Role of Artificial Intelligence in Identifying Vital Biomarkers with Greater Precision in Emergency Departments During Emerging Pandemics
Abstract
:1. Introduction
2. Results
3. Discussion
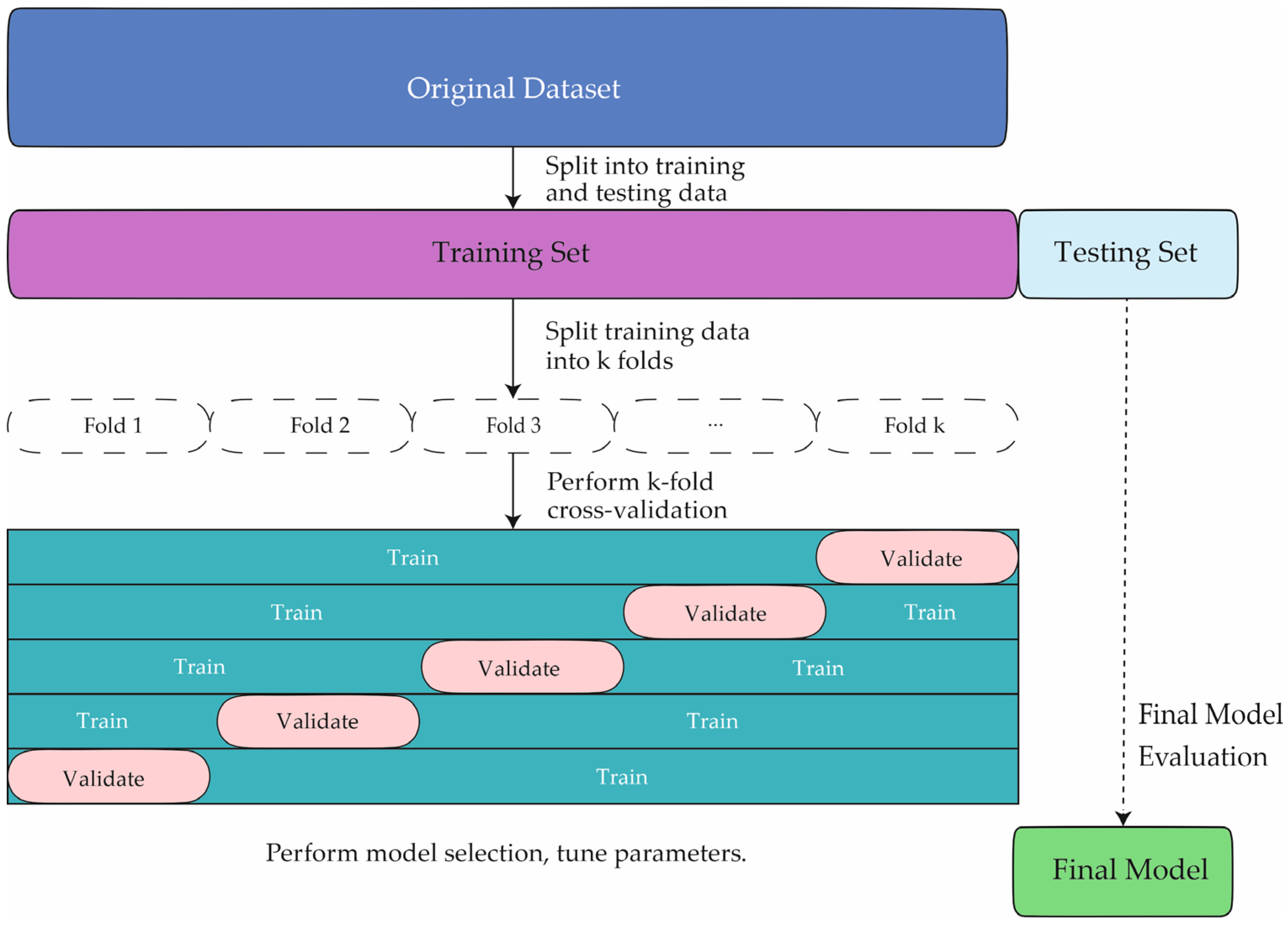
4. Materials and Methods
4.1. Patients
4.2. Artificial Intelligence Method
- Bootstrapping (Sampling with Replacement): From a training dataset containing n observations and pp features, a subset Di is generated by randomly selecting n samples with replacement from the original dataset. This technique allows certain data points to appear multiple times in Di, while others may not appear at all.
- Random Feature Selection: At each node of each tree, instead of evaluating all pp features, a random subset of kk features is selected, where k = . This reduces correlation between individual trees, enhancing the model’s generalization capability.
- Node Splitting Criterion: Each node is split based on an impurity reduction criterion, such as entropy or the Gini index, in classification tasks. In this study, the Gini index was used. The impurity GG of a node with class proportions pk is defined as:
- Tree Aggregation: Once the trees are trained, RF predictions are obtained through aggregation. For a set of tree {T1, T2, …, Tm}, the final prediction is determined by majority voting:
- Feature Importance: The importance of each feature is measured by evaluating the change in the splitting criterion when the feature is randomly permuted in the dataset. For Gini index-based importance, a feature is considered important if permuting it increases node impurity across the trees.
- Model Evaluation: The model’s performance was assessed using metrics such as accuracy, sensitivity, specificity, and AUC in classification problems or mean squared error (MSE) in regression tasks.
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yuan, Y.; Jiao, B.; Qu, L.; Yang, D.; Liu, R. The development of COVID-19 treatment. Front. Immunol. 2023, 14, 1125246. [Google Scholar] [CrossRef] [PubMed]
- Ao, D.; He, X.; Liu, J.; Xu, L. Strategies for the development and approval of COVID-19 vaccines and therapeutics in the post-pandemic period. Signal Transduct. Target. Ther. 2023, 8, 466. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Sharma, L.; Chang, D. Pathophysiology and clinical management of coronavirus disease (COVID-19): A mini-review. Front. Immunol. 2023, 14, 1116131. [Google Scholar] [CrossRef]
- Anand, U.; Jakhmola, S.; Indari, O.; Jha, H.C.; Chen, Z.-S.; Tripathi, V.; de la Lastra, J.M.P. Potential therapeutic targets and vaccine development for SARS-CoV-2/COVID-19 pandemic management: A review on the recent update. Front. Immunol. 2021, 12, 658519. [Google Scholar] [CrossRef]
- Zhang, H.-P.; Sun, Y.-L.; Wang, Y.-F.; Yazici, D.; Azkur, D.; Ogulur, I.; Azkur, A.K.; Yang, Z.; Chen, X.; Zhang, A.; et al. Recent developments in the immunopathology of COVID-19. Allergy 2022, 78, 369–388. [Google Scholar] [CrossRef] [PubMed]
- Franco-Moreno, A.I.; Bustamante-Fermosel, A.; Ruiz-Giardin, J.M.; Muñoz-Rivas, N.; Torres-Macho, J.; Brown-Lavalle, D. Utility of probability scores for the diagnosis of pulmonary embolism in patients with SARS-CoV-2 infection: A systematic review. Rev. Clin. Esp. 2022, 223, 40–49. [Google Scholar] [CrossRef] [PubMed]
- Franco-Moreno, A.; Palma-Huerta, E.; Fernández-Vidal, E.; Madroñal-Cerezo, E.; Marco-Martínez, J.; Romero-Pareja, R.; Izquierdo-Martínez, A.; Carpintero-García, L.; Ruiz-Giardín, J.M.; Torres-Macho, J.; et al. External validation of the CHEDDAR score for suspected pulmonary embolism in patients with SARS-CoV-2 infection in an independent cohort. J. Thromb. Thrombolysis 2024, 57, 352–357. [Google Scholar] [CrossRef] [PubMed]
- Vielhauer, J.; Benesch, C.; Pernpruner, A.; Johlke, A.L.; Hellmuth, J.C.; Muenchhoff, M.; Scherer, C.; Fink, N.; Sabel, B.; Schulz, C.; et al. How to exclude pulmonary embolism in patients hospitalized with COVID-19: A comparison of predictive scores. Thromb. J. 2023, 21, 51. [Google Scholar] [CrossRef] [PubMed]
- Mousavi Aghdam, M.; Crowley, Q. Application of GIS and spatiotemporal analyses in viral infection modelling using multiple datasets—A case study on the SARS-CoV-2 epidemic. Semergen 2024, 50, 102159. [Google Scholar] [CrossRef] [PubMed]
- Marín-Benesiu, F.; Chica-Redecillas, L.; Arenas-Rodríguez, V.; de Santiago, E.; Martínez-Diz, S.; López-Torres, G.; Cortés-Valverde, A.I.; Romero-Cachinero, C.; Entrala-Bernal, C.; Fernandez-Rosado, F.J.; et al. The T-cell repertoire of Spanish patients with COVID-19 as a strategy to link T-cell characteristics to the severity of the disease. Hum. Genom. 2024, 18, 94. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Li, X.X.; Yuan, N.; Li, C.; Yang, J.G.; Cheng, L.M.; Lu, Z.-X.; Hou, H.-Y.; Zhang, B.; Hu, H.; et al. T cell receptor β repertoires in patients with COVID-19 reveal disease severity signatures. Front. Immunol. 2023, 14, 1190844. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Diz, S.; Morales-Álvarez, C.M.; Garcia-Iglesias, Y.; Guerrero-González, J.M.; Romero-Cachinero, C.; González-Cabezuelo, J.M.; Fernandez-Rosado, F.J.; Arenas-Rodríguez, V.; Lopez-Cintas, R.; Alvarez-Cubero, M.J.; et al. Analyzing the role of ACE2, AR, MX1 and TMPRSS2 genetic markers for COVID-19 severity. Hum. Genom. 2023, 17, 50. [Google Scholar] [CrossRef] [PubMed]
- Meseldžić, N.; Prnjavorac, B.; Dujić, T.; Malenica, M.; Glamočlija, U.; Prnjavorac, L.; Bedak, O.; Kadrić, S.I.; Marjanović, D.; Bego, T. Association of ACE2 and TMPRSS2 genes variants with disease severity and most important biomarkers in COVID-19 patients in Bosnia and Herzegovina. Croat. Med. J. 2024, 65, 220. [Google Scholar] [CrossRef] [PubMed]
- Alam, N.; Lodhi, G.M.; Khan, U.A.; Zia, A.; Azam, M.; Khan, J.; Shah, T.A.; Okla, M.K.; Ali younous, Y.; Bourhia, M. Association of ACE2 and TMPRSS2 towards COVID-19 susceptibility. Discov. Life 2024, 54, 6. [Google Scholar] [CrossRef]
- Vera-Lastra, O.; Mora, G.; Lucas-Hernández, A.; Ordinola-Navarro, A.; Rodríguez-Chávez, E.; Peralta-Amaro, A.L.; Medina, G.; Cruz-Dominguez, M.P.; Jara, L.J.; Shoenfeld, Y. New onset autoimmune diseases after the Sputnik vaccine. Biomedicines 2023, 11, 1898. [Google Scholar] [CrossRef] [PubMed]
- Barajas Galindo, D.E.; Ramos Bachiller, B.; González Roza, L.; García Ruiz de Morales, J.M.; Sánchez Lasheras, F.; González Arnáiz, E.; Ariadel Cobo, D.; Ballesteros Pomar, M.D.; Rodríguez, I.C. Increased incidence of Graves’ disease during the SARS-CoV2 pandemic. Clin. Endocrinol. 2022, 98, 730–737. [Google Scholar] [CrossRef]
- Sandeep, F.; Kiran, N.; Rahaman, Z.; Devi, P.; Bendari, A. Pathology in the age of artificial intelligence (AI): Redefining roles and responsibilities for tomorrow’s practitioners. Cureus 2024, 16, e56040. [Google Scholar] [CrossRef]
- Lu, H.; Li, L.; Ong, K.; Wang, Y.; Jiao, Y.; Wang, X.; Cai, C.; Zhang, J.; Hou, J.; Zhao, H.; et al. AI-Based Computational Pathology and Its Contribution to Precision Medicine. In Frontiers in Bioimage Informatics Methodology; World Scientific: Singapore, 2024; pp. 167–193. [Google Scholar]
- Casado, P.; Cutillas, P.R. Proteomic characterization of acute myeloid leukemia for precision medicine. Mol. Cell. Proteom. MCP 2023, 22, 100517. [Google Scholar] [CrossRef]
- Balzanelli, M.G.; Distratis, P.; Dipalma, G.; Vimercati, L.; Catucci, O.; Amatulli, F.; Cefalo, A.; Lazzaro, R.; Palazzo, D.; Aityan, S.K.; et al. Immunity profiling of COVID-19 infection, dynamic variations of lymphocyte subsets, a comparative analysis on four different groups. Microorganisms 2021, 9, 2036. [Google Scholar] [CrossRef]
- Galli, F.; Bindo, F.; Motos, A.; Fernández-Barat, L.; Barbeta, E.; Gabarrús, A.; Ceccato, A.; Bermejo-Martin, J.F.; Ferrer, R.; Riera, J.; et al. Procalcitonin and C-reactive protein to rule out early bacterial coinfection in COVID-19 critically ill patients. Intensive Care Med. 2023, 49, 934–945. [Google Scholar] [CrossRef]
- Balzanelli, M.G.; Distratis, P.; Lazzaro, R.; Cefalo, A.; Catucci, O.; Aityan, S.K.; Dipalma, G.; Vimercati, L.; Inchingolo, A.D.; Maggiore, M.E.; et al. The Vitamin D, IL-6 and the eGFR markers a possible way to elucidate the lung–heart–kidney cross-talk in COVID-19 disease: A foregone conclusion. Microorganisms 2021, 9, 1903. [Google Scholar] [CrossRef]
- Vranic, S.; Gatalica, Z. PD-L1 testing by immunohistochemistry in immuno-oncology. Biomol. Biomed. 2023, 23, 15–25. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hao, L.; Li, S.; Deng, J.; Li, N.; Yu, F.; Jiang, Z.; Zhang, J.; Shi, X.; Hu, X. The current status and future of PD-L1 in liver cancer. Front. Immunol. 2023, 14, 1323581. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Thomas, J.; Klebanov, A.; John, S.; Miller, L.S.; Vegesna, A.; Amdur, R.L.; Bhowmick, K.; Mishra, L. CEACAMS 1, 5, and 6 in disease and cancer: Interactions with pathogens. Genes Cancer 2023, 14, 12–29. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ma, R.X.; Wei, J.R.; Hu, Y.W. Characteristics of Carcinoembryonic Antigen-Related Cell Adhesion Molecules and Their Relationship to Cancer. Mol. Cancer Ther. 2024, 23, 939–948. [Google Scholar] [CrossRef] [PubMed]
- Gan, P.; Li, P.; Xia, H.; Zhou, X.; Tang, X. The application of artificial intelligence in improving colonoscopic adenoma detection rate: Where are we and where are we going. Gastroenterol. Hepatol. 2023, 46, 203–213, (In English & Spanish). [Google Scholar] [CrossRef] [PubMed]
- Chow, K.W.; Bell, M.T.; Cumpian, N.; Amour, M.; Hsu, R.H.; Eysselein, V.E.; Srivastava, N.; Fleischman, M.W.; Reicher, S. Long-term impact of artificial intelligence on colorectal adenoma detection in high-risk colonoscopy. World J. Gastrointest. Endosc. 2024, 16, 335. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, L.M.; Chahla, R.E.; Vega, I.M.; Ortega, E.S.; Barrenechea, G.G.; Contreras, M.F. Artificial Intelligence: Accuracy for the diagnosis of precancerous lesions of the cervix. Medicina 2024, 84, 459–467. (In Spanish) [Google Scholar] [PubMed]
- Kim, S.; An, H.; Cho, H.W.; Min, K.J.; Hong, J.H.; Lee, S.; Song, J.Y.; Lee, J.K.; Lee, N.W. Pivotal Clinical Study to Evaluate the Efficacy and Safety of Assistive Artificial Intelligence-Based Software for Cervical Cancer Diagnosis. J. Clin. Med. 2023, 12, 4024. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Zhuo, Y.; Wang, T.Q.; Lv, C.E.; Yao, L.H.; Zhang, S.Y. Procalcitonin and C-reactive protein as diagnostic biomarkers in COVID-19 and Non-COVID-19 sepsis patients: A comparative study. BMC Infect. Dis. 2024, 24, 45. [Google Scholar] [CrossRef] [PubMed]
- Al-Janabi, G.; Al-Fahham, A.; Alsaedi, A.N.N.; Al-Amery, A.Y.K. Correlation between hepcidin and procalcitonin and their diagnostic role in patients with COVID-19. Wiad. Lek. 2023, 76, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Gugo, K.; Tandara, L.; Juricic, G.; Pavicic Ivelja, M.; Rumora, L. Effects of Hypoxia and Inflammation on Hepcidin Concentration in Non-Anaemic COVID-19 Patients. J. Clin. Med. 2024, 13, 3201. [Google Scholar] [CrossRef]
- Julián-Jiménez, A.; García, D.E.; Merinos-Sánchez, G.; de Guadiana-Romualdo, L.G.; del Castillo, J.G. Diagnostic accuracy of procalcitonin for bacteremia in the emergency department: A systematic review. Rev. Esp. Quimioter. 2023, 37, 29–42. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Liu, J.; Hu, S.; Mao, Y. LDH and NLR, as inflammatory markers, the independent risk factors for COVID-19 complicated with respiratory failure in elderly patients. Pak. J. Med. Sci. 2024, 40, 2112–2117. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Murdoch, B. Privacy and artificial intelligence: Challenges for protecting health information in a new era. BMC Med. Ethics 2021, 22, 122. [Google Scholar] [CrossRef] [PubMed]
- Gerke, S.; Minssen, T.; Cohen, G. Ethical and legal challenges of artificial intelligence-driven healthcare. In Artificial Intelligence in Healthcare; Academic Press: Cambridge, MA, USA, 2020; pp. 295–336. [Google Scholar]
- Linkeviciute, A.; Curigliano, G.; Peccatori, F.A.; Pakutinskas, P. The regulatory impact of a harmonized artificial intelligence regulation proposal on the clinical research landscape in the European Union. BioLaw J.-Riv. BioDiritto 2022, 509–524. [Google Scholar] [CrossRef]
- Gaïffas, S.; Merad, I.; Yu, Y. WildWood: A new random forest algorithm. IEEE Trans. Inf. Theory 2023, 69, 6586–6604. [Google Scholar] [CrossRef]
- Chen, X.; Yu, D.; Zhang, X. Optimal weighted random forests. arXiv 2023, arXiv:2305.10042. [Google Scholar]
- Rothacher, Y.; Strobl, C. Identifying informative predictor variables with random forests. J. Educ. Behav. Stat. 2024, 49, 595–629. [Google Scholar] [CrossRef]
- Fife, D.A.; D’Onofrio, J. Common, uncommon, and novel applications of random forest in psychological research. Behav. Res. Methods 2023, 55, 2447–2466. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Pei, J.; Tong, H. Data Mining: Concepts and Techniques; Morgan Kaufmann: San Francisco, CA, USA, 2022. [Google Scholar]
- Gil-Rojas, S.; Suárez, M.; Martínez-Blanco, P.; Torres, A.M.; Martínez-García, N.; Blasco, P.; Torralba, M.; Mateo, J. Application of Machine Learning Techniques to Assess Alpha-Fetoprotein at Diagnosis of Hepatocellular Carcinoma. Int. J. Mol. Sci. 2024, 25, 1996. [Google Scholar] [CrossRef] [PubMed]
- Escobar-Ipuz, F.A.; Torres, A.M.; García-Jiménez, M.A.; Basar, C.; Cascón, J.; Mateo, J. Prediction of patients with idiopathic generalized epilepsy from healthy controls using machine learning from scalp EEG recordings. Brain Res. 2023, 1798, 148131. [Google Scholar] [CrossRef] [PubMed]
- Soria, C.; Arroyo, Y.; Torres, A.M.; Redondo, M.Á.; Basar, C.; Mateo, J. Method for classifying schizophrenia patients based on machine learning. J. Clin. Med. 2023, 12, 4375. [Google Scholar] [CrossRef]
- Mora, D.; Nieto, J.A.; Mateo, J.; Bikdeli, B.; Barco, S.; Trujillo-Santos, J.; Soler, S.; Font, L.; Bosevski, M.; Monreal, M.; et al. Machine learning to predict outcomes in patients with acute pulmonary embolism who prematurely discontinued anticoagulant therapy. Thromb. Haemost. 2022, 122, 570–577. [Google Scholar] [CrossRef]
- Suárez, M.; Gil-Rojas, S.; Martínez-Blanco, P.; Torres, A.M.; Ramón, A.; Blasco-Segura, P.; Torralba, M.; Mateo, J. Machine Learning-Based Assessment of Survival and Risk Factors in Non-Alcoholic Fatty Liver Disease-Related Hepatocellular Carcinoma for Optimized Patient Management. Cancers 2024, 16, 1114. [Google Scholar] [CrossRef] [PubMed]
- Johnson, K.B.; Wei, W.Q.; Weeraratne, D.; Frisse, M.E.; Misulis, K.; Rhee, K.; Zhao, J.; Snowdon, J.L. Precision medicine, AI, and the future of personalized health care. Clin. Transl. Sci. 2021, 14, 86–93. [Google Scholar] [CrossRef] [PubMed]
- Suárez, M.; Martínez, R.; Torres, A.M.; Ramón, A.; Blasco, P.; Mateo, J. Personalized risk assessment of hepatic fibrosis after cholecystectomy in metabolic-associated steatotic liver disease: A machine learning approach. J. Clin. Med. 2023, 12, 6489. [Google Scholar] [CrossRef] [PubMed]
- Probst, P.; Wright, M.N.; Boulesteix, A.L. Hyperparameters and tuning strategies for random forest. Wiley Interdiscip. Rev. Data Min. Knowl. Discov. 2019, 9, e1301. [Google Scholar] [CrossRef]
- Zhu, M.; Xia, J.; Jin, X.; Yan, M.; Cai, G.; Yan, J.; Ning, G. Class weights random forest algorithm for processing class imbalanced medical data. IEEE Access 2018, 6, 4641–4652. [Google Scholar] [CrossRef]
- Valkenborg, D.; Rousseau, A.J.; Geubbelmans, M.; Burzykowski, T. Support vector machines. Am. J. Orthod. Dentofac. Orthop. 2023, 164, 754–757. [Google Scholar] [CrossRef]
- Rajaguru, H.; Kumar Prabhakar, S. Bayesian Linear Discriminant Analysis for Breast Cancer Classification. In Proceedings of the 2017 2nd International Conference on Communication and Electronics Systems (ICCES), Coimbatore, India, 19–20 October 2017; pp. 266–269. Available online: https://ieeexplore.ieee.org/abstract/document/8321279 (accessed on 23 January 2024).
- Kourou, K.; Exarchos, T.P.; Exarchos, K.P.; Karamouzis, M.V.; Fotiadis, D.I. Machine learning applications in cancer prognosis and prediction. Comput. Struct. Biotechnol. J. 2014, 13, 8–17. [Google Scholar] [CrossRef] [PubMed]
- Chaplot, N.; Pandey, D.; Kumar, Y.; Sisodia, P.S. A Comprehensive Analysis of Artificial Intelligence Techniques for the Prediction and Prognosis of Genetic Disorders Using Various Gene Disorders. Arch. Comput. Methods Eng. 2023, 30, 3301–3323. [Google Scholar] [CrossRef]
- Uddin, S.; Haque, I.; Lu, H.; Moni, M.A.; Gide, E. Comparative performance analysis of K-nearest neighbour (KNN) algorithm and its different variants for disease prediction. Sci. Rep. 2022, 12, 6256. [Google Scholar] [CrossRef]

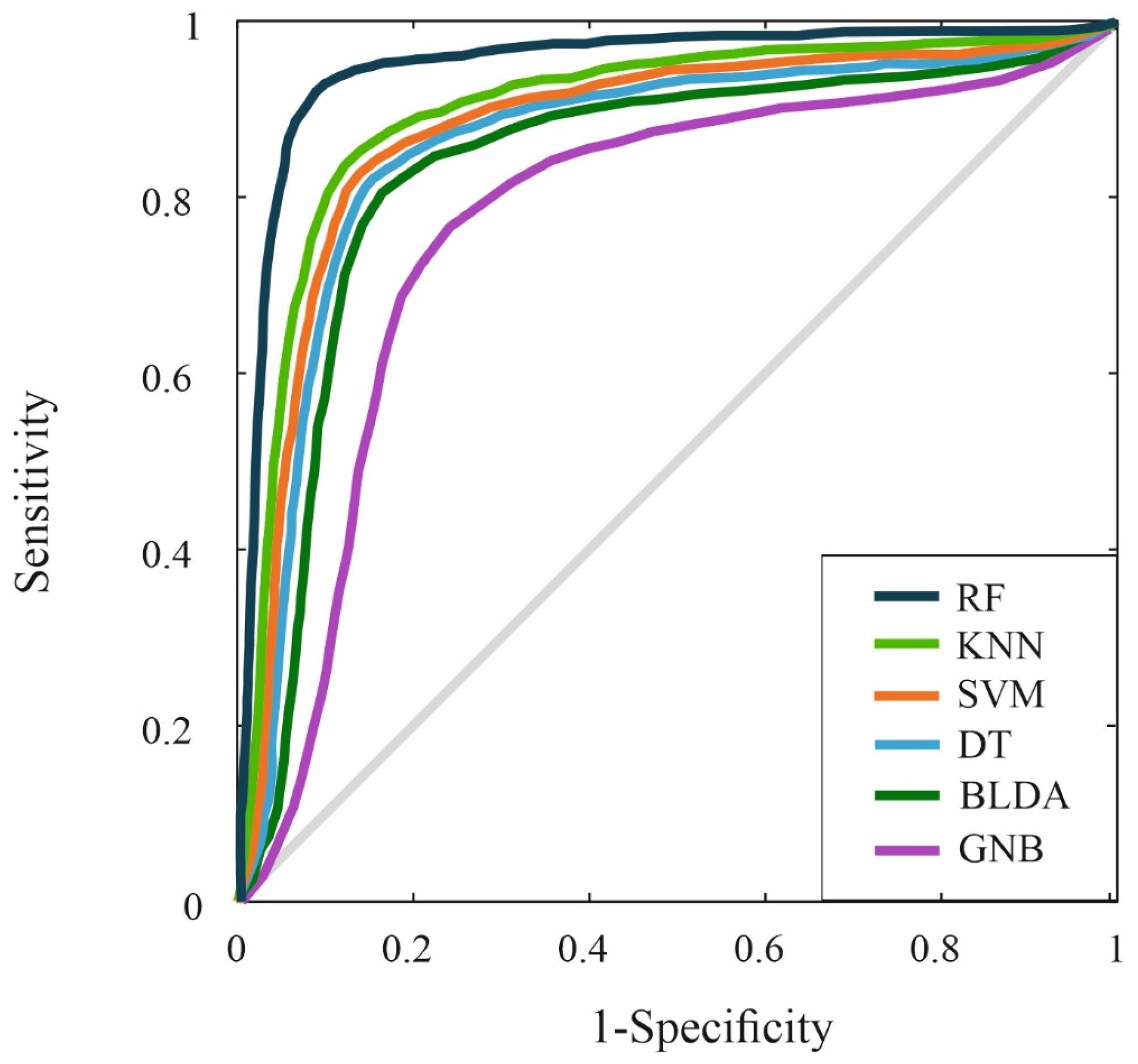
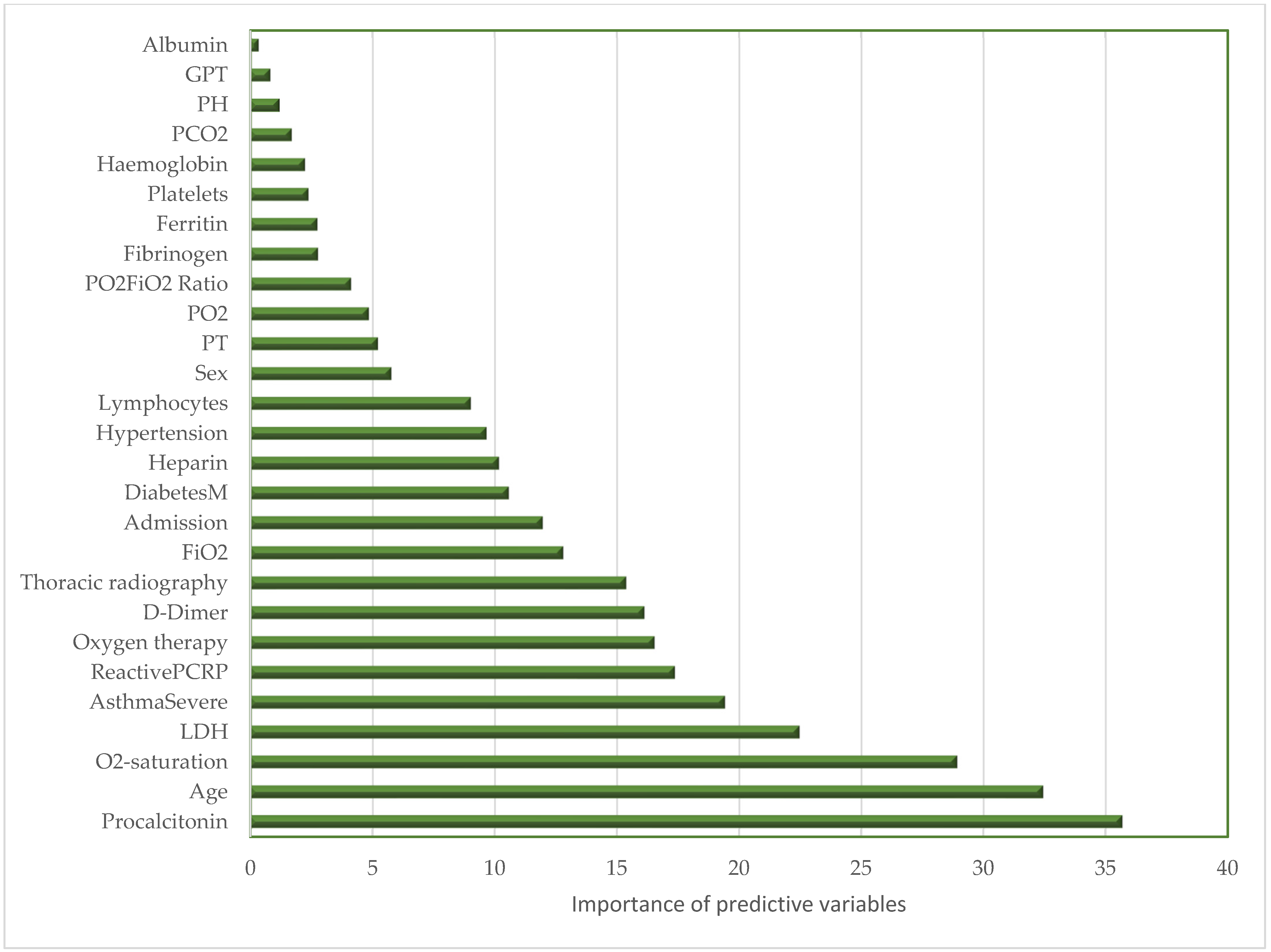
| Recall | Specificity | MCC | AUC1 | F1 Score | |
|---|---|---|---|---|---|
| SVM | 84.85 ± 0.87 | 84.65 ± 0.85 | 75.20 ± 0.82 | 0.85 ± 0.02 | 84.49 ± 0.88 |
| BLDA | 82.03 ± 0.96 | 81.83 ± 1.03 | 72.70 ± 0.85 | 0.82 ± 0.02 | 81.68 ± 1.01 |
| DT | 83.84 ± 0.91 | 83.64 ± 0.93 | 74.31 ± 0.92 | 0.84 ± 0.02 | 83.49 ± 0.93 |
| GNB | 77.16 ± 1.08 | 76.98 ± 1.10 | 68.39 ± 1.05 | 0.77 ± 0.02 | 76.84 ± 1.04 |
| KNN | 87.28 ± 0.74 | 87.07 ± 0.75 | 77.35 ± 0.78 | 0.87 ± 0.01 | 86.91 ± 0.73 |
| RF | 92.72 ± 0.51 | 92.50 ± 0.48 | 82.18 ± 0.45 | 0.93 ± 0.01 | 92.34 ± 0.49 |
| Accuracy | Precision | Kappa | DYI | |
|---|---|---|---|---|
| SVM | 84.75 ± 0.83 | 84.14 ± 0.84 | 75.45 ± 0.81 | 84.75 ± 0.82 |
| BLDA | 81.93 ± 0.99 | 81.35 ± 0.98 | 72.94 ± 0.95 | 81.93 ± 1.02 |
| DT | 83.74 ± 0.92 | 83.15 ± 0.90 | 74.55 ± 0.89 | 83.74 ± 0.91 |
| GNB | 77.07 ± 1.05 | 76.52 ± 1.03 | 68.61 ± 1.02 | 77.07 ± 1.06 |
| KNN | 87.17 ± 0.76 | 86.55 ± 0.74 | 77.61 ± 0.76 | 87.17 ± 0.75 |
| RF | 92.61 ± 0.49 | 91.95 ± 0.48 | 82.45 ± 0.46 | 92.61 ± 0.48 |
| Recall | Specificity | MCC | AUC1 | F1 Score | |
|---|---|---|---|---|---|
| SVM | 82.95 ± 0.82 | 82.67 ± 0.80 | 73.51 ± 0.83 | 0.82 ± 0.02 | 82.76 ± 0.83 |
| BLDA | 79.83 ± 1.02 | 79.89 ± 1.05 | 71.02 ± 0.91 | 0.79 ± 0.02 | 79.76 ± 1.03 |
| DT | 81.56 ± 0.96 | 81.48 ± 0.97 | 69.86 ± 0.95 | 0.81 ± 0.02 | 81.37 ± 0.94 |
| GNB | 74.99 ± 1.09 | 74.87 ± 1.12 | 66.59 ± 1.07 | 0.74 ± 0.02 | 74.78 ± 1.06 |
| KNN | 85.23 ± 0.76 | 85.19 ± 0.76 | 76.52 ± 0.72 | 0.85 ± 0.01 | 85.31 ± 0.78 |
| RF | 91.84 ± 0.52 | 91.35 ± 0.51 | 81.09 ± 0.49 | 0.91 ± 0.01 | 91.57 ± 0.51 |
| Accuracy | Precision | Kappa | DYI | |
|---|---|---|---|---|
| SVM | 82.89 ± 0.89 | 82.67 ± 0.91 | 73.58 ± 0.88 | 82.54 ± 0.89 |
| BLDA | 79.90 ± 1.02 | 79.96 ± 1.03 | 71.01 ± 0.97 | 79.87 ± 1.04 |
| DT | 81.39 ± 0.96 | 81.42 ± 0.94 | 70.13 ± 0.93 | 81.37 ± 0.95 |
| GNB | 75.06 ± 1.08 | 75.48 ± 1.06 | 66.53 ± 1.04 | 75.02 ± 1.07 |
| KNN | 85.34 ± 0.78 | 85.27 ± 0.79 | 75.93 ± 0.77 | 85.22 ± 0.78 |
| RF | 91.75 ± 0.52 | 91.47 ± 0.51 | 81.34 ± 0.51 | 91.57 ± 0.52 |
| Method | Parameters |
|---|---|
| SVM | Kernel function: Gaussian Sigma = 0.5 C = 1.0 Numerical tolerance = 0.001 Iteration limit = 100 |
| DT | Minimum number of instances in leaves = 4 Minimum number of instances in internal nodes = 6 Maximum depth = 100 |
| BLDA | Kernel: Bayesian |
| GNB | Usekernel: False fL = 0 Adjust = 0 |
| KNN | Number of neighbours = 20 Distance metric: Euclidean Weight: Uniform |
| RF | Number of estimators: 120, Maximun_depth: 20, Minimum_samples_split: 10, Minimum_samples_leaf: 4, Maximun _features: ‘sqrt’ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garrido, N.J.; González-Martínez, F.; Torres, A.M.; Blasco-Segura, P.; Losada, S.; Plaza, A.; Mateo, J. Role of Artificial Intelligence in Identifying Vital Biomarkers with Greater Precision in Emergency Departments During Emerging Pandemics. Int. J. Mol. Sci. 2025, 26, 722. https://doi.org/10.3390/ijms26020722
Garrido NJ, González-Martínez F, Torres AM, Blasco-Segura P, Losada S, Plaza A, Mateo J. Role of Artificial Intelligence in Identifying Vital Biomarkers with Greater Precision in Emergency Departments During Emerging Pandemics. International Journal of Molecular Sciences. 2025; 26(2):722. https://doi.org/10.3390/ijms26020722
Chicago/Turabian StyleGarrido, Nicolás J., Félix González-Martínez, Ana M. Torres, Pilar Blasco-Segura, Susana Losada, Adrián Plaza, and Jorge Mateo. 2025. "Role of Artificial Intelligence in Identifying Vital Biomarkers with Greater Precision in Emergency Departments During Emerging Pandemics" International Journal of Molecular Sciences 26, no. 2: 722. https://doi.org/10.3390/ijms26020722
APA StyleGarrido, N. J., González-Martínez, F., Torres, A. M., Blasco-Segura, P., Losada, S., Plaza, A., & Mateo, J. (2025). Role of Artificial Intelligence in Identifying Vital Biomarkers with Greater Precision in Emergency Departments During Emerging Pandemics. International Journal of Molecular Sciences, 26(2), 722. https://doi.org/10.3390/ijms26020722





