Mousa, A.; Zhou, Y.; Ando, D.; Sun, G.; Fukuuchi, T.; Iwasaki, Y.; Xiang, J.; Byrne, S.R.; et al. Translational response to mitochondrial stresses is orchestrated by tRNA modifications. bioRxiv 2024. [Google Scholar] [CrossRef]
- Deng, W.; Babu, I.R.; Su, D.; Yin, S.; Begley, T.J.; Dedon, P.C. Trm9-Catalyzed tRNA Modifications Regulate Global Protein Expression by Codon-Biased Translation. PLoS Genet. 2015, 11, e1005706. [Google Scholar] [CrossRef]
- Chan, C.T.Y.; Pang, Y.L.J.; Deng, W.; Babu, I.R.; Dyavaiah, M.; Begley, T.J.; Dedon, P.C. Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins. Nat. Commun. 2012, 3, 937. [Google Scholar] [CrossRef]
- Guo, H.; Xia, L.; Wang, W.; Xu, W.; Shen, X.; Wu, X.; He, T.; Jiang, X.; Xu, Y.; Zhao, P.; et al. Hypoxia induces alterations in tRNA modifications involved in translational control. BMC Biol. 2023, 21, 39. [Google Scholar] [CrossRef]
- Pinkard, O.; McFarland, S.; Sweet, T.; Coller, J. Quantitative tRNA-sequencing uncovers metazoan tissue-specific tRNA regulation. Nat. Commun. 2020, 11, 4104. [Google Scholar] [CrossRef]
- Yu, P.; Zhou, S.; Gao, Y.; Liang, Y.; Guo, W.; Wang, D.O.; Ding, S.; Lin, S.; Wang, J.; Cun, Y. Dynamic landscapes of tRNA transcriptomes and translatomes in diverse mouse tissues. Genom. Proteom. Bioinform. 2022, 21, 834–849. [Google Scholar] [CrossRef]
- Oerum, S.; Meynier, V.; Catala, M.; Tisne, C. A comprehensive review of m6A/m6Am RNA methyltransferase structures. Nucleic Acids Res. 2021, 49, 7239–7255. [Google Scholar] [CrossRef] [PubMed]
- van den Born, E.; Vagbo, C.B.; Songe-Moller, L.; Leihne, V.; Lien, G.F.; Leszczynska, G.; Malkiewicz, A.; Krokan, H.E.; Kirpekar, F.; Klungland, A.; et al. ALKBH8-mediated formation of a novel diastereomeric pair of wobble nucleosides in mammalian tRNA. Nat. Commun. 2011, 2, 172. [Google Scholar] [CrossRef]
- Kawarada, L.; Suzuki, T.; Ohira, T.; Hirata, S.; Miyauchi, K.; Suzuki, T. ALKBH1 is an RNA dioxygenase responsible for cytoplasmic and mitochondrial tRNA modifications. Nucleic Acids Res. 2017, 45, 7401–7415. [Google Scholar] [CrossRef]
- Yamamoto, T.; Fujimura, A.; Wei, F.Y.; Shinojima, N.; Kuroda, J.I.; Mukasa, A.; Tomizawa, K. 2-Methylthio Conversion of N6-Isopentenyladenosine in Mitochondrial tRNAs by CDK5RAP1 Promotes the Maintenance of Glioma-Initiating Cells. iScience 2019, 21, 42–56. [Google Scholar] [CrossRef]
- Berg, M.D.; Brandl, C.J. Transfer RNAs: Diversity in form and function. RNA Biol. 2021, 18, 316–339. [Google Scholar] [CrossRef]
- Avery, J.C.; Hoffmann, P.R. Selenium, Selenoproteins, and Immunity. Nutrients 2018, 10, 1203. [Google Scholar] [CrossRef]
- Gao, L.; Behrens, A.; Rodschinka, G.; Forcelloni, S.; Wani, S.; Strasser, K.; Nedialkova, D.D. Selective gene expression maintains human tRNA anticodon pools during differentiation. Nat. Cell Biol. 2024, 26, 100–112. [Google Scholar] [CrossRef]
- Rashad, S.; Byrne, S.R.; Saigusa, D.; Xiang, J.; Zhou, Y.; Zhang, L.; Begley, T.J.; Tominaga, T.; Niizuma, K. Codon Usage and mRNA Stability are Translational Determinants of Cellular Response to Canonical Ferroptosis Inducers. Neuroscience 2022, 501, 103–130. [Google Scholar] [CrossRef]
- Tumu, S.; Patil, A.; Towns, W.; Dyavaiah, M.; Begley, T.J. The gene-specific codon counting database: A genome-based catalog of one-, two-, three-, four- and five-codon combinations present in Saccharomyces cerevisiae genes. Database 2012, 2012, bas002. [Google Scholar] [CrossRef]
- Liu, Q.; Shvarts, T.; Sliz, P.; Gregory, R.I. RiboToolkit: An integrated platform for analysis and annotation of ribosome profiling data to decode mRNA translation at codon resolution. Nucleic Acids Res. 2020, 48, W218–W229. [Google Scholar] [CrossRef]
- Benisty, H.; Hernandez-Alias, X.; Weber, M.; Anglada-Girotto, M.; Mantica, F.; Radusky, L.; Senger, G.; Calvet, F.; Weghorn, D.; Irimia, M.; et al. Genes enriched in A/T-ending codons are co-regulated and conserved across mammals. Cell Syst. 2023, 14, 312–323.e3. [Google Scholar] [CrossRef] [PubMed]
- Müller, M.; Legrand, C.; Tuorto, F.; Kelly, V.P.; Atlasi, Y.; Lyko, F.; Ehrenhofer-Murray, A.E. Queuine links translational control in eukaryotes to a micronutrient from bacteria. Nucleic Acids Res. 2019, 47, 3711–3727. [Google Scholar] [CrossRef] [PubMed]
- Dixit, S.; Kessler, A.C.; Henderson, J.; Pan, X.; Zhao, R.; D’Almeida, G.S.; Kulkarni, S.; Rubio, M.A.T.; Hegedusova, E.; Ross, R.L.; et al. Dynamic queuosine changes in tRNA couple nutrient levels to codon choice in Trypanosoma brucei. Nucleic Acids Res. 2021, 49, 12986–12999. [Google Scholar] [CrossRef]
- Wu, X.; Yuan, H.; Wu, Q.; Gao, Y.; Duan, T.; Yang, K.; Huang, T.; Wang, S.; Yuan, F.; Lee, D.; et al. Threonine fuels glioblastoma through YRDC-mediated codon-biased translational reprogramming. Nat. Cancer 2024, 5, 1024–1044. [Google Scholar] [CrossRef]
- Presnyak, V.; Alhusaini, N.; Chen, Y.H.; Martin, S.; Morris, N.; Kline, N.; Olson, S.; Weinberg, D.; Baker, K.E.; Graveley, B.R.; et al. Codon optimality is a major determinant of mRNA stability. Cell 2015, 160, 1111–1124. [Google Scholar] [CrossRef]
- Bae, H.; Coller, J. Codon optimality-mediated mRNA degradation: Linking translational elongation to mRNA stability. Mol. Cell 2022, 82, 1467–1476. [Google Scholar] [CrossRef]
- Buschauer, R.; Matsuo, Y.; Sugiyama, T.; Chen, Y.H.; Alhusaini, N.; Sweet, T.; Ikeuchi, K.; Cheng, J.; Matsuki, Y.; Nobuta, R.; et al. The Ccr4-Not complex monitors the translating ribosome for codon optimality. Science 2020, 368, eaay6912. [Google Scholar] [CrossRef]
- Hanson, G.; Coller, J. Codon optimality, bias and usage in translation and mRNA decay. Nat. Rev. Mol. Cell Biol. 2018, 19, 20–30. [Google Scholar] [CrossRef]
- Bornelov, S.; Selmi, T.; Flad, S.; Dietmann, S.; Frye, M. Codon usage optimization in pluripotent embryonic stem cells. Genome Biol. 2019, 20, 119. [Google Scholar] [CrossRef]
- Guimaraes, J.C.; Mittal, N.; Gnann, A.; Jedlinski, D.; Riba, A.; Buczak, K.; Schmidt, A.; Zavolan, M. A rare codon-based translational program of cell proliferation. Genome Biol. 2020, 21, 44. [Google Scholar] [CrossRef]
- Allen, S.R.; Stewart, R.K.; Rogers, M.; Ruiz, I.J.; Cohen, E.; Laederach, A.; Counter, C.M.; Sawyer, J.K.; Fox, D.T. Distinct responses to rare codons in select Drosophila tissues. Elife 2022, 11, e76893. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Zhang, J. Preferred synonymous codons are translated more accurately: Proteomic evidence, among-species variation, and mechanistic basis. Sci. Adv. 2022, 8, eabl9812. [Google Scholar] [CrossRef] [PubMed]
- Sabi, R.; Volvovitch Daniel, R.; Tuller, T. stAIcalc: tRNA adaptation index calculator based on species-specific weights. Bioinformatics 2017, 33, 589–591. [Google Scholar] [CrossRef]
- Sharp, P.M.; Li, W.H. The codon Adaptation Index-a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987, 15, 1281–1295. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Alias, X.; Benisty, H.; Schaefer, M.H.; Serrano, L. Translational efficiency across healthy and tumor tissues is proliferation-related. Mol. Syst. Biol. 2020, 16, e9275. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, L.; Lin, A.; Xu, C.; Li, Z.; Liu, K.; Liu, B.; Ma, X.; Zhao, F.; Jiang, H.; et al. Algorithm for optimized mRNA design improves stability and immunogenicity. Nature 2023, 621, 396–403. [Google Scholar] [CrossRef]
- Delaunay, S.; Pascual, G.; Feng, B.; Klann, K.; Behm, M.; Hotz-Wagenblatt, A.; Richter, K.; Zaoui, K.; Herpel, E.; Munch, C.; et al. Mitochondrial RNA modifications shape metabolic plasticity in metastasis. Nature 2022, 607, 593–603. [Google Scholar] [CrossRef]
- Hayes, P.; Fergus, C.; Ghanim, M.; Cirzi, C.; Burtnyak, L.; McGrenaghan, C.J.; Tuorto, F.; Nolan, D.P.; Kelly, V.P. Queuine Micronutrient Deficiency Promotes Warburg Metabolism and Reversal of the Mitochondrial ATP Synthase in Hela Cells. Nutrients 2020, 12, 871. [Google Scholar] [CrossRef]
- RajBhandary, U.L.; Ghosh, H.P. Studies on polynucleotides. XCI. Yeast methionine transfer ribonucleic acid: Purification, properties, and terminal nucleotide sequences. J. Biol. Chem. 1969, 244, 1104–1113. [Google Scholar] [CrossRef]
- Goodman, H.M.; Abelson, J.; Landy, A.; Brenner, S.; Smith, J.D. Amber suppression: A nucleotide change in the anticodon of a tyrosine transfer RNA. Nature 1968, 217, 1019–1024. [Google Scholar] [CrossRef]
- Doctor, B.P.; Loebel, J.E.; Sodd, M.A.; Winter, D.B. Nucleotide sequence of Escherichia coli tyrosine transfer ribonucleic acid. Science 1969, 163, 693–695. [Google Scholar] [CrossRef] [PubMed]
- Harada, F.; Nishimura, S. Possible anticodon sequences of tRNAHis, tRNAAsm, and tRNAAsp from Escherichia coli B. Universal presence of nucleoside Q in the first postion of the anticondons of these transfer ribonucleic acids. Biochemistry 1972, 11, 301–308. [Google Scholar] [CrossRef] [PubMed]
- Fergus, C.; Al-Qasem, M.; Cotter, M.; McDonnell, C.M.; Sorrentino, E.; Chevot, F.; Hokamp, K.; Senge, M.O.; Southern, J.M.; Connon, S.J.; et al. The human tRNA-guanine transglycosylase displays promiscuous nucleobase preference but strict tRNA specificity. Nucleic Acids Res. 2021, 49, 4877–4890. [Google Scholar] [CrossRef]
- Marks, T.; Farkas, W.R. Effects of a diet deficient in tyrosine and queuine on germfree mice. Biochem. Biophys. Res. Commun. 1997, 230, 233–237. [Google Scholar] [CrossRef] [PubMed]
- Farkas, W.R. Effect of diet on the queuosine family of tRNAs of germ-free mice. J. Biol. Chem. 1980, 255, 6832–6835. [Google Scholar] [CrossRef] [PubMed]
- Richard, P.; Kozlowski, L.; Guillorit, H.; Garnier, P.; McKnight, N.C.; Danchin, A.; Maniere, X. Queuine, a bacterial-derived hypermodified nucleobase, shows protection in in vitro models of neurodegeneration. PLoS ONE 2021, 16, e0253216. [Google Scholar] [CrossRef]
- Hernandez-Alias, X.; Katanski, C.D.; Zhang, W.; Assari, M.; Watkins, C.P.; Schaefer, M.H.; Serrano, L.; Pan, T. Single-read tRNA-seq analysis reveals coordination of tRNA modification and aminoacylation and fragmentation. Nucleic Acids Res. 2023, 51, e17. [Google Scholar] [CrossRef]
- Su, D.; Chan, C.T.; Gu, C.; Lim, K.S.; Chionh, Y.H.; McBee, M.E.; Russell, B.S.; Babu, I.R.; Begley, T.J.; Dedon, P.C. Quantitative analysis of ribonucleoside modifications in tRNA by HPLC-coupled mass spectrometry. Nat. Protoc. 2014, 9, 828–841. [Google Scholar] [CrossRef]
- Cozen, A.E.; Quartley, E.; Holmes, A.D.; Hrabeta-Robinson, E.; Phizicky, E.M.; Lowe, T.M. ARM-seq: AlkB-facilitated RNA methylation sequencing reveals a complex landscape of modified tRNA fragments. Nat. Methods 2015, 12, 879–884. [Google Scholar] [CrossRef]
- Pichot, F.; Hogg, M.C.; Marchand, V.; Bourguignon, V.; Jirstrom, E.; Farrell, C.; Gibriel, H.A.; Prehn, J.H.M.; Motorin, Y.; Helm, M. Quantification of substoichiometric modification reveals global tsRNA hypomodification, preferences for angiogenin-mediated tRNA cleavage, and idiosyncratic epitranscriptomes of human neuronal cell-lines. Comput. Struct. Biotechnol. J. 2023, 21, 401–417. [Google Scholar] [CrossRef]
- Li, G.; Manning, A.C.; Bagi, A.; Yang, X.; Gokulnath, P.; Spanos, M.; Howard, J.; Chan, P.P.; Sweeney, T.; Kitchen, R.; et al. Distinct Stress-Dependent Signatures of Cellular and Extracellular tRNA-Derived Small RNAs. Adv. Sci. 2022, 9, e2200829. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Rashad, S.; Tominaga, T.; Niizuma, K. Dynamic mRNA stability changes buffer transcriptional activation during neuronal differentiation and are regulated by RNA binding proteins. bioRxiv 2023. [Google Scholar] [CrossRef]
- Vu, L.P.; Pickering, B.F.; Cheng, Y.; Zaccara, S.; Nguyen, D.; Minuesa, G.; Chou, T.; Chow, A.; Saletore, Y.; MacKay, M.; et al. The N(6)-methyladenosine (m(6)A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat. Med. 2017, 23, 1369–1376. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Akhmedov, M.; Martinelli, A.; Geiger, R.; Kwee, I. Omics Playground: A comprehensive self-service platform for visualization, analytics and exploration of Big Omics Data. NAR Genom. Bioinform. 2020, 2, lqz019. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Albers, J.; Danzer, C.; Rechsteiner, M.; Lehmann, H.; Brandt, L.P.; Hejhal, T.; Catalano, A.; Busenhart, P.; Gonçalves, A.F.; Brandt, S.; et al. A versatile modular vector system for rapid combinatorial mammalian genetics. J. Clin. Investig. 2015, 125, 1603–1619. [Google Scholar] [CrossRef]
- Chan, K.Y.; Jang, M.J.; Yoo, B.B.; Greenbaum, A.; Ravi, N.; Wu, W.L.; Sánchez-Guardado, L.; Lois, C.; Mazmanian, S.K.; Deverman, B.E.; et al. Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat. Neurosci. 2017, 20, 1172–1179. [Google Scholar] [CrossRef] [PubMed]
- Challis, R.C.; Ravindra Kumar, S.; Chan, K.Y.; Challis, C.; Beadle, K.; Jang, M.J.; Kim, H.M.; Rajendran, P.S.; Tompkins, J.D.; Shivkumar, K.; et al. Systemic AAV vectors for widespread and targeted gene delivery in rodents. Nat. Protoc. 2019, 14, 379–414. [Google Scholar] [CrossRef] [PubMed]
- Rohart, F.; Gautier, B.; Singh, A.; KA, L.C. mixOmics: An R package for ’omics feature selection and multiple data integration. PLoS Comput. Biol. 2017, 13, e1005752. [Google Scholar] [CrossRef] [PubMed]



| Abbreviation | Full Name | Occurrence in Small (<200nt) RNAs |
|---|---|---|
| ac4C | N4-acetylcytidine | tRNA |
| acp3U | 3-(3-amino-3-carboxypropyl)uridine | tRNA |
| Am | 2′-O-methyladenosine | rRNA, snRNA, snoRNA |
| Cm | 2′-O-methylcytidine | rRNA, snRNA, tRNA |
| cm5U | 5-carboxymethyluridine | tRNA |
| D | 5-carboxymethyluridine | rRNA, tRNA |
| f5C | 5-formylcytidine | tRNA |
| f5Cm | 5-formyl-2′-O-methylcytidine | tRNA |
| Gm | 2′-O-methylguanosine | rRNA, snRNA, tRNA |
| hm5C | 5-hydroxymethylcytidine | tRNA |
| hm5Cm | 2′-O-methyl-5-hydroxymethylcytidine | tRNA |
| I | Inosine | tRNA |
| i6A | N6-isopentenyladenosine | tRNA |
| m1A | 1-methyladenosine | rRNA, tRNA |
| m1G | 1-methylguanosine | rRNA, tRNA |
| m1I | 1-methylinosine | tRNA |
| m1Y | 1-methylpseudouridine | unknown |
| m227G | N2,N2,7-trimethylguanosine | rRNA? |
| m22G | N2,N2-dimethylguanosine | tRNA, rRNA |
| m2G | N2-methylguanosine | rRNA, snRNA, tRNA |
| m3C | 3-methylcytidine | tRNA |
| m3U | 3-methyluridine | rRNA |
| m5C | 5-methylcytidine | tRNA, rRNA |
| m5U | 5-methyluridine | tRNA, rRNA |
| m5Um | 5,2′-O-dimethyluridine | tRNA |
| m66A | N6,N6-dimethyladenosine | rRNA |
| m6A | N6-methyladenosine | rRNA, snRNA |
| m6Am | N6,2′-O-dimethyladenosine | snRNA |
| m6t6A | N6-methyl-N6-threonylcarbamoyladenosine | tRNA |
| m7G | 7-methylguanosine | rRNA, tRNA, miRNA |
| man Q | Mannosyl-queuosine | tRNA |
| gal Q | Galactosyl-queuosine | tRNA |
| mchm5U_R | 5-(carboxyhydroxymethyl)uridine methyl ester (R) | tRNA |
| mchm5U_S | 5-(carboxyhydroxymethyl)uridine methyl ester (S) | tRNA |
| mcm5s2U | 5-methoxycarbonylmethyl-2-thiouridine | tRNA |
| mcm5U | 5-methoxycarbonylmethyluridine | tRNA |
| mcm5Um | 5-methoxycarbonylmethyl-2′-O-methyluridine | tRNA |
| ms2i6A | 2-methylthio-N6-isopentenyladenosine | tRNA |
| ms2t6A | 2-methylthio-N6-threonylcarbamoyladenosine | tRNA |
| ncm5U | 5-carbamoylmethyluridine | tRNA |
| ncm5Um | 5-carbamoylmethyl-2′-O-methyluridine | tRNA |
| oHyW | Hydroxywybutosine | tRNA |
| Q | Queuosine | tRNA |
| t6A | N6-threonylcarbamoyladenosine | tRNA |
| Um | 2′-O-methyluridine | rRNA, snRNA, snoRNA, tRNA |
| Y | Pseudouridine | rRNA, snRNA, snoRNA, tRNA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ando, D.; Rashad, S.; Begley, T.J.; Endo, H.; Aoki, M.; Dedon, P.C.; Niizuma, K. Decoding Codon Bias: The Role of tRNA Modifications in Tissue-Specific Translation. Int. J. Mol. Sci. 2025, 26, 706. https://doi.org/10.3390/ijms26020706
Ando D, Rashad S, Begley TJ, Endo H, Aoki M, Dedon PC, Niizuma K. Decoding Codon Bias: The Role of tRNA Modifications in Tissue-Specific Translation. International Journal of Molecular Sciences. 2025; 26(2):706. https://doi.org/10.3390/ijms26020706
Chicago/Turabian StyleAndo, Daisuke, Sherif Rashad, Thomas J. Begley, Hidenori Endo, Masashi Aoki, Peter C. Dedon, and Kuniyasu Niizuma. 2025. "Decoding Codon Bias: The Role of tRNA Modifications in Tissue-Specific Translation" International Journal of Molecular Sciences 26, no. 2: 706. https://doi.org/10.3390/ijms26020706
APA StyleAndo, D., Rashad, S., Begley, T. J., Endo, H., Aoki, M., Dedon, P. C., & Niizuma, K. (2025). Decoding Codon Bias: The Role of tRNA Modifications in Tissue-Specific Translation. International Journal of Molecular Sciences, 26(2), 706. https://doi.org/10.3390/ijms26020706






